Abstract
To define features of the B cell response to HIV that may be translated to vaccine development, we have isolated a panel of monoclonal antibodies (MAbs) from HIV-infected patients. These MAbs are all highly reactive to HIV envelope (Env) from multiple clades, and include gp120 and gp41 specificities. Three of the MAbs exhibit substantial homology to previously described VH1-69, VH3-30, and VH4-59 HIV broadly neutralizing antibody lineages. An inherently autoreactive VH4-34 encoded MAb was reactive to diverse Env despite its minimal mutation from germline. Its isolation is consistent with our previous observation of increased VH4-34+antibodies in HIV-infected patients. These results suggest that conserved developmental processes contribute to immunoglobulin repertoire usage and maturation in response to HIV Env and that intrinsically autoreactive VH genes, despite the absence of mutation, could serve as effective templates for maturation and development of protective antibodies. These results also bear significant implications for the development of immunogens.
Introduction
The induction of protective humoral response to HIV envelope (Env) is a primary objective of preventive vaccination strategies against HIV infection. Approximately 20% of HIV-infected patients develop serum antibodies that have the ability to neutralize a broad range of HIV isolates; the appearance of these broadly neutralizing antibodies (bNAbs) often does not occur until several years after infection,(1,2) and typically they do not neutralize autologous contemporary HIV isolates from the patient.(3,4) The frequent induction of these bNAbs demonstrates the capability of the immune response that if re-capitulated by vaccination would be an attractive strategy to prevent HIV infection. The characterization of HIV Env-reactive MAbs generated from HIV-infected patients has provided substantial insight into the humoral response, including identifying MAbs that have potent and broad neutralizing activity, and enabling the further characterization of other antibody-mediated effector functions. Additionally, molecular characterization of these MAbs has provided insight into their developmental process and identified common features, including extensive mutation from germline, long immunoglobulin (Ig) heavy chain complementarity determining region 3 (HCDR3), and increased usage of specific Ig variable genes.(5–7)
The number of HIV broadly neutralizing MAb (bNMAbs) isolated from HIV-infected patients has grown substantially in recent years and currently exceeds two dozen.(6,7) These bNMAbs have importantly identified regions of Env that may be of particular value for targeting through vaccination. The CD4 binding site of gp120, recognized by several bNMAbs including b12, VRC01/03, and CH103, is an attractive region due in part to its conserved nature and participation in viral entry; however, adequate recognition frequently requires long HCDR3 for effective binding. Quaternary epitopes of gp120 that include the V1 and V2 regions are recognized by PG9/PG16 bNMAbs, and 2G12 and PGT recognizes glycan-dependent epitopes on gp120. The highly conserved membrane proximal external region (MPER) of gp41 is recognized by the bNAbs 4E10, 2F5, CAP205-CH12, and 10E8. Many gp41-directed MAbs exhibit poly-reactivity, including reactivity to self- and bacterial antigens, which suggests that their induction may be subject in part to regulation by tolerance mechanisms.
To further define the B cell response to HIV infection, we have isolated a panel of novel HIV Env-specific MAbs from HIV-infected patients. Our analysis revealed the presence of gp120- and gp41-specific MAbs, including those with cross-clade HIV Env binding, and neutralizing activity in HIV-infected patients, including those receiving anti-retroviral therapy (ART) with suppressed viremia.
Materials and Methods
Patient samples
Peripheral blood samples were obtained from HIV-1 infected patients at the University of Rochester Medical Center. All subjects provided signed written informed consent. PBMC were isolated within 2 h of sampling using CPT tubes (Becton Dickinson, Franklin Lakes, NJ). Tubes were immediately inverted 8 to 10 times and processed according to the manufacturer's instructions. All procedures and methods were approved by the University of Rochester Research Subjects Review Board.
