Abstract
Nanopores have recently emerged as powerful tools in single-molecule investigations. Biological nanopores however have drawbacks, including a fixed size and limited stability in lipid bilayers. Inspired by the great success of directed evolution approaches in tailoring enzymes properties, in this work we evolved Cytolysin A from Salmonella typhi (ClyA) to a high level of soluble expression and desired electrical properties in lipid bilayers. Evolved ClyA nanopores remained open up to −150 mV applied potential, which allowed the detailed characterization of folded proteins by ionic current recordings. Remarkably, we also found that ClyA forms several nanopore species; among which we could isolate and characterize three nanopore types most likely corresponding to the 12mer, 13mer and 14mer oligomeric forms of ClyA. The ability to employ nanopores with identical amino acid composition but different size is a new feature in the biological nanopore field. Our experiments show that sub-nanometer variations in the diameter of nanopores greatly affect the recognition of analyte proteins.
Keywords: single-molecule sensing, folded protein translocation, enhanced stability, permissive oligomerization
INTRODUCTION
The transport of ions or molecules across a biological membrane is a fundamental process in cellular life and is tightly regulated by ion channels, transporters and pores. Recently, researchers have adopted biological,1-3 solid-state,4,5 DNA origami6-9 and hybrid6,7,10 nanopores in single-molecule analysis.11 Biological nanopores have advantages compared to their synthetic counterparts, mostly because they can be reproducibly fabricated and modified with an atomic level precision that cannot yet be matched by artificial nanopores. Biological nanopores, however, also have drawbacks. The mechanical stability of biological nanopores depends on individual cases. Alpha hemolysin from Staphylococcus aureus (αHL) and porin A from Mycobacterium smegmatis (MspA) nanopores remain open in lipid bilayers at high-applied potentials and can cope surprisingly well with extreme conditions of temperature,12,13 pH13-16 and denaturant concentrations.13,17,18 However, most of other porins and channels are much less robust. Arguably, still, the biggest disadvantage of biological nanopores is their fixed size.
A significant number of studies sampled the translocation of proteins through artificial nanopores.19-24 Nonetheless, the investigation of proteins with solid-state nanopores is difficult because proteins have a non-uniform charge distribution, they often adsorb to the nanopores surface and they translocate too quickly to be properly sampled.21 Furthermore, because proteins have a compact folded structure, the diameter of the nanopore should be similar to that of the protein.20 Biological nanopores have also been used to characterize proteins. However, the small internal diameter (~1 nm) of the most common nanopores such as αHL, MspA or aerolysin, although allowed the analysis of small peptides or un-folded proteins,25-34 precludes the direct investigations of folded proteins. Among the larger biological nanopores, the connector protein in the bacteriophage phi29 DNA-packaging motor has been modified to insert into artificial bilayers to form nanopores of ~3.5 nm diameter (phi29 nanopores).35 Despite their suitable size, however, phi29 nanopores have not yet been used to investigate proteins. More recently, bacterial ferric hydroxamate uptake component A (FhuA) has been engineered to produce a stable nanopore, which open pore ionic current can be transiently blocked by folded protein analytes.7
Recently, we have introduced Cytolysin A from Salmonella typhi (ClyA) as the first biological nanopore that allows the direct investigation of natively folded proteins.14 The ClyA structure is ideal for this task because proteins such as thrombin (37 kDa) or malate dehydrogenase (35 kDa, monomer)14 can be electrophoretically trapped between the wide cis entrance and the narrower trans exit, and can therefore be sampled for several minutes. Ionic currents through ClyA are so sensitive to the vestibule environment that blockades of human and bovine thrombin can be easily distinguished.14 Our work was based on a ClyA construct where the two native cysteine residues of ClyA wild type (ClyA-WT) were replaced by serine (ClyA-SS) so that an additional cysteine residue could be incorporated and chemically modified at the nanopore surface.14 ClyA nanopores, however, showed several drawbacks. ClyA-WT displayed a very heterogeneous unitary conductance distribution and ClyA-WT and ClyA-SS monomers mostly expressed in inclusion bodies (see below). Further, ClyA-SS monomers showed low water solubility and low hemolytic activity (Figure S1, Supplementary Information). Finally in planar lipid bilayers ClyA-SS spontaneously opened and closed (gated) at applied potentials higher than +60 mV or lower than −90 mV.14 Therefore we aimed to obtain a ClyA variant amenable to site–specific chemical modifications (not including cysteine residues) with desired properties of solubility, activity and stability in planar lipid bilayers.
