Abstract
Innate lymphoid cells (ILCs) are increasingly recognised as an innate immune counterpart of adaptive TH cells. In addition to their similar effector cytokine production, there is a strong parallel between the transcription factors that control the differentiation of TH1, TH2 and TH17 cells and ILC Groups 1, 2 and 3, respectively. Here, we review the transcriptional circuit that specifies the development of a common ILC progenitor and its subsequent programming into distinct ILC groups. Notch, GATA-3, Nfil3 and Id2 are identified as early factors that suppress B and T cell potentials and are turned on in favour of ILC commitment. Natural killer cells, which are the cytotoxic ILCs, develop along a pathway distinct from the rest of the helper-like ILCs that are derived from a common progenitor to all helper-like innate lymphoid cells (CHILPs). PLZF− CHILPs give rise to lymphoid tissue inducer cells while PLZF+ CHILPs have multi-lineage potential and could give rise to ILCs 1, 2 and 3. Such lineage specificity is dictated by the controlled expression of T-bet, RORα, RORγt and AHR. In addition to the type of transcription factors, the developmental stages at which these factors are expressed are crucial in specifying the fate of the ILCs.
Introduction
Transcriptional programming of immune cell fate and lineage specificity is essential for the commitment and development of the hematopoietic system1-3. The recent discovery of innate lymphoid cells (ILCs) has sparked an intriguing question relating to their ontogeny – ie. where do these cells come from? The ILCs are characterised by their lymphoid origin and hence their requirement for the common cytokine receptor gamma chain4. Like other innate immune cells, the ILCs lack somatically rearranged antigen-specific receptors and can respond rapidly to stimuli. However, the ILCs mediate their immune effector functions through the secretion of key effector cytokines that were previously primarily associated with a T helper cell (TH) response. Three groups of ILCs have been assigned. Group 1 ILCs (ILC1s) are defined by their production of the signature type 1 cytokine interferon gamma (IFNγ), Group 2 ILCs (ILC2s) produce the type 2 cytokines interleukin 4 (IL-4), IL-5 and/or IL-13, and Group 3 ILCs (ILC3s) produce the TH17-associated cytokines IL-17 and/or IL-224. The ILCs include the previously discovered natural killer cells (NK)5,6 and lymphoid tissue inducer cells (LTi)7,8 and these cells are now reclassified as Group 1 and 3 ILCs, respectively4. Importantly, functionally equivalent populations of human ILCs have been identified4,9-11.
ILCs have been implicated in immune protective functions and tissue homeostasis, but their release of potent pro-inflammatory cytokines has also been shown to contribute to inflammatory conditions such as allergic asthma and inflammatory bowel diseases (IBD)10,11. It is noteworthy that genes required for ILC2 growth and differentiation have been associated with differences in asthma severity in large-scale genome wide association studies12,13. ILC3s in mice were first linked to colitis14 but subsequent studies have implicated human Group 1 ILC- and Group 3 ILC-like cells in Crohn’s disease as well15,16. ILC3s are IL-23-responsive cells, and the reported association between polymorphism in the IL-23 receptor with IBD re-affirms the pathological role of ILC3s in IBD17. ILC2 and NCR+ ILC3 have also been recently implicated in atopic dermatitis and psoriasis, respectively, after these cells were shown to accumulate in the skin lesion of these patients18,19.
With the discovery of the ILCs, immune functions and pathologies once assumed to be TH cell-dependent are now being revisited to determine ILC involvement and this may allow development of more targeted therapies tailored to the ILCs. Understanding the cues for ILC development has therefore become a focus of interest and major advances have been made within a relatively short period of time. Reviews on the biology of ILCs and its cytokine effector functions have been published elsewhere4,10,11. This review will thus focus on the developmental programming of the ILCs and is aimed at consolidating current information on known transcription factors that regulate the development of a common ILC progenitor and its subsequent differentiation into the distinct ILC groups. We will begin with an overview of the development of the three ILC groups, followed by a discussion of some key transcription factors that are required for the functional differentiation/maturation of ILCs.
Development of the different ILC groups
A common ILC progenitor?
The notion of a common ILC progenitor arose from various early observations that the deletion of the transcription factor inhibitor of DNA binding 2 (Id2) resulted in the ablation of all recognised ILC groups3,20-22, suggesting that all the ILCs are developmentally related. Significant progress towards our understanding of the relatedness of the ILCs was made with the description of an Id2+ progenitor that was termed the common progenitor to all helper-like ILCs (CHILPs). CHILPs have multi-ILC lineage potential, and with the exception of NK cells, CHILPs give rise to members of all three ILC groups following adoptive transfer4,23 (Figure 1). Transcription factor profiling of the CHILP revealed that it was made up of heterogenous populations of cells and could be bisected into those that expressed the transcription factor promyelocytic leukaemia zinc finger (PLZF+), and those that did not (PLZF−). While all the ILCs could arise from the PLZF+ population, LTi cells appears to be derived from the PLZF− precursor24 (Figure 1). This showed that all the other ILCs are more closely related to each other than to LTi cells and even less so to NK cells. However, the heterogeneity of the CHILPs suggests that greater refinement is required to discriminate the ILC progenitors. Indeed, the involvement of GATA-34,25 and Nfil37,8,26 at these stages of commitment and the regulation of the downstream ILCs 1, 2 and 3 defining factors remains to be defined fully.
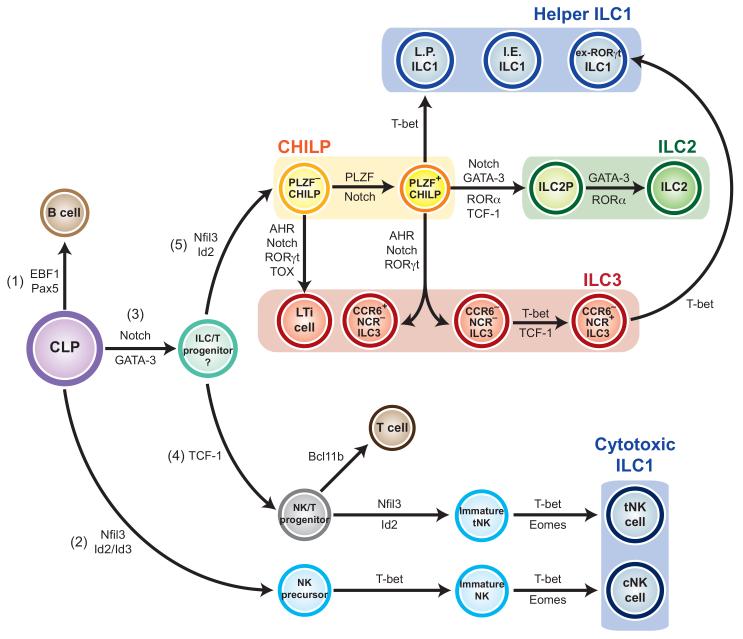
Figure 1. Schematic of the proposed transcriptional circuit regulating ILC development.
