Summary
Dynamics of the nucleosome and exposure of nucleosomal DNA play key roles in many nuclear processes but local dynamics of the nucleosome and its modulation by DNA sequence are poorly understood. Using single-molecule assays we observed that the nucleosome can unwrap asymmetrically and directionally under force. The relative DNA flexibility of the inner quarters of nucleosomal DNA controls the unwrapping direction such that the nucleosome unwraps from the stiffer side. If the DNA flexibility is similar on two sides, it stochastically unwraps from either side. The two ends of the nucleosome are orchestrated such that the opening of one end helps to stabilize the other end, providing a mechanism to amplify even small differences in flexibility to a large asymmetry in nucleosome stability. Our discovery of DNA flexibility as a critical factor for nucleosome dynamics and mechanical stability suggests a novel mechanism of gene regulation by DNA sequence and modifications.
Introduction
The fundamental unit for genome compaction in eukaryotic cells is the nucleosome, in which ∼147 base pairs of DNA wrap ∼1.7 turns around a histone octamer core (Kornberg, 1974). Nucleosome dynamics regulates replication, repair, and transcription (Andrews and Luger, 2011; Bintu et al., 2012; Kulaeva et al., 2013; Li et al., 2007; Nag and Smerdon, 2009). Nucleosomal DNA can be invaded either passively due to spontaneous fluctuations (Hodges et al., 2009; Koopmans et al., 2007; Li et al., 2005; Li and Widom, 2004) or actively by forces generated by polymerases and chromatin remodelers (Sirinakis et al., 2011; Yin et al., 1995). In addition, highly dynamic chromatin anchored to various subcellular structures is likely to experience tension. Nucleosomal DNA under tension has been proposed to unwrap in two major stages; the outer turn unwraps at low force followed by unwrapping of the inner turn at higher force (Brower-Toland et al., 2002; Mack et al., 2012; Mihardja et al., 2006). However, previous mechanical studies relied on end-to-end distance detection of the DNA tethers, interpretation of which can be indirect, and is unable to report on local conformational changes of different parts of the nucleosome.
Understanding the physical basis of how DNA sequence and modifications affect nucleosome dynamics will help elucidate how genomic and epigenetic modifications regulate cellular functions. In the nucleosome, DNA of about one persistence length (147 bp) has to be bent and twisted to form ∼1.7 turns around the histone octamer (Chua et al., 2012; Kulaeva et al., 2013; Luger et al., 1997). DNA sequence may affect the strength of DNA-histone interactions through formation of specific DNA-histone interactions or by affecting the static curvature, dynamic flexibility, permanent or dynamic twist (Widom, 2001). These mechanical properties of DNA are affected by sequence composition and a variety of modifications (Hagerman, 1988; Mirsaidov et al., 2009; Rief et al., 1999; Severin et al., 2011; Vafabakhsh and Ha, 2012; Widom, 2001). The DNA sequence has a profound effect on nucleosome positioning, structure and stability (Chua et al., 2012; North et al., 2012; Tóth et al., 2013; Widom, 2001), but how it affects nucleosome dynamics is poorly understood.
Here, we employ a single-molecule assay which combines fluorescence with optical tweezers (Hohng et al., 2007; Maffeo et al., 2014; Zhou et al., 2011) to simultaneously manipulate an individual nucleosome under force and probe its local conformational transitions.
Results
Probing Local Conformational Dynamics of the Nucleosome under Tension
In order to obtain clearly interpretable data on local nucleosome dynamics we chose the nucleosome positioning sequence 601 (Lowary and Widom, 1998), which has been used for previous high resolution single molecule studies (Bintu et al., 2011,2012; Böhm et al., 2011; Brower-Toland et al., 2002; Deindl et al., 2013; Gansen et al., 2009; Hall et al., 2009; Hodges et al., 2009; Kruithof and van Noort, 2009; Mack et al., 2012; Mihardja et al., 2006; North et al., 2012; Sheinin et al., 2013; Shundrovsky et al., 2006; Sudhanshu et al., 2011; Tóth et al., 2013). A nucleosome was anchored to a PEG-coated glass surface on one end of the DNA and pulled via a λ-DNA tethered to the other end by an optical trap (Figure 1A). A fluorescence resonance energy transfer (FRET) dye pair, a donor and an acceptor, attached to various positions on the DNA enable the measurement of conformational changes of defined locality.
Figure 1. Observation of Local Conformational Changes of Nucleosome under Tension.
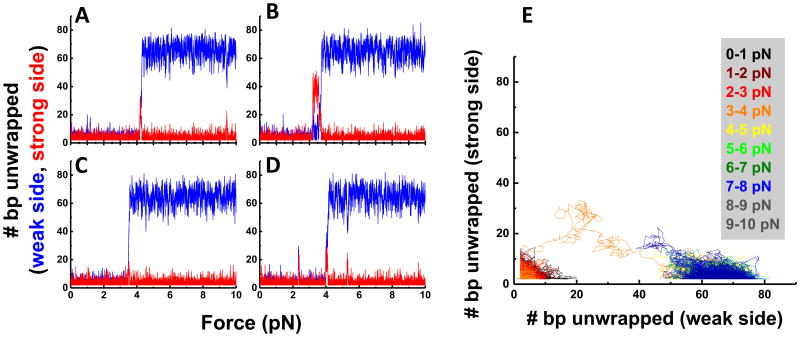
(A) Experimental scheme: a nucleosome was immobilized on a microscope slide via a 14 bp dsDNA handle beyond the nucleosome core sequence. The other end was connected to a micron-diameter bead through a λ-DNA linker which was held in place by an optical trap which applies force. Local conformational changes were recorded by FRET between the donor (green) and the acceptor (red) on the DNA. (B) Positions of donor and acceptor fluorophores in the ED1-labeling scheme superposed on two different views of the nucleosome structure (Protein Data Bank [PDB] file 3MVD). (C and D) Single-molecule time traces of the ED1 construct recorded during stretching and relaxing at a stage speed of 455 nm/s at a set maximum force of ∼ 6 pN (C) and ∼ 20 pN (D): force (black), donor signal (green), acceptor signal (red), and FRET efficiency (blue). (E and F) The average FRET versus force when the maximum force was set to ∼ 6 pN (E): average of 26 traces and ∼ 20 pN (F): average of 25 traces. See also Figure S1.