Env-specific B cell isolation
To isolate gp140-specific B cells, PBMC were stained with purified oligomeric HIV-1 SF162 (clade B) and KNH1144 (clade A) gp140 directly conjugated to AlexaFluor660 and AlexaFluor 647, respectively, and Tetanus Toxoid conjugated to FITC (Calbiochem, San Diego, CA), in addition to anti-CD19-PE-Cy7, anti-CD20-APC-Cy7, anti-IgD-PE, anti-IgM-PerCP-Cy5.5, anti-CD3-PE-Cy5, anti-CD14-PE-Cy5, 7AAD for dead cell exclusion, and biotinylated 9G4 MAb/streptavidin Qdot800 (Invitrogen, Carlsbad, CA) at 4°C for 60 min, as previously described.(8) Single 7AAD- CD3- CD14- CD19+Tetanus Toxoid – gp140+ cells were directly sorted into 96-well PCR plates (Bio-Rad, Hercules, CA) containing 4 μL/well 0.5X PBS with 10 mM DTT (Invitrogen), 8 U RNAsin (Promega, Madison, WI), and 0.4 U 5′-3′ Prime RNAse Inhibitor (Eppendorf, Hamburg, Germany). Plates were sealed with Microseal F Film (Bio-Rad) and immediately frozen on dry ice before storage at −80°C until used for RT-PCR.
Alternatively, single cells were sorted into 384-well tissue culture plates containing irradiated CD40L-expressing fibroblasts, 10 ng/mL IL-2 (Peprotech, Rocky Hill, NJ), 2.5 μg/mL CpG2006 (IDT, Coralville, IA), 2.5 μg/mL R848 (Invitrogen, San Diego, CA), and 1:100,000 Staphylococcus aureus Cowan (SAC, EMD Millipore, Darmstadt, Germany) and cultured for 7–10 days. Following culture, qualitative 384-well ELISA was performed to identify wells containing cells secreting gp140-reactive IgG. Cells from positive wells were collected and resuspended in PCR buffer described above.
MAb generation and production
cDNA was synthesized and semi-nested RT-PCR for IgH, Igλ, and Igκ V gene transcripts was performed as previously described.(9) Purified PCR2 products were sequenced at Genewiz Sequences and analyzed by IgBlast (www.ncbi.nlm.nih.gov/igblast) and IMGT/V-QUEST (www.imgt.org/IMGT_vquest) to identify germline V(D)J gene segments with highest identity and determine sequence properties including mutation, CDR3 length, and charge. Expression vector cloning and transfection of HEK293T cells (ATCC, Manassas, VA) were performed as previously described(10) using expression vectors kindly provided by Dr. Eric Meffre (Yale University, New Haven, CT). MAbs were purified using Aspire Protein G tips (Thermo Scientific, Waltham, MA).
ELISA
ELISA plates were coated with recombinant HIV-1 Env proteins at 0.5 μg/mL, MAbs were diluted in PBS+0.01% Tween-20, and binding detected with horseradish peroxidase–conjugated anti-human IgG (Jackson ImmunoResearch, West Grove, PA). Env proteins tested included: oligomeric SF162 gp140, oligomeric PVO.4 gp140, oligomeric X1936 gp140, oligomeric KNH1144 gp140, and oligomeric YU2 gp140 ΔV1V2V3 provided by the UR CFAR Recombinant Protein Production Core; monomeric MN gp120 (Protein Sciences, Meriden, CT); monomeric CN54 gp120 (NIH AIDS Reagent Repository), monomeric Con of Cons gp120 (Immune Technology, New York, NY); and YU2 gp41 (ImmunoDx, Woburn, MA). b12, 17b, and 4E10 MAbs were obtained from NIH AIDS Reagent Repository. Polyclonal human IgG was obtained from Jackson ImmunoResearch. Anti-Nuclear Antigen (ANA) IgG ELISA (Inova Diagnostics, San Diego, CA) was performed, testing samples in duplicate using kits according to the manufacturer's recommendations. Anti-cardiolipin ELISA was performed as previously described.(8)
HIV-1 neutralization activity
The neutralizing activity of sera was determined as previously described.(11) Briefly, pseudotyped viruses were incubated with heat-inactivated sera and then added to TZM-bl indicator cells. Neutralizing antibody titers were determined as the MAb concentration (μg/mL) at which luciferase expression was reduced by 50% (ID50).