ILC1s are grouped in blue and are divided into the helper ILC1s and cytotoxic ILC1s (NK cells), ILC2s are grouped in green, ILC3s in red and CHILPs in yellow. I.E. ILC1 = intraepithelial ILC1; L.P. ILC1 = lamina propria ILC1.
B cells, T cells and all ILCs are derived from a multipotent CLP, with the decision to differentiate into any of these immune cell types dependent on the transcription factor that is turned on. Expression of EBF1 and Pax5 for example allows differentiation into a B cell (1). Expression of Nfil3 and Id2/Id3 (2) leads to an NK precursor that then progresses onto a mature cNK cell via a pathway that requires T-bet and Eomes. If Notch and GATA-3 are switched on instead (3), a multipotent ILC/T progenitor may be generated. Expression of TCF-1 by this progenitor (4) leads to a more restricted NK/T progenitor that continues to develop in the thymus. If Bcl11b is switched on, a T cell is generated, but sequential expression of Nfil3, Id2, T-bet and Eomes gives rise to tNK cells instead. ILC/T progenitors that otherwise express Nfil3 and Id2 become CHILPs (5). PLZF− CHILPs develop into LTi cells, while PLZF+ CHILPs give rise to the remaining helper ILC1s, ILC2s and NCR+/− ILC3s via the expression of lineage-specific transcription factors. These are T-bet for the ILC1s, GATA-3 and RORα for ILC2s and AHR and RORγt for ILC3s. CCR6− NCR+ ILC3s demonstrate plasticity and can further differentiate to become an ILC1 by expressing T-bet. The origin of the I.E. ILC1 has not been determined.
Group 1 ILCs (ILC1s)
Group 1 ILCs (ILC1s) are defined by their production of IFNγ and their requirement for the T-box transcription factor T-bet (Tbx21)4,9-11. To date, members of the group include the conventional NK (cNK) and thymic NK (tNK) cells10, ex-ILC3s (described later) that have developed the ability to secrete IFNγ and express T-bet 27-29, intestinal and tonsillar intraepithelial ILC130 and a recently identified subset of lamina propria resident ILC123. Although T-bet is considered the signature transcription factor of the ILC1s, NK cells are also dependent on another T-box transcription factor Eomes for their terminal maturation31 and Eomes is also expressed by intraepithelial ILC1s30.
All the identified ILC1s produce IFNγ in response to the pro-inflammatory cytokine IL-1223,29,30,32,33, but a combination of IL-12 and IL-18 has been suggested to act synergistically on NK cells32. With the exception of the intraepithelial ILC1s30, the development of all the other ILC1s are dependent on IL-1523,29,34-36. In fact, IL-15, IL12 and IL-18 induce T-bet expression37 and in the case of the ILC3-derived ILC1s, IL-15 and IL-12 are important for the downregulation of the ILC3-defining transcription factor RORγt29 (described later) in order for these cells to acquire an ILC1 phenotype.
NK cells posses potent cytolytic activity towards virally-infected cells and tumours through their production of perforin and granzymes38. They are activated via surface natural cytotoxicity receptors (NCR), which are important for both tumour 39 and viral antigen recognition40. While NKp44 (NCR2) is only present in human, NKp46 (NCR1) is conserved in human and mice41,42. Similar to NK cells, the remaining members of the Group 1 ILCs also express NCRs but confer immune protection against non-viral intracellular pathogens such as Salmonella enterica28 and Toxoplasma gondii23, although they have also been implicated in immune pathology such as colitis28-30. While intraepithelial ILC1s express perforin and granzymes which is suggestive of their potential cytotoxicity30, lamina propria ILC1s23 and ex-ILC3s27,29 do not and these latter cells are considered helper-like ILCs rather than cytotoxic ILCs.
Of all members of the Group 1 ILCs, the developmental pathways of the NK cells are best characterised. The bone marrow is the primary site of NK cell development at steady state in the adult human and mice, but the liver43,44, thymus33,45 and lymph nodes46,47 also serve as sites of NK-poiesis. However, it has been suggested that NK cell development in the bone marrow differs from that in the liver – both in the temporal acquisition of the mature NK markers NKp46 and DX548, and their dependence on the transcription factor Eomes31.
In mice, mature cNK cells develop from a bone marrow-derived common lymphoid progenitor (CLP) that progressively undergoes three major stages of development, each characterised by a sequential change in the expression of distinct cell surface markers2,49,50. The expression of surface NK1.1 and DX5 are commonly used to distinguish these different intermediary stages51-54. The earliest progenitor committed to the NK lineage is a CD122+ NK precursor (NK1.1− DX5−), which then progresses through an immature NK cell stage (NK1.1+ DX5−) before becoming a mature NK cell (NK1.1+ DX5+). In human, cNK cells develop from a CD34+ hematopoietic progenitor that undergoes four major stages of development characterised by variable expression of the surface markers CD34, CD94 and CD11755. Mature human cNK cells are then further divided into two subsets based on their levels of CD56 expression56.
In contrast, tNK cells are derived from a bipotent NK/T cell progenitor found in the thymus of humans and mice57,58. In mice, tNK cells are distinguished from the BM-derived cNK based largely on the expression of CD12733 but T cell receptor gamma (TCRγ) rearrangement was also suggested as a unique marker of these cells59. The decision of thymocytes to commit into either a T or NK cell is dependent on the expression of the transcription factor Bcl11b at the double negative 2a (DN2a) stage (Figure 1). Bcl11b was shown to promote the expression of a panel of genes associated with T cell development, but repress NK-cell associated genes such as Id2, Nfil3, Tbx21 and Eomes, leading to a commitment to the T cell lineage. Consistent with this, Bcl11b-deficient thymocytes beyond the DN2 stage acquire NK-like properties with a concomitant loss of T cell features60,61. Following their development, NK cells migrate to seed other anatomical sites including the lung, liver, peripheral blood and secondary lymphoid organs, although the contribution of NK cells of different origin to these sites may differ33,62.
The development of other ILC1s is distinct from the NK cells. For example, a subset of ILC1 are derived from the re-differentiation of an ILC3 and were defined by being RORγt fate-map positive27,29. Intraepithelial ILC1 and lamina propria ILC1 are RORγt fate-map negative23,30, indicating that they are not of an ILC3 origin. The lamina propria ILC1s develop directly from the CHILPs23. However, CHILPs did not give rise to Eomes+ progeny23 and hence may not be the progenitor to intraepithelial ILC1. Both intraepithelial ILC1 and NK cells are T-bet+ Eomes+, suggesting that they are more closely related, but unlike the NK cells, intraepithelial ILC1 are independent of IL-15 signalling30. The developmental pathway for this unique class of ILC1 remains to be fully determined.
Group 2 ILCs (ILC2s)
Members of the Group 2 ILCs (ILC2s) were discovered almost concurrently by three independent research groups and were initially termed natural helper cells (NHC)20, nuocytes63 and innate type 2 helper cells (Ih2)64. These cells were first reported within the fat-associated lymphoid clusters (FALC), mesenteric lymph nodes, intestines, spleen and liver20,63,64, but were later shown to reside within the lung65,66 and skin18,19 as well. The different ILC2s show variation in their surface markers and cytokine secretion, but these variations may simply reflect their different anatomical location or activation state.
