Abstract
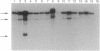
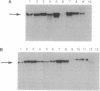
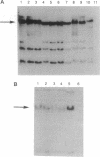
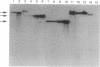
We tested the hypothesis that different genes can have different abilities to be amplified after transfection under comparable selection conditions. DNA from human lymphoid or choriocarcinoma cell lines was transfected into L cells. Transfectants for CD5, CD8A, TROP1, and TROP2, genes expressed on lymphocytes or trophoblast and carcinomas, were selected by fluorescence-activated cell sorting. To select for amplification of the transfected gene we cloned twice by fluorescence-activated cell sorting the transfectants with the highest expression. We analyzed a total of 38 families (1768 clones) derived from the original transfectants. We then analyzed by Southern blotting the clones with the highest increase in surface expression and determined the copy number of each transfected gene. CD5, CD8A, and TROP2 were amplified with high frequency and progressively, whereas TROP1 essentially was not amplified at all. We examined the hypothesis that DNA methylation prevents the amplification of the TROP1 gene by treating JAR choriocarcinoma cells with 5-azacytidine to decrease DNA methylation. DNA extracted at different times after the treatment was used for transfection. When DNA that showed demethylation of the TROP1 gene was used, 16 Trop-1 transfectants were obtained and 6 of them were found to contain up to 40 copies of the TROP1 gene per haploid genome. Thus, we showed that transfectants obtained from a demethylated TROP1 gene were amplified efficiently and progressively. We propose that DNA methylation affects DNA amplification either by altering the recognition of methylated DNA sequences or by changing the conformation of the chromatin of methylated segments. We speculate that DNA methylation is a determinant of gene amplification in vivo, for example in tumor cells.
Full text
PDF
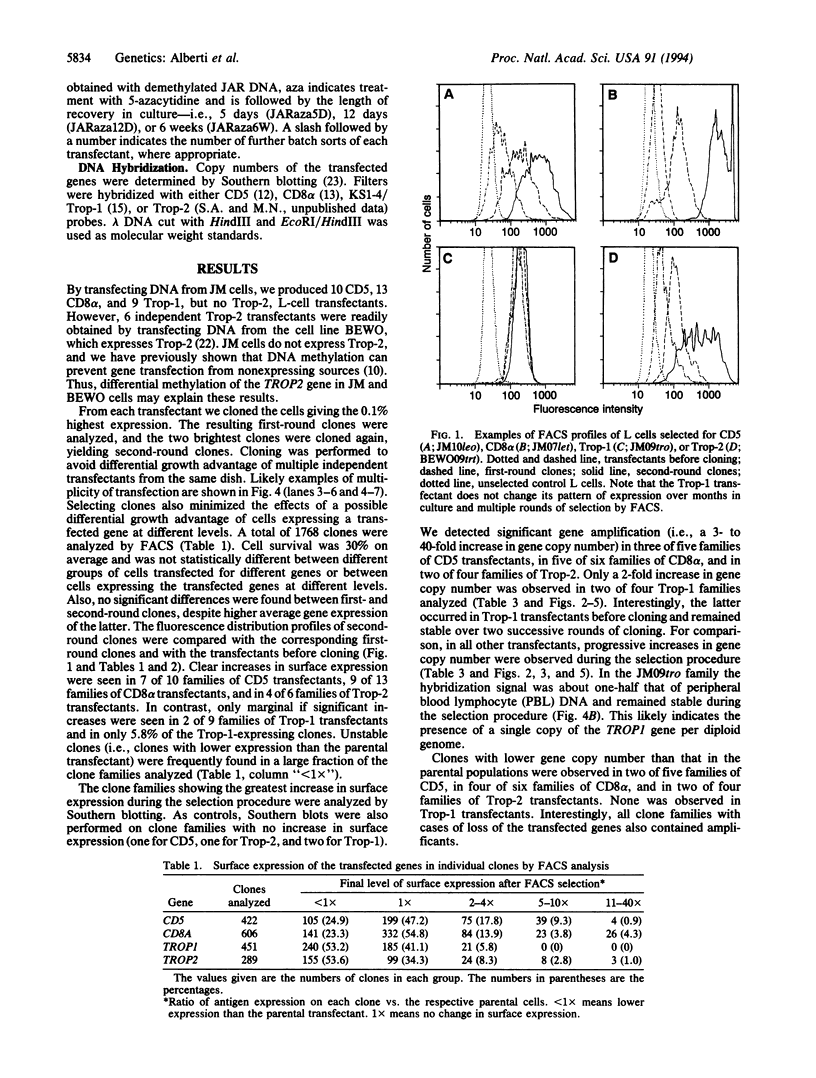


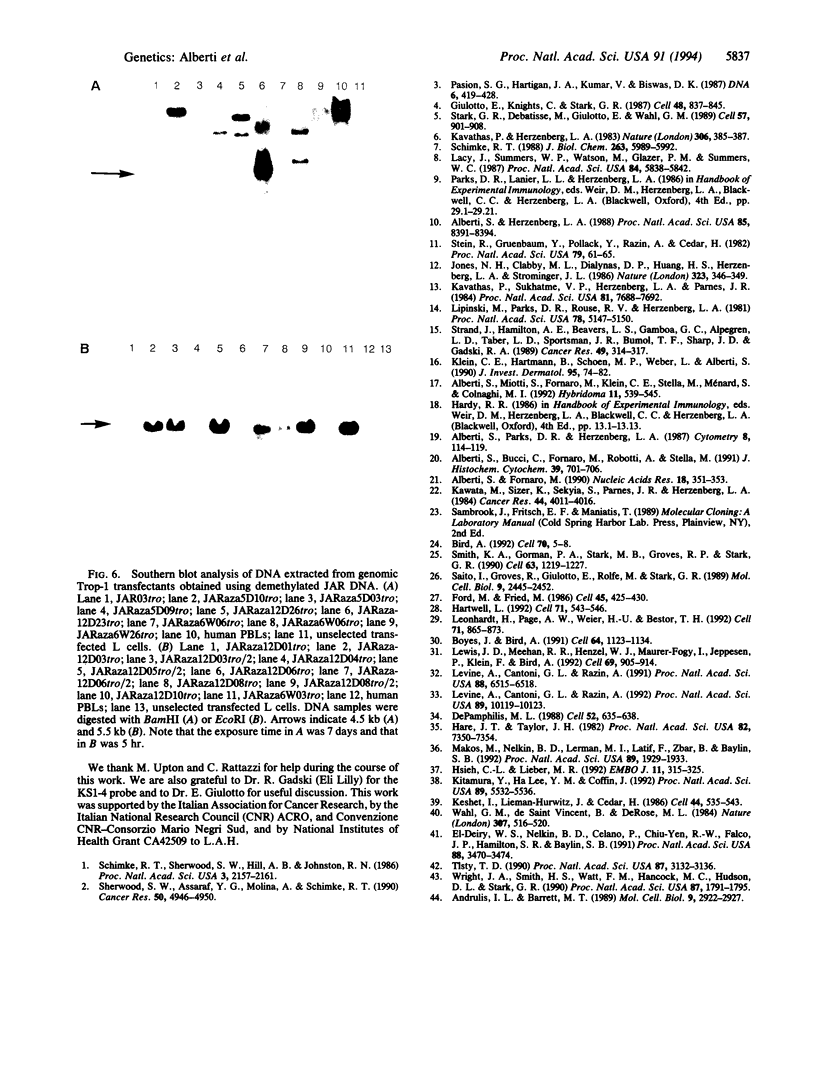
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberti S., Bucci C., Fornaro M., Robotti A., Stella M. Immunofluorescence analysis in flow cytometry: better selection of antibody-labeled cells after fluorescence overcompensation in the red channel. J Histochem Cytochem. 1991 May;39(5):701–706. doi: 10.1177/39.5.1901878. [DOI] [PubMed] [Google Scholar]
- Alberti S., Fornaro M. Higher transfection efficiency of genomic DNA purified with a guanidinium thiocyanate-based procedure. Nucleic Acids Res. 1990 Jan 25;18(2):351–353. doi: 10.1093/nar/18.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Herzenberg L. A. DNA methylation prevents transfection of genes for specific surface antigens. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8391–8394. doi: 10.1073/pnas.85.22.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Miotti S., Stella M., Klein C. E., Fornaro M., Menard S., Colnaghi M. I. Biochemical characterization of Trop-2, a cell surface molecule expressed by human carcinomas: formal proof that the monoclonal antibodies T16 and MOv-16 recognize Trop-2. Hybridoma. 1992 Oct;11(5):539–545. doi: 10.1089/hyb.1992.11.539. [DOI] [PubMed] [Google Scholar]