RESULTS AND DISCUSSION
Tuning the properties of ClyA by directed evolution
Inspired by the great success of directed evolution approaches in tailoring enzymes with desired properties, we used directed evolution to improve the activity of ClyA-SS, reasoning that mutations compensating for the deleterious effects of C87S and C285S substitutions would also increase the soluble expression and stability of ClyA. Random libraries were generated on the background of ClyA-SS by error prone PCR (approximately 1-4 mutations per gene per round) and screened for hemolytic activity (Figure S1, Supplementary Information). The most active variants were then purified by Ni-NTA affinity chromatography and tested for oligomer formation by blue native polyacrylamide gel electrophoresis (BN-PAGE, Figure S2, Supplementary Information). Selected nanopore variants were finally screened in planar lipid bilayers for the desired behavior (uniformity of formed channels, low electrical noise and ability to remain open at high applied potentials), which served as final and critical criteria for selection. After just four rounds of screening, we isolated ClyA-CS variant (Figure 1 and Table 1) that showed low electrical noise and remained open in planar lipid bilayers from +90 mV to −150 mV (Figure S3 Supplementary Information). ClyA-CS displayed five mutations relative to the ClyA-SS sequence: S87C, L99Q, E103G, F166Y and K294R (Figure 1). All of the accumulated mutations correspond to positions that are located outside the lumen of the nanopore according to the E. coli ClyA crystal structure36 (Figure 1) and do not have an obvious role in the stability or hemolytic activity of the nanopore; with the only possible exception of phenylalanine 166, which is located next to the cluster of hydrophobic residues implicated in the pre-pore to pore transition of ClyA.36 Remarkably, the serine at position 87 converted back to cysteine, the original residue in the wild-type gene (Figure 1). Since we desired to obtain a cysteine-less ClyA variant amenable to site-specific chemical modification, we subjected ClyA-CS to an additional round of directed evolution (Supplementary Information), from which we selected ClyA-AS with desired electrical properties. In ClyA-AS cysteine 87 was converted to alanine and isoleucine 203 to valine (Figure 1), the latter being the same residue as in E.coli ClyA-WT. Contrary to ClyA-SS and ClyA-WT, evolved ClyA variants expressed in E.coli cells in the soluble fraction (Figure S1, Supplementary Information) and could be purified without the use of detergents, allowing a ~10-fold increase in the purification yield (~0.6 mg pure ClyA-CS per 10 ml culture).
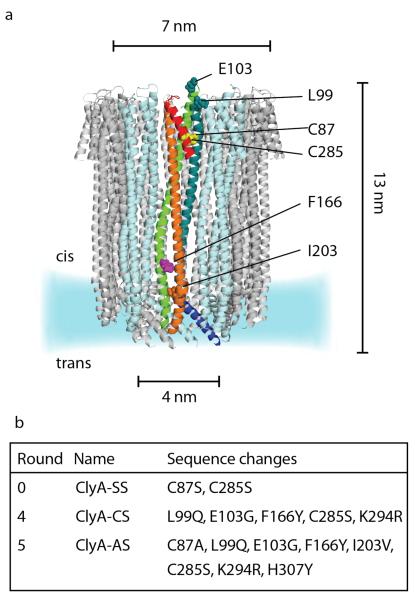
Figure 1.
(a) Ribbon representation of a S. typhi ClyA nanopore constructed by homology modeling from the E. coli ClyA structure (PDB: 2WCD, 90% sequence identity).36 One protomer is highlighted, with the secondary structure elements coloured from blue to red from N to C termini; other protomers are shown alternating in pale blue and grey. The amino acids side chains changed by the directed evolution experiments are displayed as spheres. The two native cysteine residues are colored in yellow, while Phenylalanine 166 is colored in cyan. K294 and H307 are not displayed because they are not shown in the ClyA crystal structure. (b) Sequence changes in ClyA-SS, ClyA-CS and ClyA-AS relative to ClyA-WT.
Table 1. Parameters for ClyA-SS and ClyA-CS nanopores.
The diameters of Type I ClyA-SS and ClyA-CS were taken from the crystal structure of E. coli ClyA (PDB: 2WCD). The diameters of Type II and Type III ClyA-CS were measured from the models shown in Figure 4d. The diameters were determined including the Van der Waals radii of the atoms. The values of GO are from the gel purified ClyA nanopores (Figure 3). Errors are given as standard deviations.
| Type I ClyA-SS* | Type I ClyA-CS | Type II ClyA-CS | Type III ClyA-CS | |
|---|---|---|---|---|
| Trans diameter, nm | 3.3 | 3.3 | 3.7 | 4.2 |
| Cis diameter, nm | 5.5 | 5.5 | 5.9 | 6.5 |
| GO at −35 mV, nS | 1.8 ± 0.1 | 1.79 ± 0.04 | 2.21 ± 0.08 | 2.64 ± 0.08 |
| GO at +35 mV, nS | 2.0±0.1 | 2.03 ± 0.20 | 2.42 ± 0.16 | 2.88 ± 0.08 |
| GO from equation 2 | 2.06 | 2.06 | 2.47 | 3.07 |
| HT Occupancy of L2 at −35 mV, % | 22 ± 5 | 30 ± 10 | 96 ± 2 | 100 |
| HT Level 1 at −35 mV, IRES% | 56 ± 1 | 56 ± 1 | 68 ± 1 | NA |
| HT Level 2 at −35 mV, IRES% | 23±1 | 23 ± 3 | 31 ± 1 | 32±9 |