The ILC2s are dependent on IL-7 for development and in response to IL-25, IL-33, thymic stromal lymphopoietin (TSLP) and TL1A, they secrete primarily IL-5 and IL-13, although their secretion of IL-4 and IL-6 have also been reported20,63,64,66-68. Interestingly, in addition to these type 2 cytokines, ILC2s also produce the growth factor amphiregulin69. ILC2s are indispensable for conferring immune defence against helminth infection63,64 and through the release of amphiregulin, ILC2s promote tissue remodelling following influenza virus-induced airway damage69. However, ILC2s also mediate type 2 immune pathology and have been shown to be the key players in viral-70-72 and allergen-induced airway inflammation65,66. Functionally equivalent ILC2s have also been identified in the human lung69 and gut and were enriched in the nasal polyps of patients with chronic rhinosinusitis73.
ILC2s could be derived from the PLZF+ fraction of CHILPs23,24 and require the transcription factors GATA-367 and RORα74,75 for their commitment. A GATA-3high ILC2 precursor (ILC2P) that has a transcriptional profile similar to that of ILC2s but lacks surface expression of KLRG1 that is found on mature ILC2s has also been identified67. Conditional deletion of GATA-3 in Id2+ cells demonstrated the requirement of GATA-3 for ILC2 development67. In addition to GATA-3, ILC2s are also dependent on the transcription factor retinoic acid receptor-related orphan nuclear receptor alpha (RORα)74,75. RORα-deficient mice lack ILC2s and bone marrow progenitors from these mice fail to develop into ILC2 both in vivo and in vitro. Although GATA-3high ILC2P has been reported to express lower levels of RORα compared to mature ILC2s67, possibly suggesting that RORα is required for the maturation of ILC2s from an ILC2P stage, experimental evidence is still required for the relationship of RORα and GATA-3 in ILC2 commitment.
GATA-3 also drives human ILC2 development and function, and human ILC2s similarly express high amounts of RORα76. Thus, the transcription factors GATA-3 and RORα can be used to define the mature ILC2s. However, it was shown recently that early ablation of GATA-3 is detrimental to the development of all ILC groups as well25, indicating that GATA-3 is required for the development of the CHILPs.
Group 3 ILCs (ILC3s)
Group 3 ILCs (ILC3s) represent the most developmentally diverse group of ILCs. The hallmarks of the ILC3s are their requirement for the transcription factor ROR gamma t (RORγt) and their release of the TH17-associated cytokines IL-17 and/or IL-22 upon IL-23 and IL-1β stimulation14,77-81. In fact, ILC3s are the main producers of host-protective IL-17 following Candida albicans infection82 and very recently, both ILC2s and ILC3s were shown to be expanded in the peripheral blood of filarial-infected patients83. Similar to the ILC2s, ILC3s express IL-7Rα and are dependent on IL-7 for their development14,21,78-80,84-86. Members of the Group 3 ILCs include the lymphoid tissue inducer (LTi) cells, and two subsets of ILC3s distinguished by their expression of NCR – the NCR+ ILC3s and NCR− ILC3s.
As their name suggests, LTi cells are the key drivers of secondary lymphoid organogenesis throughout life: in fetal development of lymph nodes and Peyer’s patches7,8, postnatal development of cryptopatches and isolated lymphoid follicles87,88, and in adults, for maintaining the integrity of the secondary lymphoid organs89. LTi cells mediate such functions by expressing surface lymphotoxin LTα1β2 that engage and activate lymphoid stromal cells expressing the corresponding LTβ receptor90. In addition to these established roles, LTi cells were later discovered to contribute to immune defence. CD4+ and CD4− LTi-like cells were shown to secrete IL-17 and IL-22 upon IL-2381 and IL-1β stimulation77 and mediate the expulsion of the intestinal bacterial pathogen Citrobacter rodentium by providing an early source of the effector cytokine IL-2291. Human LTi cells that similarly express LTα and LTβ and produce IL-17 and IL-22 have also been discovered92.
This area of cytokine producing innate lymphoid cells underwent a rapid burst of expansion with concurrent reports of the identification RORγt-dependent ILCs that are now collectively referred to as NCR+ ILC3s78-80. The NCR+ ILC3s reside in the lamina propria of mice and were identified as RORγt+ NKp46+ NK1.1−/lo cells that produce only IL-22. Equivalent IL-22 producers were also discovered in human tonsils, Peyer’s patches and uteri84,93-95. In earlier reports, the NCR+ ILC3s were referred to as NCR-2221, NK-2284,93, NKR-LTi29 and ILC22 77,96 cells. IL-22 has previously been shown to mediate the expulsion of the attaching and effacing pathogen C.rodentium by triggering the release of anti-microbial peptides97. Thus, similar to the LTi cells, mouse NCR+ ILC3s were identified to be important sources of IL-22 for defence against C.rodentium infection80. Notably, although NCR+ ILC3s express NKp46, they do not exhibit the cytotoxicity characteristic of the NK cells78-80.
In addition to the NCR+ ILC3s, NCR− ILC3s were also described. This innate population of CD4− LTi-like cells accumulate in the inflamed intestines of Helicobacter hepaticus-infected mice. In addition to IL-17 and IL-22, these cells secrete IFNγ and express the transcription factor T-bet14. Like the LTi cells, these innate CD4− cells express CCR6, but not the NKp46 NCR that is associated with NK cells despite exhibiting an NK-like phenotype. To distinguish them from the LTi and NK cells, they were initially referred to as NCR− ILC3s4. However, to be able to specifically distinguish this subset from the other NCR− ILC3s (discussed below), we refer to them here as CCR6+ NCR− ILC3s (Figure 1).
The many similarities between NCR+ ILC3s and LTi cells led to initial speculation that LTi cells were precursors to NCR+ ILC3s29,98. However, more elaborate analysis in mice suggested that NKp46− RORγt+ cells that were thought to be representative of the LTi cells reported in earlier studies actually consisted of a heterogenous mix of cell types. Using CCR6, CD4 and NKp46 surface staining, Sawa et al.99 and Klose et al.28 described a more comprehensive subgrouping of the RORγt+ cells where the cells were first divided into two main fractions based on CCR6 expression. The CCR6+ fraction included the classical CD4+ LTi cells (LTi4) and CD4− LTi cells (LTi0) both of which were NKp46−, while the CCR6− fraction was invariably CD4− but could be further divided into NKp46+ and NKp46− cells. By adoptive transfer, Klose et al. demonstrated that intestinal NCR+ ILC3s arise from a CCR6− CD4− NKp46− precursor, but not from CCR6+ CD4+/− NKp46− LTi cells28. Similar observations were also confirmed in vitro where cultures of LTi4 and LTi0 cells failed to give rise to NKp46+ cells99. The CCR6− precursors to NCR+ ILC3s were also not derived from CCR6+ LTi cells28. Moreover, depletion of CD4+ cells led to a reduction in only the numbers of CD4+ LTi cells but not NCR+ ILC3s, indicating that NCR+ ILC3s originated from a CD4− parent99. This was confirmed by Rankin et al. who reported that only cultures of CD4− NKp46− cells differentiated into CD4− NKp46+ progenies100. However, in the absence of CCR6 staining, the population of CD4− NKp46− precursors identified by Rankin et al. would have included the CCR6− cells described by Klose et al. Taken together, all these findings suggest that in mice, NCR+ ILC3s are developmentally distinct from the LTi cells (Figure 1).
