Abstract
In glioblastoma (GBM), the EGF receptor (EGFR) and Src family kinases (SFKs) contribute to an aggressive phenotype. EGFR may be targeted therapeutically; however, resistance to EGFR-targeting drugs such as Erlotinib and Gefitinib develops quickly. In many GBMs, a truncated form of the EGFR (EGFRvIII) is expressed. Although EGFRvIII is constitutively active and promotes cancer progression, its activity is attenuated compared with EGF-ligated wild-type EGFR, suggesting that EGFRvIII may function together with other signaling receptors in cancer cells to induce an aggressive phenotype. In this study, we demonstrate that in EGFRvIII-expressing GBM cells, the urokinase receptor (uPAR) functions as a major activator of SFKs, controlling phosphorylation of downstream targets such as p130Cas and Tyr-845 in the EGFR in vitro and in vivo. When EGFRvIII expression in GBM cells was neutralized, either genetically or by treating the cells with Gefitinib, paradoxically, the cells demonstrated increased cell migration. The increase in cell migration was explained by a compensatory increase in expression of urokinase-type plasminogen activator, which activates uPAR-dependent cell-signaling. GBM cells that were selected for their ability to grow in vivo in the absence of EGFRvIII also demonstrated increased cell migration, due to activation of the uPAR signaling system. The increase in GBM cell migration, induced by genetic or pharmacologic targeting of the EGFR, was blocked by Dasatinib, highlighting the central role of SFKs in uPAR-promoted cell migration. These results suggest that compensatory activation of uPAR-dependent cell-signaling, in GBM cells treated with targeted therapeutics, may adversely affect the course of the disease by promoting cell migration, which may be associated with tumor progression.
Keywords: uPAR, EGF Receptor, EGFRvIII, Src Family Kinase, cell migration, glioblastoma
INTRODUCTION
Glioblastoma (GBM) is a typically lethal malignancy in which c-Src and other members of the Src family kinase (SFK) gene family are frequently highly activated, contributing to tumor aggressiveness.1–4 SFKs are non-receptor tyrosine kinases with broad regulatory activity in cell survival, proliferation, cell migration, and angiogenesis.5,6 SFKs function independently and in conjunction with receptors in the plasma membrane, including receptor tyrosine kinases (RTKs), integrins, G protein-coupled receptors (GPCRs), and the erythropoietin receptor, to activate downstream cell-signaling pathways that include the Ras-ERK1/2 pathway, the PI3K-Akt-mTOR1 pathway, and the Tiam1-Rac1 pathway.5–11 By phosphorylating Tyr-845 in the activated EGF receptor (EGFR), SFKs promote activation of the transcription factor, STAT5b, which supports cancer cell proliferation and survival.12–16 Phospho-Tyr-845 also binds cytochrome C oxidase subunit II, which may be involved in cancer cell survival when the EGFR translocates to mitochondria.17
A second gene product that plays a central role in GBM pathogenesis is the EGF receptor (EGFR), which is frequently amplified or over-expressed and in many cases, may be truncated to form a constitutively active mutant called EGFR variant III (EGFRvIII).18–21 Although EGFRvIII expression promotes an aggressive phenotype in GBM cells, the catalytic activity of EGFRvIII is attenuated compared with EGF-ligated wild-type (wt) EGFR.22 Thus, EGFRvIII may require novel interactions with other receptors and cell-signaling proteins to most robustly affect cancer cell physiology.
