SUMMARY
Neisseria gonorrhoeae is an obligate human pathogen and the causative agent of the sexually-transmitted disease gonorrhea. The control of this disease has been compromised by the increasing proportion of infections due to antibiotic-resistant strains, which are growing at an alarming rate. N. gonorrhoeae MtrF is an integral membrane protein, which belongs to the AbgT family of transporters for which no structural information is available. Here we describe the crystal structure of MtrF, revealing a dimeric molecule with architecture distinct from all other families of transporters. MtrF is a bowl-shaped dimer with a solvent-filled basin extending from the cytoplasm to halfway across the membrane bilayer. Each subunit of the transporter contains nine transmembrane helices and two hairpins, posing a plausible pathway for substrate transport. A combination of the crystal structure and biochemical functional assays suggests that MtrF is an antibiotic efflux pump, mediating bacterial resistance to sulfonamide antimetabolite drugs.
INTRODUCTION
Neisseria gonorrhoeae is a gram-negative diplococcus, which is found only in humans and causes the sexually transmitted disease gonorrhea. Since it is a strictly human pathogen and can colonize both male and female genital mucosal surfaces and other sites, it has developed mechanisms to overcome antimicrobial systems of the host’s innate defense. One major mechanism that N. gonorrhoeae uses to resist antimicrobial agents is the expression of multidrug efflux pumps that recognize and actively export a variety of structurally unrelated toxic compounds from the bacterial cell, including antibacterial peptides, long-chain fatty acids, and several clinically important antibiotics (Lee and Shafer, 1999; Rouquette-Loughlin et al., 2003; Shafer et al., 1998; Shafer et al., 2001; Unemo and Shafer, 2014).
The best characterized efflux system in N. gonorrhoeae is the MtrC-MtrD-MtrE multidrug efflux system (Delahay et al., 1997; Lucas et al., 1995; Hagman et al., 1995; Hagman et al., 1997; Veal and Shafer, 2003; Warner et al., 2008). This system is similar to other efflux pumps of the resistance-nodulation-cell division (RND) superfamily (Tseng et al., 1999) possessed by many Gram-negative bacteria. MtrD (Bolla et al., 2014; Hagman et al., 1995; Hagman and Shafer, 1995; Hagman et al., 1997) is the inner membrane transporter component of the tripartite RND pump. The complex is formed by interactions between MtrD, the periplasmic membrane fusion protein MtrC (Hagman et al., 1995; Hagman et al., 1997; Veal et al., 2002; Janganan et al., 2013), and the outer membrane channel MtrE (Delahay et al., 1997; Lei et al., 2014; Janganan et al., 2011; Janganan et al., 2013). This powerful efflux complex mediates the export of several structurally diverse hydrophobic antimicrobial agents, such as antibiotics, nonionic detergents, antibacterial peptides, bile salts, and gonadal steroidal hormones (Delahay et al., 1997; Hagman et al., 1995; Hagman et al., 1997; Shafer et al., 1998). In addition, the Mtr efflux system includes another inner membrane protein MtrF (Veal and Shafer, 2003; Folster and Shafer, 2005), which belongs to the AbgT family of transporters (Prakash et al., 2003). It has been proposed that MtrF cooperates with the MtrC-MtrD-MtrE complex to export certain antimicrobials by a yet unknown mechanism (Veal and Shafer, 2003).
To date, approximately 13,000 putative transporters of the AbgT family have been identified. AbgT-type proteins are commonly found in Gram-negative bacteria such as Salmonella enterica, Gram-positive bacteria such as Staphylococcus aureus, as well as yeasts such as Saccharomyces arboricola. Surprisingly, among proteins in this diverse family, only Escherichia coli AbgT (Hussein et al., 1998; Carter et al., 2007) and N. gonorrhoeae MtrF (Folster and Shafer, 2005; Veal and Shafer, 2003) have been partially characterized. Thus far, there is no structural information available for this family of membrane proteins, obscuring the details of their function and mechanism.
The products of the E. coli abg genes have been shown to catalyze the uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate (Carter et al., 2007). Particularly, E. coli AbgT has been demonstrated to import the catabolite p-aminobenzoyl-glutamate for de novo folic acid synthesis (Carter et al., 2007). As E. coli AbgT and N. gonorrhoeae MtrF belong to the same family of transporters, there is a chance that the MtrF protein may also act as an importer to uptake p-aminobenzoyl-glutamate and related small molecules for the synthesis of the essential folate vitamin. However, since MtrF is needed for the high-level resistance of gonococci to hydrophobic antimicrobials, including erythromycin and TX-100 (Folster and Shafer, 2005), it seems more likely that it participates in drug efflux. To understand the transport functions of members of the AbgT family, we here present the crystal structure of the N. gonorrhoeae MtrF transporter. Importantly, we show that N. gonorrhoeae MtrF is capable of exporting the folate metabolite p-aminobenzoic acid (PABA) from cells. A combination of the three-dimensional structure and genetic analysis allows us to identify key MtrF residues that are important for the function of this membrane protein. Finally, we demonstrate that MtrF behaves as an antibiotic efflux pump, which is responsible for removing sulfonamides from the cell and mediating bacterial resistance to this class of antimetabolites.
RESULTS AND DISCUSSION
Overall structure of N. gonorrhoeae MtrF
The N. gonorrhoeae MtrF transporter is composed of 522 amino acids, sharing 38% identity with E. coli AbgT (Fig. S1). The crystal structure of this membrane protein was determined to a resolution of 3.95 Å (Table S1). Two molecules of MtrF, which assemble as a dimer, are found in the asymmetric unit (Fig. S2 and Fig. 1). Superimposition of these two MtrF molecules gives an RMSD of 0.5 Å over 506 Cα atoms, indicating that their conformations are nearly identical to each other. The dimeric form of MtrF in the crystal lattice is in good agreement with the oligomerization state of this membrane protein in detergent solution in which it also assembles as a dimer (Fig. S3).
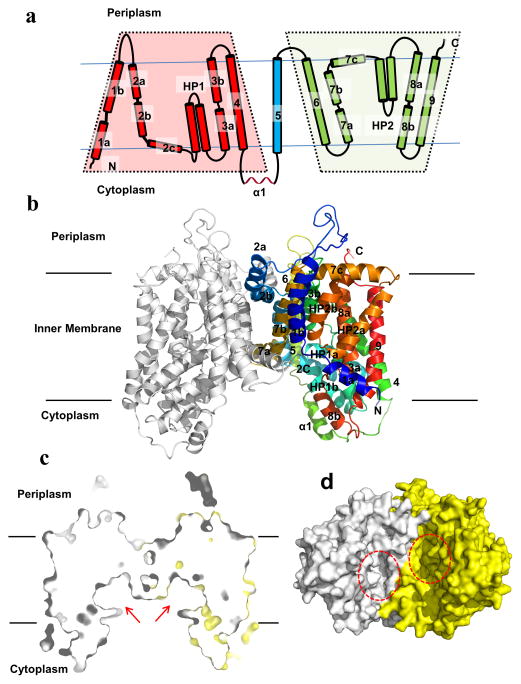
Fig. 1.
Structure of the N. gonorrhoeae MtrF transporter. (a) Transmembrane topology of N. gonorrhoeae MtrF. The transporter contains nine transmembrane helices (TMs) and two hairpins (HPs). (b) Ribbon diagram of a dimer of MtrF viewed in the membrane plane. The right subunit of the dimer is colored using a rainbow gradient from the N-terminus (blue) to the C-terminus (red), whereas the left subunit is colored gray. The MtrF dimer forms a bowl-shaped structure with a concave aqueous basin facing the intracellular solution. (c) Surface representation of a cross section of the MtrF dimer sliced through the middle of the protein. Each protomer forms an internal cavity (red arrow), which is accessible to the cytoplasm. (d) Bottom view of a surface representation of the MtrF dimer, indicating a solvent accessible cavity (red circle) from each protomer of the protein. The two protomers are colored gray and yellow.