Results and Discussion
Patient characteristics and MAb generation
Peripheral blood B cells were isolated from a diverse group of HIV-infected patients, including three patients receiving ART with suppressed viral loads of <50 copies/mL; the remaining two patients had viral loads of <1000 copies/mL (Table 1). The inclusion of patients with and without detectable viremia allows the exploration of the necessity of ongoing viremia for the persistence of HIV-specific memory B cells. HIV Env-reactive B cells were identified by binding to fluorescently conjugated oligomeric gp140 and isolated by flow cytometric sorting. Single gp140-reactive B cells were either directly subjected to single cell RT-PCR for Ig heavy and light chain variable (V) region sequencing and cloning or single cells cultured (SCC) briefly and pre-screened by ELISA for gp140-reactive antibody production prior to RT-PCR (Table 2). Heavy and light chain variable regions were cloned and expressed as human IgG1 MAbs.
Table 1.
HIV+ Subject Demographics
| Subject | MAb | Gender | Age (yr) | Race | Time since Dx (yr) | ART status | CD4 (cells/μL) | Viral Load (copies/mL) |
|---|---|---|---|---|---|---|---|---|
| HIV018 | KS18-5 | F | 51 | Black | 7 | On | 344 | <50 |
| HIV037 | KS37-6 | F | 33 | Black | 12 | Off | 447 | 470 |
| HIV041 | KS41-2 | M | 52 | Hispanic | 12 | On | 235 | <50 |
| KS41-11 | ||||||||
| KS41-22 | ||||||||
| HIV059 | KS59-5 | F | 49 | Black | 1 | On | 479 | <50 |
| KS59-12 | ||||||||
| HIV074 | KS74-11 | F | 48 | Black | 2 | Never | 900 | 624 |
Table 2.
Molecular Characteristics of MAbs
| Heavy chain | Light chain | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject | mAb | Selection Strategy | V | D | J | AA mutation (%) | CDR3 length (AA) | V | J | AA mutation (%) | CDR3 length (AA) |
| HIV018 | KS18-5 | gp140+IgD- | 3-15*05 | 3-22*01 | 5*02 | 27 | 13 | κ1-NL1*01 | κ4*01 | 20.9 | 9 |
| HIV037 | KS37-6 | gp140+9G4+ | 4-34*01 | 2-2*02 | 6*02 | 0 | 26 | λ9-49*01 | λ1*01 | 1 | 14 |
| HIV041 | KS41-2 | gp140+SCC | 4-4*02 | 1-20*01 | 6*02 | 17.3 | 20 | κ1-27*01 | κ1*01 | 7.7 | 9 |
| KS41-11 | gp140+SCC | 4-59*08 | 3-3*01 | 6*02 | 11.3 | 20 | κ3-20*01 | κ2*01 | 4.8 | 10 | |
| KS41-22 | gp140+SCC | 1-18*01 | 3-10*01 | 3*02 | 21.6 | 16 | κ1-27*01 | κ1*01 | 3.8 | 9 | |
| HIV059 | KS59-5 | gp140+IgG+ | 3-30*02 | 3-3*01 | 3*02 | 16.3 | 23 | κ2-30*02 | κ1*01 | 7.7 | 9 |
| KS59-12 | gp140+IgG+ | 3-23*01 | 1-26*01 | 3*02 | 14.3 | 17 | λ2-11*01 | λ1*01 | 7.1 | 12 | |
