Abstract
Motivation: MicroRNAs (miRNAs) play critical roles in gene regulation. Although it is well known that multiple miRNAs may work as miRNA modules to synergistically regulate common target mRNAs, the understanding of miRNA modules is still in its infancy.
Results: We employed the recently generated high throughput experimental data to study miRNA modules. We predicted 181 miRNA modules and 306 potential miRNA modules. We observed that the target sites of these predicted modules were in general weaker compared with those not bound by miRNA modules. We also discovered that miRNAs in predicted modules preferred to bind unconventional target sites rather than canonical sites. Surprisingly, contrary to a previous study, we found that most adjacent miRNA target sites from the same miRNA modules were not within the range of 10–130 nucleotides. Interestingly, the distance of target sites bound by miRNAs in the same modules was shorter when miRNA modules bound unconventional instead of canonical sites. Our study shed new light on miRNA binding and miRNA target sites, which will likely advance our understanding of miRNA regulation.
Availability and implementation: The software miRModule can be freely downloaded at http://hulab.ucf.edu/research/projects/miRNA/miRModule.
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: haihu@cs.ucf.edu or xiaoman@mail.ucf.edu.
1 Introduction
MicroRNAs (miRNAs) play critical roles in gene regulation (Bartel, 2004, 2009). MiRNAs are a family of small (∼22 nucleotides) non-coding RNAs. They can bind mRNAs at 5′ untranslated regions (UTRs), coding sequences (CDSs), and 3′ UTRs. The binding is traditionally thought to be through base-pairing of the seed regions in miRNAs with the partially complementary sequences in target mRNAs (Bartel, 2009). The seed region refers to the 5′ end of miRNAs from position 2 to position 7 (Lewis et al., 2003, 2005). Depending on the pairing quality, miRNA target sites are classified into two categories: canonical sites and non-canonical sites. The former are target sites that are perfect complementary to the seed regions, while the latter are target sites with imperfect seed complementarity (G:U wobbles or mismatches). With the advance of biotechnology, it is accepted that base-pairing can involve both seed regions and non-seed regions (Hafner et al., 2010; Helwak et al., 2013; Wang 2014). That is, other types of target sites exist in addition to the canonical and non-canonical target sites. We define unconventional sites as target sites other than the canonical sites. Regardless of the types of target sites, the binding of miRNAs to their target mRNAs during diverse cellular processes may degrade target mRNAs, and/or repress the translation of target mRNAs to proteins (Bartel, 2004, 2009; Wang et al., 2011). Due to such pivotal roles in gene regulation, it is critical to study miRNAs and their target sites.
miRNAs often form modules to regulate target mRNAs (Doench and Sharp, 2004; Krek et al., 2005; Saetrom et al., 2007; Vella et al., 2004; Wu et al., 2010). In this study, a miRNA module is defined as a group of miRNAs that co-bind a significant number of mRNAs and repress the expression of these common mRNAs significantly more than individual miRNAs in the module (Sections 2.2–2.3). Several studies show that miRNA modules synergistically control the expression of their common target mRNAs (Doench and Sharp, 2004; Krek et al., 2005; Saetrom et al., 2007; Vella et al., 2004; Wu et al., 2010). For instance, miR-124, miR-375, and let-7b form a miRNA module that coordinately regulates the gene Mtpn in a murine pancreatic cell line (Krek et al., 2005). The distance between adjacent target sites of miRNAs in the same miRNA modules may play critical roles in target mRNA downregulation (Doench and Sharp, 2004; Saetrom et al., 2007). According to multiple past experiments, the distance of miRNA target sites for optimal downregulation of target mRNAs is between 13 and 35 nucleotides (Brennecke et al., 2005; Doench and Sharp, 2004; Kloosterman et al., 2004; Vella et al., 2004). Saetrom et al. computationally showed that miRNA target sites within 130 nucleotides are more likely conserved than more distant sites in 3′ UTRs (Saetrom et al., 2007), suggesting that target sites of miRNAs may need to be within certain ranges to be functional. Several studies also defined miRNA modules by harnessing predicted miRNA target sites in 3′ UTRs and the co-expression relationship of target mRNAs of the same miRNAs (Bryan et al., 2013; Jayaswal et al., 2011; Zhang et al., 2011).
Despite various studies mentioned above, our understanding of miRNA modules is still rudimentary. To our knowledge, all published large-scale studies on miRNA modules so far are based on computationally predicted miRNA target sites in 3′ UTRs. However, 3′ UTRs only account for a small portion of potential miRNA target site residing regions (Hafner et al., 2010; Helwak et al., 2013). Moreover, even the most well-known target site prediction methods currently produce a significant fraction of false positive target sites (Witkos et al., 2011). In addition, in defining miRNA modules, few computational studies require the higher downregulation of target gene expression by miRNA modules than that by subsets of miRNAs contained in the modules (Wu et al., 2010). Therefore, although we have gained basic insight into miRNA modules from previous studies, our understanding of miRNA modules may be biased and limited.
