Abstract
Since 1980, the global prevalence of obesity has doubled; in the United States, it has almost tripled. Billions of people are overweight and obese; the WHO reports that >65% of the world’s population die of diseases related to overweight rather than underweight. Obesity is a complex disease that can be studied from “metropolis to metabolite”—that is, beginning at the policy and the population level through epidemiology and intervention studies; to bench work including preclinical models, tissue, and cell culture studies; to biochemical assays; and to metabolomics. Metabolomics is the next research frontier because it provides a real-time snapshot of biochemical building blocks and products of cellular processes. This report comments on practical considerations when conducting metabolomics research. The pros and cons and important study design concerns are addressed to aid in increasing metabolomics research in the United States. The link between metabolism and inflammation is an understudied phenomenon that has great potential to transform our understanding of immunometabolism in obesity, diabetes, cancer, and other diseases; metabolomics promises to be an important tool in understanding the complex relations between factors contributing to such diseases.
Keywords: metabolomics, obesity, diabetes, high-fat diet, macrophage
Obesity and Inflammation
Over the past 30 y, the prevalence of obesity has increased epidemically in both Western nations and in developing countries, with little hope for effective treatments on the horizon (1, 2). More than one-third of adults and almost 1 in 5 children in the United States are obese. Globally, the WHO estimates that 500 million adults and almost 43 million children <5 y are obese (3). Childhood obesity is of particular concern because health care professionals are increasingly treating children for conditions that were traditionally considered to be adult diseases such as type 2 diabetes, high blood pressure, insulin resistance, and dyslipidemia (4, 5).
Importantly, obesity is a preventable disease. In the United States, the American Medical Association recently classified obesity as a disease (6). There are various complications associated with obesity, including coronary artery disease, type 2 diabetes, certain cancers (7), hypertension, dyslipidemia, stroke, liver and gallbladder disease, sleep apnea and respiratory problems, osteoarthritis, and gynecologic disorders such as abnormal menses and infertility. The underlying causes of obesity represent a complex web of interactions including inherited genetic traits, low physical activity levels, environmental factors such as access to affordable healthy food, cultural identity, socioeconomic status, and others.
At the cellular level, work over the past decade has increasingly linked obesity and inflammation (2). It has been demonstrated that immune cells infiltrate adipose tissue at the onset of weight gain and directly contribute to and perpetuate the inflammatory state of fat, systemic insulin resistance, and the promotion of obesity (8–14). One inflammatory cell, the macrophage, accumulates in obese adipose tissue and quantities directly correlate with adipocyte size, age, female sex, and numerous inflammatory mediators (15). Clear associations exist between obesity, metabolic syndrome, and macrophage-mediated inflammation (16). Classically activated macrophages infiltrate adipose tissue at the onset of weight gain, and contribute to the chronically inflamed state that leads to insulin resistance and diabetes. In contrast, alternatively activated macrophages safeguard insulin sensitivity and tissue homeostasis in metabolically active tissues (2). Many questions remain about how macrophage phenotypes are regulated within the adipose microenvironment along the classical-alternative polarization spectrum. Our group focuses on the relation between the tissue microenvironment and macrophage metabolism in regulating obesity-associated inflammation in diseases such as diabetes, atherosclerosis, and breast and ovarian cancers. This review focuses on the preclinical and clinical considerations of using metabolomics as a tool to aid in understanding how metabolism regulates the inflammatory response.
How Is Obesity Studied?
Preclinical models are commonly used to mimic human obesity, albeit with the understanding that mice do not always perfectly model human physiology. There are 2 methods for modeling obesity in preclinical rodent-based systems: by using genetic manipulation approaches, such as gene mutation or overexpression, or by using custom diets to induce obesity. The first knockout mouse model was created in 1989 (17). Since then, the use of genetic models in the laboratory where a single gene is deleted or overexpressed is a common method that is widely used for modeling and understanding the etiology and progression of an array of diseases, including obesity. Various model systems have been developed that are aimed at studying the underlying biochemical mechanisms that play a role in the onset and progression of obesity, as well as the metabolic pathologies associated with obesity. Existing mouse models are available through companies such as The Jackson Laboratory or Charles River Laboratories, or models can be made in the laboratory or at many mouse core facilities at most research institutes. Examples of genetic models of obesity include ob/ob mice, which lack leptin; db/db mice, which lack leptin receptor; Agouti yellow mice; melanocortin 4 receptor–deficient mice; and Zucker fa/fa obese rats. Such monogenic approaches have proven to be powerful tools providing abundant information into the contribution of single pathways to obesity. However, they are limited in terms of how this information can be translated to the condition of human obesity because it is exceedingly rare that humans have a single gene mutation that leads to obesity (18). The reality is that obesity is a complex genetic disorder involving multiple single nucleotide polymorphisms, epigenetic patterns, and multiple, cumulative exposures beginning in utero and continuing over an entire life span. The environmental exposures at play include physical activity, environmental chemicals, our microbiome, and importantly, our diet—all of which contribute to obesity onset, progression, and severity.
