Abstract
Background
A rapid surge of female breast cancer has been observed in young women in several East Asian countries. The BIM deletion polymorphism, which confers cell resistance to apoptosis, was recently found exclusively in East Asian people with prevalence rate of 12%. We aimed to evaluate the possible role of this genetic alteration in carcinogenesis of breast cancer in East Asians.
Method
Female healthy volunteers (n = 307), patients in one consecutive stage I-III breast cancer cohort (n = 692) and one metastatic breast cancer cohort (n = 189) were evaluated. BIM wild-type and deletion alleles were separately genotyped in genomic DNAs.
Results
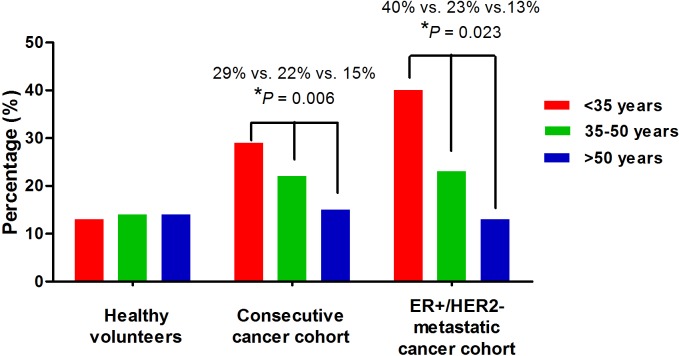
Both cancer cohorts consistently showed inverse associations between the BIM deletion polymorphism and patient age (≤35 y vs. 36-50 y vs. >50 y: 29% vs. 22% vs. 15%, P = 0.006 in the consecutive cohort, and 40% vs. 23% vs. 13%, P = 0.023 in the metastatic cohort). In healthy volunteers, the frequencies of the BIM deletion polymorphism were similar (13%-14%) in all age groups. Further analyses indicated that the BIM deletion polymorphism was not associated with specific clinicopathologic features, but it was associated with poor overall survival (adjusted hazard ratio 1.71) in the consecutive cohort.
Conclusions
BIM deletion polymorphism may be involved in the tumorigenesis of the early-onset breast cancer among East Asians.
Introduction
The incidence of breast cancer among Asian women is in general lower than that in Western countries. However, all health statistics indicated that breast cancer has been rapidly increasing in recent decades in East Asia, including Singapore, Korea, Japan, and Taiwan [1–4]. Compared to Caucasian American women, the age-period-cohort analyses consistently revealed a much stronger birth cohort effect on the breast cancer incidence of Singaporean, Japanese, and Taiwanese women [1,3,4]. This strong birth cohort effect correlated directly with a rapid increase in the incidence of early-onset breast cancer in these countries. Intuitively, Westernized lifestyle is thought to be the major cause of this rapidly increasing young female breast cancer (YFBC) in Asia [5]. However, our recent study demonstrated a major discrepancy of molecular subtype distributions between Taiwanese and Caucasian YFBC. In contrast to their Western counterpart, Taiwanese YFBCs are characterized by a luminal A subtype (defined as estrogen receptor [ER] and/or progesterone receptor [PR] positive and human epidermal growth factor receptor 2 [HER2] negative) prevalence, and low basal-like subtype prevalence [6]. These findings implied that the emerging YFBCs in Taiwan might not just be a mirror image of their Western counterparts. We hypothesize that some unique genetic factors or interactions between genetic factors and environmental factors may play a role in East Asian YFBC carcinogenesis.