In this study, we used experimentally determined instead of computationally predicted target sites to study miRNA modules. We predicted 181 miRNA modules and 306 potential miRNA modules. We analyzed binding energy, location, and distances of target sites in these predicted miRNA modules. We observed that target sites of these predicted modules were in general weaker compared with target sites not bound by miRNA modules. We also discovered that miRNAs in predicted modules preferred to bind only unconventional target sites, instead of only canonical sites, or a mixture of canonical and unconventional sites. Contrary to a previous study (Saetrom et al., 2007), we noticed that most target sites of miRNAs from the same modules were not within the range of 10–130 nucleotides. Interestingly, the distance of target sites bound by miRNAs in the same modules was shorter when miRNA modules bound unconventional instead of canonical sites. Our study sheds new light on miRNA binding, which will likely advance our understanding of miRNA regulation.
2 Material and methods
2.1 Target sites and gene expression data
We downloaded the experimentally determined miRNA target sites from the crosslinking, ligation and sequencing of hybrids (CLASH) experiments (Helwak et al., 2013). In these experiments, a miRNA and one of its interacting mRNA target sites were ligated and sequenced as a chimeric segment. The sequenced chimerical segments for multiple miRNAs and their target mRNAs were then separated into segments from miRNAs and segments from their target mRNAs, which provided the information of which miRNAs targeted which region of their target mRNAs. We obtained 18 514 high-confidence miRNA–mRNA interactions in HEK293 cells. These interactions involved 399 miRNAs, 7390 mRNAs, 4130 canonical sites, 10 300 non-canonical sites and 14 384 unconventional sites. Among these target sites, 1034 (5.6%), 11 367 (61.4%) and 6096 (32.9%) of them were within 5′ UTRs, CDSs and 3′ UTRs, respectively. The remaining 17 target sites were not within any annotated mRNA.
We also downloaded mRNA expression data from (Schmitter et al., 2006). The expression data before and after AGO2 protein knocking down in HEK293 were used to measure the downregulation of miRNAs and miRNA modules. This was because the knock-down of the AGO2 protein basically prevented the functions of all miRNAs, as AGO2 is an essential component of the RNA-induced silencing complex that recognizes target mRNAs and loads miRNAs to target mRNAs (Bartel, 2009).
2.2 The pipeline to predict miRNA modules
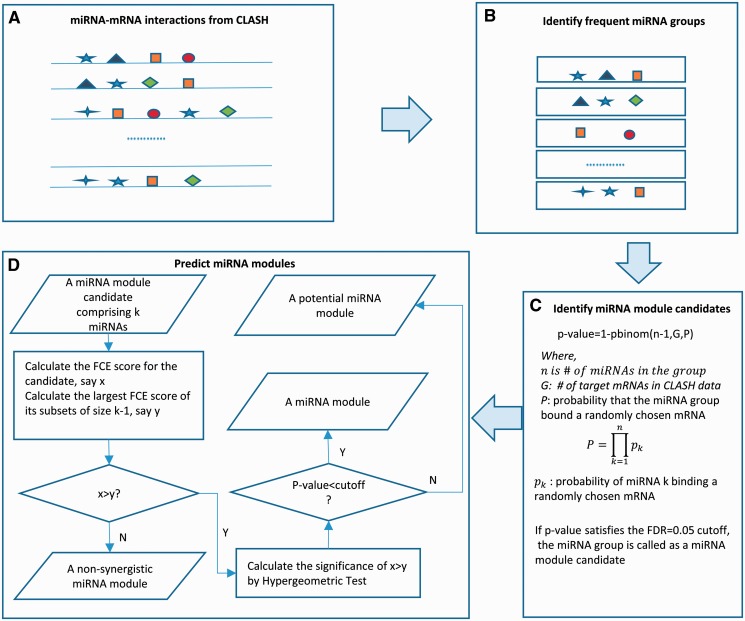
We developed the following pipeline to predict miRNA modules (Fig. 1): starting from 18 514 experimentally validated target sites, we modified the ChIPModule approach (Ding et al., 2013) to discover groups of miRNAs that co-bind at least S mRNAs; next, we applied the binomial test to assess the statistical significance of every group of miRNAs identified above. Each significant group of miRNAs was called a miRNA module candidate; finally, we predicted miRNA modules based on hypergeometric testing. A miRNA module was defined as a module candidate that significantly decreased the expression of their common mRNA targets than individual miRNAs contained in this candidate did. The details were in the following two sections.
Fig. 1.
The pipeline to predict miRNA modules. (A) MiRNA-mRNA interaction data from CLASH. Each line represents a target mRNA, each box represents a miRNA target site, with different shapes representing different miRNAs. (B). Identify miRNA groups with their target sites frequently co-occurring in common mRNAs. (C) Identify miRNA module candidates by binomial tests. (D). Predict miRNA modules based on hypergeometric tests
2.3 Predicting miRNA module candidates
We modified ChIPModule (Ding et al., 2013) to discover miRNA module candidates. ChIPModule was developed to discover significantly co-occurring binding sites of a group of transcription factors (TFs) in input sequences. In brief, with all known motifs in a database, ChIPModule defines putative TF binding sites (TFBSs) in input sequences. It then identifies TF groups of variable sizes with frequently co-occurring TFBSs in input sequences by an effective tree-based approach. Finally, ChIPModule assesses the statistical significance of each TF group with frequent co-occurring TFBSs by Poisson clumping heuristic (Hu et al., 2008) and output significant TF groups as TF modules. Because of its superior performance to other methods in TF module discovery (Ding et al., 2013), we applied a modified version of ChIPModule to miRNA module candidate discovery.