| HIV074 | KS74-11 | gp140+SCC | 1-69*13 | 3-3*01 | 6*03 | 14.3 | 12 | κ1-5*03 | κ2*02 | 7.7 | 10 |
HIV envelope binding activity
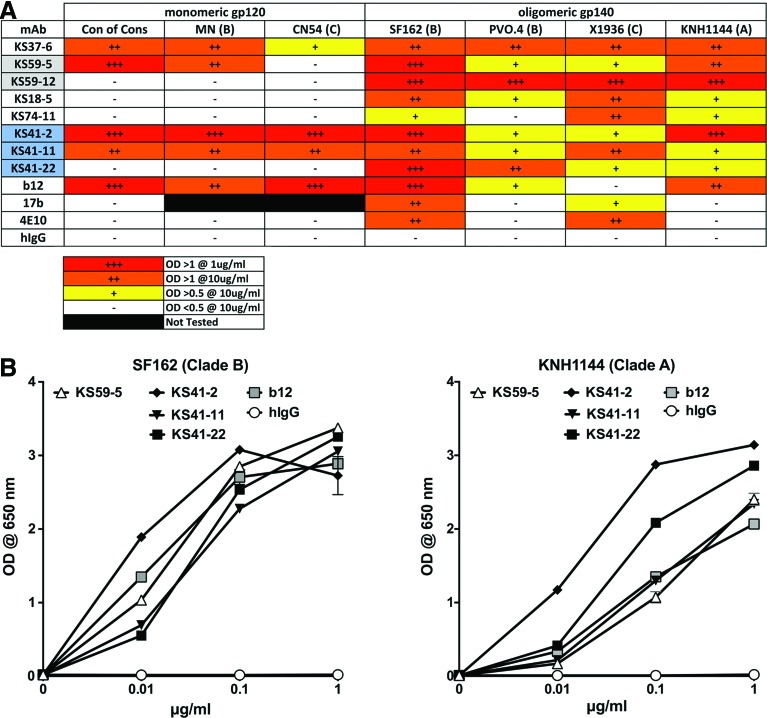
Eight MAbs that exhibited HIV binding activity during a preliminary screen on oligomeric HIV SF162 gp140 (data not shown) were tested against a comprehensive panel of monomeric HIV gp120 and oligomeric HIV gp140 proteins by ELISA (Fig. 1A). Four of the MAbs (KS37-6, KS59-5, KS41-2, and KS41-11) were reactive to monomeric gp120, suggesting that their binding does not depend on quaternary structure or gp41. All MAbs as expected based on screening were reactive to oligomeric gp140; additionally all of the MAbs displayed multi-clade reactivity. Further titration of the highest binding MAbs demonstrated significant (p<0.001) multi-clade binding to SF162 gp140 (clade B) and KNH1144 (clade A) at 0.01 μg/mL for KS41-2, KS41-22, KS41-11, and KS59-5 as compared to control human IgG (Fig. 1B). KS41-2 displayed the highest binding to SF162 gp140 and KNH1144, which was significantly greater (p<0.0001) at even 0.01 μg/mL compared to the other MAbs tested.
FIG. 1.
HIV Env binding profiles of MAbs. Human recombinant MAbs were added in triplicate to ELISA plates coated with 0.5 μg/mL of indicated HIV Env proteins and binding detected with anti-human IgG. (A) MAbs were tested at 10 and 1 μg/mL and relative binding indicated; shading indicates MAbs were isolated from the same subject. (B) MAbs were serially diluted and binding to indicated oligomeric gp140 determined, mean±SEM presented. MAbs b12, 17b, and 4E10 included as positive controls. Polyclonal human IgG included as negative control.
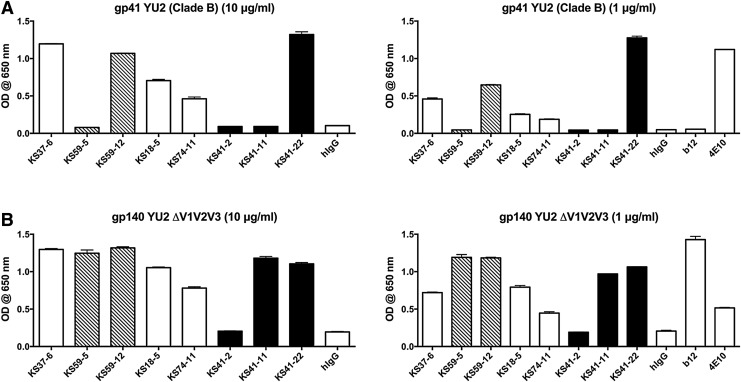
The MAbs KS41-22, KS59-12, KS37-6, KS18-5, and KS74-11 displayed reactivity to gp41 (Fig. 2A). The MAb KS37-6 was reactive to both gp120 and gp41, suggesting it may be binding in a poly-reactive manner, which is consistent with its VH4-34 usage, a subset of immunoglobulin with inherent poly-reactivity recognized by the 9G4 anti-idiotype antibody.(9,12) Its polyreactive nature is also consistent with its reactivity to anti-nuclear antigen (data not shown), in a similar fashion to VH4-34 antibodies isolated from healthy controls previously published by our laboratory.(9) None of the other MAbs exhibited any reactivity to anti-nuclear antigen or cardiolipin (data not shown), suggesting they were not poly-reactive.