To probe unwrapping of the outer DNA turn, we constructed the ED1 (Entry-Dyad 1) labeling scheme consisting of a donor close to the dyad and an acceptor close to an entry. ED1 nucleosomes displayed a single high FRET population due to close proximity of the probes (Figure S1A) as expected from the nucleosome crystal structure (Makde et al., 2010) (Figure 1B). In the absence of force, FRET time traces were stable within our temporal resolution of 30 ms (Figure S1B). The same DNA reconstituted with the (H3/H4)2 tetramer produced a very different distribution with low FRET values attributed to the tetrasome (Figure S1A).
We increased the applied force starting from a low value (typically between 0.4 – 1.0 pN) to a predetermined higher value and then returned it to the low value. FRET gradually decreased as the force increased followed by fast fluctuations and finally a sharp decrease in FRET (Figures 1C and 1D). Upon relaxation, the nucleosome reformed, retracing the dynamics observed during stretching if the force was held below 6 pN to limit the extent of unwrapping (Figures 1C and 1E) or displaying hysteresis when we extended the force range to 20 pN (Figures 1D and 1F). The initial gradual FRET decrease indicates that DNA unwraps steadily without going through a major energy barrier at low tension. The FRET fluctuation that follows likely represents a bistable hopping behavior reported previously (Mihardja et al., 2006). Subsequent stretching/relaxation cycles reproduced the same behavior, suggesting that each cycle brings the nucleosome back to the initial state.
To probe inner turn unwrapping, we attached FRET probes to a region approximately 40 bp from the dyad (INT) (Figure 2G). As with ED1, the INT nucleosome showed a single narrow FRET peak at zero force and was distinguishable from the tetrasome species which displayed a broad range of FRET (Figures S1C and S1D). At low forces, the INT nucleosome maintained a stable high FRET value with occasional hopping to an intermediate FRET state (Figure 2H). As the force increased to higher values (10–15 pN), FRET suddenly dropped to a final low value (Figure 2H). As an additional control, the INT-tetrasome showed a distinct FRET vs. force stretching pattern, unraveling at much lower force (3-5 pN), thus confirming that INT nucleosome contained the histone octamer (Figure S1E).
Figure 2. Nucleosome Unwraps Directionally under Tension.
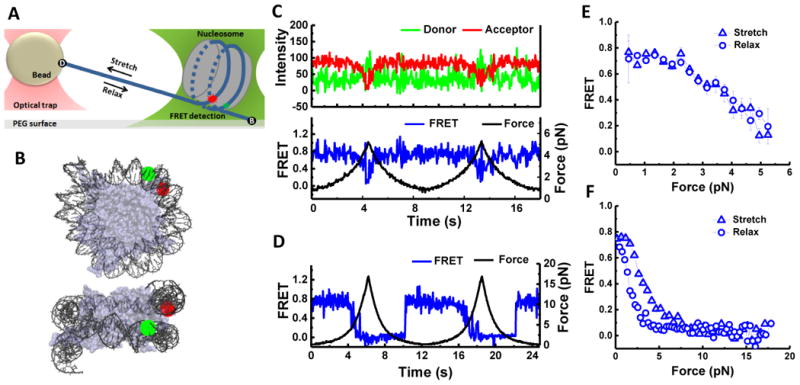
(A-H) FRET vs. force during stretching for various FRET pairs spanning two sides of the nucleosome illustrated in (G) (see Figure S2 for labeling positions). Representative data for single cycles are shown in gray. The averaged curves are in blue for the weak side, in red for the strong side, and in black for the inner turn probes. Error bars are SEM of 25 traces for ED1 (A), 15 traces for ED1.5 (B), 8 traces for ED1.7 (C), 20 traces for ED2 (D), 7 traces for ED2.5 (E), 40 traces for ED2.8 (F), and 22 traces for INT (H). (I) Overlay of ED1, ED2, and INT stretching curves. Substeps, which may arise from progressive unwrapping, could be seen for ED1.7 both in the averaged trace and in individual traces (three out of eight cycles). See also Figure S2.
Taken together, our nucleosome stretching data with probes at ED1 and INT positions are consistent with previous studies on the effect of force on global nucleosome dynamics (Brower-Toland et al., 2002; Kruithof and van Noort, 2009; Mihardja et al., 2006; Sheinin et al., 2013); the outer turn unwraps at low force (3-5 pN) and the inner turn unwraps at higher force (12-15 pN). In addition, ED1 probe allows us to observe gradual unwrapping before an abrupt transition of the initial DNA end segment at the low force range (< 3 pN).
Nucleosome Unwrapping is Asymmetric
Previous investigations of nucleosome unwrapping (Brower-Toland et al., 2002; Kruithof and van Noort, 2009; Mack et al., 2012; Mihardja et al., 2006; Sheinin et al., 2013) assumed that two nucleosomal DNA ends respond similarly to the applied force since unwrapping of the two DNA ends was not separately observable. Our assay, which is sensitive to local conformational changes, enables the examination of two sides separately by comparing the FRET-Force response on the two ends. We designed a construct termed ED2 with a FRET pair placed at the opposite entry/dyad region—the “left” end (Figures 2G and S2A). Surprisingly, the FRET-Force pattern of ED2 displayed a pattern very different from ED1 (Figure 2D). FRET remained stable at low forces and did not decrease until higher force (15 – 20 pN) was reached, in contrast to the decrease below 5 pN observed for ED1 on the “right” end. This result indicates that a significant asymmetry exists in the DNA unwrapping behavior.