We modified ChIPModule in two aspects. One was that we considered experimentally determined miRNA target sites instead of putative TFBSs (Ding et al., 2013). The other was that we calculated statistical significance of the co-occurrence of a group of miRNAs differently. Assume we observed a group of k miRNAs, all of which bound n of the 7390 mRNAs. We assessed the statistical significance of this miRNA group as follows: first, for each miRNA, we calculated its probability to have a target site in a randomly chosen mRNA, which was the ratio of the number of mRNAs containing the CLASH target sites of this miRNA to 7390. Second, we calculated the probability that this group of miRNAs bound a randomly chosen mRNA by assuming each miRNA bound the mRNA independently. That is, this probability was measured as the product of the k probabilities that each of the k miRNAs bound the mRNA. Finally, we calculated the binomial tail probability of observing n of the 7390 mRNAs containing target sites of all k miRNAs in this group as the statistical significance of this group of miRNAs. With the tail probabilities for all groups of miRNAs that frequently co-bind their common mRNA targets, we applied the Q-value software (Storey and Tibshirani, 2003) to output significant miRNA groups so that the false discovery rate (FDR) was controlled. The significant miRNA groups were considered as miRNA module candidates (Fig. 1).
We applied this modified approach to output miRNA module candidates, with S = 10 and FDR = 0.05. S = 10 referred to the requirement that all miRNAs in a module candidate shared at least 10 common target mRNAs. We also tried S = 20 to predict candidates. All predicted candidates with S = 20 were a subset of the predicted candidates with S = 10, which suggested S = 20 may be too stringent. We thus reported our analyses with S = 10.
2.4 Identifying miRNA modules
Given a module candidate comprising k miRNAs, all of which bound the same n of the 7390 mRNAs, we determined whether it was a miRNA module by the following procedure (Fig. 1). First, we calculated a fold change extent (FCE) score for the candidate and all its subsets with k − 1 miRNAs. The FCE score of a group of miRNAs was defined as the fraction of their common target mRNAs with fold changes larger than a pre-defined cutoff D, when the expression levels of genes before and after AGO2 protein knockdown were compared. We used three cutoffs of D, corresponding to the 99, 95 and 90% quartile of the distribution of the fold changes of the 7390 mRNAs. Second, we checked whether the FCE score of this miRNA module candidate was larger than that of any subset of size k − 1. Third, if this candidate had larger FCE scores than any subset, we assessed its significance of higher downregulation of target genes than the k subsets by the following hypergeometric testing. Without loss of generality, we assumed that (i) the subset with the largest FCE score had N common target mRNAs, among which M mRNAs had fold changes larger than D; and (ii) the fold changes of m out of the n target mRNAs of this candidate were larger than D. Under these assumptions, the significance of observing higher downregulation of targets of the module than that of any subset was measured by a hypergeometric testing tail probability of observing at least m of n targets with large fold changes randomly chosen from the population of N targets with M targets of large fold changes. Finally, we assigned the module type of this candidate. If the significance at step three satisfied the required FDR of 0.05 (Storey and Tibshirani, 2003), this candidate was claimed as a synergistic miRNA module. Otherwise, if the FCE score of the candidate was larger than that of any subset at step two, this candidate was a potential synergistic miRNA module. In all remaining cases, this candidate was considered as a non-synergistic miRNA module. The synergistic, potential and non-synergistic miRNA modules were abbreviated as miRNA modules, potential modules or non-synergistic modules below, respectively.
With the above procedure, we predicted 193, 181 and 190 miRNA modules, using the 99, 95 and 90% quartile of the distribution of fold changes as D, respectively. More than 80% of the predicted miRNA modules with the three choices of D were shared by the three sets. For convenience, we reported our results based on the second choice of D (95%).
2.5 Validating predicted candidates especially the predicted miRNA modules
To assess whether the predicted candidates especially the miRNA modules were likely functional, we studied the overlap of their target genes with known pathway genes or gene sets of known functions. For known pathway genes, we used pathways at http://www.broadinstitute.org/gsea/index.jsp. To evaluate the overlap significance, we used hypergeometric testing (Boyle et al., 2004). We also checked the order of target sites of different miRNAs in a predicted candidate as in previous studies (Cai et al., 2010; Ding et al., 2014). We also searched literature to see whether a predicted module was supported as well. For each candidate, we searched in Google scholar to retrieve top 20 hits. For every hit, we manually checked whether all miRNAs contained in this candidate were reported to (i) bind the same targets; (ii) be active under the same experimental conditions (e.g. highly co-upregulated in specific cancers); or (iii) be found in co-transfection experiments. If at least one type of evidence was found, this candidate was considered to be supported by literature.
2.6 Comparing the strength of target sites
We compared the strength of target sites bound by predicted modules with that by individual miRNAs. The strength was measured by target site binding energy downloaded from the CLASH paper (Helwak et al., 2013). The binding energy approximated the interaction strength of a miRNA and one of its target sites. The higher the energy was, the weaker the target site was. In brief, we treated the binding energy of target sites bound by predicted miRNA modules as samples of a random variable X1. We also obtained the samples of another random variable Z, which was the binding energy of the sites in mRNAs that were not bound by any predicted candidate. We then applied Wilcoxon rank-sum test to test the null hypothesis Z > X1 with the alternative hypothesis as Z <= X1 (Wilcoxon, 1945). Similarly, we compared the strength of target sites of the potential modules with that of individual miRNAs. We also compared the strength of target sites of individual miRNAs in individual modules with the strength of target sites of the same miRNAs but not bound by any candidate.