We previously demonstrated that in GBM cells, phosphorylation of Tyr-845 in EGFRvIII is dependent on the urokinase receptor (uPAR),15 a GPI-anchored membrane protein that interacts with integrins, FPR-like receptor-1 (FPRL1), and various RTKs to form a multiprotein complex with potent cell-signaling activity.23 The role of uPAR in phosphorylation of Tyr-845 in EGFRvIII may reflect a physical interaction between uPAR and EGFRvIII that increases availability of Tyr-845, as has been reported previously for uPAR and wt-EGFR.24 Alternatively, in EGFRvIII-expressing GBM cells, uPAR may control SFK activity in general. We and others have shown that SFKs play an essential role in the cell-signaling pathways by which uPAR promotes cell survival and stimulates cell migration;25–29 however, the importance of uPAR in controlling SFK activation in comparison with other receptors has not been explored. This problem is particularly important in GBM because SFKs are activated by numerous RTKs implicated in GBM progression, including wt-EGFR and EGFRvIII.1,30,31
In this study, first we examined the role of uPAR in activation of SFKs in GBM cells by measuring phosphorylation of Tyr-416, which reports the fully activated form of SFKs,32 and two SFK substrates, Tyr-845 in the EGFR and p130Cas. Our results demonstrate that in EGFRvIII-expressing GBM cells and in cells in which EGFRvIII expression was neutralized, uPAR functions as a general activator of SFKs, affecting substrates in addition to EGFR Tyr-845. Next, we examined the effects of EGFRvIII neutralization on GBM cell migration. We previously demonstrated that reversing EGFRvIII gene expression in GBM cells, in vitro or in vivo, induces expression of increased levels of urokinase type-plasminogen activator (uPA), stimulating uPAR-dependent cell-signaling and promoting cell survival.15 We now report that blocking EGFRvIII gene expression paradoxically promotes cell migration. This response was entirely attributable to activation of the uPA-uPAR signaling system. Similarly, EGFRvIII-expressing GBM cells that were treated with the EGFR tyrosine kinase inhibitor (TKI), Gefitinib, demonstrated increased uPA expression and accelerated cell migration. Gefitinib has been evaluated in clinical trials for GBM.33,34 We propose that signaling systems, which are activated as a compensatory response to support cancer cell survival, may also stimulate processes such as cell migration, involved in cancer progression. Importantly, the uPA-dependent increase in GBM cell migration that accompanied EGFRvIII blockade was reversed by the SFK-targeting cancer therapeutic, Dasatinib.4,35,36
Results
uPAR controls SFK activation in EGFRvIII-expressing GBM cells
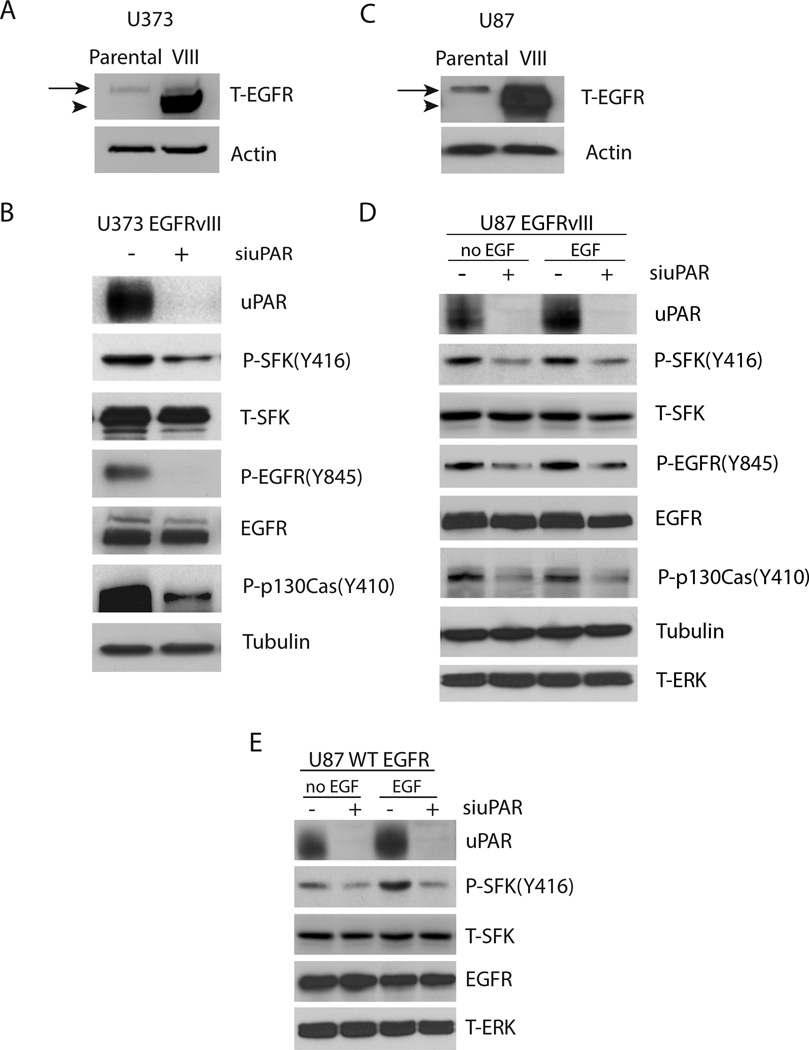
To compare the role of EGFRvIII and uPAR in SFK activation, first EGFRvIII was expressed in U373MG GBM cells. Fig. 1A shows that U373MG parental cells express low levels of wt-EGFR and that EGFRvIII is robustly expressed in transduced cells. The mobility of EGFRvIII is slightly increased compared with wt-EGFR due to receptor truncation.37 When SDS-PAGE was conducted for an extended time, to allow resolution of the bands for wt-EGFR and EGFRvIII, expression of wt-EGFR did not appear to be regulated by EGFRvIII (results not shown).