Recently, Ng KP et al. discovered a common germline polymorphism (deletion of intron 2 of BIM gene) which was uniquely detected in East-Asian people (12.3% carrier frequency) and was absent in Africans and Caucasians. BIM deletion polymorphism conferred an inferior response to tyrosine kinase inhibitors in patients with chronic myeloid leukemia and epidermal growth factor receptor mutated non-small cell lung cancer [7–9]. The BIM gene encodes B-cell lymphoma 2 interacting mediator of cell death (BIM) protein, which is a member of the Bcl-2 family. Wild type BIM protein, which contains a BCL2-homology domain 3 (BH3), which functions as an apoptosis facilitator and has been shown to mediate apoptosis in response to stimuli such as cytokine deprivation, deregulated calcium flux and microtubule perturbation. Thus, BIM is considered a protector of tissue homeostasis [10,11]. The BIM deletion polymorphism switched BIM splicing from exon 4 to exon 3, and resulted in expression of BIM isoforms lacking the pro-apoptotic BH3 domain and conferred intrinsic resistance to tyrosine kinase inhibitors [7].
Since BIM deletion polymorphism is unique in East Asian people, and its product (BIM isoforms) may be linked to tumorigenesis, it is crucial to clarify whether this genetic change plays a role in the carcinogenesis of YFBC in East Asian women.
Materials and Methods
Patients and sample collection
All participants in this study gave written informed consent. The study received approval from the National Taiwan University Hospital (NTUH) ethics committee (201307001RINA). The study included 307 female healthy volunteers, 692 patients with stage I-III breast cancer in one consecutive cohort and 189 patients with ER+/HER2- breast cancer in one metastatic cohort with available germline or tumor DNAs (S1–S3 Datasets). The healthy volunteers participated in the prior study exploring the association of breast cancer and gene polymorphism [12]. The consecutive cancer cohort was obtained from a prospectively collected database which included stage I-III breast cancer consecutively newly diagnosed at NTUH between 2004 and 2006 [6]. The metastatic cancer cohort was obtained from a retrospectively collected database which includes patients with ER+/ HER2- metastatic breast cancer patients diagnosed at NTUH between 2001 and 2006. To avoid bias by double counting, we excluded 19 patients from the consecutive cohort because these patients were included in the consecutive cohort and had distant metastasis between 2004 and 2006. The methods and definitions of ER and HER2 positivity were previously described [6]. For ER and PR, Tumors with ≥10% positively-staining nuclei were considered positive. The HER2 status was considered positive if score 3+ by immunohistochemical analysis or 2+ with gene amplification on fluorescence in situ hybridization.
Evaluation of BIM deletion polymorphism
The DNAs from healthy volunteers were extracted from blood specimens. The DNAs from patients in consecutive and metastatic cancer cohorts were extracted from formalin-fixed paraffin-embedded tumor specimens. The genomic DNAs of blood and tumor specimens were isolated using the QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA, USA). For DNAs from each blood or tumor specimen, we performed two separate polymerase chain reaction reactions to determine the presence of the wild-type and deletion alleles as previously described [7]. The forward and reverse primers for the deletion allele were CCACCAATGGAAAAGGTTCA and GGCACAGCCTCTATGGAGAA, respectively. The forward and reverse primers for the wild-type allele were CCACCAATGGAAAAGGTTCA and CTGTCATTTCTCCCCACCAC, respectively. The resulting PCR products from the deletion (1,323 bp) and the wild-type (4,226 bp) alleles were analyzed on 1% agarose gels. For each PCR, we used genomic DNAs from KCL22 and PC-9 cells as positive and negative control, respectively.
Statistical analysis
Data on clinicopathological features between wild and deleted BIM groups were compared using chi-square test (or two tailed Fisher’s exact test if expected number of each cell was less than five cases). The Mantel-Haenszel chi-square test was used to verify age, histologic grade, tumor size, lymph node status and American Joint Committee on Cancer (AJCC) stage-related trend. For survival analysis in the consecutive cancer cohort, only patients with stage I-III breast cancer were included, and distant metastasis free survival (DMFS) and overall survival (OS) were used as the endpoints. DMFS was defined as the duration from diagnosis to confirmation of distant recurrences. OS was defined as the duration from breast cancer diagnosis to death from any cause. Survival curves were constructed using the Kaplan-Meier method. The associations between each of the categorical variables and survival were analyzed using the log-rank test. Cox proportional hazards analysis was used to determine the relative contribution of various factors to survival. The backward stepwise variable selection procedure was applied to obtain the best candidate final Cox’s proportional hazards model. A P value ≤0.05 was used to indicate statistical significance; all tests were two-tailed. All statistical analyses were performed using the statistical package SPSS for Windows (Version 17.0).