2.7 Analyzing the preferred target site combinations of miRNA modules
We investigated the combinations of target sites a (potential) miRNA module preferred to bind. We had three possible combinations: all canonical sites (type 1), a mixture of canonical and unconventional sites (type 2) and all unconventional sites (type 3). For a (potential) miRNA module comprising k different miRNAs, the probability of the type 1 combination of target sites was pk, where p is the fraction of the number of canonical sites in the 18 514 target sites. Similarly, the probability of the type 3 combination was (1 − p)k. The probability of the type 2 combination was 1 − pk − (1 − p)k. We assume that we observed this (potential) module targeting n mRNAs, m1, m2 and m3 of which had the type 1, type 2 and type 3 target site combinations, respectively. We calculated the significance of this (potential) module preferring the type 1 combination as the binomial tail probability of observing at least m1 successful experiments in n experiments, each of which had a success rate of Pk. Similarly, we calculated the significance of this (potential) module preferring other types of combinations.
We analyzed five different location combinations of target sites: all sites in CDSs (type 1), all sites in 3′ UTRs (type 2), all sites in 5′ UTRs (type 3), at least one 3′ UTR site and another site not from 3′ UTR (type 4) and all other sites (type 5). We did similar tests to determine whether a (potential) miRNA module preferred a specific combination of site locations.
2.8 Inferring preferred distance ranges of adjacent target sites of miRNA modules
We defined preferred distance ranges of adjacent target sites of a miRNA module as follows: First, we divided the distances of adjacent target sites of a miRNA module into 10 nucleotides long bins. Second, we calculated the P-value of the enrichment of distances in each bin, by assuming the distances were evenly distributed across bins. If the P-value is small (FDR = 0.05), the bin was considered as significantly enriched. Third, we extended each significant bin to obtain a region with the smallest P-value of enrichment and defined this region as a preferred region. More precisely, for a significant bin, say A, we considered combining A with its left neighbouring bin. We calculated the P-value of the enrichment of distances in these two bins under the same uniform distribution assumption. Similarly, we considered the two bins comprising A and its right neighbouring bin. We then chose the extension with the smaller P-value, for instance, the extension to the left. If this smaller P-value is small (FDR = 0.05) and smaller than the P-value of A, we extended A into a preferred region comprising two bins. We repeated this procedure until no more extension could be made. Finally, we reported all non-overlapping preferred regions as the preferred distance ranges of adjacent target sites of this miRNA module. Similarly, we defined the preferred distance ranges of adjacent target sites of other types of candidates.
3 Results
3.1 Predicted miRNA modules were supported by functional evidence
With FDR = 0.05 (Storey and Tibshirani, 2003), we discovered 507 miRNA module candidates (Supplementary File S1). Each candidate consisted of 2 5 miRNAs, with an average of 2.72 miRNAs. The number of candidates with 2, 3, 4 and 5 miRNAs was 174, 300, 32 and 1, respectively. All miRNAs in a candidate shared at least 10 common mRNA targets.
We investigated whether the 507 candidates significantly downregulated their target mRNAs more than any subset of the contained miRNAs (Section 2). We found 181 candidates downregulated their target mRNAs significantly more than their contained subsets (FDR = 0.05). We considered these 181 candidates miRNA modules. We also noticed that 306 candidates downregulated their target mRNAs more than their contained subsets, with or without satisfying the required FDR of 0.05. We considered these 306 candidates potential miRNA modules, which included the above 181 modules. The remaining 201 candidates, which did not downregulate their target genes more than their subsets, were defined as non-synergistic modules.
We explored whether the 507 candidates, especially the 181 miRNA modules, were functional (Supplementary File S2). We studied the overlap of target mRNAs of a candidate with genes in a known pathway or annotated with a common gene ontology (GO) term, as in previous studies (Ambros, 2004; Xu et al., 2011). The rationale was that if a candidate was functional, its target genes likely significantly overlap with genes in a known pathway, or genes annotated with a specific GO term (Ambros, 2004; Xu et al., 2011). We found that the function of the majority of the predicted candidates, especially the predicted miRNA modules, was supported (Table 1). For instance, the target mRNAs of 68.4% of the 507 candidates significantly shared at least one GO term. To assess the statistical significance of the pathway and GO support, we generated 507 random miRNA groups, each of which consisted of miRNAs randomly chosen from the 399 miRNAs mentioned in the CLASH paper and contained the same number of miRNAs as the corresponding predicted candidate. We found that target mRNAs of only 18 random miRNA groups significantly overlap with genes in a pathway, and target mRNAs of only six random miRNA groups shared a GO term (Table 1).
Table 1.