Although rodent genetic models of obesity have provided valuable insight into the biochemical underpinnings of obesity, for the reasons discussed above diet-induced obesity (DIO6) models more accurately represent obesity observed in humans. Thus, the use of animal models exposed to obesogenic diets best model the human condition (19). There are many diets that cause obesity. Rodents readily consume high-fat diets and can be weaned onto these diets. Obesity onset is usually rapid, with significant weight gain observed within weeks to a few months from the initiation of the diet. DIO experiments can be extremely well controlled due to the ability to use littermates, which ensures that study animals are genetically identical and exposed to similar in utero, maternal, and home cage environments.
Humans consume, on average, 34–45% of energy from fat (20), which must be taken into consideration in the laboratory setting, where scientists aim for the most human-relevant high-fat intake through DIO models. There are several DIO diets that are regularly used, each with its own set of advantages and shortcomings; however, first, it is important to understand unpurified diets, especially when considering the most appropriate “control” for the DIO diet to be used. Rodents in animal facilities are commonly maintained on a feed pellet diet, consisting of ground oats, barley, wheat, and corn (a diet high in fiber and phytoestrogens). Importantly, unpurified diets are undefined, and although the composition of macro- and micronutrients in unpurified diets is always constant, the source of those nutrients can vary from lot to lot. Recognizing this fact is important when analyzing and interpreting DIO model data if unpurified diets are chosen as the “control” diet.
Purified or defined diets are diets that are mass produced by companies wherein all diet components are known. Choline-deficient and methyl-donor–deficient diets, which lack choline, folate, and methionine, or high-sucrose diets are 3 obesogenic diets that can be used in DIO studies. However, by far the most commonly used diets are the high-fat diets. The most widely used diet is a lard-based van Heek series, which includes a range of high-fat diets with 45% or 60% of energy derived from fat, typically lard; therefore, these diets have a high saturated fat content. They are commonly paired with a diet with 10% of energy derived from fat (a “low fat” control diet) that does not induce obesity. The pitfall with this 10% control diet is that to be isocaloric to the high-fat diets that contain 45% or 60% of energy from fat, the diet containing 10% of energy from fat is usually high in sucrose, which can induce fatty liver. Another alternative diet is the Western diet, which also derives its fat from lard, as with the van Heek series, and contains added cholesterol to induce atherosclerosis. In addition, specialized diets are available, including the Surwit diet which is high in coconut oil (medium-chain fats), for specific scientific questions. As discussed above, a complicating factor for many of these diets is that to remain isocaloric, sucrose is added to the low-fat diet to match the caloric content of the high-fat diet. Sucrose is a combination of glucose and fructose, which can induce weight gain through multiple pathways (21), although to a lesser extent than high-fat diets. To overcome this limitation, sucrose-free diets are available in which sucrose has been replaced with a more complex carbohydrate such as cornstarch.
Finally, the diet model that may best mimic human obesity is called the Cafeteria (CAF) diet. The CAF diet is a labor-intensive smorgasbord feeding model in which rodents are provided the choice of unpurified diets or highly palatable, calorically dense, nutrient-deficient human “junk” foods purchased from a grocery store. Therefore, the food exposure more closely parallels what humans are eating. Indeed, Piernas et al. (22) reported that in preschool-aged kids, snack foods account for 27% of total caloric intake. Why is this important? Snack foods are energy dense, providing lots of calories with little nutritional value. For example, 100 g of fresh corn has 83 kcal, a corn tortilla contains 210 kcal, and fully processed, energy-dense tortilla chips contain 493 kcal per serving.