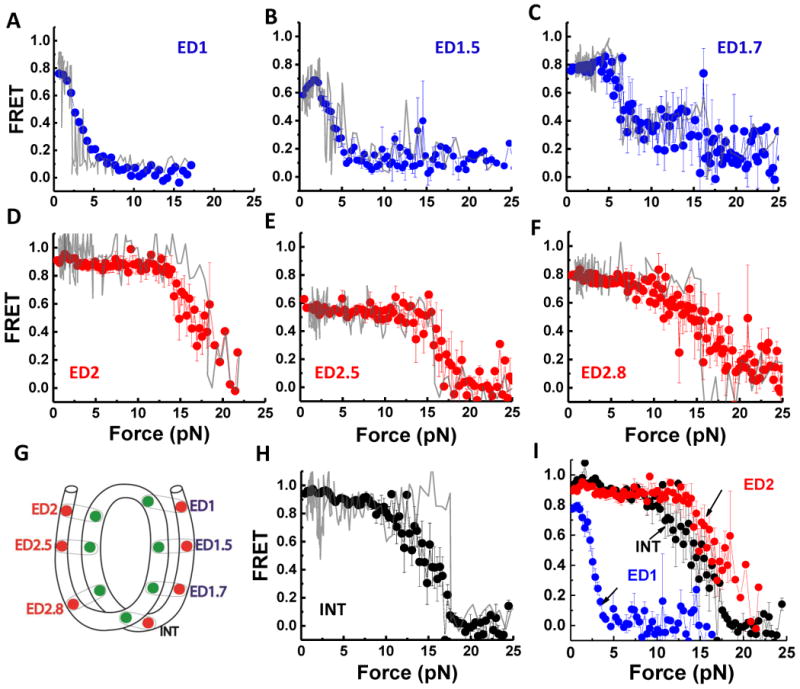
We performed various control experiments to confirm the unwrapping asymmetry result and to rule out alternative explanations. First, additional constructs with probes at symmetric locations on the DNA handles outside the core sequence confirmed that the nucleosome is not mispositioned on the 601 sequence (Figure S3). Second, we swapped the orientation of surface tethering and pulling via the lambda DNA tether and found that the strong side unwrapped at high forces for both configurations (with essentially identical FRET versus force curves), ruling out surface tethering via a particular end as the reason for the asymmetry (Figure 3A). Third, replacing the first 10 bp of the left handle with the corresponding region on the right handle showed that the sequence difference just outside the core region is not responsible for the asymmetry (Figure 3B).
Figure 3. Unwrapping Force Is Not Affected by Pulling Configuration or Extra-Nucleosomal Handle Sequence.
(A) Switched pulling configurations for the same labeling position ED2. In the ED2 scheme, the 5′ end of the bottom J strand (the right end) is biotinylated. In the ED2B scheme, the 5′ end of the top I strand (the left end) is biotinylated. Averaged stretching traces for both ED2 pulling configurations show identical high force required for unwrapping (ED2: average of 20 traces, ED2B: average of 4 traces). (B) Changing the handle sequence on the left side does not alter the high force range required to open nucleosomal DNA on this side. Averaged stretching curves show identical high force required for unwrapping for 601-ED2 (average of 20 traces) and RRH-1-10-ED2 (average of 15 traces). See also Figure S3.
To examine if the observed asymmetry may be induced by position-specific perturbations caused by the fluorophores, we designed four additional constructs for comparison of the two sides: ED1 versus ED2, ED1.5 versus ED2.5, and ED1.7 versus ED2.8 (Figure 2G and Figure S2A). Generally, the force required for a significant FRET decrease was lower for ED1 (Figure 2A), ED1.5 (Figure 2B) and ED1.7 (Figure 2C) than for those labeled at symmetrically related sites, ED2 (Figure 2D), ED2.5 (Figure 2E) and ED2.8 (Figure 2F), respectively, showing that the asymmetry is highly unlikely due to position-dependent perturbations by the fluorophores, and indicating that one side of the nucleosome is indeed weaker than the other when the DNA is under tension.
Strikingly, the force needed for a major unwrapping signal was larger for the ED2 end (16.8±0.4 pN) than the DNA inner turn (14.7 ± 0.5 pN) (Figure 2D and 2H) (the errors represent the SEM). This effect was even clearer when the pulling rate was halved to 233 nm/sec (14.2 ± 0.5 pN vs. 11.2 ± 0.9 pN, Figure S2). Thus, the data suggest that DNA unwrapping occurs directionally, starting from the ‘weak’ end (ED1) at the lowest unwrapping force, followed by the inner turn, and then to the ‘strong’ end (ED2). However, the small difference between the INT and ED2 unwrapping force would allow the inner turn to unwrap later than the strong end in some cases.
Such mechanical asymmetry may influence gene expression by affecting DNA exposure or transcriptional pausing. In fact, an in vitro transcription study (Bondarenko et al., 2006) observed that nucleosomes can form a polar barrier to transcriptional elongation. Specifically, our “strong” side (ED2) corresponds to the 601R transcription orientation where polymerases face a higher outer turn barrier (the +15 barrier).
Unwrapping of the Nucleosome on One End Stabilizes the Other End
In the low force range, FRET of the strong outer turn ED2 is stable and remains unchanged until the final drop at high force (16.8±1.5 pN) (Fig. 2D). When the pulling rate is lowered 2-fold (Figure S2), we observed a small decrease in FRET followed by a FRET recovery in the low force range for some stretching traces. Therefore, we probed the earliest unwrapping process of the strong (ED2) side by moving the probes to either one (ED2-1, Figure 4C) or twelve (ED2-12, Figure S4A) nucleotides beyond the nucleosome core sequence on the strong side. At low forces, ED2-1 and ED2-12 probes on the strong side showed the same stretching pattern as ED1 probe on the weak side: FRET decreased gradually at low force followed by fluctuations at 3-6 pN (Figures 4A and S4). However, on the weak side the FRET dropped entirely after 6 pN, while on the strong side, FRET recovered and did not fully drop until much higher force was reached. Our force-fluorescence spectroscopy approach allows detection of unwrapping/rewrapping of a specific side. In contrast, in previous studies measuring the overall end-to-end distance (Mihardja et al., 2006; Sheinin et al., 2013), simultaneous rewrapping of the strong end and unwrapping of the weak end may not have given a detectable change in overall length. Coordination in FRET-force patterns of ED1 and ED2-1 indicate that two extreme ends of the nucleosome are slightly unwrapped at low forces but once the weak end significantly unwraps, the strong end rewraps and stays stable until much higher forces are applied.
Figure 4. Coordinated Dynamics of the Two Nucleosomal DNA Ends.