Figure 1.
Regulation of SFK activation by uPAR in EGFRvIII-expressing U373MG and U87MG cells. (a) Cell extracts from parental and EGFRvIII-expressing U373MG cells were subjected to immunoblot analysis to detect total EGFR (T-EGFR) and actin as a loading control. The mobility of wild-type EGFR is shown with an arrow and the mobility of EGFRvIII is shown with an arrowhead. (b) EGFRvIII-expressing U373MG cells were transfected with uPAR-specific (+) or NTC siRNA (−), allowed to recover, and then cultured in SFM for 24 h. Cell extracts were prepared and subjected to immunoblot analysis to detect uPAR, phospho-Tyr-416 in SFKs (P-SFK(Y416)), total SFKs (T-SFK), phosphorylated Tyr-845 in the EGFR (P-EGFR(Y845)), total EGFR, phosphorylated Tyr-410 in p130Cas (P-p130Cas(Y410)) and tubulin. (c) Cell extracts from parental and EGFRvIII-expressing U87MG cells were subjected to immunoblot analysis to detect total EGFR (T-EGFR) and actin as a loading control. (d) EGFRvIII-expressing U87MG cells were transfected with uPAR-specific (+) or NTC siRNA (−), allowed to recover, and then cultured in SFM for 24 h. The cells were then treated with EGF (2 ng/mL) for 10 min as specified (the two right-hand lanes). Immunoblot analysis was performed. (e) wt-EGFR-over-expressing U87MG cells and EGFRvIII-expressing U87MG cells were transfected with uPAR-specific (+) or NTC (−) siRNA, transferred to SFM, and treated with EGF (2 ng/mL) or vehicle for 10 min. Immunoblot analysis was performed.
We previously reported that uPAR gene-silencing decreases phosphorylation of Tyr-845 in EGFRvIII in U373MG cells.15 Tyr-845 is a well described SFK substrate.12,38 Fig. 1B confirms our original result, showing that Tyr-845 phosphorylation in EGFRvIII is substantially decreased by uPAR gene-silencing. To test whether the decrease in phospho-Tyr-845 reflects decreased SFK activity, or a distinct mechanism, such as altered availability of the substrate, first we examined phosphorylation of SFK Tyr-416, which was decreased by uPAR gene-silencing. Tyr-416 is known to be phosphorylated in maximally activated SFKs; however, active forms of SFKs also exist in which Tyr-416 is not phosphorylated.6,32,39 We therefore examined phosphorylation of a second SFK substrate, p130Cas, and demonstrated that uPAR gene-silencing decreases phospho-p130Cas as well. The total level of SFKs remained approximately unchanged in uPAR gene-silenced cells. Although in EGFRvIII-expressing U373MG cells, uPAR gene-silencing most substantially affected Tyr-845 phosphorylation, the effects on phosphorylation of Tyr-416 in SFKs and p130Cas suggest that uPAR functions as an general SFK activator in these cells, even though EGFRvIII expresses constitutive activity and also activates SFKs.1,30,31
To test the role of uPAR in SFK activation in a second model system, we expressed EGFRvIII in U87MG GBM cells. Fig. 1C shows that EGFRvIII was robustly expressed in these cells. Again in U87MG cells, EGFRvIII did not have an apparent effect on wt-EGFR expression, as determined by immunoblot analysis when the time of SDS-PAGE was extended. uPAR gene-silencing reduced phosphorylation of SFK Tyr-416 in EGFRvIII-expressing U87MG cells and phosphorylation of the SFK substrates: EGFR Tyr-845 and p130Cas, without having a major effect on the total level of SFKs (Fig. 1D). In the U87MG model system, the effects of uPAR gene-silencing on phosphorylation of SFK Tyr-416, EGFRvIII Tyr-845, and p130Cas were similar in magnitude. Thus, again in this cell line, uPAR functions as a general regulator of SFK activation. The previously reported activity of uPAR in controlling phosphorylation of Tyr-845 in EGFRvIII15 probably reflects, at least in part, the general activity of uPAR in SFK activation in EGFRvIII-expressing cells, although a direct interaction between uPAR and EGFRvIII cannot be ruled out by the studies presented here.
Because the U373MG and U87MG cell lines express low levels of wild-type EGFR, as a control, we tested whether EGF treatment affects SFK activation in EGFRvIII-expressing U87MG cells. As shown in Fig. 1D, phospho-Tyr-416, phospho-Tyr-845, and phospho-p130Cas were unchanged in cells that were treated with 2 ng/ml EGF for 10 min.