Our laboratory was interested in comparing the obesogenic and inflammatory effect of various diets including the CAF diet. We provided a choice of 3 human foods that varied from day to day, including cookies, salami, chips, chocolate, muffins, cereals, and cheese, among other options. Detailed information on the diets is available in Sampey et al. (21, 23). Although rats fed this junk-food diet ate ∼17% of calories from feed pellets, overall we observed hyperphagia resulting in rapid weight gain in the CAF diet–fed rats above and beyond what we observed in the rats fed diets that contained 45% of energy from fat and 10% of energy from fat and an unpurified diet–only control diet. Rats fed the control diets consumed ∼100 kcal/d, whereas rats fed the CAF diet consumed 130–150 kcal/d (21). The CAF diet is a model to broadly mimic human snacking, because a variety of options and the palatability of snack foods lead to overeating (21). CAF diet–fed rats developed prediabetes characterized by elevated insulin, fasting blood glucose, and circulating nonesterified FFAs, with concurrent impaired glucose and insulin tolerance. Both the liver and adipose tissue from CAF diet–fed rats were severely inflamed, and markers of pancreatic islet dysfunction were evident. Of note, when we compared rats fed the low-fat (10% of energy from fat) and high-fat (45% of energy from fat)–defined diets, we did not see significant divergence between the groups at the same time point at which we saw dramatic CAF diet–induced metabolic dysfunction. Given the remarkable effects of the CAF diet, we turned to metabolomics to provide insight into why the CAF diet induced inflammation that was much more severe than the typical 45% high-fat experimental diet. Using metabolomics and bioinformatics analysis, we identified mitochondrial dysfunction and a specific acylcarnitine, lauroylcarnitine, a metabolite of FA oxidation that was elevated in adipose tissue of CAF diet–fed rats compared with high-fat-diet–fed or control rats (21, 23). Lauroylcarnitine significantly correlated with crown-like structures in adipose tissue, markers of inflammation and insulin resistance. We then demonstrated that lauroylcarnitine drove proinflammatory macrophage polarization and cytokine release, therefore providing a potential mechanism linking obesity, lipid metabolism, and inflammation to be tested in future studies (21, 23).
How Does a Molecular Biologist Incorporate Metabolomics into Nutrition Research?
Substantial progress has been made in understanding the inflammatory phenotype of macrophages in the microenvironment of tissues such as adipose, liver, and the vessel wall in atherosclerosis. However, how metabolism regulates macrophage polarization, thus influencing tissue inflammation and insulin resistance within metabolically active tissue microenvironments, is poorly understood (2). Addressing this question is essential because cellular and molecular pathways regulated by metabolites, such as bioactive lipid mediators, have yet to be explored in great detail and will have tremendous potential as novel therapeutic targets. The ability to harness control of the “immune-metabolic” milieu could break the proinflammatory cycle and would control insulin resistance, compensatory hyperinsulinemia, and hyperglycemia of diabetes. Our previous work on cytosolic FA binding protein aP2 (FABP4) lends support to the concept of fuel metabolism and inflammation being interdependent; we demonstrated that the integration of lipid trafficking and signaling in the macrophage contributes significantly to the pathogenesis of atherosclerosis, insulin resistance, and obesity through multiple pathways such as the nuclear hormone receptor PPAR-γ, modulation of reverse cholesterol transport, the inflammatory inhibitor of kappa B kinase (Iκk)–NF-κB kinase pathway, and endoplasmic reticulum stress (2, 21, 23–33). Furthermore, Vats et al. (34) established that generation of alternatively activated macrophages is an FA oxidation-dependent process. We recently demonstrated that metabolic reprogramming of macrophages is possible in vitro through overexpressing glucose transporter 1 (GLUT1) (35). Using metabolomics, we showed that forced glucose metabolism and activation of the pentose phosphate pathway was sufficient to polarize macrophages to the classically activated macrophage phenotype (35). Therefore, an important modifier of macrophage plasticity and the inflammatory tone contributing to the onset of diabetes is the availability of fuel substrates to the macrophages in the microenvironment. Biochemists recognize that glucose- and lipid-involving metabolic pathways, in fact all metabolic pathways, work in concert to maintain cellular and tissue homeostasis. Therefore, altering some aspect of glucose metabolism, for example, will undoubtedly have effects on lipid and protein metabolic pathways. Classic radiotracer methods are useful for determining flux through a specific pathway, but to get at the heart of how a cell or tissue is responding to a metabolic change in a more holistic way, metabolomics can be used.