- Alberti S., Parks D. R., Herzenberg L. A. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987 Mar;8(2):114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- Andrulis I. L., Barrett M. T. DNA methylation patterns associated with asparagine synthetase expression in asparagine-overproducing and -auxotrophic cells. Mol Cell Biol. 1989 Jul;9(7):2922–2927. doi: 10.1128/mcb.9.7.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Giulotto E., Knights C., Stark G. R. Hamster cells with increased rates of DNA amplification, a new phenotype. Cell. 1987 Mar 13;48(5):837–845. doi: 10.1016/0092-8674(87)90080-8. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992 Nov 13;71(4):543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- Hsieh C. L., Lieber M. R. CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO J. 1992 Jan;11(1):315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. H., Clabby M. L., Dialynas D. P., Huang H. J., Herzenberg L. A., Strominger J. L. Isolation of complementary DNA clones encoding the human lymphocyte glycoprotein T1/Leu-1. 1986 Sep 25-Oct 1Nature. 323(6086):346–349. doi: 10.1038/323346a0. [DOI] [PubMed] [Google Scholar]
- Kavathas P., Herzenberg L. A. Amplification of a gene coding for human T-cell differentiation antigen. Nature. 1983 Nov 24;306(5941):385–387. doi: 10.1038/306385a0. [DOI] [PubMed] [Google Scholar]
- Kavathas P., Sukhatme V. P., Herzenberg L. A., Parnes J. R. Isolation of the gene encoding the human T-lymphocyte differentiation antigen Leu-2 (T8) by gene transfer and cDNA subtraction. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7688–7692. doi: 10.1073/pnas.81.24.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M., Sizer K., Sekiya S., Parnes J. R., Herzenberg L. A. Limitation of differential expression of HLA-A,B,C antigens on choriocarcinoma cell lines by messenger RNA for HLA heavy chain but not by beta 2-microglobulin. Cancer Res. 1984 Sep;44(9):4011–4016. [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Lee Y. M., Coffin J. M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C. E., Hartmann B., Schön M. P., Weber L., Alberti S. Expression of 38-kD cell-surface glycoprotein in transformed human keratinocyte cell lines, basal cell carcinomas, and epithelial germs. J Invest Dermatol. 1990 Jul;95(1):74–82. doi: 10.1111/1523-1747.ep12873988. [DOI] [PubMed] [Google Scholar]
- Lacy J., Summers W. P., Watson M., Glazer P. M., Summers W. C. Amplification and deregulation of MYC following Epstein-Barr virus infection of a human B-cell line. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5838–5842. doi: 10.1073/pnas.84.16.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H., Page A. W., Weier H. U., Bestor T. H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992 Nov 27;71(5):865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Methylation in the preinitiation domain suppresses gene transcription by an indirect mechanism. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10119–10123. doi: 10.1073/pnas.89.21.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lipinski M., Parks D. R., Rouse R. V., Herzenberg L. A. Human trophoblast cell-surface antigens defined by monoclonal antibodies. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5147–5150. doi: 10.1073/pnas.78.8.