The acquisition of NKp46 by the NCR+ ILC3s is accompanied by their expression of the ILC1-defining transcription factor T-bet. In fact, following the expression of T-bet, NCR+ ILC3s develop the ability to secrete IFNγ and they downregulate the expression of RORγt over time28,29. Such progressive change in functional ability and transcriptional control marks the transition of the cell from an ILC3 to an ILC1 and eventually gives rise to a population of ILC1 that is identified as RORγt-fate-map positive29, as described earlier. To date, this functional plasticity has only been observed in the NCR+ ILC3s.
The CHILP progenitors of ILCs are characterised as being Lineage− Id2+ α4β7+ RORγt− cells23. Prior to the discovery of CHILPs, a population of α4β7+ RORγ+ progenitors downstream of the CLP was already shown to generate LTi4 and LTi0 cells, while it was proposed that α4β7− RORγt+ progenitors generated the NCR+ ILC3s99,101. However, ablation of Id2 in mice resulted in marked reduction in the frequency of the α4β7+ RORγt+ cells and hence LTi cells, but did not perturb the frequency of α4β7− RORγt+ cells although a severe reduction in NCR+ ILC3s was observed21,101. Thus, Id2 is required for the development of α4β7+ RORγt+ cells and the subsequent generation of NCR+ ILC3s, but not for the generation of α4β7− RORγt+ precursors. These data also highlighted that the α4β7− RORγt+ cells were not derived from α4β7+ RORγt+ cells, and may represent an independent branching from the CLP.
Common transcription factors for multiple ILC lineages
In the last section, we have provided an overview of the progenitor-progeny relationship between members of the different ILC groups. Here, we discuss the transcription factors that are needed to programme the CLP into a common ILC precursor and the lineage-defining factors that then induce differentiation into the various ILC branches.
Id2 (Inhibitor of DNA binding 2)
All ILC groups are dependent on the transcription factor Id2 for development20-22,52. Id2 is highly expressed in NK precursors (NKPs)102,103 and CHILPs23 but not in hematopoietic stem cells (HSCs) and CLPs67, indicating that the expression of Id2 downstream of the CLP is needed for subsequent ILC development (Figure 1). In addition to the ILCs, Id2 has also been implicated for the development of dendritic cells104,105.
The earliest evidence for the importance of Id2 was demonstrated by Id2-deficient mice having significantly reduced NK and LTi cell populations. As a consequence, these mice failed to develop lymph nodes and Peyer’s patches22. Subsequent studies then found that Id2−/− mice also lacked ILC2s20 and NCR+ ILC3s21. While the loss of ILC2s and 3s could be attributed to the loss of CHILPs, loss of NK cells was not due to failure to generate the NKPs. Indeed, Id2-deficient mice have normal proportions of NKPs and immature NK (iNK) cells despite a significant reduction in mature NK (mNK) cells52. This ability to generate NKPs in an Id2−/− background was attributed to the compensatory role of Id3, another member of the Id family of transcription factors. The expression patterns of Id2 and Id3 were found to be inversely correlated. While Id3 is highly expressed in CLPs and NKPs, the expression of Id2 is low in CLPs but increases significantly in NKPs and mNK cells. The expression of Id3 was also found to be doubled in Id2-deficient NKPs52. Nevertheless, despite the compensatory role of Id3 in early NKP development, Id2 appears to be indispensable for subsequent NK maturation. Ectopic expression of Id3 was also shown to drive the development of human NK cells from a CD34+ bipotent NK/T cell progenitor. This was accompanied by a concomitant block in TCR gene rearrangement and a loss of T cell potential106.
Id2 acts by sequestering the E box protein transcription factors, thus preventing their binding to DNA and the induction of E protein target genes107. Deletion of one of the major E proteins E2A in Id2-deficient mice restored both NK and LTi cell numbers and lymph node and Peyer’s patch development52. E2A proteins are essential for B cell development by inducing the B cell-defining transcription factor Pax5108,109 and are crucial for early thymocyte commitment to become T cell precursors110. Taken together, Id2 appears to drive ILC development by suppressing intrinsic B and T cell lineage potentials to allow for the expression of ILC-specific factors.
GATA-3 (GATA binding protein 3)
The role of the zinc finger transcription factor GATA-3 in immunity was first implicated in T cells. GATA-3 is essential for thymocytes to develop beyond the earliest double negative 1 (DN1) CD4− CD8− stage111, and in a mature T cell, GATA-3 is the key driver of TH2 differentiation and to induce the expression of the type 2 cytokines IL-4, IL-5 and IL-13112.
Consistent with its TH2-associated role, GATA-3 was initially reported as an ILC2 lineage-defining transcription factor67. However, GATA-3 also regulates NK cell function. Although GATA-3 deficiency did not affect the frequency of total CD3− NK1.1+ NK cells, the resulting Gata3−/− NK cells displayed an immature phenotype and were characterised by lower T-bet expression and defective IFNγ production in response to IL-12 and/or IL-18 stimulation113. Indeed, collective data derived from various GATA-3 deletion models suggest that GATA-3 plays niche roles at various stages of ILC development and that these differ between ILC groups. Deletion of GATA-3 from all hematopoietic cells using Gata3fl/fl-Vav-Cre mice resulted in the most widespread effect on the ILCs. These mice exhibited a marked reduction in all CD127+ ILCs including thymic NK cells (ILC1), ILC2s and RORγt+ ILC3s, but not CD127− cNK cells25. A reduction in RORγt+ ILC3s was also observed following the adoptive transfer of GATA-3-deficient hematopoietic precursors into irradiated recipient mice, and the residual Gata3−/− ILC3s that are formed in these mice failed to produce IL-22 upon IL-23 stimulation114. This suggest that ILC3s require GATA-3 for both early development and for effector function. However, in another deletion model, GATA-3 ablation in Id2-expressing cells markedly reduced the size of the ILC2 pool but spared the RORγt+ ILC3s67, suggesting that unlike the ILC2s, RORγt+ ILC3s no longer require GATA-3 for development at the stage where Id2 is turned on. In addition, when GATA-3 was deleted in NKp46-expressing cells, only lamina propria resident ILC1 were reduced in numbers but the cNK cell and NCR+ ILC3 pools remained intact23.