The fold of MtrF is unique and composed of a number of unusual structural elements. Each subunit of MtrF comprises nine α-helical transmembrane segments and two helical hairpins: TM1 (a (12–22) and b (26–47)), TM2 (a (78–92), b (94–112) and c (114–125)), HP1 (a (128–145) and b (147–164)), TM3 (a (168–182) and b (191–205)), TM4 (218–240), TM5 (269–292), TM6 (310–334), TM7 (a (341–353), b (356–374) and c (376–391)), HP2 (a (396–413) and b (417–434)), TM8 (a (438–451) and b (462–471)) and TM9 (480–506). It should be noted that five of these TMs (TM1, TM2, TM3, TM7 and TM8) are broken into segments within the membrane. In addition, HP1 and HP2 are formed by relatively short helices, which are only long enough to span half of the membrane (Fig. 1a and b). Interestingly, the intramembrane loops of several of these TMs and HPs are right next to each other, allowing the transporter to form an internal cavity within the membrane (Fig. 1c and d). Noticeably, our structure indicates that the N- and C-terminal ends of MtrF are located at different sides of the inner membrane (Fig. 1a and b). As there was no signaling peptide added when expressing the MtrF protein, the N-terminal end of this protein should be located at the cytoplasm. Thus, the C-terminal end of MtrF should be positioned within the periplasmic space. This is in good agreement with the predicted E. coli AbgT topology that the C-terminal end of this protein is located at the periplasm (Daley et al., 2005).
Each protomer of MtrF contains a relatively small periplasmic domain. This domain is made up of two long loops formed between TMs 1 and 2, and TMs 5 and 6, respectively. Below the inner leaflet of the membrane, a small cytoplasmic domain links TMs 4 and 5 together. This domain is comprised by a relatively long random loop and helix (α1) (Fig. 1a).
Viewed in parallel to the membrane, the MtrF dimer is bowl-shaped with a concave aqueous basin facing the intracellular solution (Fig. 1b). The dimer is about 75 Å tall, 80 Å wide and 50 Å thick, with the transmembrane portion of the transporter lying approximately in the middle. The rim of the basin is as large as 45 Å. The bowl-shaped structure is 25 Å in depth and deeply penetrates into the inner leaflet of the cytoplasmic membrane (Fig. 1c). This basin probably allows aqueous solution to reach the midpoint of the membrane bilayer. Interestingly, a similar bowl-shaped structure has also been found in the glutamate transporter (Yernool et al., 2004). However, the bowl-shape, which deeply penetrates into the membrane, is formed within the trimer of this transporter (Yernool et al., 2004).
The structure of the MtrF dimer can be roughly divided into the inner and outer cores (Fig. 2). The inner core contains TMs 1, 2, 5, 6, and 7, creating a frame-like structure to house the outer core of the protein (Fig. 2a). A portion of this inner core is responsible for forming a dimerization domain of the transporter. Viewed along the membrane normal, TMs 1b, 2a, 2b, 6, 7a, and 7b, as well as their counter parts from the next subunit, contribute to this distinct dimerization domain.
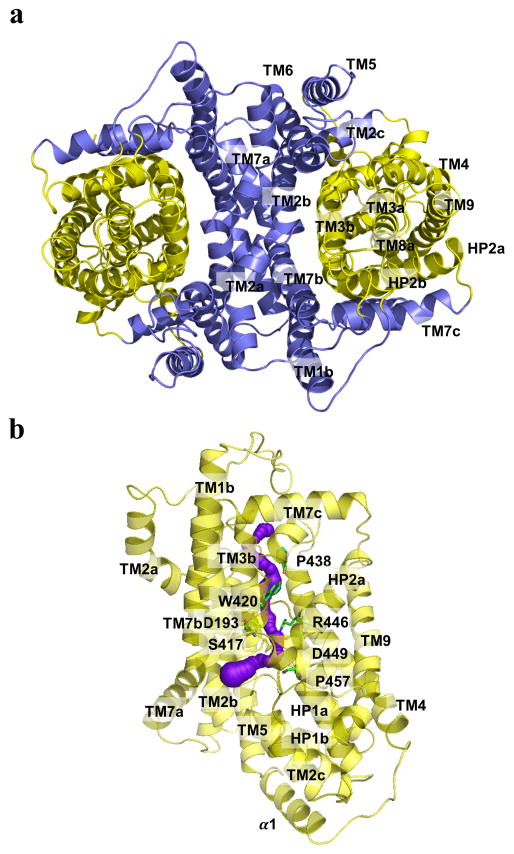
Fig. 2.
Inner and Outer cores of MtrF. (a) The inner core of MtrF, comprising TMs 1, 2, 5, 6 and 7 (colored slate), contributes to dimerization as well as formation of a frame-like structure housing the outer core of the protomer. The outer core of MtrF is composed of TMs 3, 4, 8, 9 as well as HPs 1 and 2 (colored yellow). (b) The outer core of MtrF forms a channel (colored purple) spanning approximately from the middle of the inner membrane up to the periplasmic space. This channel was calculated using the program CAVER (http://loschmidt.chemi.muni.cz/caver). The secondary structural elements of the MtrF protomer are in yellow. Residues D193, S417, W420, P438, R446, D449, and P457 are in green sticks.
Noticeably, the outer core of the MtrF protomer, comprising TMs 3, 4, 8, and 9, as well as HPs 1 and 2, folds into a cylindrical structural feature (Fig. 2a). It is most likely that this outer core cylinder forms a substrate-binding site and transport pathway. Based on the crystal structure, the outer core cylinder contributes to form a tunnel spanning approximately from the middle of the inner membrane up to the periplasmic space (Figs. 2b and S4). This tunnel is surrounded with HPs 1 and 2, as well as TMs 3 and 8. Interestingly, this tunnel is connected to the cytoplasmic space through an opening located at the basin of the bowl-shaped structure. The loop regions of HP1, HP2, TM3, and TM8 form this opening. Several conserved residues, including D193, S417, W420, P438, R446, D449, and P457, are found to line the wall of the channel (Fig. 2b). These residues may play an important role for the function.
MtrF is capable of exporting p-aminobenzoic acid from the cell
As E. coli AbgT has been shown to enable uptake of the folate catabolite p-aminobenzoyl-glutamate (Carter et al., 2007), we investigated if E. coli cells expressing N. gonorrhoeae MtrF can grow in liquid minimal medium (containing 90.4 mM Na2HPO4, 22.0 mM KH2PO4, 8.5 mM NaCl, 0.1 mM CaCl2, 1.0 mM MgSO4, 20.0 mM NH4Cl, and 22.2 mM glucose) supplemented with p-aminobenzoyl-glutamate. Based on several studies on folic acid transporters (Eudes et al., 2010; Klaus et al., 2005; Salcedo-Sora et al., 2011), we made an E. coli knockout strain BL21(DE3)ΔabgTΔpabA that lacks the abgT gene (Hussein et al., 1998; Carter et al., 2007) to impede uptake of the catabolite p-aminobenzoyl-glutamate and pabA gene (Kaplan and Nichols, 1986) to impair intracellular synthesis of the metabolite PABA. We then transformed these double knockout cells with pET15bΩmtrF, expressing N. gonorrhoeae mtrF, or the empty vector pET15b. Surprisingly, the double knockout E. coli BL21(DE3)ΔabgTΔpabA cells, transformed with either pET15bΩmtrF or pET15b, could not grow in this liquid medium supplemented with p-aminobenzoyl-glutamate up to 1 mM. However, these cells were capable of growing in liquid minimal medium when supplemented with 30 nM PABA. Within the E. coli abg operon, there are two additional genes, abgA and abgB, located upstream of abgT. The products of these two genes are the AbgA and AbgB aminoacyl aminohydrolases, which cleave the catabolite p-aminobenzoyl-glutamate imported by the AbgT transporter to form PABA for folic acid synthesis (Carter et al., 2007). In contrast, the mtr operon of N. gonorrhoeae does not possess abgA or abgB related genes (www.genome.ou.edu). Thus, our data suggest that MtrF and AbgT may function differently even though these two membrane proteins belong to the same family.