What Are the Options for Metabolomics for Clinicians or Bench Scientists?
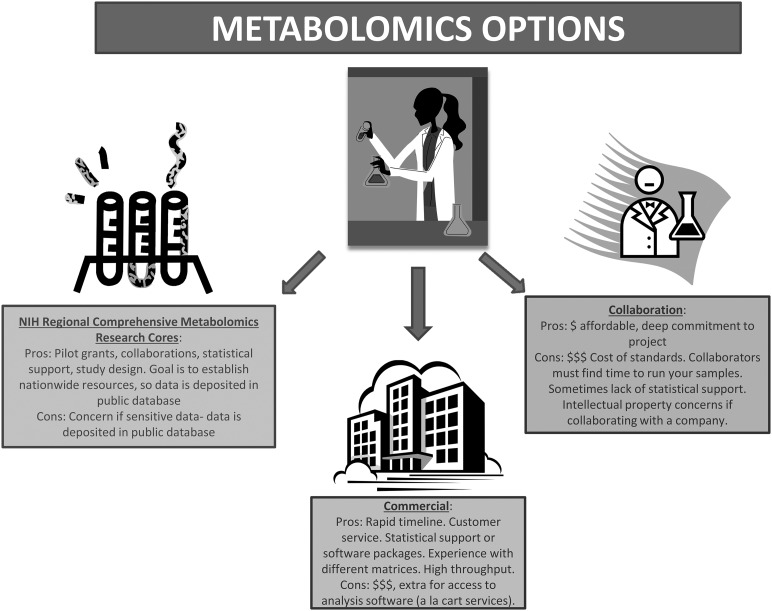
As biochemists who want to understand the metabolic underpinnings of processes but do not have the chemistry, informatics, or programming background to answer such questions alone, we have to turn to others to conduct metabolomics analyses (Figure 1). When designing a metabolomics study, one must carefully consider the hypothesis to be tested or question asked while at the same time designing studies that are sufficiently powered to detect differences considering the expected biological variation and keeping within budgetary constraints. One may conduct targeted investigations to detect a few important metabolites or run samples through multiple platforms in an unbiased discovery-based approach. As with other technologies, cell culture studies are inherently less variable than mouse samples, which are, in turn, less variable than human samples. Controlling for as many variables as possible is imperative. In our experience, 50–200 mg for most tissues, 50–100 μL of plasma, or ∼1 million cells are sufficient for most metabolomics analyses (23, 24, 35–37). Sample size should be as great as possible because multiple metabolites are measured, which is similar to a microarray study. Clinical considerations include sample size and proper controls as in any study; however, one must consider the metabolic state of patients (fasted, taking medications, etc.) and the time to capture the sample from the operating room or clinic to the freezer. Some studies use a “stop watch” approach and place a stop watch with the sample when the clinician isolates it. The stop watch only gets turned off when the sample is frozen in liquid nitrogen. Depending on the isolation of the sample, availability of the research associate, and transport to the laboratory, sample freezing times can vary from minutes to many hours. This is highly relevant when analyzing metabolites. Furthermore, because clinical samples are precious, it is best to optimize on practice samples. Finally, a benefit of using metabolomics rather than other biomarkers (e.g., serum or urine proteins such as a cytokine) is that metabolites do not vary by species. There can be extensive cross-species validation.
FIGURE 1.
The pros and cons of metabolomics.
There are several approaches for initiating metabolomics research—with pros and cons for each—and every laboratory must decide what fits best. First, for those “do it yourself-ers” with the proper machinery and training, metabolomics is a matter of having the right authentic standards to quantitate specific metabolites of interest and the right people to prepare samples and operate your machines. One caveat to conducting your own metabolomics is that it is often cost-prohibitive to purchase the hundreds of authentic standards necessary to conduct comprehensive discovery work. Furthermore, without the proper training in programming and statistics, one is often left “eye-balling” the data for significant findings. Of course, there are plenty of outstanding researchers who have excellent metabolomics facilities. Collaborating with such experts is the second route to pursue metabolomics research as a novice.