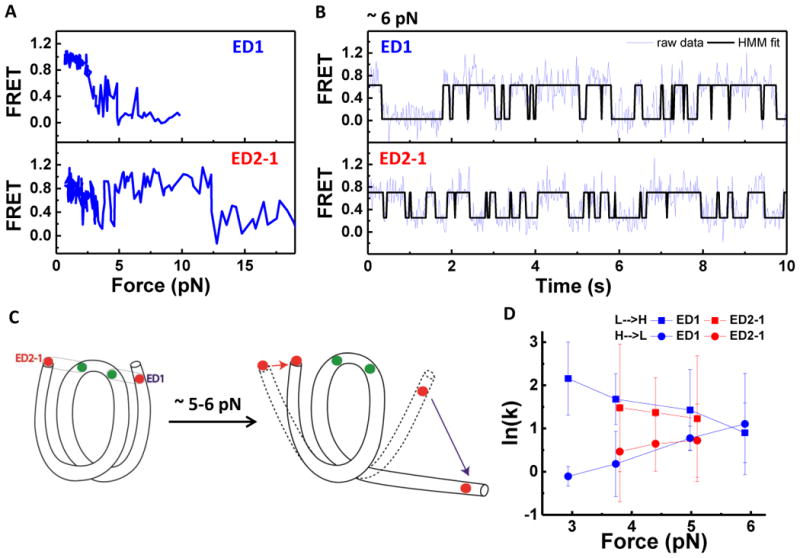
(A) Representative single-molecule stretching traces of ED1 and ED2-1 as indicated in C. (B) Representative time traces of FRET efficiency at a constant force of 6 pN, showing hopping between high and low FRET states. Fits from Hidden Markov modeling are overlaid. (C) Illustration of how major unwrapping of one side of the nucleosome facilitates rewrapping on the other end. Initially, two extreme ends of the nucleosome synchronously unwrap and rewrap at forces below ∼ 5pN (dashed shape). Once the ED1 side majorly unwraps (blue arrow), this facilitates the rewrapping of the ED2 side (red arrow). (D) Rates of transition between high and low FRET states vs. force. Unwrapping rates (high to low FRET transitions) in circles and rewrapping rates (low to high FRET transitions) in squares. See also Figure S4.
At constant forces, FRET time traces of both ED1 and ED2-1 constructs (Figure 4B) showed two-state hopping between wrapped and partially unwrapped state, respectively. Hidden Markov Modeling (McKinney et al., 2006) was used to determine the transition rates between the two states (Figure 4D). As the force increased, the unwrapping rate increased and the wrapping rate decreased, consistent with a previous report (Mihardja et al., 2006), and the rates on the two DNA ends were similar. Our observation that the two ends of the nucleosome are orchestrated such that the opening of one end helps stabilize the other end raises the possibility that even relatively small asymmetry between the two sides may result in one side winning reliably (cartoon in Figure 4C).
Asymmetry of Nucleosome Unwrapping Is Directed by DNA Local Flexibility
We propose that the observed asymmetry in mechanical stability originates from the DNA sequence differences between the two sides of the 601 sequence for the following reasons. First, the protein core structure is symmetric around the dyad axis (Chua et al., 2012; Luger et al., 1997) whereas the DNA sequence is nonpalindromic. Second, unzipping of the nucleosomal DNA under certain experimental configurations shows a higher off-dyad barrier on one side (“strong” side in our study) than the other (Hall et al., 2009). Third, symmetrization of certain sequence features can affect the overall thermodynamic stability as measured by salt titration (Chua et al., 2012).
Since the unwrapping asymmetry is observed for the outer turn, we first symmetrized DNA content at the entry regions by replacing the AT-rich region (nucleotide 8-24 from the right end) on the weak side with the corresponding GC-rich segment on the strong side (Figure S5A). This construct, termed LL8-24, exhibited the same asymmetry as the 601 nucleosome (Figure S5C), ruling out the differences in AT/GC-content of the entry region as the source of asymmetry.
Because DNA has to be bent and deformed to wrap around the histone octamer, the intrinsic DNA flexibility may influence DNA-histone binding affinity (Lowary and Widom, 1998; Widom, 2001). Therefore, we hypothesized that the more flexible sequences would unravel at higher forces by better tolerating the sharply bent DNA conformation. To test this hypothesis, we examined the relative flexibility of the two 73 bp DNA fragments flanking the dyad in the 601 sequence using a single molecule DNA cyclization assay (Vafabakhsh and Ha, 2012). The ‘strong’ side (LH for left half) yielded a cyclization time of 26 minutes while the ‘weak’ side (RH for right half) took 189 minutes to cyclize, indicating that the left side of the 601 is more flexible than the right side by a factor of 7 according to our measurement (Figure 5C). Thus, the asymmetry in DNA flexibility appears to correlate with asymmetric unwrapping - the more flexible DNA side unwraps at higher force and vice versa. Here, ‘flexibility’ is an operational definition equivalent to ‘cyclizability’ in our assay because we do not yet know whether a static bend or dynamic flexibility (represented by lower bending energy) determines the apparent flexibility.
Figure 5. Asymmetric Nucleosome Unwrapping Controlled by DNA Local Flexibility.
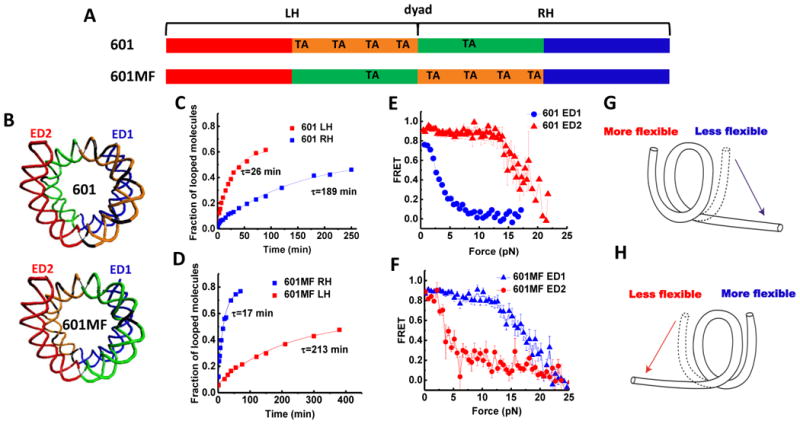
(A) Variations of the 601 the sequence where the inner quarters are colored in orange and green and the outer quarters are colored in red and blue. TA steps are indicated. (B) Nucleosomal DNA structures are shown in the same color scheme with corresponding scheme of the sequence. (C and D) Single exponential fits to the looped DNA fraction versus time yield the average looping time τ measured using single DNA cyclization assay for the 73 bp left or right halves (LH and RH, respectively). (E and F) Averaged stretching time traces of FRET efficiency versus force for nucleosomes in ED1 and ED2 labeling schemes. Error bars denote SEM of 25 traces for 601 ED1, 15 traces for 601 ED2, 29 traces for 601MF ED1, 19 traces for 601MF ED2. (G and H) Illustrations of the relationship between the direction of nucleosome unwrapping and the DNA flexibility of the two halves of the nucleosomal DNA sequence. The nucleosome unwraps from the stiffer side (single-headed arrows) if the DNA flexibility differs significantly between the two sides. See also Figure S5.