As PABA is an important precursor for folic acid synthesis, we thought that N. gonorrhoeae MtrF might enable uptake of PABA. It should be noted that PABA is capable of diffusing into bacterial cells through the membrane, participating as an intermediate in de novo synthesis of the essential vitamin folic acid. Nonetheless, we compared the radioactive PABA content over time in cells transformed with either pET15bΩmtrF or the empty vector pET15b. Surprisingly, E. coli cells producing N. gonorrhoeae MtrF showed a significant decrease in the level of [3H]-PABA, in contrast to cells transformed with the empty pET15b vector (Fig. 3a). Instead of an importer, the data strongly suggested to us that MtrF may act as an exporter, capable of expelling the intracellular PABA metabolite from the cell.
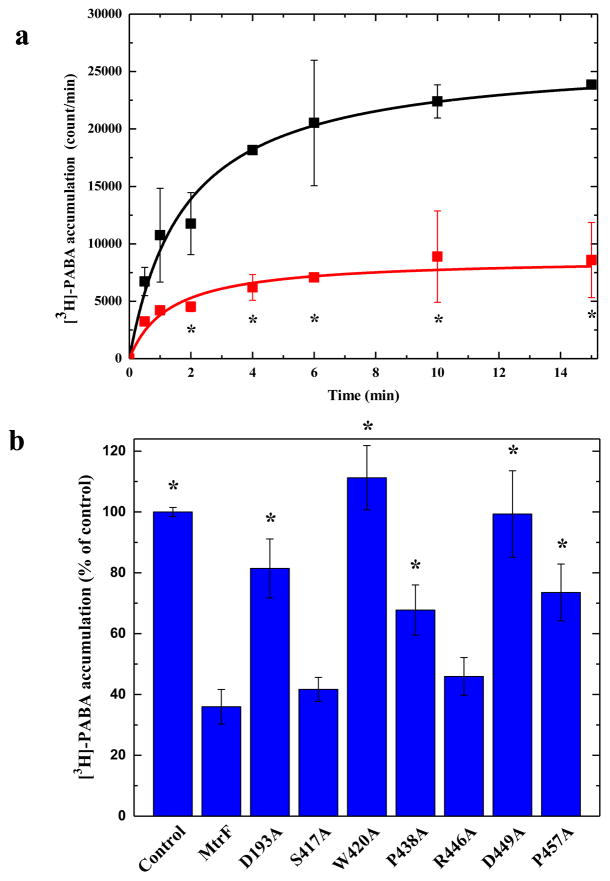
Fig. 3.
Accumulation of radioactive p-aminobenzoic acid. (a) Time course of [3H]-PABA accumulation by E. coli BL21(DE3)ΔabgTΔpabA double knockout cells transformed with pET15bΩmtrF or pET15b. Cells expressing mtrF (red curve) show a significant decrease in [3H]-PABA accumulation when compared with cells carrying the empty vector (black curve). “*” indicates values of BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells that are significantly different from the control (BL21(DE3)ΔabgTΔpabA/pET15b) values (P < 0.05). (b) Mutants of the MtrF transporter. Cells possessing the mutant transporter D193A, W420A, P438A, D449A and P457A show a significant increase in the level of [3H]-PABA accumulations compared with cells expressing wild-type MtrF. However, cells expressing S417A and R446A only show a modest change on the [3H]-PABA concentration when compared with cells carrying wild-type MtrF. “*” indicates values of BL21(DE3)ΔabgTΔpabA/pET15b and BL21(DE3)ΔabgTΔpabA cells expressing the mutant transporters that are significantly higher than that of BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF expressing wild-type MtrF (P < 0.009). The data showed in (a) and (b) are the cumulative average of three successive recordings.
In order to determine whether the conserved MtrF residues, D193, S417, W420, P438, R446, D449, and P457, lining the inner wall of the tunnel formed by each protomer are important for the function of the transporter, we mutated each residue to alanine, individually (Table S2). We expressed these mutant transporters in BL21(DE3)ΔabgTΔpabA. Western analysis suggested that the expression levels of these mutant transporters are comparable with that of wild-type MtrF (Fig. S5). In addition, Western analysis on the crude cell lysates indicates that the level of expression of the wild-type MtrF transporter was ~200 copies per cell (Fig. S6). In comparison with the expression level of the AcrB pump, which is ~500 copies per cell (Tikhonova and Zgurskaya, 2004), the expression level of our MtrF proteins should be low enough for physiological measurement. We then measured the accumulation of [3H]-PABA in cells harboring the mutant transporters, D193A, S417A, W420A, P438A, R446A, D449A, and P457A. The results showed a significant increase in the levels of [3H]-PABA accumulation in cells possessing the mutant transporter D193A, W420A, P438A, D449A, or P457A, when compared with cells expressing wild-type N. gonorrhoeae MtrF (Fig. 3b). However, cells expressing S417A and R426A only indicated a modest change in the [3H]-PABA concentration when compared with cells carrying wild-type MtrF (Fig. 3b). On the contrary, the levels of [3H]-PABA accumulation in cells expressing the mutants W420A and D449A were nearly the same as those transformed with the empty vector, indicating that these mutant transporters are not functional and cannot export PABA from cells. The data suggest that MtrF acts as an efflux pump, and these amino acids are important for its function.
Expression of the MtrF transporter decreases intracellular folic acid concentration
We hypothesized that N. gonorrhoeae MtrF is capable of catalyzing the efflux of the folate metabolite PABA. If this is the case, then cells expressing MtrF should show a lower level of intracellular folic acid concentration. Therefore, we measured the intracellular folic acid content of these cells microbiologically using Lactobacillus casei (Wilson and Horne, 1982). Cells were grown in liquid minimal medium supplemented with 30 nM PABA. These cells were harvested when the optical density (OD600 nm) reached 0.5. Cells were disrupted with a French pressure cell. The crude cell lysates were then assayed microbiologically using L. casei to obtain their folic acid concentrations. Consistent with the results from PABA accumulation, the folic acid concentration in cells producing MtrF were markedly reduced in comparison with cells transformed with the empty vector (Fig. 4). These data further support our hypothesis that MtrF acts as an efflux pump and participates in exporting PABA from the cell.