| HT Occupancy of L2 at −150 mV, % | NA | 100 | 100 | NA |
| HT Level 2 (−150 mV), IRES, % | NA | 23 ± 1 | 29 ± 2 | NA |
| HT Dwell time at −150 mV, ms | NA | 0.8 ± 0.4 | 172 ± 108 | NA |
Data taken from reference 14
Isolation of ClyA nanopores with different conductance
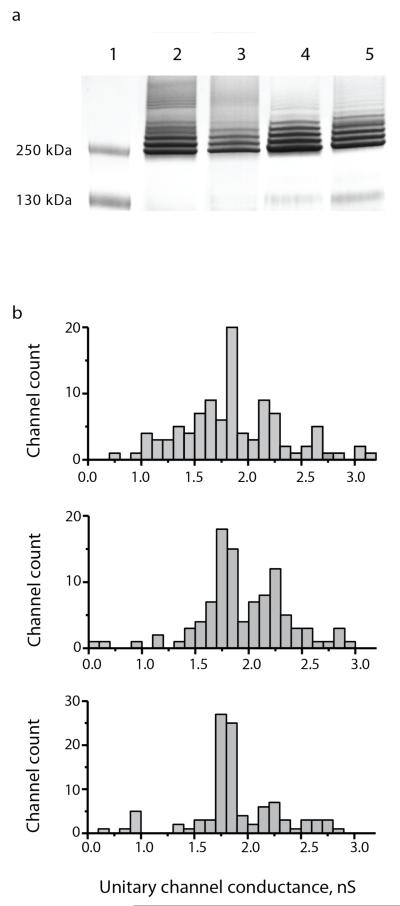
ClyA-WT, ClyA-SS and evolved ClyA oligomers, formed by incubation of purified ClyA monomers with 0.5% w/v β-dodecyl maltoside (DDM), migrated as multiple, closely spaced bands on a blue-native gradient PAGE (Figure 2a). Because on such gels the electrophoretic mobility is related to the size of the protein, the observed gel migration pattern strongly suggests that purified ClyA monomers assemble into several oligomeric forms with different size. Furthermore, in lipid bilayers pre-assembled ClyA-WT nanopores showed a wide distribution of open nanopore conductance (GO), spanning approximately 2 nS (Figure 2b, top), suggesting that in lipid bilayers ClyA-WT might assemble into nanopores of different size and/or geometry. The major peak in the distribution of unitary conductance corresponded to nanopores with an average conductance of 1.81±0.04 nS that we named Type I (GO range from 1.7 to 1.9 nS, at −35 mV, 15 mM Tris.HCl pH 7.5 and 150mM NaCl, Table 1 and Figure 2b, top). Interestingly, evolved ClyA nanopores showed an altered pattern of conductance distributions. Pre-assembled ClyA-CS showed two major peaks: the first corresponded to Type I (1.79±0.05 nS) and the second to a novel nanopore type with an average conductance of 2.23±0.08 nS that we named Type II (GO range from 2.1 to 2.4 nS, −35 mV, Table 1 and Figure 2b, middle). By contrast, ClyA-AS showed a relatively uniform distribution of mainly Type I nanopores (GO= 1.80±0.05, Figure 2b, bottom).
Figure 2.
Oligomerization pattern and nanopore conductance of pre-oligomerised ClyA nanopores. (a) Oligomerisation of ClyA nanopores examined by a 4-20% BN-PAGE. Proteins (1 mg/ml) were pre-incubated with 0.5% DDM for 30 min at 37 °C before loading into the gel (40 μg/lane). Lane 1: Protein ladder, lane 2: ClyA-WT, lane 3: ClyA-SS, lane 4: ClyA-AS and lane 5: ClyA-CS(b) Unitary nanopore conductance distribution obtained from 100 nanopores reconstituted in planar lipid bilayers for ClyA-WT (top), ClyA-CS (middle) and ClyA-AS (bottom) nanopores after pre-oligomerization in 0.5% DDM. Top: Type I ClyA-WT (GO = 1.81±0.04 nS) and Type II ClyA-WT (GO=2.21±0.07 nS) represented 24% and 19% of the inserted nanopores, respectively. Middle: ClyA-CS open pore conductance showed two clearly visible peaks. Type I ClyA-CS (GO=1.79±0.05 nS) included 35% and Type II ClyA-CS (2.23±0.08 nS) 23% of the reconstituted nanopores. Bottom: ClyA-AS nanopore conductance was relatively uniform with 52% of the reconstituted nanopores corresponding to Type I ClyA-AS (GO=1.80±0.05 nS) and 16% to Type II ClyA-AS (GO=2.24 ±0.09 nS).Recordings were carried out at −35 mV in 15 mM Tris.HCl, pH 7.5, 150mM NaCl and the temperature was 28°C.
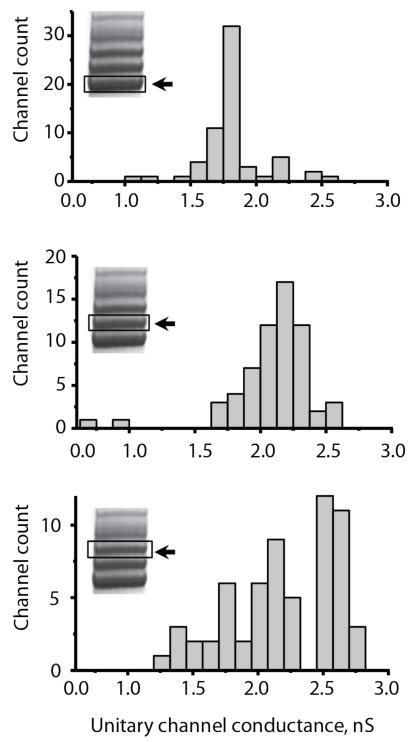
To establish whether the ClyA species with different electrophoretic mobility on BN-PAGE form nanopores with different conductance, we gel-extracted ClyA-CS from the three lowest oligomeric bands and measured the unitary conductance of 62 nanopores derived from each band within two days from gel extraction. Remarkably, the majority of ClyA-CS oligomers from the lowest band (band 1) formed Type I ClyA-CS nanopores (1.79±0.04 nS, Figure 3, top), while most of nanopores extracted from the second lowest band (band 2) reconstituted as Type II ClyA-CS nanopores (GO = 2.21 ± 0.08 nS, Figure 3, middle). Interestingly, a large fraction of the nanopores extracted from the third lowest band (band 3) reconstituted in lipid bilayers as a third nanopore type showing an average conductance of 2.64 ± 0.08 nS that we named Type III (GO range from 2.5 to 3.0 nS, at −35 mV, Table 1 and Figure 3, bottom). In order to check whether the oligomers remain intact after gel purification, extracted ClyA oligomers were re-loaded on a BN-PAGE. Despite the presence of faint additional bands probably originating from contamination during gel extraction or reversible oligomer to monomer transitions, gel purified ClyA-CS oligomers migrated mainly as single bands, indicating that the nanopores remained largely intact after gel purification (Figure S2c-d, Supplementary Information).