FIG. 2.
HIV gp41 and V1V2V3 dependent binding of MAbs. Human recombinant MAbs were added in triplicate at 10 and 1 μg/mL to ELISA plates coated with (A) 2 μg/mL of YU2 gp41 or (B) 0.5 μg/mL of oligomeric YU2 gp140 ΔV1V2V3 and binding detected with anti-human IgG.
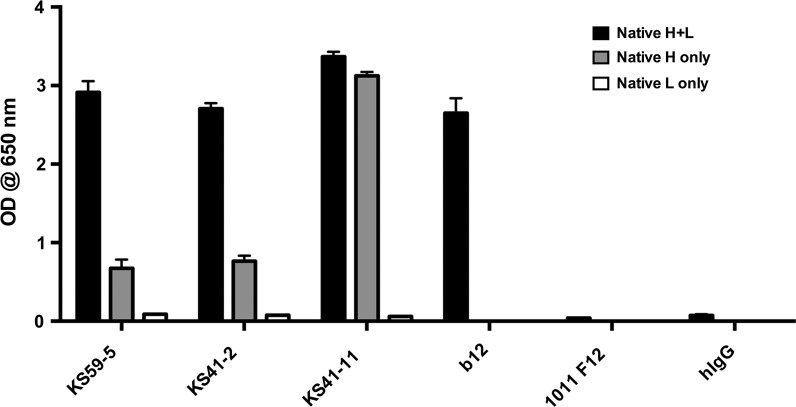
All MAbs, except KS41-2, were reactive to an HIV gp140 that lacks the V1, V2, and V3 variable loops (Fig. 2B), suggesting that only KS41-2 binding was dependent on the presence of one or a combination of these variable loops. These results indicate that we have identified a diverse panel of MAbs recognizing gp41- and gp120-dependent epitopes and that HIV Env-specific memory B cells persist in patients whose viremia is suppressed by ART. To determine the relative dependence on heavy or light chains on binding activity, KS59-5, KS41-2, and KS41-11 light chains were replaced with an irrelevant light chain, resulting in a 69%, 72%, and 7% reduction in binding to SF162 gp140, respectively (Fig. 3). This indicates that native KS59-5 and KS41-2 light chains are substantially contributing to its ability to bind to gp140, although pairing of these light chains with an irrelevant heavy chain did not confer binding to SF162 gp140 (Fig. 3), indicating that the light chain reactivity alone is not sufficient for gp140 binding. Native (paired heavy and light chain) b12 was included for comparison. The substantial contribution of the light chain to the binding of gp140 for KS59-5 and KS41-2 does highlight the importance of studying light chain interactions to further dissect the molecular basis of HIV Env binding.
FIG. 3.
Immunoglobulin light chain contribution to HIV gp140 binding. KS59-5, KS41-2, and KS41-11 MAbs were generated either as their native Ig heavy and light chain pairing or by replacing their native heavy or light chain with that of the irrelevant 1011 F12 MAb. MAbs were tested in triplicate at 10 μg/mL by ELISA for binding to SF162 gp140. b12 and polyclonal hIgG, both only tested in native H+L format.