In order to test the correlation further, we modified the 601 sequence so as to locally switch the DNA flexibility on the two sides by flipping the middle 73 bp (601MF) (Figures 5A and 5B). The single molecule cyclization showed that the right side of 601MF has now become more flexible (17 min. looping time) than the left side (213 min. looping time) by a factor of 12 (Figure 5D), reversing the relation found in the original 601 sequence, and correspondingly, the left side of the 601MF nucleosome (now containing stiffer DNA sequence) unwrapped at a lower force than the right side (Figure 5F). This implies that the direction of outer turn unwrapping can be controlled by the relative flexibility of internal regions of DNA such that the nucleosome first unwraps from the DNA side connected to a less flexible inner turn DNA (Figures 5G and 5H).
We further tested how nucleosome unwrapping is affected when the DNA sequence is similar in flexibility on both sides. We were guided by the 10 bp TA steps rule suggested by Widom (Lowary and Widom, 1998; Widom, 2001) to construct this DNA sequence. Chua et al. (2012) confirmed by crystallography that TA dinucleotides accommodate the highest degree of distortion of the DNA structure within nucleosome. The 601 sequence is nonpalindromic with 10 bp TA steps situated only on the left (strong) side. Therefore, we pseudo-symmetrized the flexibility of the sequence by adding three copies of TA dinucleotides spaced 10 bp apart to the right (weak) side (601RTA) (Figures 6A and 6B). The resulting 601RTA right half (RH) became more flexible (cyclization time decreased from 189 to 63 min) (Figure 6C) and closer to the left half (26 min.). We ensured that the nucleosome positioning is maintained on all three sequences (601, 601MF and 601RTA), as nucleosomes reconstituted from all three sequences show the same electrophoretic mobility on a 5% native PAGE gel and displayed similar single-molecule FRET histograms (Figure S6). Strikingly, instead of one side winning the match every time, which side unwraps at low forces became stochastic (Figure 6F). The fraction of traces unwrapped at low force and high force was 37% and 67% for the left half (601RTA-ED2) and 44% and 56% for the right half (601RTA-ED1), respectively (Figures 6G and 6H). Averaging over all stretching traces produced almost identical FRET-force patterns for these two constructs (Figure 6D). These results imply that when the flexibility of DNA on the two sides of the nucleosome is similar, each side of the nucleosome unwraps stochastically at either low force or high force (Figure 6E).
Figure 6. Stochastic Unwrapping of Nucleosome on the Sequence with Similar Flexibility on Two Sides.
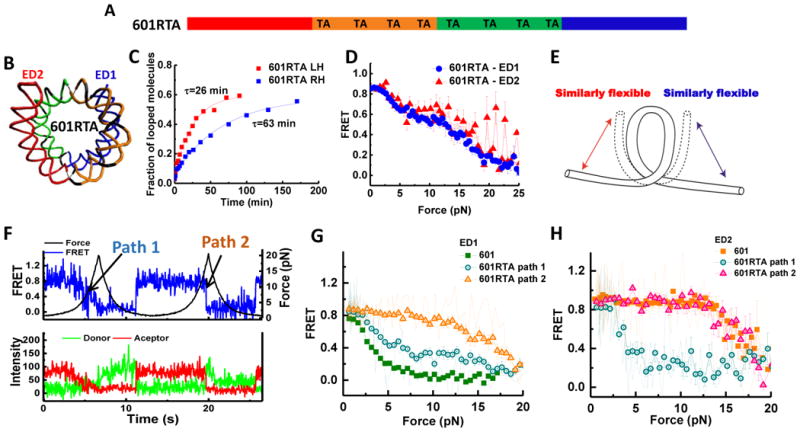
(A) Scheme of the 601RTA sequence which is derived from the 601 sequence by substitution of three dinucleotides on the right side by three TA steps. (B) Nucleosomal DNA structures are shown in the same color scheme with the scheme of the sequence. (C) Single exponential fits to the looped DNA fraction versus time yield the average looping time τ measured using single DNA cyclization assay for the 73 bp left or right halves (LH and RH, respectively) for the 601RTA sequence. (D) Averaged stretching time traces of FRET efficiency versus force for nucleosomes in ED1 (average of 57 traces) and ED2 (average of 7 traces) labeling schemes for the 601 RTA sequence. Error bars denote SEM. (E) A cartoon illustrating stochastic unwrapping of nucleosome from either side when the DNA flexibility on the two sides is made similar on the 601RTA sequence. (F) Representative single-molecule fluorescence-force time trace for 601-RTA nucleosome reconstituted with the ED1 labeling scheme. Two unwrapping paths are shown. Path 1 is gradual FRET decrease at low force (similar to original weak side), while path 2 is sudden FRET decrease at high force (similar to original strong side). (G and H) Averaged FRET versus force stretching curves for 601-RTA-ED1 (25 traces for path 1 and 32 traces for path 2) nucleosomes (G) and 601-RTA-ED2 (four traces for path 1 and three traces for path 2) nucleosomes (H), comparing to that of ED1 and ED2 of the 601 sequence. Representative single-molecule stretching traces are shown in lighter color lines. See also Figure S6.
Monte Carlo Simulation of Asymmetric Unwrapping of Nucleosomal DNA
In order to model the asymmetric nucleosome dynamics under tension, we adopted a continuum model of symmetric nucleosomal DNA unwrapping developed by Sudhanshu et al. (2011) and extended it to a more general, asymmetric case where m and n base pairs can be unwrapped from the weak and strong side, respectively. The only modification to the energy function used by Sudhanshu et al. (2011) was a reduction in the binding energy of the inner quarter of the weak side (see Supplemental Information for details). With this energy function, we performed Monte Carlo simulations starting from 0.1 pN and increasing the force in 0.1 pN increments every 2000 time steps until 10 pN of force was reached. Four representative trajectories of m and n values, the number of base pairs unwrapped from the weak and strong sides, respectively, are shown in Figure 7 (blue for m and red for n). At ~3-5 pN of force, we observed major unwrapping of the weak side (m values reaching around 65 bp). In three cases (Figures 7A, 7B, and 7D), initial unwrapping of the strong side (transient increase in n, i.e., unwrapping of the strong side) precedes rewrapping of the strong side and major unwrapping of the weak side. Figure 7E shows an example trajectory in (m, n) space (corresponding to Figure 7B). A transient unwrapping of the strong side is seen in the force range 3-4 pN before the systems moves to the asymmetrically unwrapped state.