Support of the predicted miRNA module candidates and the random miRNA groups
| Module types | No. (%) of modules supported by Pathway | No. (%) of modules supported by GO | No. (%) of modules supported by literature | No. (%) of modules supported by order | Total no. (%) of modules supported |
|---|---|---|---|---|---|
| 181 synergistic modules | 125 (69.0%) | 165 (91.2%) | 32 (17.7%) | 62 (34.3%) | 178 (98.3%) |
| 306 possible modules | 211 (69.0%) | 274 (89.5%) | 57 (16.8%) | 103 (33.7%) | 298 (97.4%) |
| 201 non-synergistic modules | 136 (67.7%) | 174 (86.6%) | 42 (20.9%) | 44 (21.9%) | 194 (96.5%) |
| 507 predicted candidates | 347 (68.4%) | 448 (88.4%) | 99 (19.5%) | 147 (29.0%) | 492 (97.0%) |
| 507 random miRNA groups | 18 (3.55%) | 6 (1.18%) | 11 (2.1%) | 0 (0.0%) | 27 (5.3%) |
In addition, we studied the order of target sites of miRNAs in the same predicted candidates (Supplementary File S2). We found that 34.3% of the 181 miRNA modules, 33.7% of the 306 potential modules and 29% of the 507 candidates contained miRNA pairs with statistically significant orders (Table 1, FDR = 0.05). For instance, three miRNAs MIR-222, LET-7B and MIR-615-3P formed a miRNA module. MIR-615-3P preferred to bind at the 5′ of the target sites of MIR-222 (FDR = 0.0286), which preferred to bind at the 5′ of the target sites of LET-7B (FDR = 6.1E-4). In contrast, no random miRNA group had preferred orders.
We also did a literature search to check whether the predicted candidates were supported (Supplementary File S2). We found that 99 of the 507 candidates and 32 of the 181 miRNA modules were supported by literature (Table 1). By comparison, we did literature search for the 507 random miRNA groups and found that 11 groups were supported. The supporting rate of 11/100 was likely over-estimated, as the 399 miRNAs were known to be active under the same conditions and may actually play certain roles together. Even given the over-estimated supporting rate of random miRNA combinations, the chance of observing the number of supported candidates was 0 (Table 1).
3.2 Target sites of miRNA modules were weaker compared with other target sites
We compared the strength of target sites bound by predicted (potential) miRNA modules with that by individual miRNAs. Target sites of the predicted miRNA modules had significantly higher energy than target sites bound by individual miRNAs (Wilcoxon rank-sum P-value 3.0 E −19). Similarly, this was true for target sites of the potential miRNA modules (P-value 4.6 E −15). Target sites of (potential) miRNA modules were thus weaker than target sites not bound by any of the 507candidates.
We also compared the strength of target sites of the predicted modules and the potential modules with that of non-synergistic modules. Target sites of the 181 miRNA modules had significantly higher energy than target sites bound by the 201 non-synergistic modules (P-value 1.6 E −56). Similarly, target sites of the 306 potential modules had significantly higher energy than those bound by the 201 non-synergistic modules (P-value 1.7 E −87).Therefore, target sites of the (potential) miRNA modules were weaker than those of the 201 non-synergistic modules, which implied that there was a difference between candidates that downregulated target mRNAs more than their subsets and candidates that did not.
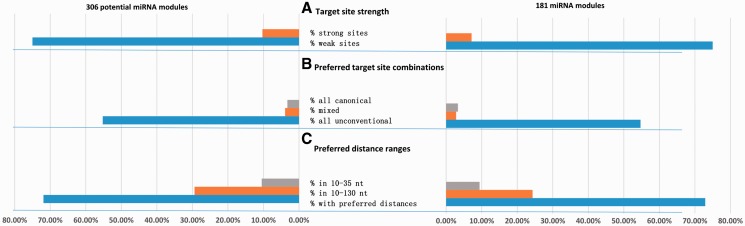
We further compared the strength of target sites of individual miRNAs bound by (potential) miRNA modules with that of target sites of the same miRNAs not bound by any of the 507 candidates. For 42 of the 56 miRNAs in the 181 miRNA modules, their target sites were significantly weaker compared with their target sites not bound by any of the 507 candidates (FDR = 0.05). On the contrary, for only 4 of the 56 miRNAs, their target sites were significantly stronger than those not bound by any candidate (FDR = 0.05), suggesting that for the majority of miRNAs, target sites bound by modules were not as strong as target sites not bound by any candidate. A similar observation was made for the 306 potential modules, for which target sites of 51 of the 68 miRNAs bound by modules were weaker and target sites of only 7 of the 68 miRNAs bound by modules were stronger (FDR = 0.05). Therefore, consistent with the above pooled analyses of target sites of all (potential) modules, target sites of the majority of miRNAs bound by individual (potential) modules were weaker (Figure 2).
Fig. 2.
The target site strength, site combinations, and distances of adjacent sites preferred by miRNA modules. (A) Percentages of miRNAs in modules preferring sites of different strengths. (B) Percentages of preferred target site combinations. (C) Percentages of preferred distance ranges
3.3 Most miRNA modules preferred to bind unconventional target sites
We investigated the preferred combinations of canonical and unconventional target sites of a (potential) miRNA module. For the 181 miRNA modules, 99, 5 and 6 miRNA modules preferred target sites composed of all unconventional sites (type 1), a mixture of canonical and unconventional sites (type 2) and all canonical sites (type 3), respectively (FDR = 0.05). Similarly, for the 306 potential miRNA modules, 169, 12 and 10 potential miRNA modules preferred target sites of types 1, 2 and 3, respectively (FDR = 0.05). It was thus evident that more than half of the (potential) miRNA modules preferred to bind unconventional sites instead of a mixture of canonical and unconventional sites.