Results
Frequencies of BIM deletion polymorphism by age groups among healthy volunteers and breast cancer patients
The healthy volunteers had a median age of 48 (range 25–80) years at enrollment. The patients in consecutive and metastatic cancer cohorts had a median age of 49 (range 23–86) years and 50 (range 26–82) years at initial diagnosis of breast cancer, respectively. Among the 307 healthy volunteers, BIM deletion polymorphism was detected in 48 (14%) subjects and the frequencies were similar among the three age groups (≤35 vs. 36–50 vs. >50 years, 13% vs. 14% vs. 14%, P = 0.974). Compared with healthy volunteers, patients in the consecutive cancer cohort (19% vs. 14%, P = 0.018) had significantly higher frequencies of BIM deletion polymorphism and patients in the metastatic cancer cohort (19% vs. 14%, P = 0.089) had a trend toward higher frequencies of BIM deletion polymorphism (Table 1).
Table 1. Comparison of BIM deletion polymorphism in women benign breast disease and two breast cancer cohorts with healthy volunteers among age groups.
| No. (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤35 years | 36–50 years | >50 years | Whole | |||||||||
| BIM polymorphism | wild | deleted | P | wild | deleted | P | wild | deleted | P | wild | deleted | P |
| Healthy (reference) | 42 (88) | 6 (13) | 140 (86) | 22 (14) | 125 (86) | 20 (14) | 307 (86) | 48 (14) | ||||
| Consecutive cohort | 37 (71) | 15 (29) | 0.045 | 254 (78) | 71 (22) | 0.029 | 267 (85) | 48 (15) | 0.640 | 558 (81) | 134 (19) | 0.018 |
| Metastatic cohort | 9 (60) | 6 (40) | 0.028* | 62 (78) | 18 (23) | 0.079 | 82 (87) | 12 (13) | 0.820 | 153 (81) | 36 (19) | 0.089 |
* two tailed Fisher’s exact test.
Both cancer cohorts consistently showed inverse association of BIM deletion polymorphism with patient age (≤35 years vs. 36–50 years vs. >50 years: 29% vs. 22% vs. 15%, P = 0.006 in consecutive cohort, and 40% vs. 23% vs. 13%, P = 0.023 in metastatic cohort). Among women ≤50 years, both cancer cohorts consistently showed higher frequencies of BIM deletion polymorphism than healthy volunteers (consecutive cancer cohort, 23% vs.13%, P = 0.005; metastatic cancer cohort, 25% vs. 13%, P = 0.010). In contrast, the frequencies of BIM deletion polymorphism were quite similar (healthy volunteer, 14%; consecutive cancer cohort, 15%; metastatic cancer cohort, 13%) among the three study groups of women >50 years (Table 1 and Fig 1).
Fig 1. Inverse association of BIM deletion polymorphism with age in young breast cancer patients.
Patient clinicopathological characteristics by BIM status in two breast cancer cohorts
Among the 692 patients with stage I-III breast cancer in the consecutive cohort, BIM deletion polymorphism was detected in 140 (19%) subjects. The BIM deletion polymorphism rate was significantly higher in young patients. Other clinicopathological variables including histology, histologic grade, AJCC stage, ER status, PR status, HER2 status and molecular subtype were not significantly associated with this polymorphism (Table 2).