Comparison of SFK activation in wt-EGFR and EGFRvIII-expressing GBM cells
EGFRvIII is constitutively active despite its inability to bind EGF; however, the catalytic activity of EGFRvIII is substantially decreased compared with EGF-ligated wt-EGFR.22,37 To compare the role of uPAR in SFK activation in wt-EGFR- and EGFRvIII-expressing cells, U87MG cells were transduced to over-express wt-EGFR. Fig. 1E shows that EGF increased phosphorylation of Tyr-416 in SFKs in wt-EGFR over-expressing cells; however, uPAR gene-silencing decreased phospho-Tyr-416 both before and after EGF treatment. Thus, uPAR plays a role controlling SFK activation in GBM cells that express EGFRvIII or wt-EGFR.
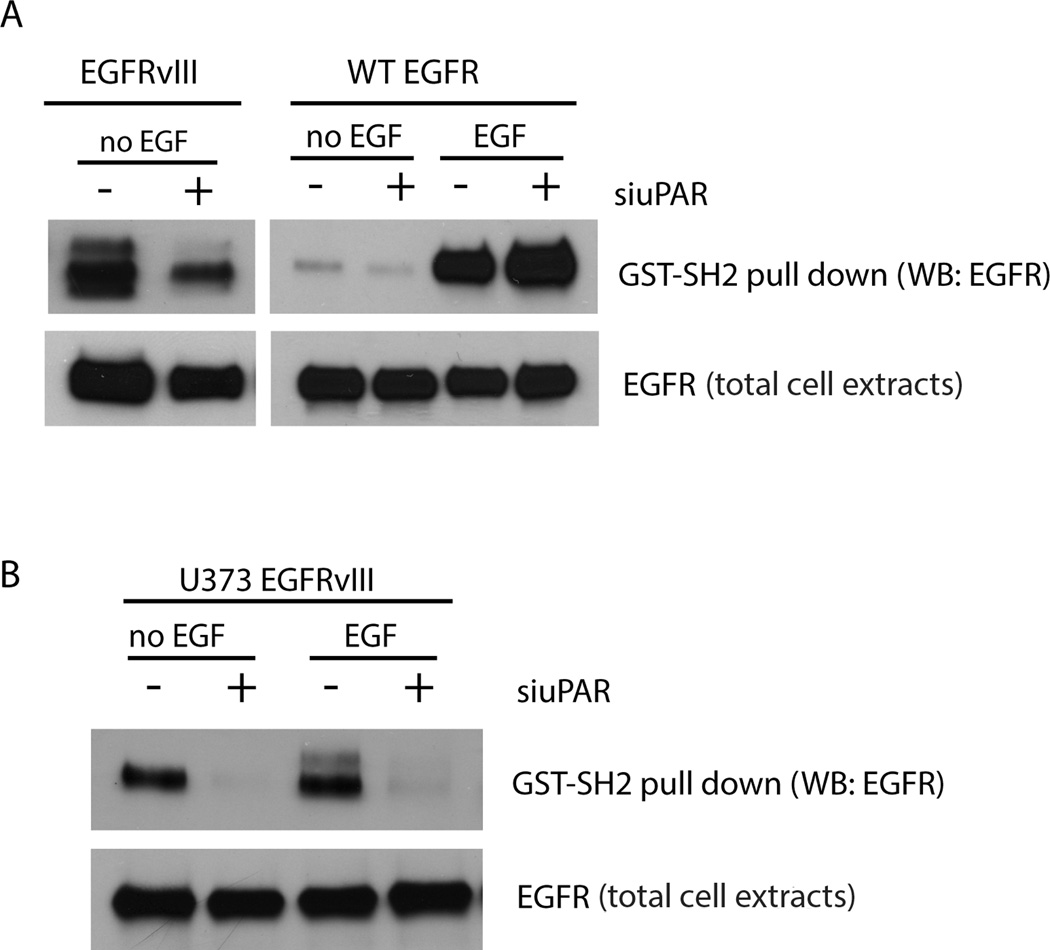
To further compare the activity of uPAR in controlling SFK activity in wt-EGFR- and EGFRvIII-expressing GBM cells, we performed affinity precipitation experiments with a GST fusion protein that contains the SH2 domain of c-Src (GST-SH2). This fusion protein binds phospho-Tyr residues, which in the EGFR, may be generated by auto-phosphorylation or through the activity of SFKs.5,6,12 In EGFRvIII-expressing U87MG cells, EGFRvIII readily affinity-precipitated with GST-SH2; however, when uPAR was silenced, EGFRvIII precipitation with GST-SH2 was substantially decreased (Fig. 2A). These results suggest a major role for uPAR, and by extension, SFKs, in generating the phospho-Tyr residues that serve as SH2-binding sites in EGFRvIII in U87MG cells.