These findings highlight that: (1) GATA-3 is dispensable for cNK cell commitment, and more importantly, (2) GATA-3 is crucial for the generation of a common ILC progenitor and appears to be needed upstream of Id2 (Figure 1). Once a GATA-3-dependent ILC progenitor is formed from the CLP, Id2 becomes crucial for the maintenance of this ILC progenitor while the role of GATA-3 becomes dispensable until needed again for the development of the lamina propria ILC1 and ILC2, or for the proper functioning of the ILC3s. Thus, the need for GATA-3 occurs in two waves; first during early progenitor development, and later during ILC differentiation. The GATA-3-dependent progenitor may represent a multipotent ILC/T cell progenitor, with the decision to become an ILC or a T cell dependent on the transcriptional programming that follows. Notch, GATA-3 and T cell factor 1 (TCF-1) are the three transcription factors that are crucial in the early stages of T cell commitment115. As will be discussed in the next section, Notch signalling is similarly required for early ILC development. Thus, it seems likely that Notch and GATA-3 may give rise to an ILC/T cell progenitor but while TCF-1 would then specify a T cell lineage, Id2 specifies an ILC lineage.
Notch signalling
Unlike most of the other transcription factors, Notch signalling is not cell intrinsic but is dependent on the engagement of the Notch receptors with extracellular Notch ligands expressed by neighbouring cells. In vertebrates, four Notch receptors (Notch 1, 2, 3 and 4,) and five Notch ligands (Delta-like (DL)-1, -3 and -4 and Jagged-1 and -2) have been identified116.
Notch signalling is essential for silencing the early B cell factors, EBF1 and Pax5, allowing commitment to a T cell fate3. In vitro cultures of CLPs in the absence of Notch ligands generates mostly B cells, but in the presence of Notch ligands, T cells, ILC2s and ILC3s could be derived75,101,117. These observations suggest that Notch acts in parallel with GATA-3, being required for facilitating ILC- and T-potential and repressing the B cell programme (Figure 1). However, Notch signalling is dispensable for cNK96,101 and tNK118 cell development and indeed, CLPs cultured in the absence of Notch ligands develop NK potential101,117. These data suggest that cNK cells may have already branched from the other ILC lineages early in development, and that a subpopulation of tNK cells may also develop independently of a T/NK progenitor.
Notch signalling is crucial for the development of ILC2s75,119, NCR+ ILC3s96,100 and LTi cells96. As mentioned previously, the development of NCR+ ILC3s is accompanied by T-bet expression, which is dependent on the presence of Notch100. Nevertheless, the overall role of Notch signalling in LTi cells appears less straightforward. Notch signalling was reported to be crucial for the generation of an α4β7+ RORγt− LTi progenitor from fetal liver CLPs in vitro, but continued Notch engagement retarded the development of mature LTi cells101. However, LTi cells were reduced following the ablation of Notch effector protein RBP-Jκ in mice96, indicating that Notch is needed for LTi cell development. This discrepancy between the need for Notch in the fetal and adult stages may be related to LTi cell’s requirement for aryl hydrocarbon receptor (AHR), a key transcription factor that is required for ILC3 development. Notch1 has been identified as a target gene of AHR96 and fetal LTi cells are AHR-independent, and therefore Notch-independent, while postnatal LTi cells are AHR- and Notch – dependent96,120,121. Such change in the requirement for Notch was also consistent with another study that showed that Notch signalling was important only for the development of RORγt+ ILC3s from adult bone marrow-derived CLPs but not from fetal liver CLPs117. These different requirements for Notch may suggest an adaptation of the cells for the differential expression of Notch ligands in the adult and fetal hematopoietic microenvironments.
TCF-1 (T cell factor 1)
TCF-1 is induced by Notch signalling and is required for T cell development3. TCF-1 is encoded by the Tcf7 gene and is highly expressed in early thymocytes, NK cells, ILC2s and both NCR+ and NCR− ILC3s122. However, Tcf7−/− mice do not show impairment in the frequency of NK cells or NCR− ILC3s, but they lack ILC2s and NCR+ ILC3s119,122. As a result of the loss of ILC2s, Tcf7−/− mice mount a delayed immune response to helminth infection119 and allergen-induced airway challenge119,122. GATA-3 and RORα expression was also decreased in heterozygous Tcf7+/− mice, suggesting that TCF-1 may be required for their expression122. Indeed, overexpression of TCF-1 upregulated the expression of GATA-3, and GATA-3 was required for TCF-1-mediated upregulation of ILC2-associated genes such as Il17rb and Il2ra119.
Despite the high levels of Tcf7 expression in NCR− ILC3s, this pool of cells developed normally in TCF-1-deficient mice. This may be indicative of the role of TCF-1 in inducing the transition of NCR− ILC3s to NCR+ ILC3s, consistent with the loss of the latter population in Tcf7−/− mice122. The diminished numbers of NCR+ ILC3s resulted in increased susceptibility to C.rodentium infection. Thus, while TCF-1 is important to switch off ILC potential early in development, it is required again during ILC2 and NCR+ ILC3 differentiation.
Nfil3 (Nuclear factor interleukin-3)
Nfil3, known also as E4-binding protein 4 (E4bp4), is essential for NK cell development. Nfil3−/− mice develop normal frequencies of NKPs, but have decreased iNK cells and almost undetectable mNK cells53,54 and tNK cells103. However, the NKP cells in these early reports were defined according to an initial definition of Lin− CD3− CD122+ NK1.1− DX5− cells that were enriched for, but did not exclusively contain NKP50. Using a more refined definition of NKP proposed by Fathman et al.123 which segregates the NKPs into pre-NKP (pNKP) and refined NKP (rNKP) based on distinct surface markers, Male et al. showed that deletion of Nfil3 resulted in a significant reduction of pNKP and rNKP cells124. Consistent with this, deletion of Nfil3 also resulted in decreased proportions of Id2+ NKP, demonstrating that Nfil3 is required at the NKP stage103.
The arrest of the majority of NK cells at the immature stage in Nfil3−/− mice may be related to a reduced expression of Eomes which is crucial for NK cell maturation31. Nfil3−/− bone marrow progenitor cells had lower expression of Id2 and Eomes and ectopic expression of Id2 and Eomes in Nfil3−/− cells were shown to overcome the need for Nfil3 and restored NK cell development53,95,103. Nfil3 binding sites have been found both in the transcriptional regulatory region of the Id2 and Eomes genes124. However, in contrast to the Nfil3−/− bone marrow hematopoietic progenitors, residual NK cells that can develop in an Nfil3-deficient background expressed Id2 at comparable levels with wild type NK cells. Thus, Id2 expression may be sustained through Nfil3-independent means following NK lineage commitment. Apart from Id2 and Eomes, lower GATA-3 expression was also observed in Nfil3−/− hematopoietic progenitor cells53. However, overexpression of GATA-3 into Nfil3−/− bone marrow cells did not rescue NK cell development124.