The above analyses only showed that most (potential) miRNA modules preferred to bind mRNAs containing only unconventional sites. It was not clear whether target sites in the majority of mRNA targets of a (potential) miRNA module were composed of only unconventional sites. We thus checked the (potential) miRNA modules which had more than 70% of mRNA targets that contained only unconventional sites. We found that 116 of the 181 (64.1%) miRNA modules had more than 70% of their mRNA targets with only unconventional sites, and 198 of the 306 (64.7%) potential miRNA modules had more than 70% of mRNA targets with only unconventional sites. Therefore, the combination of all unconventional target sites was the most dominant combination found in target mRNAs of most (potential) miRNA modules.
Since unconventional sites included non-canonical sites and other sites, we explored whether miRNA modules prefer to bind non-canonical sites. For the 181 significant modules, only 52 (28.7%) modules significantly preferred all non-canonical sites. Moreover, only four modules had more than 70% of mRNA target with only non-canonical sites. Similarly, for 306 potential synergistic modules, only 82 (26.8%) modules significantly preferred all non-canonical sites. Only five modules had more than 70% of mRNA target with only non-canonical sites. It was thus evident that miRNA modules preferred unconventional sites instead of non-canonical unconventional sites (Figure 2).
3.4 Most miRNA modules preferred to bind the first or the last exons
We studied the location combinations of target sites of (potential) miRNA modules (Section 2). We found that most (potential) miRNA modules preferred to bind target sites in CDSs instead of a mixture of CDSs and UTRs. For the 181 miRNA modules, 127, 7, 2 and 2 miRNA modules preferred target sites all in CDSs (type 1), all in 3′ UTRs (type 2), all in 5′ UTRs (type 3), and in both 3′ UTRs and other locations (type 4), respectively (FDR = 0.05). Similarly, for the 306 potential miRNA modules, 210, 16, 3 and 2 potential miRNA modules preferred target sites of types 1, 2, 3 and 4, respectively. In contrast, 1, 0, 0 and 0 of the 507 random miRNA groups preferred target sites of types 1, 2, 3 and 4, respectively.
Because the majority of (potential) miRNA modules preferred to bind only in CDSs, we further investigated whether target sites in an mRNA target of a (potential) module were always in the same exons, adjacent exons or others. We found that the majority of the (potential) miRNA modules prefer to bind the same exons. For instance, for 150 of the 181 miRNA modules, more than 50% of their target sites in a target mRNA were in the same exon. Similarly, for 256 of the 306 potential modules, more than 50% of their target sites in a target mRNA were in the same exon. In contrast, the 507 random miRNA groups did not have exon binding preference, of which only 14 random miRNA groups had more than 50% of their target sites in the same exon in an mRNA.
Since most target sites in a target mRNA of a (potential) module were in the same exon, we also studied whether the target sites of a (potential) module preferred a specific type of exons. Indeed, we found that the first and the last exon of the target mRNAs were preferred to be bound by the (potential) miRNA modules. For instance, 64.9% of target sites of the 181 miRNA modules that were in the same exons were in either the first exons or the last exons. Similarly, 67.3% of target sites of the 306 potential miRNA modules that were in the same exons were in either the first exons or the last exons. Therefore, we concluded that most (potential) miRNA modules preferred to bind one exon in the target mRNAs, either the first or the last exons.
3.5 miRNA modules preferred target sites within certain ranges
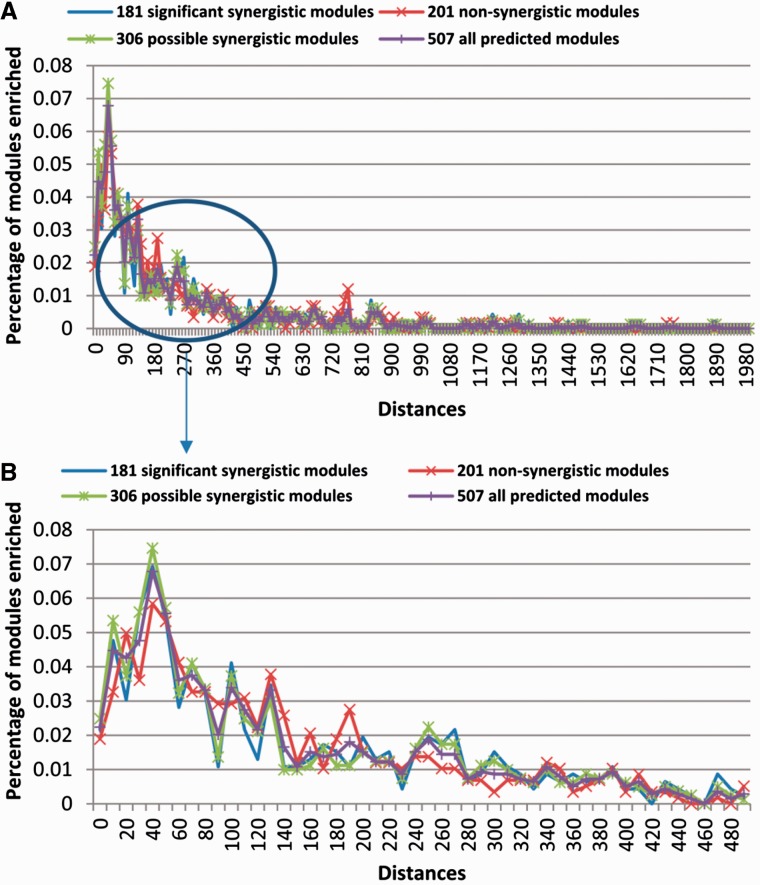
With the experimentally determined target sites, we studied the preferred distance range of the adjacent target sites of miRNA modules. We defined the preferred distance ranges of target sites of the 181 miRNA modules, 306 potential miRNA modules, 202 non-synergistic modules and the 507 module candidates, respectively (Table 2 and Supplementary File S3). For each of the four types of miRNA combinations, more than 70% of combinations had preferred distance ranges. The distribution of the preferred distance ranges of the 181 miRNA modules and that of the 306 potential modules were more similar to each other, compared with that of the 202 non-synergistic modules (Figure 3). For all four types of miRNA combinations with preferred distance ranges, more than 90% of combinations in each type had all preferred distance ranges <360 nucleotides (Figure 3 and Table 2). In contrast, 20 (3.9%) of the 507 random miRNA groups had preferred distance ranges, 10 (50.0%) of which had preferred distance ranges <360 nucleotides.