Figure 2.
uPAR regulates availability of SH2-binding sites in EGFRvIII. (a) EGFRvIII-expressing and wt-EGFR-over-expressing U87MG cells were transfected with uPAR-specific (+) or NTC (−) siRNA, transferred to SFM, and treated with 10 ng/mL EGF or with vehicle for 10 min. Cell extracts were isolated and incubated with GST-SH2 coupled to glutathione-Sepharose for 3 h at 4°C. The Sepharose beads were wash ed and re-suspended in SDS-sample buffer for SDS-PAGE. EGFR that affinity-precipitated with GST-SH2 was determined by immunoblot analysis. Total cell extracts also were subjected to immunoblot analysis as a control for load. (b) EGFRvIII-expressing U373MG cells were transfected with uPAR-specific (+) or NTC (−) siRNA, transferred to SFM, and treated with 10 ng/mL EGF or with vehicle for 10 min. Binding of EGFRvIII to GST-SH2 was determined as described in panel a.
In U87MG cells that over-express wt-EGFR, only low levels of EGFR affinity-precipitated with GST-SH2 unless the cells were pre-treated with EGF (Fig. 2A). Following EGF pre-treatment, affinity precipitation of wt-EGFR with GST-SH2 was robust and uPAR gene-silencing did not affect this result, most likely reflecting the greatly increased capacity of EGF-ligated wt-EGFR to induce auto-phosphorylation.22 We confirmed the relationship between uPAR and EGFRvIII phosphorylation in U373MG GBM cells. uPAR gene-silencing almost entirely blocked affinity precipitation of EGFRvIII with GST-SH2 in U373MG cells (Fig. 2B). This result was not affected by EGF pre-treatment, as anticipated because EGFRvIII does not bind EGF.37
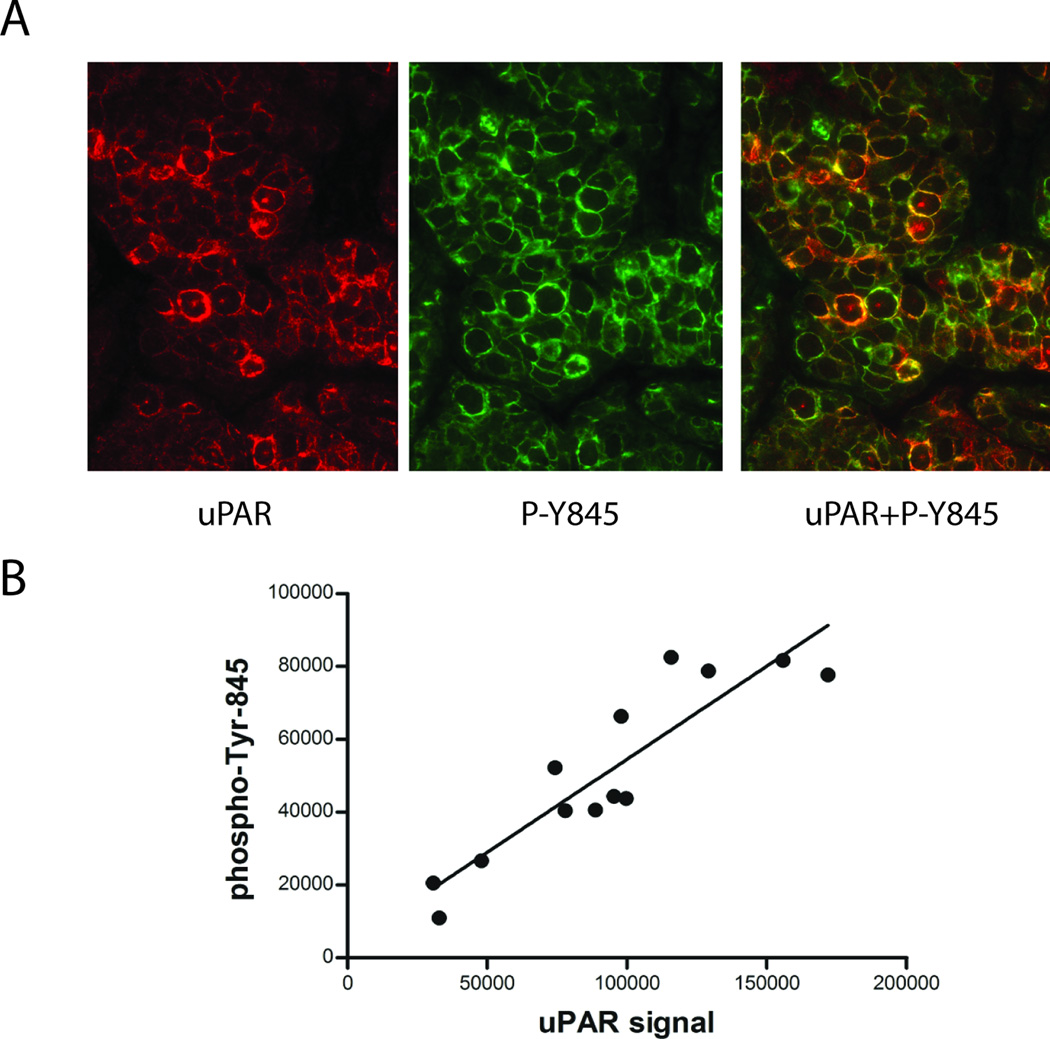
The results presented here demonstrate that the previously reported relationship between uPAR expression and phosphorylation of Tyr-845 in EGFRvIII in GBM cells may represent one component of a more general phenomenon in which uPAR plays a major role regulating SFK activation. To determine whether this relationship exists in vivo, we examined the EGFRvIII-expressing human GBM, GBM39, which has been propagated as a xenograft in mice.40 Tumor tissue was recovered from mice and analyzed by quantum dot immunofluorescence (IF) microscopy. This method allows precise quantitation of uPAR antigen