Figure 3.
Isolation of ClyA Types. Unitary conductance of 62 ClyA-CS nanopores extracted from the lowest (top), second lowest (middle) and third lowest (bottom) oligomeric band of ClyA-CS separated on 4-20% acrylamide BN-PAGE. The bands that were excised are boxed and marked with an arrow on the insets. Recordings were carried out at −35 mV, 28° C in 15 mM Tris.HCl pH 7.5 containing 150 mM NaCl. ClyA oligomers were extracted in 15 mM Tris.HCl pH 7.5, 150 mM NaCl containing 1 mM EDTA and 0.2% DDM.
These findings show that the three major ClyA bands observed on the BN-PAGE correspond to three distinct nanopore types. Although alternative folded ClyA structures cannot be excluded, the observation that ClyA oligomers with a slower electrophoretic migration form nanopores with higher conductance in lipid bilayers suggests that the three ClyA types might correspond to oligomers with different subunit stoichiometry. This is not necessarily surprising given that high order symmetrical oligomeric structures are often permissive with respect to subunit stoichiometry,37,38 and the exact stoichiometry of E. coli ClyA (~90% sequence identity to Salmonella typhi ClyA) oligomerisation is controversial: the crystal structure (PDB_ID: 2WCD) revealed a dodecamer36, while earlier cryo-EM studies revealed nanopores with 839 or 1340 subunits. We hypothesize that the major band on the BN-PAGE corresponding to Type I ClyA most likely represents the 12mer of the crystal structure, while the band corresponding to Type II ClyA might be the 13mer observed in earlier cryo-EM studies. By analogy, Type III ClyA-CS nanopores may correspond to a 14mer version of ClyA not observed before. Both Type I and Type II ClyA-CS nanopores remained open at high-applied potentials (Figure 4a and b) and showed low electrical noise (Figure 4c). Type III ClyA-CS, however, was not very stable in lipid bilayers as shown by frequent gating events especially at applied potentials lower than −40 mV and higher than +50 mV. In addition, Type III ClyA-CS, showed relatively high low-frequency noise (Figure 4c) and often gated irreversibly, (Figure S3b, Supplementary Information) precluding a routine use in planar lipid bilayers.
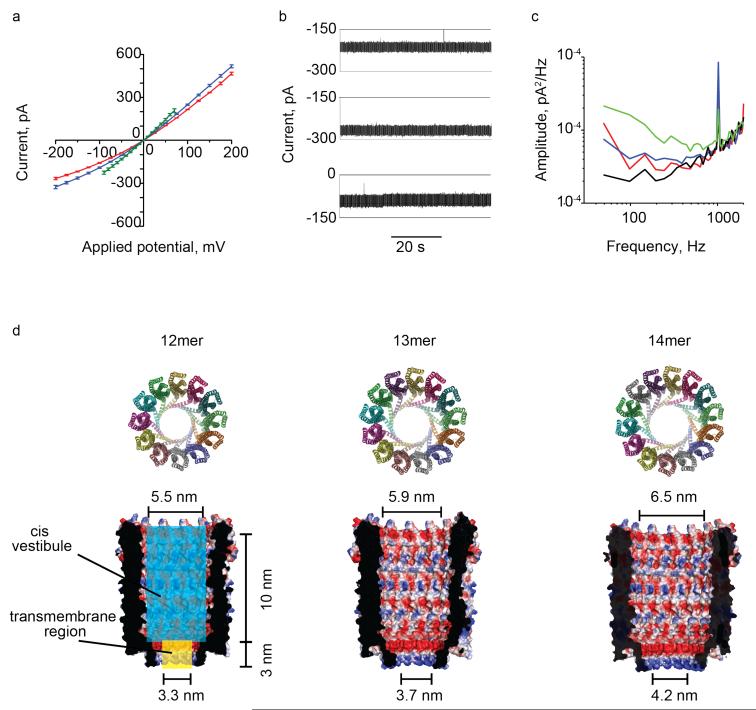
Figure 4.