As with any collaboration, there are pluses and minuses: one must discuss costs, personnel, and authorship upfront. A typical hurdle with collaborations is the timeline for study completion and the possible extended wait time for your samples to be analyzed. However, working closely with experts will help with experimental design, data interpretation, and planning of further experiments. Collaborating with a company that conducts metabolomics is also an option, but that has drawbacks as well, such as access (because not every company will want to collaborate with you) and intellectual property concerns. For example, the method of processing samples used may be proprietary and there may be substantial hurdles to clear when it is time to publish.
The third option is a fee-for-service through companies that specialize in metabolomics such as Metabolon, Lipomics (recently acquired by Metabolon), Metabolomic Discoveries, and Chenomx, among others (Table 1). There can be substantial costs associated with this approach depending on the service package purchased; services offered can range from study design and sample prep to statistical analysis, data interpretation, and figure preparation. Through their high-throughput capacity and standard operating procedures, companies often will have established protocols for your samples, including quantities necessary, tricks for preparing samples, etc., that save time and money because assay optimizations are not necessary. Because it is a fee-for-service rather than a collaboration, data are usually generated within an established time as agreed upon when a contract is signed. Companies must consider customer satisfaction and repeat business, but this is a relationship that you pay for. Although you are guaranteed a final product, the option to go back and conduct follow-up experiments is not possible without acquiring additional costs.
TABLE 1.
Location and representative publications for sites conducting metabolomic analysis1
| Commercial sites for metabolomic analysis | |||
| Name | Location | URL | Representative publications |
| Metabolon, Inc. | Durham, NC (RTP) | www.metabolon.com | (23, 36, 38) |
| Lipomics, Inc. (owned by Metabolon, Inc.) | Sacramento, CA | www.lipomics.com | (24) |
| Metabolomic Discoveries | Potsdam-Golm, Germany | www.metabolomicdiscoveries.com | (39, 40) |
| Chenomx, Inc. | Edmonton, Canada | www.chenomx.com | (41, 42) |
| RCMRCs, NIH-funded and Common Fund’s Metabolomics program | |||
| NIH West Coast Metabolomics Center at UC Davis (WC3MRC) | Davis, CA | www.metabolomics.ucdavis.edu | (43, 44) |
| Michigan Regional Comprehensive Metabolomics Research Core (MRC) | Ann Arbor, MI | www.mrc2.umich.edu | (45, 46) |
| NIH Eastern Regional Comprehensive Metabolomics Resource Core at RTI International (RTI RCMRC) | Durham (RTP), NC | www.rti.org | (47, 48) |
| Southeast Center for Integrated Metabolomics at University of Florida (SECIM) | Gainesville, FL | www.secim.ufl.edu | (49, 50) |
| Resource Center for Stable Isotope–Resolved Metabolomics at University of Kentucky (RC-SIRM) | Lexington, KY | www.bioinformatics.cesb.uky.edu | (49, 50) |
| Metabolomics Resource Core at the Mayo Clinic | Rochester, MN | www.mayo.edu | (51, 52) |
RCMRC, Regional Comprehensive Metabolomics Research Core; RTP, Research Triangle Park, NC.
A fourth and final metabolomics option is a new initiative supported by the NIH funded through the Common Fund’s Metabolomics program (Table 1). This is a national program aimed at increasing metabolomics capacity by developing centers with high-throughput technologies, training, and mentoring for metabolomics researchers. The goal is to standardize protocols in an effort to increase sensitivity and speed to identify metabolites. In addition, by providing reference standards and data sharing, the NIH hopes to build a publicly accessible national database that will propel metabolomics research forward. There are currently 6 Regional Comprehensive Metabolomics Research Cores (RCMRCs) funded by the Metabolomics program. The cores have expertise, instruments, and training programs to increase metabolomics capacity. Existing sites are as follows: 1) NIH West Coast Metabolomics Center at UC Davis (WC3MRC), 2) Michigan Regional Comprehensive Metabolomics Research Core (MRC), 3) NIH Eastern Regional Comprehensive Metabolomics Resource Core at RTI International (RTI RCMRC), 4) Southeast Center for Integrated Metabolomics (SECIM), 5) Resource Center for Stable Isotope-Resolved Metabolomics (RC-SIRM), and 6) the Metabolomics Core at the Mayo Clinic.