expression and phospho-Tyr-845 at the single-cell level.41 Representative IF micrographs showed that uPAR expression and phospho-Tyr-845 varied from cell to cell (Fig. 3A). uPAR and phospho-Tyr-845 co-localized in the plasma membranes of many cells. Well-defined cells were identified and relative antigen levels were determined using Nuance Multispectral Imaging System software. The quantitative analysis of antigen intensity is shown in Fig. 3B. As predicted by our in vitro studies, there was a tight correlation between uPAR expression and phospho-Tyr-845 (R2= 0.87) in vivo, at the single cell level, in human GBM tissue.
Figure 3.
uPAR and phospho-Tyr-845 vary concordantly in individual cells in human GBM tissue. (a) An EGFRvIII-expressing human GBM was propagated as a xenograft. Harvested tissue was subjected to quantum dot immunofluorescence microscopy using antibodies that detect human uPAR (red, left) and phospho-Tyr-845 (green, middle). The merged image (right), highlights co-localization of uPAR and phospho-Tyr-845 (yellow). (b) To quantify antigen signal intensity, multispectral imaging was performed followed by spectral intensity analysis. The signal intensity for uPAR-staining was plotted against that for phospho-Tyr-845-staining in randomly selected individual cells, revealing a tight correlation (R2=0.87).
Control of SFK activation when EGFRvIII expression is blocked in GBM
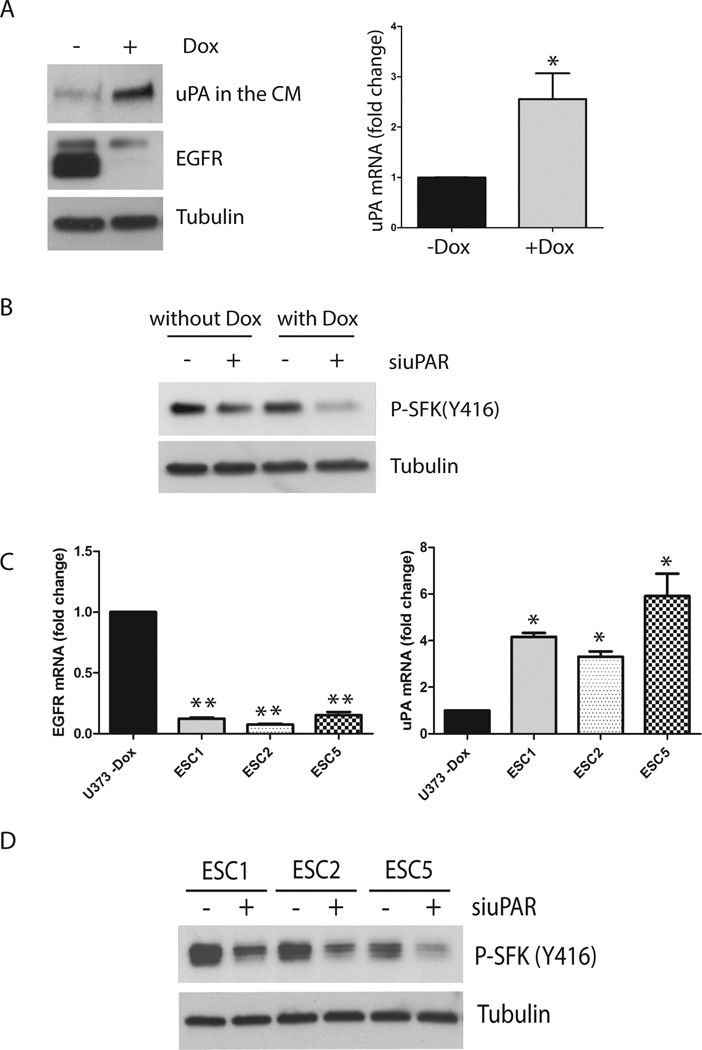
We previously described a model system in which EGFRvIII is expressed under the control of a doxycycline (Dox) repressible promoter in U373MG GBM cells.42 When EGFRvIII expression is neutralized by culturing the cells in the presence of Dox for at least 4 days, uPA expression is increased, activating compensatory uPAR-dependent pro-survival cell-signaling.15 Fig. 4A compares cells that were Dox-treated and control cells by immunoblot analysis. EGFRvIII expression was essentially absent in the Dox-treated cells, revealing a residual lower mobility band corresponding to wt-EGFR, which is expressed at low levels in these cells. When we analyzed serum-free medium (SFM), which was conditioned by the cells for 48 h, the amount of uPA recovered from Dox-treated cells was substantially increased. qPCR analysis confirmed that in Dox-treated cells, uPA expression was increased at the mRNA level (p<0.05).