Table 2. The characteristics of patients in the consecutive cancer cohort by BIM deletion polymorphism.
| Characteristics | No. | No. (%) | ||
|---|---|---|---|---|
| All (n = 692) | BIM wild (n = 558) | BIM deleted (n = 134) | P value | |
| Age at initial diagnosis | 0.006 | |||
| ≤35 years | 52 | 37 (71) | 15 (29) | |
| 36–50 years | 325 | 254 (78) | 71 (22) | |
| >50 years | 315 | 267 (85) | 48 (15) | |
| Histology type | 0.783 | |||
| Ductal carcinoma | 659 | 532 (81) | 127 (19) | |
| Others | 33 | 26 (79) | 7 (21) | |
| Histologic grade | 0.556 | |||
| I | 131 | 108 (82) | 23 (18) | |
| II | 362 | 293 (81) | 69 (19) | |
| III | 166 | 131 (79) | 35 (20) | |
| Unknown | 33 | 26 | 7 | |
| Tumor size | 0.186 | |||
| <2 cm | 300 | 237 (79) | 63 (21) | |
| 2–5 cm | 331 | 268 (81) | 68 (19) | |
| >5 cm | 61 | 53 (87) | 8 (13) | |
| Axillary lymph node | 0.494 | |||
| None or cN0 | 374 | 298 (80) | 76 (20) | |
| 1–3 or cN1 | 194 | 160 (82) | 34 (18) | |
| 4–9 or cN2 | 78 | 60 (77) | 18 (23) | |
| ≥10 or cN3 | 46 | 40 (87) | 6 (13) | |
| AJCC stage | 0.881 | |||
| I | 237 | 191 (81) | 46 (19) | |
| II | 327 | 265 (81) | 62 (19) | |
| III | 128 | 102 (80) | 26 (20) | |
| ER status | 0.434 | |||
| Negative | 221 | 182 (82) | 39 (18) | |
| Positive | 471 | 376 (80) | 95 (20) | |
| PR status | 0.921 | |||
| Negative | 395 | 318 (81) | 77 (19) | |
| Positive | 297 | 240 (81) | 57 (19) | |
| HER2 status | 0.137 | |||
| Negative | 544 | 445 (82) | 99 (18) | |
| Positive | 148 | 113 (76) | 35 (24) | |
AJCC, American Joint Committee on Cancer; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Among the 189 patients with stage ER+/HER2- metastatic breast cancer in the metastatic cohort, BIM deletion polymorphism was detected in 36 (19%) subjects. Consistent with that in consecutive cohort, the BIM deletion polymorphism rate was significantly higher in young patients and clinicopathological variables including histology, histologic grade, PR status, recurrence status and metastatic site were not significantly associated with BIM deletion polymorphism (Table 3).
Table 3. The characteristics of patients in ER+/ HER2- metastatic cancer cohort by BIM deletion polymorphism.
| Characteristics | No. | No. (%) | ||
|---|---|---|---|---|
| All (n = 189) | BIM wild (n = 153) | BIM deleted (n = 36) | P value | |
| Age at initial diagnosis | 0.023* | |||
| ≤35 years | 15 | 9 (60) | 6 (40) | |
| 36–50 years | 80 | 62 (78) | 18 (23) | |
| >50 years | 94 | 82 (87) | 12 (13) | |
| Histology | 0.374 | |||
| Ductal carcinoma | 162 | 133 (82) | 29 (18) | |
| Others | 19 | 14 (74) | 5 (26) | |
| Unknown | 8 | 6 | 2 | |
| Grade | 0.583 | |||
| I | 27 | 22 (81) | 5 (19) | |
| II | 75 | 62 (83) | 13 (17) | |
| III | 29 | 22 (76) | 7 (24) | |
| Unknown | 58 | 47 | 11 | |
| PR Status | 0.139 | |||
| Negative | 61 | 53 (87) | 8 (13) | |
| Positive | 126 | 98 (78) | 28 (22) | |
| Unknown | 2 | 2 | 0 | |
DFI, disease-free interval; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
*two tailed Fisher’s exact test.