The three ClyA-CS nanopores. (a) Current-voltage (I-V) curves for Type I (red), Type II (blue) and Type III (green) ClyA-CS pores. The I-V curve of Type III ClyA-CS shows ionic currents sampled at potentials lower than ±100mV due to the instability of the pore at higher applied potentials. (b) Current recordings of Type I (top) and Type II (middle) ClyA-CS at −150 mV and Type III ClyA-CS (bottom) at −35 mV. (c) Current power spectral densities of the Type I (red), Type II (blue) and Type III (green) ClyA-CS nanopores at −35 mV obtained from 0.5 s traces showing the low frequency noise in ClyA-CS nanopores. The current power spectral density at 0 mV is shown in black. Each line corresponds to the average of power spectra calculated from 3 recordings carried out on different single channels. (d) Section through the molecular models corresponding to the 12mer, 13mer and 14mer ClyA nanopores. The proteins are shown as surface representations and colored according to their “in vacuum” electrostatics (red for negative regions, and blue for positive regions, Pymol). The blue and yellow boxes (left) describe the theoretical cylinders corresponding to the cis vestibule and transmembrane region (trans vestibule), respectively, that are used to model the conductance of ClyA in equation 2. Recordings were carried out at −35 mV, 28° C in 15 mM Tris.HCl pH 7.5 containing 150 mM NaCl. The I-V current traces were recorded with an automated voltage protocol that applied the desired potential for 0.4 seconds. Electrical recordings were carried out at 28° C in 15 mM Tris.HCl pH 7.5 containing 150 mM NaCl.
Molecular models of the three type ClyA nanopores
We prepared molecular models of the 13mer and 14mer ClyA nanopores by adding, respectively, one and two subunits to the 12mer of the crystal structure (Figure 4d) as described in Supplementary Information. The resistance R of a cylindrical nanopore with no surface charge (or where it plays no role) is often estimated from its length (l) and cross-section area (A) using the following expression:41
| (1) |
where σ is the bulk conductivity (1.59 S m−1 at 25 °C for 150 mM NaCl42). The lumen of ClyA nanopores can be simplified by two communicating cylinders: one that opens to the cis side (cis vestibule) and the other that opens to the trans side (trans vestibule or transmembrane region, Figure 4d). Therefore, as for resistors in series, the theoretical conductance of the three ClyA nanopore types can be calculated from the inverse of the resistance by:
| (2) |
where ltrans is the length (3.0 nm) of the transmembrane region and dtrans its diameter (3.3 nm, 3.7 nm and 4.2 nm for the 12mer, 13mer and 14mer ClyA oligomers, respectively); and lcis is the length (10.0 nm) and dcis the diameter (5.5 nm, 5.9 nm and 6.5 nm for the 12 mer, 13 mer and 14 mer ClyA, respectively) of the cis vestibule (Figure 4d and Table 1). From eq. 2 the 12mer, 13mer and 14mer ClyA pores have a theoretical open pore conductance (GClyA =1/RClyA) of 2.06 nS, 2.47 nS and 3.07 nS, respectively, which compared well to the GO values measured at +35mV for Type I, II and III pores (2.03±0.20 nS, 2.42±0.16 nS and 2.88±0.08 nS, respectively, in 15 mM Tris.HCl pH 7.5 and 150mM NaCl); and they were 14%, 11% and 14% higher, respectively, than the GO values at −35 mV (Table 1, Figure 4d). Therefore, although the calculated conductance values are only simple estimates, these results are compatible with our view that the three types of ClyA nanopores correspond to three pores with different oligomer composition.
Thrombin as molecular caliper to test ClyA nanopores of different size
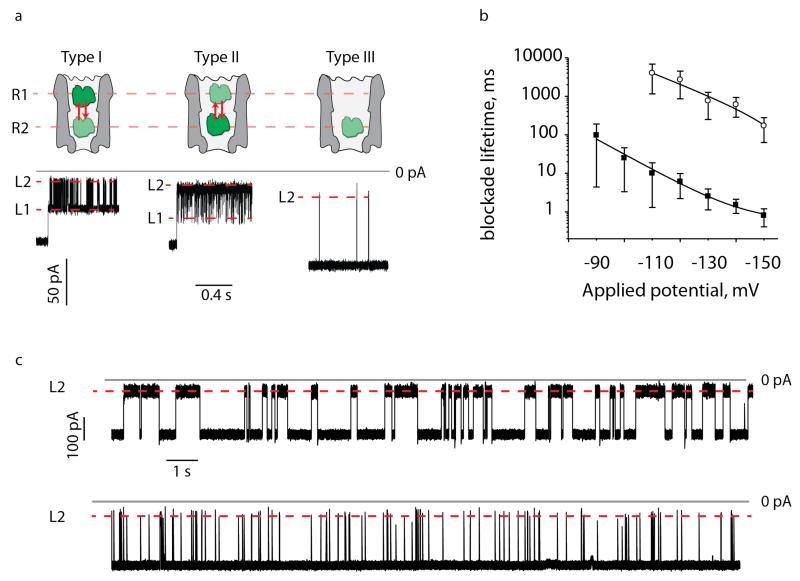
The ability to employ biological nanopores with identical amino acid composition but different size is a new feature in the biological nanopore field and is important because the size of a nanopore defines its ability to capture and study a particular molecule.20,43 We have previously shown that at −35 mV HT (human thrombin, 37kDa) inflicted well-defined current blockades to Type I ClyA nanopores that lasted for several minutes due to the entry of HT into the lumen of the pore (Figure 5a, Table 1, Figure S3, Supplementary Information).14 The current blockades switched rapidly between two blocked current levels. Level 1 [percentage of the open nanopore current (IRES%) = 56 ± 1%] and level 2 (IRES% = 23 ± 1%, Table 1, and Figure 5a). The current blockades were interpreted as two residence sites (R1 and R2) for HT within the lumen of the ClyA nanopore; where level 2 is most likely associated to HT residence at a deep more sterically constrained site R2, and level 1 is associated to the residence of HT at a site R1 located closer to the cis entrance of the nanopore (Figure 5a).14 Because thrombin provoked such well-defined pattern of current blockades we chose HT as a molecular caliper to compare the geometries of the different types of ClyA nanopores.