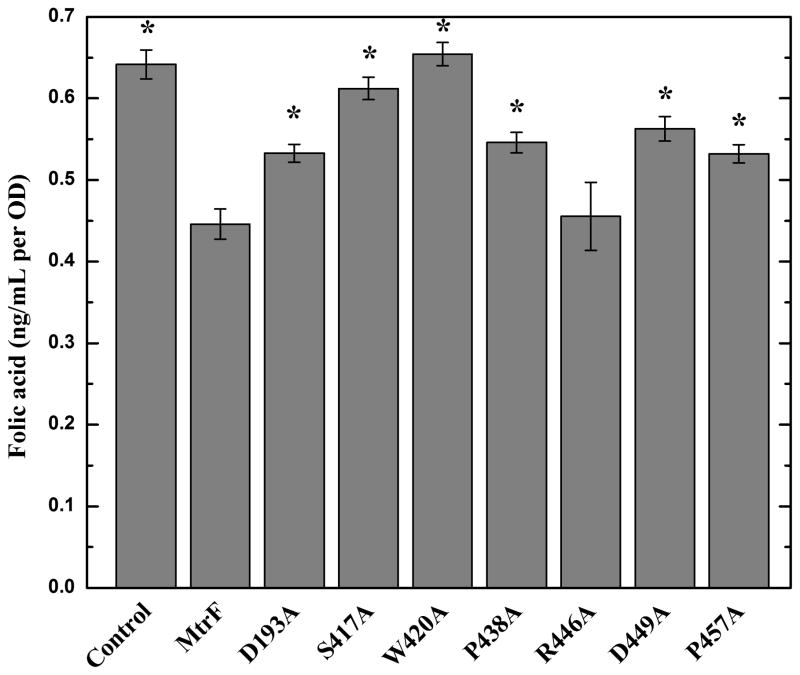
Fig. 4.
Intracellular folic acid concentration. Folic acid concentration in E. coli BL21(DE3)ΔabgTΔpabA double knockout cells expressing MtrF were markedly reduced in comparison with cells transformed with the empty vector. When transformed with plasmid expressing the mutant transporter, D193A, S417A, W420A, P438A, D449A, and P457A, folic acid production was significantly increased in these cells. However, the level of intracellular folic acid concentration in BL21(DE3)ΔabgTΔpabA cells expressing R446A was nearly identical to that of the double knockout strain carrying wild-type MtrF. Each bar represents the mean of three separate cultures. “*” indicates values of BL21(DE3)ΔabgTΔpabA/pET15b and BL21(DE3)ΔabgTΔpabA cells expressing the mutant transporters that are significantly higher than that of BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF expressing wild-type MtrF (P < 0.04).
We next investigated how residues D193, S417, W420, P438, R446, D449, and P457 affect the intracellular folic acid concentration. When transformed with plasmids expressing the mutant transporter W420A, folic acid production was restored in these cells and the level of intracellular folic acid concentration was similar to that of the double knockout strain transformed with the empty vector (Fig. 4). In addition, cells carrying the mutants, D193A, S417A, P438A, D449A, and P457A, indicated a significant increase in the level of folic acid concentration when compared with cells expressing wild-type MtrF (Fig. 4). Again, the data indicate that these residues are important for the function of the MtrF efflux pump. However, for cells expressing R446A, the intracellular folic acid content was similar to that of cells carrying the wild-type MtrF transporter.
MtrF is an antibiotic efflux pump
As PABA is an important metabolite for producing the essential folic acid in bacteria, we questioned why the function of MtrF appears to be decreasing the intracellular PABA concentration. We postulated that the answer may lie in the ability of MtrF to protect bacterial cells by extruding antimetabolites that are structurally similar to PABA, specifically sulfonamides. Sulfonamides are antimicrobial agents, sometimes referred to as antimetabolites or growth factor analogs, which are designed to specifically inhibit the essential metabolic pathway for folic acid synthesis in bacterial pathogens. At the chemical level, sulfonamides are structurally similar to the metabolite PABA, making them ideal competitive inhibitors. Sulfonamides were used in the late 1930s and early 1940s to treat gonorrhea, but the rapid emergence of strains resistant to this class of drug resulted in its removal once penicillin became available (Unemo & Shafer, 2014); resistant strains were later found to contain mutations in folP that encodes the target for sulfonamides. We therefore suspect that MtrF may be a drug efflux pump, which is capable of recognizing and extruding sulfonamide antimetabolites. In fact, we found that the minimum inhibitory concentration (MIC) of sulfanilamide for the N. gonorrhoeae strain WV16 (an mtrF knockout strain) differed from that of the N. gonorrhoeae parental strain FA140 by twofold (MIC of 500 vs. 250 μg/ml, respectively).
To determine if MtrF behaves like a sulfonamide efflux pump, we used E. coli as a surrogate host. We transformed BL21(DE3)ΔabgTΔpabA with pET15bΩmtrF or pET15b and tested for the susceptibility of these transformants to four different sulfonamide antimetabolites, sulfamethazine, sulfadiazine, sulfathiazole and sulfanilamide (Table S3). It is important to stress that in many instances expression of drug efflux pumps, including members of the RND (Tseng et al., 1999; Li et al., 1995; Rosenberg et al., 2000) and multidrug and toxic compound extrusion (MATE) superfamilies (Brown et al., 1999; Long et al., 2008; Rouquette-Loughlin et al., 2003), can have only modest changes (twofold) in bacterial susceptibility to certain antimicrobials while more significant changes in susceptibility to other agents can be observed in the same system. We found that the BL21(DE3)ΔabgTΔpabA cells producing MtrF were 32-fold less sensitive to sulfamethazine when compared with BL21(DE3)ΔabgTΔpabA cells containing the empty pET15b vector. In addition, this BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF transformant was found to be eight times more resistant to sulfanilamide in comparison to the double knockout cells transformed with pET15b. We also found that the MICs of BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells for sulfadiazine and sulfathiazole were both greater than eight times than those of BL21(DE3)ΔabgTΔpabA cells carrying the empty vector pET15b. These data indeed indicate that MtrF functions as a drug efflux pump and reduced bacterial susceptibility to sulfonamides.
The binding affinities of these four sulfonamide drugs, sulfamethazine, sulfadiazine, sulfathiazole and sulfanilamide, as well as PABA for the MtrF transporter were then determined using isothermal titration calorimetry (ITC). The data indicate that the dissociation constants, KDs, for sulfamethazine, sulfadiazine, sulfathiazole, sulfanilamide and PABA bindings are 0.33 ± 0.02, 12.74 ± 0.62, 1.52 ± 0.07, 1.14 ± 0.01 and 0.54 ± 0.02 μM, respectively (Fig. S7 and Table S4). These ligand-binding experiments indeed confirm that MtrF is capable of recognizing these ligands.
In order to further test the drug efflux capability of MtrF, we expressed the mutant transporters D193A, S417A, W420A, P438A, R446A, D449A, and P457A in BL21(DE3)ΔabgTΔpabA and evaluated for their ability to confer sulfamethazine, sulfadiazine, sulfathiazole and sulfanilamide resistance. In most cases, we found that strain BL21(DE3)ΔabgTΔpabA carrying these MtrF mutants are more sensitive to sulfamethazine and sulfanilamide in comparison with cells expressing wild-type MtrF (Table S3). With the exception of the R446 variant, our data indicate that strain BL21(DE3)ΔabgTΔpabA carrying the MtrF mutants were hypersensitive to sulfadiazine and sulfathiazole in comparison with cells expressing wild-type MtrF (Table S3). These data indeed agree with the idea that these residues are essential for the function of MtrF.