Figure 7. Monte Carlo Simulation of Nucleosome Unwrapping.
(A–D) Representative Monte Carlo simulation records show the number of base pairs unwrapped from the weak side (blue) and the strong side (red) as the force increased from 0.1 pN to 10 pN. (E) A 2D representation of unwrapping trajectory shown in (B). Different portions of the trajectory at difference forces are shown in different colors as indicated.
This simple model and simulation capture two important aspects of our data. First, asymmetric unwrapping can be obtained even when only the inner quarters are different in binding energy (presumably arising from differences in DNA flexibility where less flexible sequence has less binding energy). Second, a transient unwrapping of the strong side is often observed, and this is followed by rewrapping of the strong side and major unwrapping of the weak side in a coordinated fashion. Furthermore, our data and simulation suggest that the force-induced extension changes observed in previous studies at low forces and interpreted as symmetric unwrapping of the outer turns from both ends may need to be reinterpreted as asymmetric unraveling of the weak side only.
Discussion
Genetic information buried in nucleosome is made accessible for replication, transcription, repair and remodeling by partial unwrapping of nucleosomes (Bowman, 2010; Gansen et al., 2009; Hodges et al., 2009; Kulaeva et al., 2013; Li et al., 2005; Li and Widom, 2004; Maher et al., 2013; North et al., 2012; Tims et al., 2011). Our results provide the first demonstration of how the local flexibility of DNA governs the mechanical stability of the nucleosome and accessibility of nucleosomal DNA, and may be generalizable as a principal mechanism for regulation of DNA metabolism by nucleosomal DNA sequence and modifications.
The correlation that the more flexible the DNA sequence is, the more stable it stays bound to the histone core may aid the prediction of nucleosome positions imposed by DNA sequence. We found that this relation holds not only for DNA sequences but also for DNA modifications such as DNA mismatches, 5-methylcytosine and 5-formylcytosine (T.T.M.N., Q.Z., J. Yoo, Q. Dai, A. Aksimentiev, C. He, and T.H., unpublished data).
Stabilization of one nucleosomal DNA end upon the major opening of the other end may play a role in nucleosome integrity maintenance during transcription and nucleosome remodeling because both in vivo and in vitro studies suggest that a high fraction of nucleosomes survive after being transcribed (Bintu et al., 2011; Workman, 2006) and remodeled (Shundrovsky et al., 2006). It is also possible that such orchestration between the two nucleosome ends may help stabilize one H2A/H2B dimer during the exchange or modification of the other dimer. For example, SWR-C/SWR-1 deposits H2A.Z into only one site at a time, not both (Yen et al., 2013).
Our Monte Carlo simulations could reproduce key features of asymmetric unwrapping and coordinated dynamics of two DNA ends (Figure 7). Nevertheless, this model ignores many structural details and represents DNA sequences with the resolution of 36 bp, a quarter of the nucleosomal DNA. Other properties of the nucleosome yet to be explored may make additional contributions to the coordination of DNA ends: (1) the proposed electrostatic repulsion between two DNA turns (Mollazadeh-Beidokhti et al., 2012) where upon force-induced undocking of one end, the resulting loss of the electrostatic repulsion stabilizes the other end, (2) DNA allostery (Kim et al., 2013), and (3) the deformation of the histone octamer during unwrapping which may change charge distribution and/or contribute to the allosteric coupling. Histone deformation was suggested to govern salt-induced nucleosome dissociation (Böhm et al., 2011) and may also be involved in nucleosome remodeling by IWSI remodelers (Deindl et al., 2013). In our experiments at low tension, in addition to the early unwrapping of extreme DNA ends probed by ED1 and ED2-1, the FRET probes at ED1.5 (Figure 2B) and ED1.7 (Figure 2C) displayed an increase in FRET as a first response to applied force before a decrease, indicating possible partial DNA tightening mediated by twisting of the H2A/H2B dimer on the weak side.
We observed that nucleosomal DNA unwraps directionally under tension not only for the 601 sequence but also for the derivatives of the 601 sequence. Asymmetric unwrapping is likely to be generalizable to other sequences since the coordination of two ends would allow the system to amplify even a small difference in flexibility to cause a large asymmetry in mechanical stability.
Directionality of transcription can be ensured by the suppression of cryptic antisense (Gorman et al., 2010) through epigenetic regulation and RNA degradation (Richard and Manley, 2013). Our results linking sequence-dependent flexibility to mechanical stability of the nucleosome suggest another mechanism to maintain transcriptional direction– the possibility that nature selects for lower flexibility DNA sequences within the first half of nucleosomes in the direction of transcription. In this scenario, RNA polymerase would have greater initial access to the DNA template if it enters the nucleosomal DNA from the ‘weak’ side and would only pause when it reaches the nucleosomal dyad (Churchman and Weissman, 2011; Hodges et al., 2009). We are currently investigating DNA flexibility on a genomic scale combining sequencing and single molecule cyclization to test this possibility.
Experimental Procedures
For additional details, see the Extended Experimental Procedures.
Preparation of DNA constructs
We used PCR to amplify 181 bp ds DNA from templates which contain 147 bp 601 positioning sequence, flanked by a 14 bp linker to biotin and 20 bp spacer connected to the 12 nt COS overhang. The construct was tethered to the surface via biotin and the COS overhang was used to anneal the template to λ DNA. PCR primer oligonucleotides were designed for various templates and synthesized by Integrated DNA Technologies. The forward primer contains an amino modification (5AmMC6T) at a designated location and a biotin at the 5′ end. The reverse primer contains the same amino modification and an abasic site to create the COS overhang. The forward and reverse primers were labeled with Cy3 and Cy5 dyes respectively according to (Roy et al., 2008), and HPLC-purified when necessary to bring the labeling efficiency to >90%.