Importantly, with strong bioinformatics support, “omics”-based approaches can be used for hypothesis generating, biomarker identification, and, ideally, lead to biomarkers in vivo or in vitro to be tested as bona fide biomediators of observed effects. In summary, the link between metabolism and inflammation is an understudied phenomenon that has great potential to transform our understanding of immunometabolism in obesity, diabetes, cancer, and other diseases. One important benefit of metabolomics is that metabolites are conserved across species, as mentioned above, which makes it easy to translate preclinical findings to humans. Bioinformatics approaches provide a deeper understanding of the complex relations between these pathways that would otherwise be impossible with traditional biochemical assays. Current and future students and postdoctoral fellows should be encouraged to incorporate statistical training, computer programming, and informatics into their education in conjunction with basic molecular biology techniques and a strong understanding of metabolism to best be prepared for the next era of high-density, high-throughput data generation using metabolomics.
Acknowledgments
ARJ and LM wrote the manuscript and had responsibility for the final content. Both authors read and approved the final manuscript.
Footnotes
Abbreviations used: CAF, Cafeteria; DIO, diet-induced obesity; FABP4, fatty acid binding protein aP2; GLUT1, glucose transporter 1; IκK, inhibitor of kappa B kinase; RTP, Research Triangle Park, NC.
References
- 1.Ogden CL, Carroll MD, Flegal KM. Prevalence of obesity in the United States. JAMA 2014;312:189–90. [DOI] [PubMed] [Google Scholar]
- 2.Johnson AR, Milner JJ, Makowski L. The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev 2012;249:218–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Obesity and overweight. Geneva (Switzerland): WHO; 2011.
- 4.Ogden CL, Carroll MD, Curtin LR, Lamb MM, Flegal KM. Prevalence of high body mass index in US children and adolescents, 2007–2008. JAMA 2010;303(3):242–9. [DOI] [PubMed]
- 5.Klein JD, Dietz W. Childhood obesity: the new tobacco. Health Aff (Millwood) 2010;29(3):388–92. [DOI] [PubMed]
- 6.American Medical Association. [cited 2014 Sep 1] Available from: http://www.ama-assn.org/ama/pub/news/news/2013/2013-06-18-new-ama-policies-annual-meeting.page.
- 7.Sundaram S, Johnson AR, Makowski L. Obesity, metabolism and the microenvironment: links to cancer. J Carcinog 2013;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumeng CN, Maillard I, Saltiel AR. T-ing up inflammation in fat. Nat Med 2009;15(8):846–7. [DOI] [PubMed]
- 9.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112:1796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Divoux A, Sun J, Zhang J, Clement K, Glickman JN, Sukhova GK, Wolters PJ, Du J, Gorgun CZ, et al. Genetic deficiency and pharmacological stabilization of mast cells reduce diet-induced obesity and diabetes in mice. Nat Med 2009;15(8):940–5. [DOI] [PMC free article] [PubMed]
- 12.Ohmura K, Ishimori N, Ohmura Y, Tokuhara S, Nozawa A, Horii S, Andoh Y, Fujii S, Iwabuchi K, Onoe K, et al. Natural killer T cells are involved in adipose tissues inflammation and glucose intolerance in diet-induced obese mice. Arterioscler Thromb Vasc Biol 2010;30:193–9. [DOI] [PubMed] [Google Scholar]
- 13.Kintscher U, Hartge M, Hess K, Foryst-Ludwig A, Clemenz M, Wabitsch M, Fischer-Posovszky P, Barth TF, Dragun D, Skurk T, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol 2008;28:1304–10. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Ghosh S, Perrard XD, Feng L, Garcia GE, Perrard JL, Sweeney JF, Peterson LE, Chan L, Smith CW, et al. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation 2007;115(8):1029–38. [DOI] [PubMed]
- 15.Surmi BK, Hasty AH. Macrophage infiltration into adipose tissue: initiation, propagation and remodeling. Future Lipidol 2008;3:545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldfine AB, Fonseca V, Jablonski KA, Chen YD, Tipton L, Staten MA, Shoelson SE. Targeting inflammation using salsalate in type 2 diabetes study T. salicylate (salsalate) in patients with type 2 diabetes: a randomized trial. Ann Intern Med 2013;159:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smithies O. Science brick by brick. Nature 2010;467:S6. [DOI] [PubMed] [Google Scholar]
- 18.Farooqi IS, Matarese G, Lord GM, Keogh JM, Lawrence E, Agwu C, Sanna V, Jebb SA, Perna F, Fontana S, et al. Beneficial effects of leptin on obesity, T cell hyporesponsiveness, and neuroendocrine/metabolic dysfunction of human congenital leptin deficiency. J Clin Invest 2002;110:1093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy AJ, Ellacott KL, King VL, Hasty AH. Mouse models of the metabolic syndrome. Dis Model Mech 2010;3(3–4):156–66. [DOI] [PMC free article] [PubMed]
- 20.Agricultural Research Service, USDA. National Health and Nutrition Examination Survey. [cited 2014 Sep 1] Available from: www.ars.usda.gov/SP2UserFiles/Place/12355000/pdf/0910/Table_1_NIN_GEN_09.pdf.