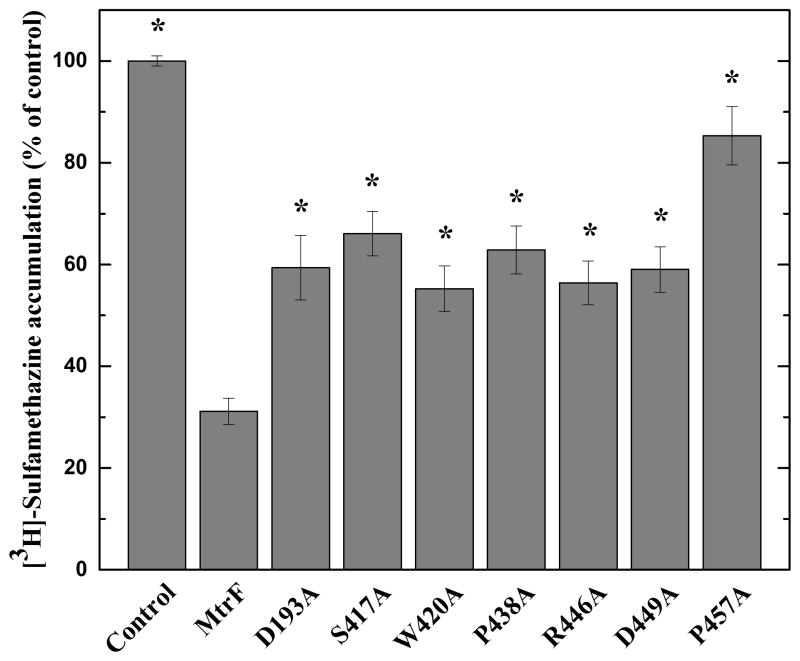
To confirm these drug susceptibility testing results, we measured the accumulation of radioactive sulfonamide in BL21(DE3)ΔabgTΔpabA cells carrying pET15bΩmtrF or pET15b. For these experiments, we compared the accumulation of [3H]-sulfamethazine over time in these cells. As shown in Fig. 5, the results indicate a much lower level of [3H]-sulfamethazine accumulation in BL21(DE3)ΔabgTΔpabA cells producing MtrF, compared to controlled cells harboring the empty pET15b vector, suggesting that MtrF is capable of exporting sulfamethazine from the cell.
Fig. 5.
Accumulation of radioactive sulfamethazine. E. coli BL21(DE3)ΔabgTΔpabA cells expressing MtrF show a significant decrease in [3H]-sulfamethazine accumulation when compared with cells carrying the empty vector. When transformed with plasmids expressing the mutant transporters, D193A, S417A, W420A, P438A, R446A, D449A, and P457A, the levels of intracellular [3H]-sulfamethazine accumulation were much higher than that of cells expressing wild-type MtrF. Each bar represents the mean of three different cultures. “*” indicates values of BL21(DE3)ΔabgTΔpabA/pET15b and BL21(DE3)ΔabgTΔpabA cells expressing the mutant transporters that are significantly higher than that of BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF expressing wild-type MtrF (P < 0.001).
When transformed with plasmids expressing the mutant transporters, D193A, S417A, W420A, P438A, R446A, D449A, and P457A, the level of intracellular sulfamethazine accumulation were much higher than that of cells expressing wild-type MtrF (Fig. 5). Again, these data indicate that residues D193, S417, W420, P438, R446, D449, and P457 are critical for the function of the MtrF efflux pump.
In addition, we determined the binding affinities of sulfamethazine, sulfadiazine, sulfathiazole, sulfanilamide and PABA for the W420A mutant transporter using ITC. The data depict that the KDs, for sulfamethazine, sulfadiazine, sulfathiazole, sulfanilamide and PABA bindings are 10.78 ± 1.17, 105.82 ± 25.60, 50.76 ± 8.90, 6.80 ± 1.49 and 41.15 ± 11.50 μM, respectively (Table S5). These bindings are much weaker than those of wild-type MtrF, suggesting that residue W420 is critical for the function of the protein.
MtrF behaves as a PMF-dependent drug efflux pump
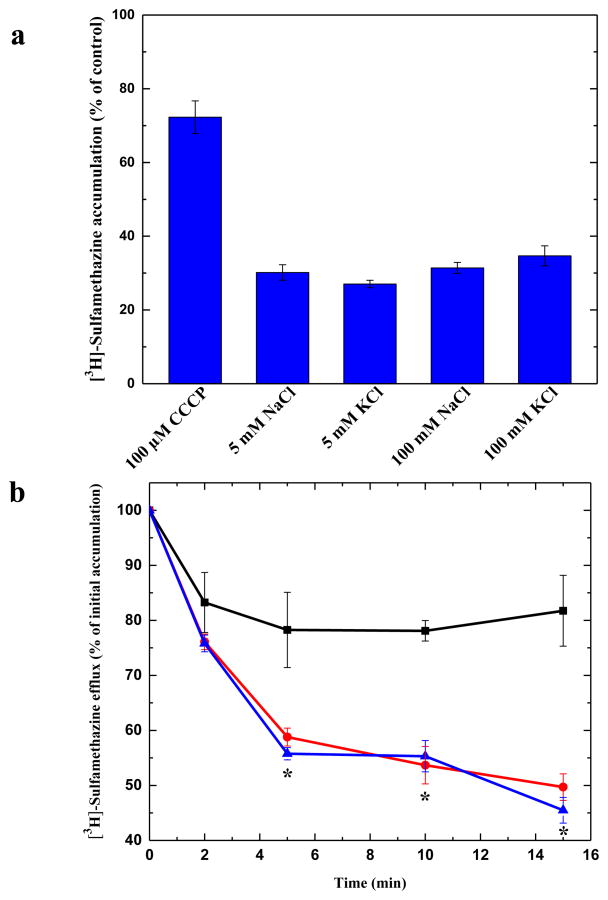
It has been suggested that members of the AbgT family use the proton-motive-force (PMF) to transport substrates across the membrane (Prakash et al., 2003). To elucidate if N. gonorrhoeae MtrF is a PMF-dependent transporter, we measured the level of intracellular sulfamethazine accumulation in the presence of carbonyl cyanide m-chlorophenylhydrazone (CCCP), a de-coupler of the membrane proton gradient. After the addition of CCCP into the assay solution, the accumulation of [3H]-sulfamethazine increased drastically in the MtrF-expressing cells (Fig. 6a), suggesting the possibility that MtrF is PMF-dependent.
Fig. 6.
Transport of sulfamethazine via MtrF. (a) Accumulation of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells with different sodium or potassium ion concentrations. The data indicate that the transport function of MtrF is independent of sodium or potassium ions. (b) Efflux of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells in the absence and presence of sodium ions. The presence of Na+ does not affect [3H]-sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells (black, controlled cells transformed with empty vector; red, 0 mM NaCl; blue, 5 mM NaCl). “*” indicates values of radioactive counts of intracellular [3H]-sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells with 0 mM NaCl (red) (P < 0.003) and 5 mM NaCl (P < 0.001) that are significantly different from those of the control (black). The data showed in (a) and (b) are the cumulative average of three successive recordings.
We then investigated the accumulation level of radioactive sulfamethazine in strain BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF in the presence of NaCl or KCl. The concentrations of these metal ions were either 5 or 100 mM. In all cases, the levels of accumulation of [3H]-sulfamethazine were similar to that in the MtrF-producing strain without the addition of any metal ions (Figs. 5 and 6a), indicating that the function of MtrF is independent of Na+ or K+.
The drug accumulation data strongly suggest that MtrF is a PMF-dependent efflux pump. To test this possibility, we next measured the efflux of [3H]-sulfamethazine that had accumulated in strain BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF over time, both in the absence and presence of Na+. Cells were first loaded with [3H]-sulfamethazine and CCCP was added to inhibit the pump. Cells were then re-energized by removing CCCP and adding glucose; thereafter, radioactive measurements were performed both in the absence and presence of 5 mM NaCl. As shown in Fig. 6b, the addition of Na+ essentially has no effect on sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF. These data suggest that MtrF is a PMF-dependent efflux pump.