HIV neutralizing activity
The neutralizing activity of the MAbs was examined by Tzm-bl assay. Only three of the eight MAbs exhibited neutralizing activity against the Tier I isolate SF162, these included KS59-5 (IC50=0.5 μg/mL), KS41-2 (IC50=2 μg/mL), KS41-11 (IC50=0.5 μg/mL) (Table 3; some data not shown). Further assessment of neutralizing activity of these MAbs revealed moderate Tier 2 neutralizing activity (10–70 μg/mL) by all three MAbs, and moderate cross-clade neutralizing activity of ZM109 (clade C) by KS41-2 (IC50=80 μg/mL) and KS41-11 (IC50=40 μg/mL). The cross-clade neutralizing activity of these MAbs is consistent with their cross-clade binding profiles. All three of these MAbs were isolated from patients on ART with suppressed viral loads indicating that memory B cells with cross-clade neutralizing activity can persist in the absence of detectable viremia, consistent with the presence of HIV neutralizing serum antibodies in HIV patients on ART that has been previously reported.(13–15)
Table 3.
HIV-1 Neutralizing Activity of MAbs
| ID50 (μg/mL) in TZM-bl cells | |||||
|---|---|---|---|---|---|
| MAb | SF162 (B) Tier 1A | BaL.26 (B) Tier 1B | SS1196.1 (B) Tier 1B | JRCSF (B) Tier 2 | ZM109 (C) Tier 1B |
| KS59-5 | 0.5 | 30 | 10 | 70 | >100 |
| KS41-2 | 2 | 10 | >100 | 10 | 80 |
| KS41-11 | 0.5 | 10 | 10 | 50 | 40 |
| b12 | ND | 0.1 | 0.5 | 0.5 | >2 |
Molecular characterization
The variable regions of the heavy and light genes of the MAbs were sequenced and analyzed to determine Ig gene usage and features (Table 2). Heavy chain VH3 and VH4 genes were most predominantly used (6/8 MAbs) consistent with their common usage in the human Ig repertoire.(16)
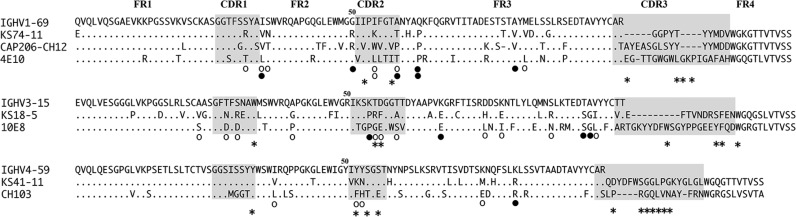
The remaining two MAbs use VH1 genes, including VH1-69 encoded KS74-11 MAb, which is also used by the bNMAbs 4E10(17,18) and CAP206-CH12,(19) which are also gp41-specific. KS74-11 shares substantial heavy chain homology with CAP206-CH12, including using VH1-69 and JH6*03; 58% of the residues mutated in KS74-11 are at the same sites that are mutated in CAP206-CH12, and 25% of the mutated residues in KS74-11 are identical to those mutated in CAP206-CH12, all of which are in FR2 or FR3 (Fig. 4). Similar to CAP206-CH12,(19) the binding of KS74-11 appears sensitive to variation in residues 674 to 677 of the MPER, as demonstrated by stronger binding of KS74-11 to X1936, weaker binding to KNH1144 and SF162, and lack of binding to PVO.4 (Fig. 1A; Supplementary Fig. 1).
FIG. 4.
Heavy chain sequence homology with HIV bNMAbs. Alignment of germline VH and relevant HIV bNMAb. ○, common residue mutation location with HIV bNMAb; •, identical residue mutation with HIV bNMAb; *, HIV Env contact site of HIV bNMAb (4E10,(17) 10E8,(20) CH103(21)). Complementarity determining regions (CDR) are shaded.