- 21.Sampey BP, Vanhoose AM, Winfield HM, Freemerman AJ, Muehlbauer MJ, Fueger PT, Newgard CB, Makowski L. Cafeteria diet is a robust model of human metabolic syndrome with liver and adipose inflammation: comparison to high-fat diet. Obesity (Silver Spring) 2011;19:1109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piernas C, Popkin BM. Trends in snacking among U.S. children. Health Aff (Millwood) 2010;29(3):398–404. [DOI] [PMC free article] [PubMed]
- 23.Sampey BP, Freemerman AJ, Zhang J, Kuan PF, Galanko JA, O'Connell TM, Ilkayeva OR, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Metabolomic profiling reveals mitochondrial-derived lipid biomarkers that drive obesity-associated inflammation. PLoS ONE 2012;7:e38812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erbay E, Babaev VR, Mayers JR, Makowski L, Charles KN, Snitow ME, Fazio S, Wiest MM, Watkins SM, Linton MF, et al. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat Med 2009;15:1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vaillancourt E, Kono K, Babaev VR, Fazio S, Linton MF, et al. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature 2007;447:959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity: macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and IkappaB kinase activities. J Biol Chem 2005;280:12888–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makowski L, Hotamisligil GS. The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Curr Opin Lipidol 2005;16:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, Hotamisligil GS. Combined adipocyte-macrophage fatty acid-binding protein deficiency improves metabolism, atherosclerosis, and survival in apolipoprotein E-deficient mice. Circulation 2004;110:1492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, Hotamisligil GS. Adipocyte fatty acid-binding protein, aP2, alters late atherosclerotic lesion formation in severe hypercholesterolemia. Arterioscler Thromb Vasc Biol 2002;22:1686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 2001;7:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotamisligil GS. Endoplasmic reticulum stress and atherosclerosis. Nat Med 2010;16(4):396–9. [DOI] [PMC free article] [PubMed]
- 32.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010;140(6):900–17. [DOI] [PMC free article] [PubMed]
- 33.Makowski L, Noland RC, Koves TR, Xing W, Ilkayeva OR, Muehlbauer MJ, Stevens RD, Muoio DM. Metabolic profiling of PPARalpha−/− mice reveals defects in carnitine and amino acid homeostasis that are partially reversed by oral carnitine supplementation. FASEB J 2009;23:586–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab 2006;4:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freemerman AJ, Johnson AR, Sacks GN, Milner JJ, Kirk EL, Troester MA, Macintyre AN, Goraksha-Hicks P, Rathmell JC, Makowski L. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a pro-inflammatory phenotype. J Biol Chem 2014;289:7884–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhatt AP, Jacobs SR, Freemerman AJ, Makowski L, Rathmell JC, Dittmer DP, Damania B. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc Natl Acad Sci USA 2012;109:11818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brauer HA, Makowski L, Hoadley KA, Casbas-Hernandez P, Lang LJ, Roman-Perez E, D'Arcy M, Freemerman AJ, Perou CM, Troester MA. Impact of tumor microenvironment and epithelial phenotypes on metabolism in breast cancer. Clin Cancer Res 2013;19:571–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makowski L, Zhou C, Zhong Y, Kuan PF, Fan C, Sampey BP, Difurio M, Bae-Jump VL. Obesity increases tumor aggressiveness in a genetically engineered mouse model of serous ovarian cancer. Gynecol Oncol 2014;133:90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pivovarova O, Gogebakan O, Kloting N, Sparwasser A, Weickert MO, Haddad I, Nikiforova VJ, Bergmann A, Kruse M, Seltmann AC, et al. Insulin up-regulates natriuretic peptide clearance receptor expression in the subcutaneous fat depot in obese subjects: a missing link between CVD risk and obesity? J Clin Endocrinol Metab 2012;97:E731–9. [DOI] [PubMed] [Google Scholar]