CONCLUSIONS
In this paper, we report the crystal structure of the N. gonorrhoeae MtrF transporter, which reveals a dimeric molecule with a fold very distinct from all other families of transporters. Our experimental data strongly suggest that N. gonorrhoeae MtrF is a drug efflux pump, capable of removing sulfonamide antimetabolites and mediating resistance to this class of drugs in bacterial cells. It is likely that many members of the AbgT family of transporters may serve as antimetabolite efflux pumps to protect cells against these noxious agents.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification of N. gonorrhoeae MtrF
Briefly, the full-length MtrF membrane protein containing a 6xHis tag at the N-terminus was overproduced in E. coli BL21(DE3)ΔacrB cells, which harbor a deletion in the chromosomal acrB gene, possessing pET15bΩmtrF. Cells were grown in 12 l of LB medium with 100 μg/ml ampicillin at 25°C. When the OD600 nm reached 0.5, the culture was treated with 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to induce mtrF expression, and cells were harvested within 15 h. The collected bacteria were resuspended in buffer containing 100 mM sodium phosphate (pH 7.2), 10% glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM phenylmethanesulfonyl fluoride (PMSF), and then disrupted with a French pressure cell. The membrane fraction was collected and washed twice with buffer containing 20 mM sodium phosphate (pH 7.2), 2 M KCl, 10% glycerol, 1 mM EDTA and 1 mM PMSF, and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM PMSF as described previously (Long et al., 2010). The membrane protein was then solubilized in 2 % (w/v) n-dodecyl-β-D-maltoside (DDM). Insoluble material was removed by ultracentrifugation at 100,000 x g. The extracted protein was purified with a Ni2+-affinity column. The purified protein was dialyzed and concentrated to 20 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM. The 6xHis tag at the N-terminus was then cleaved by adding 5 units of thrombin (GE Healthcare Bio-Sciences, Pittsburgh, PA) per mg of purified MtrF at room temperature for 20 h. The protein was subsequently passed through a Ni2+-affinity column to remove the free 6xHis tag. A final purification step was performed using a G200 size exclusion column loaded with buffer solution containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM. The purity of the MtrF protein (>95%) was judged using 10% SDS-PAGE stained with Coomassie Brilliant Blue. The purified protein was then concentrated to 20 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM.
For 6xHis selenomethionyl-substituted (SeMet)-MtrF protein expression, a 10 ml LB broth overnight culture containing E. coli BL21(DE3)ΔacrB/pET15bΩmtrF cells was transferred into 120 ml of LB broth containing 100 μg/ml ampicillin and grown at 37°C. When OD600 nm value reached 1.2, cells were harvested by centrifugation at 6000 rev/min for 10 min, and then washed two times with 20 ml of M9 minimal salts solution. The cells were re-suspended in 120 ml of M9 media and then transferred into a 12 l pre-warmed M9 solution containing 100 μg/ml ampicillin. The cell culture was incubated at 25°C with shaking. When OD600 nm reached 0.4, 100 mg/l of lysine, phenylalanine and threonine, 50 mg/l isoleucine, leucine and valine, and 60 mg/l of L-selenomethionine were added. The culture was induced with 0.2 mM IPTG after 15 min. Cells were then harvested within 15 h after induction. The procedures for purifying SeMet-MtrF were identical to those of the native protein.
Crystallization of MtrF
Crystals of the MtrF transporter were obtained using vapor diffusion. The MtrF crystals were grown at room temperature with the following procedures. A 2 μl protein solution containing 20 mg/ml MtrF protein in 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) DDM was mixed with a 2 μl of reservoir solution containing 30% PEG 400, 0.1 M sodium acetate (pH 5.0), 0.1 M magnesium acetate, 3% glycerol and 1% (w/v) octyl glucose neopentyl glycol (OGNG). The resultant mixture was equilibrated against 500 μl of the reservoir solution. The crystallization conditions for SeMet-MtrF were the same as those for native MtrF. Crystals of both MtrF and SeMet-MtrF grew to a full size in drops within a month. Typically, the dimensions of the crystals were 0.2 mm×0.2 mm×0.2 mm. Cryoprotection was achieved by raising the PEG 400 concentration to 32%.
Crystals of the Ta6Br122+ or W6(μ-O)6(μ-Cl)6Cl62− cluster derivative were prepared by incubating the crystals of MtrF in solution containing 32% PEG 400, 0.1 M sodium acetate (pH 4.6), 0.05 M magnesium acetate, 5% glycerol, 1% (w/v) OGNG, 0.05% (w/v) DDM and 0.5 mM Ta6Br12.2Br (Jena Bioscience GmbH, Jena, Germany) or 0.5 mM (NH4)2W6(μ-O)6(μ-Cl)6Cl6 for 4 hours at 25°C.
Data collection, structural determination and refinement
All diffraction data were collected at 100K at beamline 24ID-C located at the Advanced Photon Source, using a Pilatus 6M detector (Dectris Ltd., Switzerland). Diffraction data were integrated using DENZO and scaled using SCALEPACK (Otwinowski and Minor, 1997). Crystals of MtrF belong to space group P65 (Table S1). Based on the molecular weight of MtrF (56.3 kDa), two molecules per asymmetric unit with a solvent content of 71.9% were expected. Data from a native crystal, two heavy-atom derivatives, namely Ta6Br122+ and W6(μ-O)6(μ-Cl)6Cl62− clusters, and a selenomethionyl-substituted (SeMet) crystal were used for phase determination (Table S1). Four tantalum (Ta6Br122+) cluster sites were identified using SHELXD (Schneider and Sheldrick, 2002) as implemented in the HKL2MAP package (Pape and Schneider, 2004). These heavy-atom sites were refined using the program AutoSol in PHENIX (Adams et al., 2002). These phases were then used to locate six tungsten cluster sites using the corresponding W6(μ-O)6(μ-Cl)6Cl62− data set. Multiple isomorphous replacement with anomalous scattering (MIRAS), including data from the tantalum and tungsten complexes, was performed at a resolution of 5.70 Å using AutoSol implemented in PHENIX (Adams et al., 2002). These phases were then subjected to density modification, NCS averaging and phase extension to native 3.95 Å-resolution using the program RESOLVE (Terwilliger, 2001). Density modified phases were good enough to visualize the secondary structural features of the protein molecule.
After tracing the initial model manually using the program Coot (Emsley and Cowtan, 2004), molecular replacement with single wavelength anomalous diffraction (MR-SAD) was performed, utilizing the SeMet data, using the program Phaser (McCoy et al., 2007). The full-length MtrF protein contains 30 methionines (excluding the first N-terminal methionine residue), all of these corresponding 30 selenium sites were found in the difference Fourier map. The electron density map was further improved by multi-crystal averaging, including the two different crystal forms (both SeMet and wild-type) of the MtrF dimer, using PHENIX (Adams et al., 2002).
The model was then rebuilt and refined against the native data at 3.95 Å-resolution using PHENIX (Adams et al., 2002), leaving 5% of reflections in Free-R set. Feature-enhanced maps calculated using PHENIX and B-factor sharpening maps created using CCP4 were employed to ascertain loop-regions and side-chains in the structure. Iterations of refinement using PHENIX (Adams et al., 2002) and CNS (Brünger et al., 1998) and model building in Coot (Emsley and Cowtan, 2004) lead to the current structural model of the MtrF transporter (Table S1). The sulfur atom positions of all 30 methionines coincide with their corresponding Se peaks in the anomalous map.
Gel filtration
A protein liquid chromatography Superdex 200 16/60 column (GE Healthcare Bio-Sciences, Pittsburgh, PA) with a mobile phase containing 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) DDM was used in the gel filtration experiments. Blue dextran (Sigma-Aldrich, St. Louis, MO) was used to determine the column void volume. As there are no commercially available membrane protein markers, we used the well-characterized N. gonorrhoeae NorM multidrug efflux pump (Lu et al., 2013; Long et al., 2008) (which exists as a monomer in solution with the monomeric molecular weight equals 50.6 kDa) and E. coli CusC outer membrane channel (Kulathila et al., 2011; Lei et al., 2014) (which exists as a trimer in solution with the trimeric molecular weight equals 147.7 kDa) as standards. The experiments were repeated for three times. Gel filtration suggested an average molecular weight of 112.1 ± 4.1 kDa for the MtrF transporter (Fig. S3). This value is in good agreement with the theoretical value of 112.5 kDa for two MtrF protomers.