Nucleosome Reconstitution
PCR-amplified 601 templates were reconstituted with X. laevis recombinant histone octamer (purchased from Colorado State University) by salt dialysis (Dyer et al., 2004). Reconstituted nucleosomes were stored at 4°C in the dark typically at concentrations of 100–200 nM and used within 2 weeks. The efficiency of nucleosome reconstitution was measured by 5% native PAGE gel electrophoresis.
Annealing nucleosome to λ DNA
The nucleosome was annealed to λ DNA and an oligonucleotide containing digoxigenin. First, λ DNA (NEB) at 16 nM was heated in the presence of 120 mM NaCl and 1.2 mM MgCl2 at 80°C for 10 min, and then placed on ice for 5 min. Nucleosomes and BSA were added to the λ DNA at a final concentration of 8 nM and 0.1 mg/ml, respectively. The mixture was incubated with rotation in the dark at room temperature for 15 min and then for an additional 2–3 hr at 4°C. DIG oligo (see DNA Sequences in the Supplemental Information) was added to a final concentration of 200 nM and then incubated with rotation at 4°C for 1–2 hr. Samples were stored at 4°C in the dark and could be used for data acquisition for up to 2 weeks.
Sample assembly
To eliminate nonspecific surface binding, a coverslip surface was coated with polyethyleneglycol (PEG) (mixture of mPEG-SVA and Biotin-PEG-SVA, Laysan Bio) according to Roy et al. (2008). After forming an imaging chamber using the PEG coated coverslip and glass microscope slide, it was further incubated in blocking buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mg/ml BSA [NEB], 1 mg/ml tRNA [Ambion]) for 1 hr. The nucleosome sample was diluted to 10 pM in a nucleosome dilution buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM MgCl2) and immobilized on the surface via biotin-neutravidin interaction. Next, 1 μm anti-digoxigenin-coated polystyrene beads (Polysciences) diluted in nucleosome dilution buffer were added to the imaging chamber for ∼30 minutes for attachment of beads to the free end of each tether. Finally, imaging buffer (50 mM Tris-HCl pH 8, 50 mM NaCl, 1 mM MgCl2, 0.5 mg/ml BSA [NEB], 0.5 mg/ml tRNA [Ambion], 0.1% v/v Tween-20 [Sigma], 0.5% w/v D-Glucose [Sigma], 165 U/ml glucose oxidase [Sigma], 2170 U/ml catalase [Roche], and 3 mM Trolox [Sigma]) was added for data acquisition.
Fluorescence-Force Spectroscopy
We recently developed an instrument combining optical trap with fluorescence detection to monitor conformational changes of biomolecular systems under applied force (Hohng et al., 2007). The full details of this instrument can be found in our recent review (Zhou et al., 2010). Briefly, an optical trap was formed by an infrared laser (1064 nm, 800 mW, EXLSR-1064-800-CDRH, Spectra-Physics) through the back port of the microscope (Olympus) by expanding the laser beam 8-fold using two telescopes and focusing on the sample plane with a 100x oil immersion objective (Olympus). Force was applied on the sample tethers by moving the microscope slide using a piezo stage (Physik Instrument). Applied force was determined by position detection of the tethered beads using a QPD (UDT/SPOT/9DMI) and stiffness calibration as described (Hohng et al., 2007). The confocal excitation laser (532 nm, 30 mW, World StarTech) was coupled through the right port of the microscope. The excitation laser was scanned by a piezo-controlled steering mirror (S-334K.2SL, Physik Instrument). The fluorescence emission was filtered from the infrared laser by a band pass filter (HQ580/60 m, Chroma) and separated from excitation by a dichroic mirror (HQ680/60 m, Chroma) before detection by two avalanche photodiodes.
Data acquisition
Single molecule data acquisition was performed according to Hohng et al. (2007). In summary, after a bead was trapped, the origin of the tether was determined by stretching the tether in two opposite directions along both x and y axis. Then the confocal laser was scanned to locate the fluorescence spot on the tether after separating the trapped bead from its origin by 14 μm. Unless specified otherwise, the nucleosome unwrapping experiment was carried out by moving the stage between 14 μm and 16.8-17.2 μm at the speed of 455 nm/sec-1. The confocal excitation was scanned concurrently with the stage movement. Fluorescence emission was detected for 20 ms after each step in stage movement. Force-fluorescence data was obtained in the imaging buffer (50 mM Tris-HCl pH 8, 50 mM NaCl, 1 mM MgCl2, 0.5 mg/ml BSA [NEB], 0.5 mg/ml tRNA [Ambion], 0.1% v/v Tween-20 [Sigma], 0.5% w/v D-Glucose [Sigma], 165 U/ml glucose oxidase [Sigma], 2170 U/ml catalase [Roche] and 3 mM Trolox [Sigma]).
Single molecule DNA cyclization Assay
A single-molecule DNA cyclization assay was recently developed in our laboratory to quantify the flexibility of a short double stranded DNAs (< 100 bps) (Vafabakhsh and Ha, 2012). A total of six 601 DNA fragment regions listed in DNA Templates and Labeling Schemes in the Supplemental Information are generated by slow annealing (90°C for 10 min) of appropriate oligonucleotides (see DNA Sequences in the Supplemental Information) followed by slow cooling to room temperature over 4 hr. DNA fragments were immobilized on a PEG-coated microscope slide via biotin-neutravidin linkage. A FRET pair (Cy3 and Cy5) was incorporated at the two 10 nt long 5′ overhangs that are complementary to each other so that loop formation via annealing of the two overhangs was detected as a FRET increase. Data acquisition was performed in a buffered solution (10 mM Tris-HCl pH 8.0, 1 M NaCl, 0.5% w/v D-Glucose [Sigma], 165 U/ml glucose oxidase [Sigma], 2170 U/ml catalase [Roche], and 3 mM Trolox [Sigma]). Time courses of generation of high FRET population allowed us to quantify the fraction of looped molecules versus time after the high salt buffer was introduced to the chamber containing low salt buffer (10 mM NaCl) of otherwise identical composition. Here, the rate of loop formation was used as a measure of DNA flexibility. The faster the looping occurs, the more flexible the sequence is.
All single molecule measurements were performed at ∼ 22°C.