Figure 5.
Current blockades provoked by HT to the different ClyA-CS nanopores types. (a) Cartoon representation (top) of the physical interpretation of the HT blockades (below) to Type I (left), Type II (middle) or Type III (right) ClyA-CS nanopores at −35 mV. HT current blockades to Type I and Type II ClyA-CS switched between L1 (IRES% =56±1 and 68±1, respectively) and L2 (IRES% =23±3 and 31±1, respectively) current levels, most likely corresponding to R1 and R2 residence sites within the ClyA structure. The blockades lasted for several minutes, therefore only the first second of the current traces is shown. In Type I ClyA-CS, L1 was the most represented current blockade (70%), while in Type II ClyA-CS L2 was mostly populated (96%). HT current blockades to Type III ClyA-CS nanopores only showed L2 current levels (IRES=32±9). (b) voltage dependency of HT blockade duration determined for Type I (hollow circles) and Type II (filled squares) ClyA-CS nanopores. The lines indicate single exponential fits to the experimental points. (c) HT current blockades to Type I (top) and Type II (bottom) ClyA-CS nanopores at −150 mV. The blockades showed only L2 current levels for both nanopores (IRES% =23±1 and 29±2, for Type I and Type II ClyA-CS respectively). The traces in (a) were collected applying a Bessel low-pass filter with 2000 Hz cutoff and sampled at 10 kHz. The traces in (c) were collected applying a Bessel low-pass filter with10 kHz cutoff and sampled at 50 kHz in presence of 20 nM HT. All electrical recordings were carried out in 150 mM NaCl, 15 mM Tris.HCl pH 7.5 at 28°C. Errors are given as standard deviations.
At −35 mV HT current blockades to Type I ClyA-CS nanopores were almost identical to that of Type I ClyA-SS nanopores (Table 1, Figure 5a, Figure S3c Supplementary Information), confirming that mutations accumulated in our variants most likely did not change the size and geometry of the ClyA nanopore. HT current blockades to Type II ClyA also lasted for minutes (Figure S3, Supplementary Information) and switched between the two current levels (Table 1, Figure 5a). In Type I ClyA-CS, however, HT mostly lodged at the more superficial R1 site (70% occupancy), while in Type II ClyA-CS HT preferred the deeper site R2 (96% occupancy, Table 1, Figure 5a). HT occupancy of R2 in Type II ClyA at −35 mV was the same as HT occupancy of R2 in Type I ClyA at −60mV,14 indicating that HT requires a higher driving force to populate the sterically constrained R2 site in Type I ClyA-CS. Interestingly, HT blockades to Type III ClyA-CS were fast (55±48 ms) and showed only a level 2 current block (IRES% = 32±9%, Table 1, Figure 5a), suggesting an unhindered access to the R2 site. These results further support our hypothesis that the three ClyA types correspond to oligomers with a different stochiometry that offer different steric hindrance to HT.
Protein interaction with different types ClyA nanopores
The structure of the ClyA nanopores and the characteristics of HT induced current blockades suggest a physical model for the interaction of HT to Type I and Type II ClyA at −35mV in which single folded protein molecules enter the pore from the cis side and are trapped for several minutes in the cis vestibule of ClyA (Figure 5a and Figure S3c, Supporting Information). The translocation to the trans side of the Type I and Type II ClyA-CS pores might be prevented by the small diameter of the transmembrane region of Type I and Type II ClyA-CS (3.3 nm and 3.7 nm, respectively), which is smaller than the diameter of HT (~4.2 nm, Supporting Information). To our surprise, in the voltage range from −90 mV to −150 mV the duration of HT blockades to Type I and Type II ClyA-CS exponentially decreased with the applied potential (Figure 5b). The decrease of the duration of current blockades with increasing potential is often given as strong evidence to prove the translocation of an analyte molecule through a nanopore,19,24,44-49 and suggests here that HT might translocate through Type I and Type II ClyA in this voltage regime. Previous studies showed that proteins can be unfolded and translocated through small nanopores.25,34,50 The voltage dependent translocation of those proteins occurred in a few seconds and was initiated by charged peptides or nucleic acids as leader sequences.25,34,50 Therefore, the HT current blockades at high applied potentials might reflect folded HT lodging within the cis vestibule (additional discussion in Supplementary Information), before the unfolding of C- or N-termini of HT under high-applied potential promotes the fast and undetected translocation of the protein through the pore. However, a complete unfolding of HT is unlikely because the HT structure is stabilised by four disulphide bridges and the trans diameter of the pore (3.3 nm in the 12mer ClyA nanopore) most likely does not require the complete unfolding of the protein to allow translocation. Therefore, it is possible that the applied potential and/or electroosmotic flow induce the deformation or partial unfolding of HT or the ClyA transmembrane domain (additional discussion in Supporting Information), thereby allowing HT to pass through the pore. Along these lines, a larger Type II ClyA-CS pore should provide a lower barrier to HT translocation than Type I ClyA-CS, as manifested by ~200 fold faster current blockades at fixed applied potentials (Figure 5b, Table 1).