Construction of the double knockout strain
The double knockout E. coli strain BL21(DE3)ΔabgTΔpabA was produced from the BL21(DE3) strain using an RED disruption system as described by Datsenko and Wanner (Datsenko and Wanner, 2000). The ΔabgT::kan cassette, which was used to replace the chromosomal abgT gene, was produced by PCR, and then introduced into pKD46/BL21(DE3) by electroporation. The knockout BL21(DE3)ΔabgT::kan strain was selected on LB plate containing 30 μg/ml kanamycin, and verified by PCR. The kanamycin resistant gene was then released to generate the BL21(DE3)ΔabgT knockout strain. The deletion of pabA from BL21(DE3)ΔabgT was done using similar procedures as described above to generate the final BL21(DE3)ΔabgTΔpabA double knockout strain.
Site-directed mutagenesis
We performed site-directed mutagenesis on residues D193, S417, W420, P438, R446, D449, and P457 to generate single point mutants D193A, S417A, W420A, P438A, R446A, D449A, and P457A. The primers used for these mutations are listed in Table S2. All oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA) in a salt-free grade.
Accumulation assays of p-aminobenzoic acid
In brief, E. coli BL21(DE3)ΔabgTΔpabA carrying pET15bΩmtrF or pET15b were grown in LB broth with 100 μg/ml ampicillin at 37°C. When OD600 nm reached 0.5, the culture was treated with 0.2 mM IPTG to induce mtrF expression, and cells were harvested within 2 h. Cells were washed twice with buffer containing 50 mM potassium phosphate (pH 7.5), twice with buffer containing 100 mM Tris-HCl (pH 7.5), and then suspended in the same buffer to OD600 nm of 15. [3H]-PABA (Moravek Biochemicals, Brea, CA) was then added to a final concentration of 0.3 μM. Samples of 100 μl were taken at intervals, applied directly to prewetted glass-fiber filters, and washed twice with 5-ml aliquots of the same buffer; 0.5-μm glass-fiber filters (MFS, Dubbin, CA) were used with a filter apparatus. Filters were then incubated for 30 min in scintillation fluid (ScintiVerse™ BD) and counted with a Packard Tri-Carb 1600TR liquid scintillation counter (Perkin Elmer, Waltham, MA). For [3H]-PABA accumulations in cells expressing the MtrF mutants, the procedures for sample preparation were the same as above.
Assays of folic acid
The concentration of folic acid in the double knockout BL21(DE3)ΔabgTΔpabA strain transformed with pET15bΩmtrF or pET15b was measured using Lactobacillus casei based on the microbiological procedure of Wilson and Horne (1982). The concentrations of folic acid in BL21(DE3)ΔabgTΔpabA expressing the mutant transporters were measured using the same procedures.
Drug susceptibility assays
The susceptibilities to various drugs of E. coli BL21(DE3)ΔabgTΔpabA harboring pET15bΩmtrF expressing the wild-type or mutant transporters, or the pET15b empty vector were tested on agar plates. Cells were grown in Luria Broth (LB) medium with 100 μg/ml ampicillin at 37 °C. When OD600 nm reached 0.5, the cultures were induced with 0.2 mM IPTG and harvested in two hours after induction. The MICs to sulfamethazine, sulfadiazine, sulfathiazole and sulfanilamide of E. coli BL21(DE3)ΔabgTΔpabA (incoculum, 500 cells/ml) harboring these vectors were then determined using LB agar containing 50 μg/ml ampicillin, 0.1 mM IPTG and different concentrations of these drugs.
For the MIC studies with N. gonorrhoeae cells, we used strains FA140 and WV16 (as FA140 but mtrF::kan) described by Veal and Shafer (2003). These strains were grown on GCB agar plates. The susceptibilities to sulfanilamide were performed using the agar dilution assay described by Hagman et al. (1995).
Isothermal Titration Calorimetry
We used ITC to examine the binding of a variety of ligands to the purified MtrF transporter. Measurements were performed on a VP-Microcalorimeter (MicroCal, Northampton, MA) at 25 °C. Before titration, the protein was thoroughly dialyzed against buffer containing 20 mM Tris-HCl pH 7.5 and 0.03% DDM. The protein concentration was determined using the Bradford assay. The protein sample was then adjusted to a final monomeric concentration of 40 μM. Ligand solution consisting of 1.0 mM sulfamethazine, sulfadiazine, sulfathiazole, sulfanilamide or PABA in 20 mM Tris-HCl pH 7.5 and 0.03% DDM was prepared as the titrant. The protein and ligand samples were degassed before they were loaded into the cell and syringe. Binding experiments were carried out with the protein solution (1.5 ml) in the cell and the ligand as the injectant. Ten microliter injections of the ligand solution were used for data collection.
Injections occurred at intervals of 300 s, and the duration time of each injection was 20 s. Heat transfer (μcal/s) was measured as a function of elapsed time (s). The mean enthalpies measured from injection of the ligand in the buffer were subtracted from raw titration data before data analysis with ORIGIN software (MicroCal). Titration curves were fitted by a nonlinear least squares method to a function for the binding of a ligand to a macromolecule. Nonlinear regression fitting to the binding isotherm provided us the equilibrium binding constant (KA = 1/KD). Calorimetry trials were also carried out in the absence of MtrF in the same experimental conditions. No change in heat was observed in the injections throughout the experiment.
Accumulation assays of sulfamethazine
The procedures for [3H]-sulfamethazine accumulation were the same as those for [3H]-PABA accumulation. Cells were incubated with 75 nM [3H]-sulfamethazine (Moravek Biochemicals, Brea, CA) for 15 min, applied directly to prewetted glass-fiber filters, and washed immediately twice with 5-ml aliquots of the same buffer. Filters were then incubated for 30 min in scintillation fluid (ScintiVerse™ BD) and counted with a Packard Tri-Carb 1600TR liquid scintillation counter (Perkin Elmer, Waltham, MA). For metal ion dependent experiments, cells were incubated with 75 nM [3H]-sulfamethazine in the presence of NaCl or KCl (5 or 100 mM) for 15 min, then applied directly to prewetted glass-fiber filters, and washed immediately twice with 5-ml aliquots of buffer containing 100 mM Tris-HCl (pH 7.5) and 5 or 100 mM salt (NaCl or KCl).
Efflux assays of sulfamethazine
E. coli BL21(DE3)ΔabgTΔpabA carrying pET15bΩmtrF or pET15b were grown in LB broth with 100 μg/ml ampicillin at 37°C. When OD600 nm reached 0.5, the culture was induced with 0.2 mM IPTG, and cells were harvested within 2 h. Cells were washed twice with buffer containing 50 mM potassium phosphate (pH 7.5), twice with buffer containing 100 mM Tris-HCl (pH 7.5), and then suspended in the same buffer to OD600 nm of 15. CCCP was added to the cell suspension at a final concentration of 100 μM. Cells were then incubated for 10 min, pelleted, washed twice and resuspended in the same buffer to OD600 nm of 10. [3H]-sulfamethazine was then added to a final concentration of 37 nM. Cells were incubated for 15 min, and a final concentration of 0.2% glucose was then added to the cell suspension. When needed, a final concentration of 5 mM NaCl was also added at the same time. At various time points, samples of 100 μl were applied directly to prewetted glass-fiber filters, and washed twice with 5-ml aliquots of buffer containing 100 mM Tris-HCl (pH 7.5). Filters were then incubated for 30 min in scintillation fluid (ScintiVerse™ BD) and counted with a Packard Tri-Carb 1600TR liquid scintillation counter (Perkin Elmer, Waltham, MA).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R37AI021150 (W.M.S.) and R01GM086431 (E.W.Y.) and a VA Merit Award (W.M.S.) from the Medical Research Service of the Department of Veterans Affairs. W.M.S. is the recipient of a Senior Research Career Scientist from the Medical Research Service of the Department of Veterans Affairs. We are grateful to Louis Messerle (University of Iowa) for providing us the (NH4)2W6(μ-O)6(μ-Cl)6Cl6 complex used in this study. We are very thankful to Marit Nilsen-Hamilton (Iowa State University) who generously made her radioactivity counter available for us. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by an award GM103403 from the National Institutes of General Medical Sciences. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Accession Code
Atomic coordinates and structure factors for the structure of MtrF have been deposited at the RCSB Protein Data Bank with an accession code 4R1I.