Growing evidence now indicates that Nfil3 contibutes to all ILC groups. Indeed, intraepithelial ILC1, lamina propria ILC1, ILC2s, LTi cells and both the NCR+ and NCR− ILC3s are all dependent on Nfil323,26,30,125. Consequently, Nfil3−/− mice display defective formation of Peyer’s patches, increased susceptibility to C.rodentium infection and an inability to mediate allergen-induced eosinophil migration to the lung 26,125. Thus, like Id2, Nfil3 is important for the generation of a common ILC precursor and as T cells and B cells develop normally in Nfil3-deficient mice26,53,54, Nfil3 appears to be downstream of the GATA-3, but upstream of Id2.
PLZF (Promyelocytic leukaemia zinc finger)
The dependence of ILCs on the NK T cell factor PLZF (Zbtb16)126 was initially discovered when several ILCs were found to be fate-mapped in PLZF fate-mapping reporter mice24. This led to the identification of a PLZF+ Lineage− IL-7Rα+ α4β7+ ILC progenitor within the fetal liver and bone marrow. In in vitro cultures, PLZF− ILC progenitors developed into PLZF+ cells in the presence of Notch signalling. Both adoptively transferred, and in vitro cultured PLZF+ progenitors gave rise to cells that phenotypically resembled ILCs 1, 2 and 3, but not cNK, LTi, T and B cells24, demonstrating for the first time that LTi cells branched from a distinct PLZF− progenitor. Indeed, the PLZF− progenitor expressed the transcription factor TOX that has already been implicated in LTi cell development24,51.
PLZF+ and PLZF− progenitors appear to make up the population of CHILPs. CHILPs were characterised by the expression of surface markers that overlapped with the PLZF+/− progenitors such as IL-7Rα, α4β7 and cKit, but more importantly, flow cytometry analysis of the CHILPs demonstrated that it consisted of PLZF+ and PLZF− subsets of cells23,24 (Figure 1). It remains to be determined whether the PLZF+ CHILPs have multi-lineage potential or whether heterogeneity exists within the PLZF+ CHILPs for specific ILC1, 2 and 3 precursors.
ILC lineage-defining transcription factors
T-bet (T-box expressed in T cells) and Eomes (Eomesodermin)
T-bet was first identified as a TH1 cell commitment factor127 but all known Group 1 ILC1s are also dependent on T-bet for their development23,28,30,31,37,100, making T-bet the defining transcription factor of the ILC1s.
In NK cells, T-bet acts in conjunction with another T-box transcription factor Eomes to promote both cell maturation and function 31,103. Consistent with the IL-15-dependence and IL-12-responsive nature of NK cells, both these cytokines were shown to induce the expression of T-bet37, and T-bet in turn promotes IFNγ expression127. Thus, T-bet-deficient NK cells are defective in carrying out immune effector functions such as IFNγ secretion and cytolysis of target cells37. A double-deficiency in Eomes and T-bet resulted in the absence of NK cells, but the NKPs could develop at frequencies comparable to wild type mice31. This shows that T-bet and Eomes are dispensable for NKP formation and but are needed for stable NK cell commitment beyond this stage. However, while Eomes-deficient mice could give rise to iNK cells, T-bet-deficient mice could not, and in fact, T-bet deficiency led to accelerated maturation of the NK cells. This was thought to be due to the increased expression of Eomes in the residual Tbx21−/− NK cells31, and suggested that T-bet is needed at an earlier developmental stage than Eomes (Figure 1). Besides NK cells, Eomes is also found to be expressed at high levels by the intraepithelial ILC1 but the role of Eomes in its development has not been examined30.
The expression of T-bet provides the drive for the differentiation of CCR6− NCR− RORγt+ ILC3s into NCR+ ILC3s and subsequently into the ex-RORγt ILC128,100. This expression of T-bet is accompanied by the expression of NKp46, the ability to secrete IFNγ and the downregulation of RORγt28. The ablation of T-bet in Tbx21−/− Rag2−/− ulcerative colitis (TRUC) mice resulted in the development of intestinal ILC3s that produce less IFNγ, but were instead poised to produce more IL-17A that exacerbates the development of colitis128. This highlights the importance of T-bet as an important regulator that maintains the balance of intestinal homeostasis. It has been suggested that gut microbiota may induce T-bet in the NCR− ILC3s precursors, since germ-free mice have lower proportions of T-bet+ NCR− RORγt+ ILC3s compared to specific pathogen-free mice28.
RORα (retinoic acid receptor-related orphan nuclear receptor alpha)
The importance of RORα in ILC2 development was demonstrated using staggerer RORαsg/sg mice that have a spontaneous deletion in RORα129. RORαsg/sg mice have low numbers of ILC2s and residual ILC2s in these mice fail to expand in vivo in response to IL-25 administration74,75. RORαsg/sg bone marrow progenitors were also impaired in their development of ILC2s, thus confirming their dependence on RORα75. RORα-deficient mice, or recipient mice transplanted with RORαsg/sg bone marrow were defective in helminthic expulsion and suffer from greater worm burden compared to wild type mice75,130.
NK cells and RORγt+ ILC3s also express notable, but lower levels of RORα compared to the ILC2s67. However, RORα-deficient mice develop similar frequencies of NK cells and RORγt+ ILC3s74, probably due to the redundant role of RORα in these cells.
RORγt (retinoic acid receptor-related orphan nuclear receptor gamma t)
The developmental requirement for RORγt in ILC3s was first observed in LTi cells131-133, before being extended to all the subsequently discovered NCR+/− ILC3s in mice14,29,78-80. Human ILC3s also express high levels of RORγt92,93,95,98.
RORγ, RORβ and the aforementioned RORα belong to the ROR superfamily of nuclear receptors. RORγt (also called RORγ2) is one of two isoforms of the RORγ subfamily encoded by the Rorc gene and was first found to be highly expressed in CD4+ CD8+ double-positive thymocytes134. Both RORα and RORγt act in a synergistic manner to promote TH17 cell function, although RORγt appears to be more critical135,136. RORγt has been implicated in inducing the expression of IL-23R in both TH17 cells and ILC3s14,137, thus facilitating the IL-23 responsiveness of these cells. IL-7, upon which both TH17 cells and ILC3s are dependent, was also shown to stabilise the expression of RORγt29, and hence the lineage maintenance of these cells.
AHR (Aryl hydrocarbon receptor)
AHR is a ligand-activated transcription factor that has been proposed as a xenobiotic sensor that is activated upon engagement of environmental antigens such as hydrocarbon pollutants and dietary phytochemicals120,138. Like RORγt, AHR is also crucial for the differentiation of TH17 cells139, and the recent descriptions of the role of AHR in ILC3s highlights the parallel between these two groups of cells.
Consistent with its activation by environmental cues, AHR is vital for the postnatal development and maintenance of ILC3s. Ahr−/− mice have normal numbers of fetal LTi cells and equally, newborn Ahr−/− mice have normal numbers of RORγt+ ILC3s compared to wild type. However, adult Ahr−/− mice lack all subsets of lamina propria RORγt+ ILC3s beginning from weaning at 3 weeks of age96,120,121. Notably, residual ILC3s that developed in the absence of AHR were also unable to produce IL-22121 and although RORγt could directly induce IL-22 expression, AHR was demonstrated to interact with RORγt and they synergise to induce IL-22 production121.