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makos M., Nelkin B. D., Lerman M. I., Latif F., Zbar B., Baylin S. B. Distinct hypermethylation patterns occur at altered chromosome loci in human lung and colon cancer. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1929–1933. doi: 10.1073/pnas.89.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion S. G., Hartigan J. A., Kumar V., Biswas D. K. DNA sequence responsible for the amplification of adjacent genes. DNA. 1987 Oct;6(5):419–428. doi: 10.1089/dna.1987.6.419. [DOI] [PubMed] [Google Scholar]
- Saito I., Groves R., Giulotto E., Rolfe M., Stark G. R. Evolution and stability of chromosomal DNA coamplified with the CAD gene. Mol Cell Biol. 1989 Jun;9(6):2445–2452. doi: 10.1128/mcb.9.6.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured cells. J Biol Chem. 1988 May 5;263(13):5989–5992. [PubMed] [Google Scholar]
- Schimke R. T., Sherwood S. W., Hill A. B., Johnston R. N. Overreplication and recombination of DNA in higher eukaryotes: potential consequences and biological implications. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2157–2161. doi: 10.1073/pnas.83.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood S. W., Assaraf Y. G., Molina A., Schimke R. T. Flow cytometric characterization of antifolate resistance in cultured mammalian cells using fluoresceinated methotrexate and daunorubicin. Cancer Res. 1990 Aug 15;50(16):4946–4950. [PubMed] [Google Scholar]
- Smith K. A., Gorman P. A., Stark M. B., Groves R. P., Stark G. R. Distinctive chromosomal structures are formed very early in the amplification of CAD genes in Syrian hamster cells. Cell. 1990 Dec 21;63(6):1219–1227. doi: 10.1016/0092-8674(90)90417-d. [DOI] [PubMed] [Google Scholar]
- Stark G. R., Debatisse M., Giulotto E., Wahl G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989 Jun 16;57(6):901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- Stein R., Gruenbaum Y., Pollack Y., Razin A., Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A. 1982 Jan;79(1):61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad J., Hamilton A. E., Beavers L. S., Gamboa G. C., Apelgren L. D., Taber L. D., Sportsman J. R., Bumol T. F., Sharp J. D., Gadski R. A. Molecular cloning and characterization of a human adenocarcinoma/epithelial cell surface antigen complementary DNA. Cancer Res. 1989 Jan 15;49(2):314–317. [PubMed] [Google Scholar]
- Tlsty T. D. Normal diploid human and rodent cells lack a detectable frequency of gene amplification. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3132–3136. doi: 10.1073/pnas.87.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Robert de Saint Vincent B., DeRose M. L. Effect of chromosomal position on amplification of transfected genes in animal cells. Nature. 1984 Feb 9;307(5951):516–520. doi: 10.1038/307516a0. [DOI] [PubMed] [Google Scholar]
- Wright J. A., Smith H. S., Watt F. M., Hancock M. C., Hudson D. L., Stark G. R. DNA amplification is rare in normal human cells. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1791–1795. doi: 10.1073/pnas.87.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry W. S., Nelkin B. D., Celano P., Yen R. W., Falco J. P., Hamilton S. R., Baylin S. B. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]