Prognostic value of BIM deletion polymorphism in patients with stage I-III breast cancer
In consecutive cancer cohort, the median follow-up duration among the 692 patients with stage I-III breast cancer was 81.7 months (95% confidence interval (CI), 80.1–83.4). The 6-year distant metastasis-free survival rate was 92% for stage I disease, 82% for stage II disease, and 70% for stage III disease. The 6-year overall survival rate was 95% for stage I disease, 90% for stage II disease, and 74% for stage III disease.
Traditional prognostic factors such as tumor size, axillary lymph node status, ER expression, PR expression and HER2 status were associated with DMFS and/or OS in univariate and/or multivariate analyses. Univariate determined that BIM deletion polymorphism was not associated with DMFS (hazard ratio [HR] = 1.11, P = 0.636) and OS (HR = 1.45, P = 0.125). Multivariate analysis determined that BIM deletion polymorphism was not associated with DMFS, but it was significantly associated with shorter OS (adjusted HR = 1.71, P = 0.027) (Table 4).
Table 4. Analyses of distant metastasis-free and overall survival in patients with stage I-III breast cancer.
| DMFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | HR (95% CI) | P | Adjusted HR (95% CI) | P | HR (95% CI) | P | Adjusted HR (95% CI) | P | |
| BIM polymorphism | 0.636 | NS | 0.125 | 0.027 | |||||
| Wild | 558 | 1.00 | 1.00 | 1.00 | |||||
| Deleted | 134 | 1.11 (0.72–1.73) | 1.45 (0.90–2.33) | 1.71 (1.06–2.77) | |||||
| Age | 0.629 | NS | 0.008 | 0.033 | |||||
| ≤35 years | 52 | 1.00 | 1.00 | 1.00 | |||||
| 36–50 years | 325 | 1.13 (0.51–2.48) | 0.53 (0.24–1.16) | 0.48 (0.22–1.06) | |||||
| >50 years | 315 | 1.32 (0.60–2.88) | 1.08 (0.51–2.27) | 0.85 (0.40–1.82) | |||||
| Histologic grade | 0.008 | NS | 0.002 | NS | |||||
| I | 131 | 1.00 | 1.00 | ||||||
| II | 362 | 1.82 (01.0–3.31) | 1.92 (0.90–4.10) | ||||||
| III | 166 | 2.80 (1.49–5.25) | 3.67 (1.70–7.93) | ||||||
| Unknown | 33 | 2.75 (1.14–6.64) | 3.38 (1.22–9.32) | ||||||
| Tumor size | <0.001 | 0.022 | <0.001 | 0.020 | |||||
| ≤ 2 cm | 300 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| 2–5 cm | 311 | 1.81 (1.19–2.74) | 1.35 (0.87–2.09) | 2.20 (1.32–3.68) | 1.49 (0.87–2.56) | ||||
| > 5 cm | 61 | 4.34 (2.55–7.38) | 2.38 (1.29–4.40) | 5.66 (3.09–10.36) | 2.69 (1.34–5.41) | ||||
| Axillary lymph node | <0.001 | <0.001 | <0.001 | 0.001 | |||||
| None or cN0 | 374 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| 1–3 or cN1 | 194 | 2.79 (1.81–4.30) | 2.47 (1.58–3.89) | 2.39 (1.41–4.06) | 2.29 (1.31–4.01) | ||||
| 4–9 or cN2 | 78 | 3.04 (1.76–5.26) | 2.26 (1.24–4.12) | 4.13 (2.29–7.44) | 3.26 (1.70–6.23) | ||||
| ≥ 10 or cN3 | 46 | 4.43 (2.46–7.99) | 2.97 (1.54–5.72) | 5.39 (2.84–10.24) | 3.67 (1.78–7.57) | ||||
| ER status | 0.008 | 0.004 | <0.001 | <0.001 | |||||
| Negative | 221 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Positive | 471 | 0.61 (0.42–0.88) | 0.58 (0.40–0.84) | 0.41 (0.27–0.62) | 0.43 (0.28–0.67) | ||||
| PR status | 0.003 | NS | <0.001 | NS | |||||
| Negative | 396 | 1.00 | 1.00 | ||||||
| Positive | 296 | 0.56 (0.38–0.82) | 0.40 (0.25–0.65) | ||||||
| HER2 status | 0.313 | NS | 0.509 | NS | |||||
| No | 544 | 1.00 | 1.00 | ||||||
| Yes | 148 | 1.24 (0.82–1.87) | 1.18 (0.73–1.90) | ||||||
DMFS, distant metastasis-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2
Discussion
High prevalence of BIM deletion polymorphism in young Taiwanese breast cancer patients suggests that this East Asian specific genetic trait is involved in the tumorigenesis of early-onset breast cancer among East Asians. This finding supports our hypothesis that, in addition to environmental and lifestyle factors, certain genetic factors may play a role in Asian young breast cancer development.