Interestingly, the dependence of LTi cells on AHR appears to vary depending on their site of residence. NCR+ ILC3s were depleted both in the lamina propria and Peyer’s patches of Ahr−/− mice, but while LTi cells were reduced in the lamina propria, LTi cells within the Peyer’s patches were comparable to wild type mice96. Complementing these findings, Ahr−/− mice have normal development of prenatally-primed secondary lymphoid organs such as the lymph nodes and Peyer’s patches but not the postnatally-primed crytopatches and isolated lymphoid follicles96,120. In fact, AHR is dispensable for fetal LTi cell development, and the expression of AHR is important for adult but not fetal intestinal RORγt+ ILC3s121. Together, this suggests a degree of heterogeneity within the LTi cells, where AHR-dependent LTi cells that develop/expand after birth are important for the postnatal development of secondary lymphoid organs, while AHR-independent LTi cells that develop at the fetal stage are important for prenatal development of secondary lymphoid organs. The AHR-dependent and -independent LTi cells may either represent different subsets of LTi cells altogether, or they may represent LTi cells that have migrated from the lymph nodes and Peyer’s patches to the lamina propria after birth and developed the requirement for AHR.
Ligands for the activation of AHR and subsequent ILC3 induction were proposed to be dietary ligands or other food catabolites generated by the gut microflora. Dietary phytochemicals from cruciferous vegetables for example were identified as a source of AHR ligands140. However, studies on the effects of dietary plant ligands revealed conflicting conclusions. While one study found that mice fed with phytochemical-free diet resulted in a loss of ILC3s similar to AHR-deficiency120, another study found that the exclusion of vegetable products in mouse diet did not result in any impairment to ILC3 development96. Similarly, the role of commensal microbiota remains debatable. Although earlier findings have demonstrated that germ free mice do not develop NCR+ ILC3s79,80, subsequent reports found intact populations of NCR+ ILC3s in germ-free mice96,99,121. Discrepancies in these studies may be due to the different models of germ-free mice used and the technical difficulties associated with ensuring their germ-free status.
Concluding remarks
The discovery of the ILCs in recent years together with a deeper understanding of their biology has led to a major paradigm shift in the study of immunity and hematopoiesis. A striking parallel between the ILCs 1, 2 and 3 and TH1, TH 2 and TH 17 cells, respectively, highlights the possibility that the ILCs may represent the evolutionarily older innate counterparts of the adaptive TH cells before somatic diversification of antigen receptors and the ability to generate immune memory took place. Many of the transcription factors that programme TH cell differentiation are also conserved in the ILCs but the cues for switching them on, and when they can be replaced or substituted by another transcription factor are not completely understood. These aspects of ILC development form exciting research questions for future investigation.
Footnotes
Conflict of interest:
The authors declare no conflict of interest.
References
- 1.Moore AJ, Anderson MK. Dendritic cell development: a choose-your-own-adventure story. Adv Hematol. 2013;2013:949513. doi: 10.1155/2013/949513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vosshenrich CAJ, Di Santo JP. Developmental programming of natural killer and innate lymphoid cells. Curr Opin Immunol. 2013;25:130–138. doi: 10.1016/j.coi.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 5.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16:230–239. doi: 10.1002/ijc.2910160205. [DOI] [PubMed] [Google Scholar]
- 6.Kiessling R, Klein E, Pross H, Wigzell H. ‘Natural’ killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 7.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3-LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida H, Honda K, Shinkura R, Adachi S, Nishikawa S, Maki K, et al. IL-7 receptor alpha+ CD3(−) cells in the embryonic intestine induces the organizing center of Peyer’s patches. Int Immunol. 1999;11:643–655. doi: 10.1093/intimm/11.5.643. [DOI] [PubMed] [Google Scholar]
- 9.Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014;124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 10.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 11.Walker JA, Barlow JL, McKenzie ANJ. nri3349. Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 12.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savenije OE, Mahachie John JM, Granell R, Kerkhof M, Dijk FN, de Jongste JC, et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. J Allergy Clin Immunol. 2014;134:170–177. doi: 10.1016/j.jaci.2013.12.1080. [DOI] [PubMed] [Google Scholar]
- 14.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geremia A, Arancibia-Cárcamo CV, Fleming MPP, Rust N, Singh B, Mortensen NJ, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, et al. Imbalance of NKp44(+)NKp46(−) and NKp44(−)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139:882–92. 892.e1–3. doi: 10.1053/j.gastro.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 17.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim BS, Siracusa MC, Saenz SA, Noti M, Monticelli LA, Sonnenberg GF, et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci Transl Med. 2013;5:170ra16. doi: 10.1126/scitranslmed.3005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teunissen MBM, Munneke JM, Bernink JH, Spuls PI, Res PCM, Velde Te A, et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol. 2014;134:2351–2360. doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 20.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 21.Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CAJ, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207:273–280. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 23.Klose CSN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 24.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The Transcription Factor GATA3 Is Critical for the Development of All IL-7Rα-Expressing Innate Lymphoid Cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernink JH, Peters CP, Munneke M, Velde te AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 28.Klose CSN, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 29.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated Expression of Nuclear Receptor RORγt Confers Distinct Functional Fates to NK Cell Receptor-Expressing RORγt+ Innate Lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauwerys BR, Renauld JC, Houssiau FA. Synergistic proliferation and activation of natural killer cells by interleukin 12 and interleukin 18. Cytokine. 1999;11:822–830. doi: 10.1006/cyto.1999.0501. [DOI] [PubMed] [Google Scholar]
- 33.Vosshenrich CAJ, García-Ojeda ME, Samson-Villéger SI, Pasqualetto V, Enault L, Richard-Le Goff O, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 34.Cooper MA, Bush JE, Fehniger TA, VanDeusen JB, Waite RE, Liu Y, et al. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrózek E, Anderson P, Caligiuri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 37.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 38.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 41.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 42.Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moroso V, Famili F, Papazian N, Cupedo T, van der Laan LJW, Kazemier G, et al. NK cells can generate from precursors in the adult human liver. Eur J Immunol. 2011;41:3340–3350. doi: 10.1002/eji.201141760. [DOI] [PubMed] [Google Scholar]
- 44.Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- 45.Sánchez MJ, Spits H, Lanier LL, Phillips JH. Human natural killer cell committed thymocytes and their relation to the T cell lineage. J Exp Med. 1993;178:1857–1866. doi: 10.1084/jem.178.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Veinotte LL, Halim TYF, Takei F. Unique subset of natural killer cells develops from progenitors in lymph node. Blood. 2008;111:4201–4208. doi: 10.1182/blood-2007-04-087577. [DOI] [PubMed] [Google Scholar]
- 48.Gotthardt D, Prchal-Murphy M, Seillet C, Glasner A, Mandelboim O, Carotta S, et al. NK cell development in bone marrow and liver: site matters. Genes Immun. 2014;8:584–587. doi: 10.1038/gene.2014.55. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Iizuka K, Kang H-SP, Dokun A, French AR, Greco S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 50.Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31:1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 51.Aliahmad P, la Torre de B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 54.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 57.Michie AM, Carlyle JR, Schmitt TM, Ljutic B, Cho SK, Fong Q, et al. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J Immunol. 2000;164:1730–1733. doi: 10.4049/jimmunol.164.4.1730. [DOI] [PubMed] [Google Scholar]