- 40.Hische M, Larhlimi A, Schwarz F, Fischer-Rosinsky A, Bobbert T, Assmann A, Catchpole GS, Pfeiffer AF, Willmitzer L, Selbig J, et al. A distinct metabolic signature predicts development of fasting plasma glucose. J Clin Bioinform 2012;2:3. [DOI] [PMC free article] [PubMed]
- 41.Chao J, Huo TI, Cheng HY, Tsai JC, Liao JW, Lee MS, Qin XM, Hsieh MT, Pao LH, Peng WH. Gallic acid ameliorated impaired glucose and lipid homeostasis in high fat diet-induced NAFLD mice. PLoS ONE 2014;9:e96969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowan TE, Palmnas MS, Yang J, Bomhof MR, Ardell KL, Reimer RA, Vogel HJ, Shearer J. Chronic coffee consumption in the diet-induced obese rat: impact on gut microbiota and serum metabolomics. J Nutr Biochem 2014;25:489–95. [DOI] [PubMed] [Google Scholar]
- 43.Nording ML, Yang J, Georgi K, Hegedus Karbowski C, German JB, Weiss RH, Hogg RJ, Trygg J, Hammock BD, Zivkovic AM. Individual variation in lipidomic profiles of healthy subjects in response to omega-3 fatty acids. PLoS ONE 2013;8:e76575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smilowitz JT, Totten SM, Huang J, Grapov D, Durham HA, Lammi-Keefe CJ, Lebrilla C, German JB. Human milk secretory immunoglobulin A and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J Nutr 2013;143:1906–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonnell SR, Hwang SR, Rolland D, Murga-Zamalloa C, Basrur V, Conlon KP, Fermin D, Wolfe T, Raskind A, Ruan C, et al. Integrated phosphoproteomic and metabolomic profiling reveals NPM-ALK-mediated phosphorylation of PKM2 and metabolic reprogramming in anaplastic large cell lymphoma. Blood 2013;122:958–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Azzouny M, Evans CR, Treutelaar MK, Kennedy RT, Burant CF. Increased glucose metabolism and glycerolipid formation by fatty acids and GPR40 receptor signaling underlies the fatty acid potentiation of insulin secretion. J Biol Chem 2014;289:13575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumner S, Snyder R, Burgess J, Myers C, Tyl R, Sloan C, Fennell T. Metabolomics in the assessment of chemical-induced reproductive and developmental outcomes using non-invasive biological fluids: application to the study of butylbenzyl phthalate. J App Toxicol 2009;29:703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sumner SJ, Burgess JP, Snyder RW, Popp JA, Fennell TR. Metabolomics of urine for the assessment of microvesicular lipid accumulation in the liver following isoniazid exposure. Metabolomics 2010;6(2):238–49. [DOI] [PMC free article] [PubMed]
- 49.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, et al. Targeting lactate dehydrogenase–A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab 2014;19:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell JM, Fan TW, Lane AN, Moseley HN. Development and in silico evaluation of large-scale metabolite identification methods using functional group detection for metabolomics. Front Genet 2014;5:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutta T, Chai HS, Ward LE, Ghosh A, Persson XM, Ford GC, Kudva YC, Sun Z, Asmann YW, Kocher JP, et al. Concordance of changes in metabolic pathways based on plasma metabolomics and skeletal muscle transcriptomics in type 1 diabetes. Diabetes 2012;61:1004–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemutlu E, Juranic N, Zhang S, Ward LE, Dutta T, Nair KS, Terzic A, Macura S, Dzeja PP. Electron spray ionization mass spectrometry and 2D 31P NMR for monitoring 18O/16O isotope exchange and turnover rates of metabolic oligophosphates. Anal Bioanal Chem 2012;403:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]