Supplemental Information
Acknowledgments
This work was supported by the US National Institutes of Health (GM065367) and by the National Science Foundation Physics Frontiers Center program (PHY 0822613 and PHY 1430124). We thank A. J. Spakowitz for providing the energy function used for Monte Carlo simulations. T.H. is an investigator with the Howard Hughes Medical Institute.
Author Contributions: T. N. designed the research, prepared the samples, conducted all force-fluorescence experiments and part of DNA looping experiments, analyzed and interpreted the data and wrote the manuscript; Q.Z. conducted part of looping experiments; R.Z contributed to the early stage of instrumentation; J. Y. oversaw the initial stage of the project and revised the manuscript; T. H. supervised the project, performed Monte Carlo simulations, and revised the manuscript. All authors discussed the results and were given opportunities to revise the manuscript.
Contributor Information
Jaya G. Yodh, Email: jyodh@illinois.edu.
Taekjip Ha, Email: tjha@illinois.edu.
References
- Andrews AJ, Luger K. Nucleosome Structure(s) and Stability: Variations on a Theme. In: Rees DC, Dill KA, Williamson JR, editors. Annual Review of Biophysics. Vol. 40. 2011. pp. 99–117. [DOI] [PubMed] [Google Scholar]
- Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nature Structural & Molecular Biology. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal Elements that Control the Topography of the Barrier to Transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Tóth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Molecular Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Current Opinion in Structural Biology. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Research. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368. doi: 10.1038/nature09652. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X. ISWI Remodelers Slide Nucleosomes with Coordinated Multi-Base-Pair Entry Steps and Single-Base-Pair Exit Steps. Cell. 2013;152:442–452. doi: 10.1016/j.cell.2012.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods in enzymology. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Tóth K, Langowski J, Seidel CA. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc Natl Acad Sci US A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nature Structural & Molecular Biology. 2010;17:932–U937. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ. Flexibility of DNA. Annual Review of Biophysics and Biophysical Chemistry. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nature Structural & Molecular Biology. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DMJ, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Broströmer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, et al. Probing Allostery Through DNA. Science. 2013;339:816–819. doi: 10.1126/science.1229223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans WJA, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. Journal of Fluorescence. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin Structure - Repeating Unit of Histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kruithof M, van Noort J. Hidden Markov Analysis of Nucleosome Unwrapping Under Force. Biophysical Journal. 2009;96:3708–3715. doi: 10.1016/j.bpj.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Malyuchenko NV, Nikitin DV, Demidenko AV, Chertkov OV, Efimova NS, Kirpichnikov MP, Studitsky VM. Molecular mechanisms of transcription through a nucleosome by RNA polymerase II. Mol Biol. 2013;47:655–667. [PubMed] [Google Scholar]
- Li G, Widom J. Nucleosomes facilitate their own invasion. Nature Structural & Molecular Biology. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature Structural & Molecular Biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mack AH, Schlingman DJ, Ilagan RP, Regan L, Mochrie SGJ. Kinetics and Thermodynamics of Phenotype: Unwinding and Rewinding the Nucleosome. Journal of Molecular Biology. 2012;423:687–701. doi: 10.1016/j.jmb.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffeo C, Ngo TTM, Ha T, Aksimentiev A. A Coarse-Grained Model of Unstructured Single-Stranded DNA Derived from Atomistic Simulation and Single-Molecule Experiment. Journal of Chemical Theory and Computation. 2014;10:2891–2896. doi: 10.1021/ct500193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher RL, Prasad A, Rizvanova O, Wallace SS, Pederson DS. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA Repair. 2013;72:964–971. doi: 10.1016/j.dnarep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–U581. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical Journal. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsaidov U, Timp W, Zou X, Dimitrov V, Schulten K, Feinberg AP, Timp G. Nanoelectromechanics of Methylated DNA in a Synthetic Nanopore. Biophysical Journal. 2009;96:L32–L34. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollazadeh-Beidokhti L, Mohammad-Rafiee F, Schiessel H. Nucleosome Dynamics between Tension-Induced States. Biophysical Journal. 2012;102:2235–2240. doi: 10.1016/j.bpj.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag R, Smerdon MJ. Altering the chromatin landscape for nucleotide excision repair. Mutation Research-Reviews in Mutation Research. 2009;682:13–20. doi: 10.1016/j.mrrev.2009.01.002. [DOI] [PubMed] [Google Scholar]
- North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. Regulation of the nucleosome unwrapping rate controls DNA accessibility. Nucleic Acids Research. 2012;40:10215–10227. doi: 10.1093/nar/gks747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. How bidirectional becomes unidirectional. Nature Structural & Molecular Biology. 2013;20:1022–1024. doi: 10.1038/nsmb.2657. [DOI] [PubMed] [Google Scholar]
- Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nature Structural Biology. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin PMD, Zou X, Gaub HE, Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Research. 2011;39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nature Communications. 2013;4 doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nature Structural & Molecular Biology. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- Sirinakis G, Ciapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. Embo Journal. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhanshu B, Mihardja S, Koslover EF, Mehraeen S, Bustamante C, Spakowitz AJ. Tension-dependent structural deformation alters single-molecule transition kinetics. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1885–1890. doi: 10.1073/pnas.1010047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of Nucleosome Invasion by DNA Binding Proteins. Journal of Molecular Biology. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, Böhm V, Seilmann C, Danner M, Hanne J, Berg M, Barz I, Gansen A, Langowski J. Histone- and DNA sequence-dependent stability of nucleosomes studied by single-pair FRET. Cytometry Part A. 2013;83:839–846. doi: 10.1002/cyto.a.22320. [DOI] [PubMed] [Google Scholar]
- Vafabakhsh R, Ha T. Extreme Bendability of DNA Less than 100 Base Pairs Long Revealed by Single-Molecule Cyclization. Science. 2012;337:1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J. Role of DNA sequence in nucleosome stability and dynamics. Quarterly Reviews of Biophysics. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes & Development. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 Chromatin Remodelers Recognize Nucleosome-free Regions Near+1 Nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- Zhou RB, Schlierf M, Ha T. Force fluorescence spectroscopy at the single-molecule level. In: Walter NG, editor. Methods in Enzymology, Vol 475: Single Molecule Tools, Pt B: Super-Resolution, Particle Tracking, Multiparameter, and Force Based Methods. San Diego: Elsevier Academic Press Inc; 2010. pp. 405–426. [DOI] [PubMed] [Google Scholar]
- Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell. 2011;146:222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.