Table 2.
Preferred distance ranges of adjacent target sites of miRNA combinations
| Module types | % of modules with preferred distance ranges | % of modules with preferred distance ranges overlapping with 10-130nt | % of modules with preferred distance ranges overlapping with 13-35nt | % of adjacent distances <130nt (%) | % of modules with preferred distance ranges>130nt | % of modules with preferred distance ranges< 360nt |
|---|---|---|---|---|---|---|
| 181 significant synergistic modules | 132/181 = 72.9% | 44/181 = 24.3% | 17/181 = 9.4% | 25.9 | 85/181 = 47.0% | 119/132 = 90.2% |
| 306 possible synergistic modules | 220/306 = 71.9% | 90/306 = 29.4% | 32/306 = 10.5% | 28.1 | 142/306 = 46.4% | 198/220 = 90.0% |
| 201 non-synergistic modules | 148/201 = 73.6% | 69/201 = 34.3% | 16/201 = 8.0% | 27.4 | 105/201 = 52.2% | 136/148 = 91.9% |
| 507 all predicted modules | 368/507 = 72.6% | 159/507 = 31.4% | 48/507 = 9.5% | 27.8 | 247/507 = 48.7% | 334/368 = 90.1% |
| 507 random miRNA groups | 20/507 = 3.9% | 4/507 = 0.8% | 2/507 = 0.4% | 10.7 | 16/507 = 3.2% | 10/20 = 50.0% |
Fig. 3.
The distribution of preferred distance ranges of different types of miRNA combinations. A. The enriched distance ranges of four types of miRNA module candidates. B. The enriched distance ranges of four types of miRNA module candidates that are shorter than 500 nucleotides.
We tested whether two previously known preferred ranges, 10–130 nucleotides and 13–35 nucleotides, were enriched. For 44 (24.3%) miRNA modules, 90 (29.4%) potential miRNA modules and 69 (34.3%) non-synergistic modules, the 10–130 range was enriched (Binomial test, FDR = 0.05). For 17 (9.4%) miRNA modules, 32 (10.5%) potential miRNA modules and 16 (8.0%) non-synergistic modules, the 13–35 range was enriched (Binomial test, FDR = 0.05). If we pooled the distances of target sites of all 181 miRNA modules together, the 10–130 and 13–35 ranges were enriched (P-value = 0 in both cases). Similarly, the two ranges were enriched in distances of target sites of the potential modules, the non-synergistic modules and the 507 module candidates.
Although the two ranges of distances were enriched, the majority of distances of adjacent target sites of modules or module candidates were not within the two ranges (Table 2). For instance, in the 181 miRNA modules, more than 74.1% of distances of adjacent target sites of miRNA modules were longer than 130 nucleotides. Moreover, 85 (47.0%) miRNA modules had preferred distance ranges larger than 130 nucleotides (Table 2 and Figure 3). Future work may need to investigate how miRNAs with target sites in such large distance ranges interact.
3.6 Adjacent unconventional target sites of miRNA modules preferred shorter distances than other types of adjacent target sites
We studied the difference between the distances of adjacent unconventional target sites of a module and those of other types of adjacent target sites of the same module. For every predicted miRNA module, we collected the distances of adjacent unconventional target sites in each target mRNA. That is, we only considered target mRNAs that contained only unconventional sites of this module. We also collected distances of other target sites of this module. For 150 of the 181 miRNA modules, we had at least five distances collected for each of the two types of distances. We tested the null hypothesis that the distances of adjacent unconventional sites were shorter than those of other types. We only rejected the null hypothesis in five cases based on Wilcoxon rank-sum test (FDR = 0.05). We further tested the null hypothesis that the distances of adjacent unconventional sites were longer than those of other types. We rejected the null hypothesis in 31 cases (FDR = 0.05). We concluded that in six times more cases, the distances of adjacent unconventional target sites seemed shorter than that of other types of adjacent target sites.
The sample size may be too small for the above analyses using individual miRNA modules. Therefore, we considered distances of adjacent unconventional sites of all miRNA modules together instead of individual miRNA modules. For the null hypothesis that the distances of adjacent unconventional sites were shorter than those of other types, we accepted the null hypothesis (P-value > 0.99). For the null hypothesis that the distances of adjacent unconventional sites were longer than those of other types, we rejected the null hypothesis (P-value = 9.7 E −14). This pooled analysis showed that the distances of adjacent unconventional sites were indeed shorter.