CONCLUSION
In this work we used a directed evolution approach to obtain biological nanopores with desired properties. Evolved ClyA monomers showed high levels of soluble expression that allowed simple purification. In lipid bilayers evolved ClyA nanopores showed uniform open pore conductance values and remained open in a wide range of applied potentials (+90 mV to −150 mV). Remarkably, we could isolate three different nanopore types, which most likely corresponded to the 12mer observed in the crystal structure,36 the 13mer described in cryo-EM studies40 and a 14mer version of ClyA not observed before. Several evidence support our interpretation: i) The three nanopore types showed different electrophoretic mobilities in blue-native PAGE and, after gel extraction, reconstituted in planar lipid bilayers as nanopores with distinct conductance. ii) The nanopore conductance of the three ClyA types compared well to the theoretical values calculated from the length and cross-section area of the 12mer, 13mer and 14mer molecular models. iii) The ionic current blockades of the HT model protein to the three ClyA pore types showed distinguishable current blockades and dwell times suggesting that larger pores imposed less steric hindrance to thrombin. The three ClyA nanopore types provide therefore a unique opportunity to sample proteins with nanopores of different diameter but identical chemical composition, and revealed that ~10% variations in the diameter of nanopores are likely to greatly affect the recognition of a target protein analyte with the nanopore.
METHODS
Screening of ClyA nanopores
As documented in the Supplementary Information, ClyA was expressed in E. cloni® EXPRESS BL21 (DE3) cells (Lucigen) by using a pT7 plasmid. Transformants were prescreened on Brucella Agar with 5% Horse Blood (BBL™, Becton, Dickinson and Company). Individual colonies were then grown and overexpressed in 96-deep-wells plates. Monomers from cell lysates were first screened for hemolytic activity on horse erythrocytes (bioMérieux) and then purified by using Ni-NTA affinity chromatography. Purified monomers were oligomerized in the presence of 0.5% β-dodecyl maltoside (GLYCON Biochemicals GmbH)40 and loaded on 4-20% blue native gel electrophoresis gels to check for oligomerisation. The electrical properties of ClyA oligomers were then screened in planar lipid bilayers.
Purification of evolved ClyA nanopores
ClyA was expressed in E. cloni® EXPRESS BL21 (DE3) cells by using a pT7 plasmid. Monomers were purified by using Ni-NTA affinity chromatography and oligomerized in the presence of 0.5% β-dodecyl maltoside (GLYCON Biochemicals GmbH).
Electrical recordings
Ionic currents were measured by recording from planar bilayers formed from diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Alabaster, AL). Currents were measured with Ag/AgCl electrodes by using a patch-clamp amplifier (Axopatch 200B, Axon Instruments, Foster City, CA) as described previously.51
Supplementary Material
ACKNOWLEDGMENT
Funding Sources
We thank the European Research Council (European Commission’s Seventh Framework Programme, project n° 260884) for funding. AB is funded by a Ph.D. grant from the Agency for Innovation by Science and Technology (IWT) Flanders.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Full documentation of material methods, additional discussion, additional Tables and Figures. This material is available free of charge via the Internet at http://pubs.acs.org.”
REFERENCES
- (1).Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci U S A. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nat Biotechnol. 2001;19:248. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- (3).Howorka S, Cheley S, Bayley H. Nat Biotechnol. 2001;19:636. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- (4).Dekker C. Nat Nanotechnol. 2007;2:209. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- (5).Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- (6).Wei R, Martin TG, Rant U, Dietz H. Angew Chem Int Ed Engl. 2012;51:4864. doi: 10.1002/anie.201200688. [DOI] [PubMed] [Google Scholar]
- (7).Bell NA, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. Nano Lett. 2012;12:512. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- (8).Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Science. 2012;338:932. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Burns JR, Stulz E, Howorka S. Nano Lett. 2013 doi: 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- (10).Hall, Scott A, Rotem D, Mehta K, Bayley H, Dekker C. Nature Nanotechnology. 2011 doi: 10.1038/nnano.2010.237. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Venkatesan BM, Bashir R. Nat Nanotechnol. 2011;6:615. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- (12).Jung Y, Bayley H, Movileanu L. J Am Chem Soc. 2006;128:15332. doi: 10.1021/ja065827t. [DOI] [PubMed] [Google Scholar]
- (13).Heinz C, Engelhardt H, Niederweis M. J Biol Chem. 2003;278:8678. doi: 10.1074/jbc.M212280200. [DOI] [PubMed] [Google Scholar]
- (14).Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. Nano Lett. 2012;12:4895. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Franceschini L, Mikhailova E, Bayley H, Maglia G. Chem Commun (Camb) 2012;48:1520. doi: 10.1039/c1cc16124e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Maglia M, Henricus M, Wyss R, Li Q, Cheley S, Bayley H. Nano Letters. 2009;9:3831. doi: 10.1021/nl9020232. [DOI] [PubMed] [Google Scholar]