BIM deletion polymorphism was found exclusively in East Asian individuals (12.3% carrier frequency) [7]. In our study, the frequency of the polymorphism in the whole healthy volunteers (14%) was close to that reported by Ng KP et al., and the frequencies were similar among ≤35 (13%), 36–50 (14%), and >50 (14%) age groups. In breast cancer patients >50 years, the frequencies of BIM deletion polymorphism in both cancer cohorts were not significantly different from healthy volunteers. In contrast, among subjects <50 years, higher BIM deletion polymorphism frequencies were consistently shown in both cancer cohorts than healthy volunteers. Among very young (≤35 years) patients, the frequencies of BIM deletion polymorphism reached up to 29% and 40% in the consecutive and metastatic cancer cohorts, respectively.
Since no significant association between BIM deletion polymorphism and other clinicopathological factors except age was observed, we hypothesize that BIM deletion polymorphism may mediate the cancer initiation rather than tumor progression. However, how and why this genetic change affects the young ladies in East Asians remains unclear. In addition, the major limitation of the present study is lack of comprehensive information of breast cancer risk factors such as menstruation history, family history, pregnancy and birth history, alcohol consumption, and weight. To confirm BIM deletion polymorphism as a susceptible gene for YFBC carcinogenesis in East Asia, the validation by a well designed case control study is mandatory.
BIM is essential for initiation of various physiological apoptotic situations, including developmentally programmed cell death and stress-induced apoptosis. BIM is considered a protector of tissue homeostasis [10,11]. The breakdown of tissue homeostasis may lead to various pathological situations including tumor formation. In the mouse model of B cells and kidney epithelial cells, the loss of single BIM sensitizes the mice to tumorigenesis [13,14]. In mammary gland, disruption of the BH3-only proapoptotic factor BIM in mice prevents induction of apoptosis in and clearing of the lumen in terminal end buds during puberty. The findings indicate that BIM is a critical regulator of luminal space formation and maintenance during mammary morphogenesis [15–17]. Since BIM deletion polymorphism is germline genetic event and we did not observe significant association between BIM deletion polymorphism and other clinicopathological factors except age, we suggest that BIM deletion polymorphism may mediate the cancer initiation rather than tumor progression in human mammary gland.