Figure 4.
Effects of EGFRvIII neutralization on SFK activation in U373MG GBM cells. (a) U373MG cells that express EGFRvIII under the control of a Dox repressible promoter were treated with Dox (1 µg/ml) (+) or with vehicle (−) for 4 days. The cells were transferred to SFM and allowed to condition medium for 2 days. The medium was concentrated 10× and subjected to immunoblot analysis to detect uPA. Cell extracts were then prepared and immunoblotted to detect EGFR and tubulin as a control for load. Equivalent incubations were conducted to isolate RNA. uPA mRNA expression was compared in Dox-treated (+) and control (−Dox) U373MG cells (*, p<0.05, Student’s t-test). (b) U373MG cells were treated with Dox or vehicle for 6 days. uPAR-specific or NTC siRNA was introduced for 12 h, beginning at day 4. The cultures were then washed and re-equilibrated in SFM with or without Dox for the final 36 h. Cell extracts were prepared and immunoblotted to detect Tyr-416 in SFKs (P-SFK(Y416)) and tubulin as a control for load. (c) EGFRvIII-expressing U373MG cells and Escaper tumor cells (ESC1, ESC2 and ESC5) were cultured in SFM for 24 h. Total EGFR mRNA (wt-EGFR + EGFRvIII) and uPA mRNA levels were determined by qPCR and standardized against the levels present in parental cells that express EGFRvIII (mean ± SEM; n=3, *, p<0.05, **, p<0.01). (d) ESC cell lines were transfected with NTC (−) or uPAR-specific siRNA (siuPAR) (+), allowed to recover, and then cultured in SFM for 24 h. Immunoblot analysis was performed to detect SFK phospho-Tyr-416.
To study the role of uPAR in SFK activation when EGFRvIII expression is neutralized, we silenced uPAR in control and Dox-treated cells. Supplementary Fig. 1A shows that uPAR gene silencing was 95% effective. The cells were allowed to recover for 12 h and then cultured in SFM for 36 h (with continued Dox or vehicle treatment). Fig. 4B shows that SFK phospho-Tyr-416 was decreased by uPAR gene-silencing in EGFRvIII-deficient Dox-treated cells, indicating that uPAR plays a significant role in maintaining SFK activation when EGFRvIII is neutralized in GBM cells.