- (17).Pastoriza-Gallego M, Oukhaled G, Mathe J, Thiebot B, Betton JM, Auvray LC, Pelta J. FEBS Lett. 2007;581:3371. doi: 10.1016/j.febslet.2007.06.036. [DOI] [PubMed] [Google Scholar]
- (18).Japrung D, Henricus M, Li QH, Maglia G, Bayley H. Biophysical Journal. 2010;98:1856. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cracknell JA, Japrung D, Bayley H. Nano Lett. 2013 doi: 10.1021/nl400560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C. Nano Lett. 2013 doi: 10.1021/nl3042678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Niedzwiecki DJ, Grazul J, Movileanu L. Journal of the American Chemical Society. 2010;132:10816. doi: 10.1021/ja1026858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pavlenok M, Derrington IM, Gundlach JH, Niederweis M. PLoS One. 2012;7:e38726. doi: 10.1371/journal.pone.0038726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Han A, Creus M, Schurmann G, Linder V, Ward TR, de Rooij NF, Staufer U. Anal Chem. 2008;80:4651. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- (24).Stefureac RI, Trivedi D, Marziali A, Lee JS. J Phys Condens Matter. 2010;22:454133. doi: 10.1088/0953-8984/22/45/454133. [DOI] [PubMed] [Google Scholar]
- (25).Rodriguez-Larrea D, Bayley H. Nat Nanotechnol. 2013;8:288. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bikwemu R, Wolfe AJ, Xing X, Movileanu L. J Phys Condens Matter. 2010;22:454117. doi: 10.1088/0953-8984/22/45/454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. Journal of the American Chemical Society. 2007;129:14034. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- (28).Movileanu L, Schmittschmitt JP, Scholtz JM, Bayley H. Biophys J. 2005;89:1030. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Payet L, Martinho M, Pastoriza-Gallego M, Betton JM, Auvray L, Pelta J, Mathe J. Anal Chem. 2012;84:4071. doi: 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- (30).Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, Betton JM, Pelta J. J Am Chem Soc. 2011;133:2923. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- (31).Oukhaled G, Mathé J, Biance A-L, Bacri L, Betton J-M, Lairez D, Pelta J, Auvray L. Phys. Rev. Lett. 2007;98:158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- (32).Stefureac RI, Lee JS. Small. 2008;4:1646. doi: 10.1002/smll.200800585. [DOI] [PubMed] [Google Scholar]
- (33).Stefureac R, Long YT, Kraatz HB, Howard P, Lee JS. Biochemistry. 2006;45:9172. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- (34).Nivala J, Marks DB, Akeson M. Nat Biotechnol. 2013;31:247. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo PX. Nature Nanotechnology. 2009;4:765. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. Nature. 2009;459:726. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- (37).Pogoryelov D, Klyszejko AL, Krasnoselska GO, Heller EM, Leone V, Langer JD, Vonck J, Muller DJ, Faraldo-Gomez JD, Meier T. Proc Natl Acad Sci U S A. 2012;109:E1599. doi: 10.1073/pnas.1120027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bayfield OW, Chen CS, Patterson AR, Luan W, Smits C, Gollnick P, Antson AA. PLoS One. 2012;7:e44309. doi: 10.1371/journal.pone.0044309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Tzokov SB, Wyborn NR, Stillman TJ, Jamieson S, Czudnochowski N, Artymiuk PJ, Green J, Bullough PA. J Biol Chem. 2006;281:23042. doi: 10.1074/jbc.M602421200. [DOI] [PubMed] [Google Scholar]
- (40).Eifler N, Vetsch M, Gregorini M, Ringler P, Chami M, Philippsen A, Fritz A, Muller SA, Glockshuber R, Engel A, Grauschopf U. EMBO J. 2006;25:2652. doi: 10.1038/sj.emboj.7601130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kowalczyk SW, Grosberg AY, Rabin Y, Dekker C. Nanotechnology. 2011;22:315101. doi: 10.1088/0957-4484/22/31/315101. [DOI] [PubMed] [Google Scholar]
- (42).McCleskey RB. Journal of Chemical & Engineering Data. 2011;56:317. [Google Scholar]
- (43).Niedzwiecki DJ, Iyer R, Borer PN, Movileanu L. ACS Nano. 2013 [Google Scholar]
- (44).Clarke J, Wu H, Jayasinghe L, Patel A, Reid S, Bayley H. Nature Nanotechnology. 2009;4:265. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- (45).Rincon-Restrepo M, Mikhailova E, Bayley H, Maglia G. Nano Lett. 2011;11:746. doi: 10.1021/nl1038874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Maglia G, Rincon Restrepo M, Mikhailova E, Bayley H. Proc Natl Acad Sci U S A. 2008;105:19720. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Meller A. J.Phys.: Condens.Matter. 2003;15:R581. [Google Scholar]
- (48).Wanunu M, Sutin J, McNally B, Chow A, Meller A. Biophys J. 2008;95:4716. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).de Zoysa RS, Krishantha DM, Zhao Q, Gupta J, Guan X. Electrophoresis. 2011;32:3034. doi: 10.1002/elps.201100216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Thoren KL, Worden EJ, Yassif JM, Krantz BA. Proc Natl Acad Sci U S A. 2009;106:21555. doi: 10.1073/pnas.0905880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Maglia G, Heron AJ, Stoddart D, Japrung D, Bayley H. Methods in Enzymology, Vol 475: Single Molecule Tools, Pt B. 2010;474:591. doi: 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.