We also did similar analyses for the 306 potential modules. We obtained similar results. That is, for the analysis based on individual potential modules, in three times more cases (41 vs. 12), the distances of adjacent unconventional sites were shorter than the distances of other types of sites. In the pooled analysis, for the null hypothesis that the distances of adjacent unconventional sites were shorter than those of other types, we accepted the null hypothesis (P-value > 0.99). For the null hypothesis that the distances of adjacent unconventional sites were longer than those of other types, we rejected the null hypothesis (P-value = 5.0 E − 16). Therefore, it was evident that adjacent unconventional target sites preferred shorter distances than other types of adjacent target sites.
4 Discussion
We studied miRNA modules based on experimentally determined miRNA target sites. We predicted 181 miRNA modules and 306 potential miRNA modules. We demonstrated that miRNA modules preferred to bind weak sites and favoured a combination of all unconventional sites. We also observed that miRNA modules preferred to bind in CDSs and favoured the first and the last exons. We confirmed that more than 70% of miRNA modules bound sites within specific ranges, with enrichment in two previously known ranges. However, many more adjacent sites bound by miRNA modules were >130 nucleotides apart. We further showed that unconventional target sites of miRNA modules were often within shorter distances than other combinations of target sites. Our study shed new light on miRNA binding.
The majority of adjacent target sites of miRNA modules were >130 nucleotides apart, which contradicted with previous observations (Brennecke et al., 2005; Doench and Sharp, 2004; Kloosterman et al., 2004; Saetrom et al., 2007; Vella et al., 2004). To understand what resulted in different observations, we focused on target sites of the 181 miRNA modules in 3′ UTRs. We found even when we considered only target sites in 3′ UTRs, more than 75% of adjacent target sites of miRNA modules were >130 nucleotides apart. We also predicted miRNA module candidates using only the 6096 CLASH target sites in 3′ UTRs and then studied the distances of adjacent target sites of these candidates. We still observed that the majority of adjacent target sites of these candidates were >130 nucleotides apart (Supplementary File S4). Therefore, the different observations were unlikely because we used target sites in entire mRNA regions while previous studies used only target sites in 3′ UTRs. Instead, it may be due to the small number of experimentally determined sites in previous experimental studies and the limited quality of predicted sites in the previous computational study, compared with the 18 514 high-quality experimentally determined sites we used.
We predicted (potential) miRNA modules on the condition that they downregulated target genes significantly more than some of their miRNA subsets. We further checked whether these (potential) modules downregulated their target genes significantly more than any subset contained in the modules. We confirmed that for all (potential) miRNA modules, their target genes were significantly more down-regulated than the target genes of any of their subsets.
We discovered 201 non-synergistic modules. The non-synergistic modules may also play important roles in regulating target genes, as supported by GO and pathway analyses, order preference, and the literature. Moreover, these non-synergistic modules may be competitive miRNA modules that are worth further investigation (Khan et al., 2009).
Supplementary Material
Acknowledgements
The authors thank the anonymous reviewers for their insightful comments.
Funding
This work is supported by the National Science Foundation [grants 1356524, 1149955, 1125676, and 1218275] and the National Institutes of Health [grant 2R01HL048044-22]. Funding for open access charge: The National Science Foundation grant 1356524.
Conflict of interest: none declared.
References
- Ambros V. (2004). The functions of animal microRNAs. Nature, 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.I., et al. (2004). GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics, 20, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., et al. (2005). Principles of microRNA-target recognition. PLoS Biol., 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan K., et al. (2013). Discovery and visualization of miRNA-mRNA functional modules within integrated data using bicluster analysis. Nucleic Acids Res., 42, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., et al. (2010). Systematic identification of conserved motif modules in the human genome. BMC Genomics, 11, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., et al. (2013). Chipmodule: systematic discovery of transcription factors and their cofactors from chip-seq data. Pac. Symp. Biocomput., 18, 320–331. [PubMed] [Google Scholar]
- Ding J., et al. (2014). SIOMICS: a novel approach for systematic identification of motifs in ChIP-seq data. Nucleic Acids Res., 42, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A. (2004). Specificity of microRNA target selection in translational repression. Genes Dev., 18, 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., et al. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell, 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., et al. (2008). MOPAT: a graph-based method to predict recurrent cis-regulatory modules from known motifs. Nucleic Acids Res., 36, 4488–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaswal V., et al. (2011). Identification of microRNA-mRNA modules using microarray data. BMC Genomics, 12, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.A., et al. (2009). Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol., 27, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., et al. (2004). Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res., 32, 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., et al. (2005). Combinatorial microRNA target predictions. Nat. Genet., 37, 495–500. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., et al. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., et al. (2003). Prediction of mammalian microRNA targets. Cell, 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Saetrom P., et al. (2007). Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res., 35, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D., et al. (2006). Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res., 34, 4801–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA, 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., et al. (2004). The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev., 18, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2014). Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics, 30, 1377–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. (2011). Transcriptional regulation of co-expressed microRNA target genes, Genomics, 98, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F. (1945). Individual comparisons by ranking methods. Biometrics Bull., 1, 80–83. [Google Scholar]
- Witkos T.M., Koscianska E., Krzyzosiak W.J. (2011). Practical aspects of microRNA target prediction. Curr. Mol. Med., 11, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., et al. (2010). Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene, 29, 2302–2308. [DOI] [PubMed] [Google Scholar]
- Xu J., et al. (2011). MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res., 39, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., et al. (2011). A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics, 27, i401–i409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.