BIM plays important roles not only in tumorigenesis but also in treatment response. Previous preclinical studies have shown that BIM plays a key role in the anoikis, an apoptosis triggered by detachment from the extracellular matrix, of various tumor cells [18–20]. Absence of wild type BIM protein has been linked to resistance to chemotherapy and tyrosine kinase inhibitors in several cancer types [14,21–30]. In breast cancer, decrease of BIM expression has been linked to resistance to estrogen deprivation and a HER2 tyrosine kinase inhibitor [31,32]. BIM deletion polymorphism is heterozygous, so it can transcribe both wild type BIM protein and BIM isoforms which lack the BH3 domain. Although wild type BIM protein from single allele may retain certain pro-apoptosis functions, the net activity of BIM protein and isoforms in BIM deletion polymorphism cells remains inadequate in certain tumor types [7]. Our multivariate analysis showed that BIM deletion polymorphism was significantly associated with shorter OS in patients with stage I-III breast cancer (Table 4). However, it was not significantly associated with DMFS. Because of the association between BIM deletion polymorphism and age group, we conducted the stratified survival analysis in the three age groups (≤35, 36–50, and >50 years). Although the association of BIM deletion polymorphism with DMFS and OS did not reach statistical significance in the three age groups, the adjusted HRs were numerically higher in younger patients. In age group ≤35 year, the adjusted HRs were 6.03 for DMFS, and 1.99 for OS (S1 Table). Therefore, the prognostic value of BIM deletion polymorphism in patients with breast cancer warrants to be validated.
Samples used for genotyping in this study were different between healthy volunteers (blood) and cancer patients (tumor). We have analyzed both the wild and deletion alleles with positive and negative controls and did not detect homozygous deletion of BIM gene in any individual sample. In addition, both cancer cohorts consistently showed inverse association of age with BIM deletion polymorphism. Among cancer patients, women ≤50 years had significantly higher frequency of BIM deletion polymorphism than patients aged >50 years (Fig 1). Therefore, the use of different types of samples is unlikely to produce bias of our findings.
In summary, we have discovered a high prevalence of BIM deletion polymorphism in young patients with breast cancer in Taiwan, and this polymorphism may be associated with patients' poor survival. As a potential susceptible genetic factor, BIM deletion polymorphism may interact with their contemporary environmental and lifestyle factors and contribute to the YFBC carcinogenesis in East Asians. Clarification of the underlying mechanisms and interaction between BIM deletion and their contemporary environmental factors is an important step toward mitigating the rapid surge of YFBC in East Asia.
Supporting Information
(XLS)
(XLS)
(XLS)
(DOCX)
Acknowledgments
We thank the members of the Office of Medical Records at National Taiwan University for their help in assessing the clinical data. We thank Dr. S Tiong Ong (from Duke National University of Singapore Graduate Medical School, Singapore) for kindly providing genomic DNA of KCL22 cell line as positive control of BIM deletion polymorphism.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the National Taiwan University, Taiwan (grant number: NTU-ICRP-103R7557, URL: (http://irice-ca.mc.ntu.edu.tw/ aboutICECR.php), and the National Center of Excellence for Clinical Trial and Research, Taiwan (grant DOH102-TD-B-001, URL: (http://www.mohw.gov.tw/CHT/TDU/DM1_P.aspx?f_list_no=740&fod_list_no=0&doc_no=29307). ALC received the grants mentioned above. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sim X, Ali RA, Wedren S, Goh DL, Tan CS, Reilly M, et al. Ethnic differences in the time trend of female breast cancer incidence: Singapore, 1968–2002. BMC Cancer. 2006;6: 261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoo KY, Kim Y, Park SK, Kang D. Lifestyle, genetic susceptibility and future trends of breast cancer in Korea. Asian Pac J Cancer Prev. 2006;7: 679–682. [PubMed] [Google Scholar]
- 3. Matsuno RK, Anderson WF, Yamamoto S, Tsukuma H, Pfeiffer RM, Kobayashi K, et al. Early- and late-onset breast cancer types among women in the United States and Japan. Cancer Epidemiol Biomarkers Prev. 2007;16: 1437–1442. [DOI] [PubMed] [Google Scholar]