- 58.Sánchez MJ, Muench MO, Roncarolo MG, Lanier LL, Phillips JH. Identification of a common T/natural killer cell progenitor in human fetal thymus. J Exp Med. 1994;180:569–576. doi: 10.1084/jem.180.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veinotte LL, Greenwood CP, Mohammadi N, Parachoniak CA, Takei F. Expression of rearranged TCRgamma genes in natural killer cells suggests a minor thymus-dependent pathway of lineage commitment. Blood. 2006;107:2673–2679. doi: 10.1182/blood-2005-07-2797. [DOI] [PubMed] [Google Scholar]
- 60.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee S-C, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grégoire C, Chasson L, Luci C, Tomasello E, Geissmann F, Vivier E, et al. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price AE, Liang H-E, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J Allergy Clin Immunol. 2012;129:191–8. e1–4. doi: 10.1016/j.jaci.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 66.Halim TYF, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 67.Hoyler T, Klose CSN, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu X, Pappu R, Ramirez-Carrozzi V, Ota N, Caplazi P, Zhang J, et al. TNF superfamily member TL1A elicits type 2 innate lymphoid cells at mucosal barriers. Mucosal Immunol. 2014;7:730–740. doi: 10.1038/mi.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CGK, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang Y-J, Kim HY, Albacker LA, Baumgarth N, McKenzie ANJ, Smith DE, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorski SA, Hahn YS, Braciale TJ. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog. 2013;9:e1003615. doi: 10.1371/journal.ppat.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong JY, Bentley JK, Chung Y, Lei J, Steenrod JM, Chen Q, et al. Neonatal rhinovirus induces mucous metaplasia and airways hyperresponsiveness through IL-25 and type 2 innate lymphoid cells. J Allergy Clin Immunol. 2014;134:429–439. doi: 10.1016/j.jaci.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 74.Halim TYF, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 75.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, et al. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mjösberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 77.Lee Y, Kumagai Y, Jang MS, Kim J-H, Yang B-G, Lee E-J, et al. Intestinal Lin- c-Kit+ NKp46- CD4- population strongly produces IL-22 upon IL-1β stimulation. J Immunol. 2013;190:5296–5305. doi: 10.4049/jimmunol.1201452. [DOI] [PubMed] [Google Scholar]
- 78.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 79.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gladiator A, Wangler N, Trautwein-Weidner K, LeibundGut-Landmann S. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J Immunol. 2013;190:521–525. doi: 10.4049/jimmunol.1202924. [DOI] [PubMed] [Google Scholar]
- 83.Boyd A, Ribeiro JMC, Nutman TB. Human CD117 (cKit)+ innate lymphoid cells have a discrete transcriptional profile at homeostasis and are expanded during filarial infection. PLoS ONE. 2014;9:e108649. doi: 10.1371/journal.pone.0108649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci USA. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meier D, Bornmann C, Chappaz S, Schmutz S, Otten LA, Ceredig R, et al. Ectopic lymphoid-organ development occurs through interleukin 7-mediated enhanced survival of lymphoid-tissue-inducer cells. Immunity. 2007;26:643–654. doi: 10.1016/j.immuni.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Schmutz S, Bosco N, Chappaz S, Boyman O, Acha-Orbea H, Ceredig R, et al. Cutting edge: IL-7 regulates the peripheral pool of adult ROR gamma+ lymphoid tissue inducer cells. J Immunol. 2009;183:2217–2221. doi: 10.4049/jimmunol.0802911. [DOI] [PubMed] [Google Scholar]
- 87.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 88.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 89.Scandella E, Bolinger B, Lattmann E, Miller S, Favre S, Littman DR, et al. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat Immunol. 2008;9:667–675. doi: 10.1038/ni.1605. [DOI] [PubMed] [Google Scholar]
- 90.Eberl G, Lochner M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2009;2:478–485. doi: 10.1038/mi.2009.114. [DOI] [PubMed] [Google Scholar]
- 91.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ Lymphoid Tissue-Inducer Cells Promote Innate Immunity in the Gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 93.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JKM, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hughes T, Becknell B, McClory S, Briercheck E, Freud AG, Zhang X, et al. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH 17 cytokine interleukin-22. Blood. 2009;113:4008–4010. doi: 10.1182/blood-2008-12-192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Male V, Hughes T, McClory S, Colucci F, Caligiuri MA, Moffett A. Immature NK cells, capable of producing IL-22, are present in human uterine mucosa. J Immunol. 2010;185:3913–3918. doi: 10.4049/jimmunol.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 98.Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med. 2010;207:281–290. doi: 10.1084/jem.20091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 100.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carotta S, Pang SHM, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 2011;117:5449–5452. doi: 10.1182/blood-2010-11-318956. [DOI] [PubMed] [Google Scholar]
- 103.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJM, et al. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192:2667–2676. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 104.Hacker C, Kirsch RD, Ju X-S, Hieronymus T, Gust TC, Kuhl C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 105.Jackson JT, Hu Y, Liu R, Masson F, D’Amico A, Carotta S, et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 2011;30:2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heemskerk MH, Blom B, Nolan G, Stegmann AP, Bakker AQ, Weijer K, et al. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 109.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 110.Bain G, Engel I, Robanus Maandag EC, Riele te HP, Voland JR, Sharp LL, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 112.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 113.Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CAJ, Colucci F, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 114.Serafini N, Klein Wolterink RGJ, Satoh-Takayama N, Xu W, Vosshenrich CAJ, Hendriks RW, et al. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Current Opinion in Immunology. 2012;24:132–138. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 117.Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, et al. Notch signaling is necessary for adult, but not fetal, development of RORγt(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 118.Ribeiro VSG, Hasan M, Wilson A, Boucontet L, Pereira P, Lesjean-Pottier S, et al. Cutting edge: Thymic NK cells develop independently from T cell precursors. J Immunol. 2010;185:4993–4997. doi: 10.4049/jimmunol.1002273. [DOI] [PubMed] [Google Scholar]
- 119.Yang Q, Monticelli LA, Saenz SA, Chi AW-S, Sonnenberg GF, Tang J, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 121.Qiu J, Heller JJ, Guo X, Chen Z-ME, Fish K, Fu Y-X, et al. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, et al. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- 123.Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 2011;118:5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Male V, Nisoli I, Kostrzewski T, Allan DSJ, Carlyle JR, Lord GM, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211:635–642. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 128.Powell N, Walker AW, Stolarczyk E, Canavan JB, Gökmen MR, Marks E, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 130.Halim TYF, Steer CA, Mathä L, Gold MJ, Martinez-Gonzalez I, McNagny KM, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eberl G, Marmon S, Sunshine M-J, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 132.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 134.He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 136.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 138.Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127:299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J-C, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 140.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]