The KS18-5 MAb shares substantial heavy chain homology with the bNMAb 10E8, which is also gp41 specific and uses VH3-15.(20) The heavy chain of KS18-5 is 27% mutated from the germline, of which 59% of the mutations are at the same sites that are mutated in 10E8; 15% of the mutated residues in KS18-5 are identical to those mutated in 10E8, including key residues shown to be gp41 contact residues for 10E8.(20)
Similar to the previously described CD4bs-specific bNMAb CH103, KS41-11 also uses VH4-59, and 45%(5) of the residues mutated in KS41-11 are at the same sites that are mutated in CH103, of which three of the residues are in FR2 or FR3 including R81K, which is identical in CH103, and K52Y within HCDR2, which is a defined CH103 gp120 contact residue (Fig. 4).(21) Additionally, similar to CH103,(21) KS41-11 is broadly cross-reactive to multiple Env and exhibits weaker reactivity to PVO.4 and KNH isolates (Fig. 1), and the V1V2V3-independent gp120 binding of KS41-11 (Figs. 1, 2) suggests it is also a CD4bs-specific antibody.
The genetic and functional homologies of (1) KS74-11 and CAP206-CH12 and (2) KS41-11 and CH103 support the previously suggested concept of repertoire convergence, positing that for some viral epitopes the responding immunoglobulin repertoire may be biased toward usage of limited VH families and follow similar maturation pathways among different individuals.(22–27) Thus, immunization strategies capable of recapitulating HIV-specific B cell lineages displaying these shared molecular properties may be an attractive strategy for HIV vaccine development.
All of the MAbs except the VH4-34 encoded KS37-6 have substantial VH amino acid mutation from germline (14.3–27%), a feature commonly observed in HIV Env-specific MAbs.(5–7) Half of the MAbs had heavy chain complementarity determining regions (HCDR3) of 20 amino acids in length or longer, also commonly associated with HIV Env-specific MAbs. Diverse κ and λ light chain gene usage was evident among the MAbs (Table 2).
The KS37-6 MAb was isolated based on staining with the anti-idiotypic antibody 9G4, which recognizes VH4-34 encoded Ig. We have previously reported increased 9G4+B cells and serum antibodies in HIV-infected patients.(8,28) This MAb did not exhibit any heavy chain mutations from germline and subsequently maintains the VH4-34 framework 1 hydrophopic patch, which we have previously demonstrated confers B cell binding and, similar to other anti-nuclear antigen reactive VH4-34 encoded MAbs,(9) has a positively charged HCDR3 (pI=4.89). The absence of mutation is in dramatic contrast to other HIV Env-specific MAbs and suggests that B cells with inherently auto-reactive germline B cell receptors, such as 9G4+BCRs, may have an enhanced ability to bind HIV Env relative to other germline BCRs, thereby providing a desirable template potentially capable of supporting the generation of protective antibodies with a more limited mutational load than typically required for broadly neutralizing antibodies.
Conclusion
The isolation of a panel of HIV Env-specific MAbs from HIV-infected individuals has provided additional resolution of the specificity and molecular characteristics of the B cell response. Homology of several of these newly described MAbs with known bNMAbs suggests the participation of a conserved repertoire and maturation process in the humoral response to HIV and suggests that targeting these common bNAb developmental pathways may be a valuable strategy for HIV vaccines.
Supplementary Material
Acknowledgments
We are grateful for the assistance and reagents provided by Lin Silver, Erin Fox, Seana Catherman, Christopher Richardson, Diana Adlowitz, Peter Bryk, Steven Morgan, Mary Adams, Carol Greisberger, Emily Cosimano, Amneris Luque, Jonelle Mattiacio, Stephen Dewhurst, the UR CFAR Recombinant Protein Production Core, and the UR Flow Cytometry Core. A special thanks is extended to the study volunteers.
This work was supported by National Institutes of Health (NIH) grants (R01AI084808 and R21AI078459, to IS); an early-stage investigator grant through the Center for HIV/AIDS Vaccine Immunology and the HIV Vaccines Trials Network (U19AI067854); and funding through the University of Rochester Center for AIDS Research (P30AI078498, to JK) and the University of Rochester HIV/AIDS Clinical Trials Unit; Units for HIV/AIDS Clinical Trials Networks (UM1AI069511, to JK and MK).
Author Disclosure Statement
The above listed funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have no financial interests to disclose.