- 4. Shen YC, Chang CJ, Hsu C, Cheng CC, Chiu CF, Cheng AL. Significant difference in the trends of female breast cancer incidence between Taiwanese and Caucasian Americans: implications from age-period-cohort analysis. Cancer Epidemiol Biomarkers Prev. 2005;14: 1986–1990. [DOI] [PubMed] [Google Scholar]
- 5. Porter P. "Westernizing" women's risks? Breast cancer in lower-income countries. N Engl J Med. 2008;358: 213–216. 10.1056/NEJMp0708307 [DOI] [PubMed] [Google Scholar]
- 6. Lin CH, Liau JY, Lu YS, Huang CS, Lee WC, Kuo KT, et al. Molecular subtypes of breast cancer emerging in young women in Taiwan: evidence for more than just westernization as a reason for the disease in Asia. Cancer Epidemiol Biomarkers Prev. 2009;18: 1807–1814. 10.1158/1055-9965.EPI-09-0096 [DOI] [PubMed] [Google Scholar]
- 7. Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18: 521–528. 10.1038/nm.2713 [DOI] [PubMed] [Google Scholar]
- 8. Zhao M, Zhang Y, Cai W, Li J, Zhou F, Cheng N, et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. 2014;120: 2299–307. 10.1002/cncr.28725 [DOI] [PubMed] [Google Scholar]
- 9. Isobe K, Hata Y, Tochigi N, Kaburaki K, Kobayashi H, Makino T, et al. Clinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutation. J Thorac Oncol. 2014;9: 483–487. 10.1097/JTO.0000000000000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286: 1735–1738. [DOI] [PubMed] [Google Scholar]
- 12. Yu JC, Hsiung CN, Hsu HM, Bao BY, Chen ST, Hsu GC, et al. Genetic variation in the genome-wide predicted estrogen response element-related sequences is associated with breast cancer development. Breast Cancer Res. 2011;13: R13 10.1186/bcr2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101: 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7: 227–238. [DOI] [PubMed] [Google Scholar]
- 15. Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev Cell. 2007;12: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whelan KA, Caldwell SA, Shahriari KS, Jackson SR, Franchetti LD, Johannes GJ, et al. Hypoxia suppression of Bim and Bmf blocks anoikis and luminal clearing during mammary morphogenesis. Mol Biol Cell. 2010;21: 3829–3837. 10.1091/mbc.E10-04-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, et al. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25: 4591–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woods NT, Yamaguchi H, Lee FY, Bhalla KN, Wang HG. Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res. 2007;67: 10744–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293: 1829–1832. [DOI] [PubMed] [Google Scholar]
- 20. Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5: 733–740. [DOI] [PubMed] [Google Scholar]
- 21. Wang J, Zhou JY, Wu GS. Bim protein degradation contributes to cisplatin resistance. J Biol Chem. 2011;286: 22384–22392. 10.1074/jbc.M111.239566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ajabnoor GM, Crook T, Coley HM. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis. 2012;3: e260 10.1038/cddis.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006;103: 14907–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67: 11867–11875. [DOI] [PubMed] [Google Scholar]
- 27. Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4: e294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rahmani M, Anderson A, Habibi JR, Crabtree TR, Mayo M, Harada H, et al. The BH3-only protein Bim plays a critical role in leukemia cell death triggered by concomitant inhibition of the PI3K/Akt and MEK/ERK1/2 pathways. Blood. 2009;114: 4507–4516. 10.1182/blood-2008-09-177881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wickenden JA, Jin H, Johnson M, Gillings AS, Newson C, Austin M, et al. Colorectal cancer cells with the BRAF(V600E) mutation are addicted to the ERK1/2 pathway for growth factor-independent survival and repression of BIM. Oncogene. 2008;27: 7150–7161. 10.1038/onc.2008.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118: 3651–3659. 10.1172/JCI35437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, et al. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97: 1746–1759. [DOI] [PubMed] [Google Scholar]
- 32. Tanizaki J, Okamoto I, Fumita S, Okamoto W, Nishio K, Nakagawa K. Roles of BIM induction and survivin downregulation in lapatinib-induced apoptosis in breast cancer cells with HER2 amplification. Oncogene. 2011;30: 4097–4106. 10.1038/onc.2011.111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.