Author contributions
C.C.S. and J.R.B. contributed equally to this work. C.C.S., J.R.B. and E.W.Y. designed research. C.C.S., J.R.B., N.K., A.R., F.L., J.A.D. and T.H.C. performed experiments. C.C.S., J.R.B., K.R.R. and E.W.Y. performed model building and refinement. C.C.S., J.R.B., W.M.S. and E.W.Y. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCroy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, Chou TH, Delmar JA, Lei HT, Rajashankar KR, Shafer WM, Yu EW. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One. 2014;9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carter EL, Jager L, Gardner L, Hall CC, Willis S, Green JM. Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J Bacteriol. 2007;189:3329–3334. doi: 10.1128/JB.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley DO, Rapp M, Granseth E, Melèn K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahay RM, Robertson BD, Balthazar JT, Ison CA. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology. 1997;143:2127–2133. doi: 10.1099/00221287-143-7-2127. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eudes A, Kunji ERS, Noiriel A, Klaus SM, Vickers TJ, Beverley SM, Gregoty JF, III, Hanson A. Identification of transport-critical residues in a folate transporter from the folate-biopterin transporter (FBT) J Biol Chem. 2010;285:2867–2875. doi: 10.1074/jbc.M109.063651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster JP, Shafer WM. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J Bacteriol. 2005;187:3713–3720. doi: 10.1128/JB.187.11.3713-3720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- Hagman KE, Shafer WM. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol. 1995;177:4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Lucas CE, Balthazar JT, Snyder LA, Nilles M, Judd RC, Shafer WM. The MtrD protein of Neisseria gonorrhoeae is a member of resistance/nodulation/division protein family constituting part of an efflux system. Microbiology. 1997;143:2117–2125. doi: 10.1099/00221287-143-7-2117. [DOI] [PubMed] [Google Scholar]
- Hussein MJ, Green JM, Nichols BP. Characterization of mutations that allow p-aminobenozyl-glutamate utilization by Escherichia coli. J Bacteriol. 1998;180:6260–6268. doi: 10.1128/jb.180.23.6260-6268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janganan TK, Zhang L, Bavro VN, Matak-Vinkovic D, Barrera NP, Burton MF, Steel PG, Robinson CV, Borges-Walmsley MI, Walmsley AR. Opening of the outer membrane protein channel in tripartite efflux pumps is induced by interaction with the membrane fusion partner. J Biol Chem. 2011;286:5484–5493. doi: 10.1074/jbc.M110.187658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janganan TK, Bavro VN, Zhang L, Borges-Walmsley MI, Walmsley AR. Tripartite efflux pumps: energy is required for dissociation, but not assembly or opening of the outer membrane channel of the pump. Mol Microbiol. 2013;88:590–602. doi: 10.1111/mmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Nichols BP. Nucleotide sequence of Escherichia coli pabA and its evolutionary relationship to trp(G)D. J. Mol. Biol. 1986;168:451–468. doi: 10.1016/s0022-2836(83)80295-2. [DOI] [PubMed] [Google Scholar]
- Klaus SMJ, Wegkamp A, Sybesma W, Hugenholtz J, Gregory JF, III, Hanson AD. A nudix enzyme removes pyrophosphate from the dihydronepterin triphosphate in the folate synthesis pathway of bacteria and plants. J Biol Chem. 2005;280:5274–5280. doi: 10.1074/jbc.M413759200. [DOI] [PubMed] [Google Scholar]
- Kulathila R, Kulathila R, Indic M, van den Berg B. Crystal Structure of Escherichia coli CusC, the Outer Membrane Component of a Heavy Metal Efflux Pump. PLoS One. 2011;6:15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EH, Shafer WM. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol. 1999;33:7753–7758. doi: 10.1046/j.1365-2958.1999.01530.x. [DOI] [PubMed] [Google Scholar]
- Lei HT, Bolla JR, Bishop NR, Su CC, Yu EW. Crystal Structures of CusC Review Conformational Changes Accompanying Folding and Transmembrane Channel Formation. J Mol Biol. 2014;426:403–411. doi: 10.1016/j.jmb.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei HT, Chou TH, Su CC, Bolla JR, Kumar N, Radhakrishnan A, Long F, Delmar JA, Do SV, Rajashankar KR, Shafer WM, Yu EW. Crystal structure of the open state of the Neisseria gonorrhoeae MtrE outer membrane channel. PLoS One. 2014;9:e97475. doi: 10.1371/journal.pone.0097475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicro Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob Agents Chemother. 2008;52:3052–3060. doi: 10.1128/AAC.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA heavy-metal efflux pump suggest methionine-mediated metal transport mechanism. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Symersky J, Radchenko M, Koide A, Guo Y, Nie RX, Koide S. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci USA. 2013;110:2099–2104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CE, Hagman KE, Levin JC, Stein DC, Shafer WM. Importance of lipooligosaccharide structure in determining gonococcal resistance to hydrophobic antimicrobial agents resulting from the mtr efflux system. Mol Microbiol. 1995;16:1001–1009. doi: 10.1111/j.1365-2958.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, amd Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. ML-PHARE, CCP4 Proc 80–88. Daresbury Laboratory; Warrington, UK: 1991. [Google Scholar]
- Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- Prakash S, Cooper G, Singhi S, Saier MH., Jr The ion transporter superfamily. Biochim Biophys Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette-Loughlin C, Dunham SA, Kuhn M, Balthazar JT, Shafer WM. The NorM efflux pump of by Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J Bacteriol. 2003;185:1101–1106. doi: 10.1128/JB.185.3.1101-1106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Sora JE, Ochong E, Beveridge S, Johnson D, Nzila A, Biagini GA, Stocks PA, O’Neill PM, Krishna S, Bray PG, Ward SA. The molecular basis of folate salvage in Plasmodium falciparum: characterization of two folate transporters. J Biol Chem. 2011;286:44659–44668. doi: 10.1074/jbc.M111.286054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Shafer WM, Qu XD, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer WM, Veal WL, Lee EH, Zarentonelli L, Balthazar JT, Rouquette C. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J Mol Microbiol Biotechnol. 2001;3:219–225. [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. 2001;D57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st Century: Past, Evolution, and Future. Clin Microbiol Rev. 2014;27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol. 2002;184:5619–24. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal WL, Shafer WM. Identification of a cell envelope protein (MtrF) involved in hydrophobic antimicrobial resistance in Neisseria gonorrhoeae. J Antimicrob Chemother. 2003;51:27–37. doi: 10.1093/jac/dkg031. [DOI] [PubMed] [Google Scholar]
- Warner DM, Shafer WM, Jerse AE. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol. 2008;70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SD, Horne WD. Use of glycerol-crypotected Lactobacillus casei for microbiological assay of folic acid. Clin Chem. 1982;28:1198–1200. [PubMed] [Google Scholar]
- Yernool D, Boudker O, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikosii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.