References
- 1.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, and Stamatatos L: Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 2011;7:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Gils MJ, Euler Z, Schweighardt B, Wrin T, and Schuitemaker H: Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. Aids 2009;23:2405–2414 [DOI] [PubMed] [Google Scholar]
- 3.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, and Moore PL: Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 2013;9:e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richman DD, Wrin T, Little SJ, and Petropoulos CJ: Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA 2003;100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong PD, and Mascola JR: Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 2012;37:412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascola JR, and Haynes BF: HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev 2013;254:225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, and Nussenzweig MC: Antibodies in HIV-1 vaccine development and therapy. Science 2013;341:1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobie JJ, Alcena DC, Zheng B, Bryk P, Mattiacio JL, Brewer M, Labranche C, Young FM, Dewhurst S, Montefiori DC, Rosenberg AF, Feng C, Jin X, Keefer MC, and Sanz I: 9G4 autoreactivity is increased in HIV-infected patients and correlates with HIV broadly neutralizing serum activity. PLoS One 2012;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson C, Chida AS, Adlowitz D, Silver L, Fox E, Jenks SA, Palmer E, Wang Y, Heimburg-Molinaro J, Li QZ, Mohan C, Cummings R, Tipton C, and Sanz I: Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J Immunol 2013;191:4926–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H: Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008;329:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, and Montefiori DC: Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol 2005;79:10103–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter KN, Li Y, Pascual V, Williams RC, Jr., Byres LC, Spellerberg M, Stevenson FK, and Capra JD: Molecular characterization of a cross-reactive idiotope on human immunoglobulins utilizing the VH4-21 gene segment. J Exp Med 1993;178:1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gach JS, Achenbach CJ, Chromikova V, Berzins B, Lambert N, Landucci G, Forthal DN, Katlama C, Jung BH, and Murphy RL: HIV-1 specific antibody titers and neutralization among chronically infected patients on long-term suppressive antiretroviral therapy (ART): a cross-sectional study. PLoS One 2014;9:e85371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira CB, Merino-Mansilla A, Llano A, Perez I, Crespo I, Llinas L, Garcia F, Gatell JM, Yuste E, and Sanchez-Merino V: Evolution of broadly cross-reactive HIV-1-neutralizing activity: therapy-associated decline, positive association with detectable viremia, and partial restoration of B-cell subpopulations. J Virol 2013;87:12227–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina-Ramirez M, Sanchez-Merino V, Sanchez-Palomino S, Merino-Mansilla A, Ferreira CB, Perez I, Gonzalez N, Alvarez A, Alcocer-Gonzalez JM, Garcia F, Gatell JM, Alcami J, and Yuste E: Broadly cross-neutralizing antibodies in HIV-1 patients with undetectable viremia. J Virol 2011;85:5804–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, and Dunn-Walters DK: High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 2010;116:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, and Wilson IA: Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 2005;22:163–173 [DOI] [PubMed] [Google Scholar]
- 18.Kunert R, Wolbank S, Stiegler G, Weik R, and Katinger H: Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses 2004;20:755–762 [DOI] [PubMed] [Google Scholar]
- 19.Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, Donathan M, Bilska M, Gray ES, Abdool Karim SS, Kepler TB, Whitesides J, Montefiori D, Moody MA, Liao HX, and Haynes BF: Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One 2011;6:e23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, and Connors M: Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012;491:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, and Haynes BF: Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 2013;496:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharf L, West AP, Jr, Gao H, Lee T, Scheid JF, Nussenzweig MC, Bjorkman PJ, and Diskin R: Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc Natl Acad Sci USA 2013;110:6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes BF, Kelsoe G, Harrison SC, and Kepler TB: B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 2012;30:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, Horowitz L, Palese P, Bhatt RR, and Lerner RA: Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci USA 2008;105:5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, and Marasco WA: Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009;16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, and Nussenzweig MC: Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011;333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, and Mascola JR: Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011;333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcena DC, Kobie JJ, Kaminski DA, Rosenberg AF, Mattiacio JL, Brewer M, Dewhurst S, Dykes C, Jin X, Keefer MC, and Sanz I: 9G4+antibodies isolated from HIV-infected patients neutralize HIV-1 and have distinct autoreactivity profiles. PLoS One 2013;8:e85098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.