Escaper (ESC) cell lines were derived from xenografts formed by U373MG GBM cells that express Dox-repressible EGFRvIII.42 These cells require EGFRvIII expression at the time of inoculation in order to form tumors in immunocompromised mice. When EGFRvIII expression was blocked in established tumors by treating the mice with Dox, the tumors entered a temporary state of dormancy and then, released from dormancy, re-establishing aggressive growth. ESC cell lines were established from tumor cells that released from dormancy.42 We previously demonstrated that ESC cell lines secrete substantially increased levels of uPA compared with parental cells. The uPA activates uPAR-dependent cell-signaling and may explain the ability of ESC cells to release from dormancy.15 Fig. 4C shows that EGFR mRNA expression was substantially decreased in three separate ESC cell lines, compared with the EGFRvIII-expressing parental cells from which the ESC cells were derived (p<0.01), as anticipated. The ESC cells also expressed increased levels of uPA mRNA, compared with parental cells (p<0.05).
To explore the role of uPAR in SFK activation in ESC cells, we silenced uPAR in the ESC1, ESC2, and ESC5 cells. Supplementary Fig. 1B shows that uPAR gene silencing was >95 effective in all three cell lines. Next, we examined activation of SFKs in the control and uPAR gene-silenced cells by performing immunoblot analysis to detect phospho-Tyr-416. In each of the ESC cell lines, uPAR gene-silencing decreased phospho-Tyr-416 (Fig. 4D), indicating a role for uPAR in controlling SFK activation in tumor cells that acquire resistance to EGFR deficiency in vivo.
EGFRvIII neutralization in GBM cells promotes cell migration
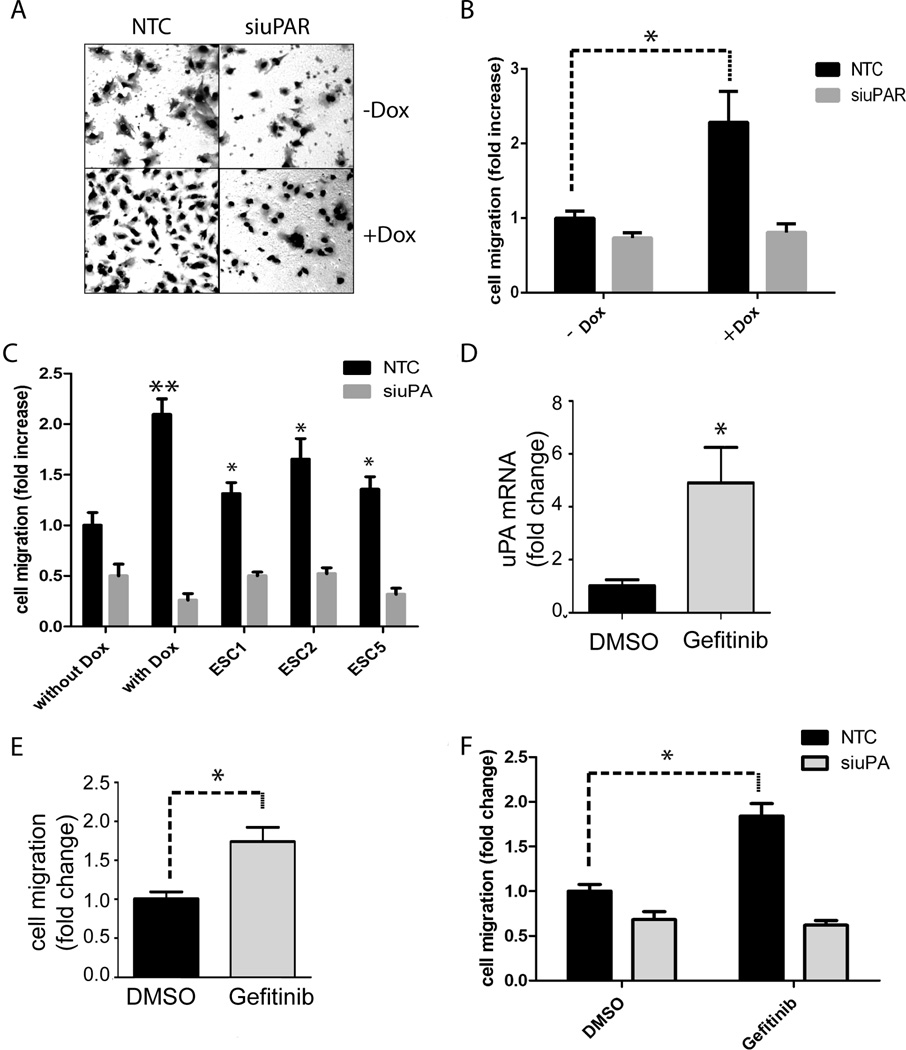
Dox-treatment to block EGFRvIII expression in U373MG cells provides a model of changes that may occur in EGFRvIII-expressing GBM cells when tumors are treated with TKIs such as Erlotinib and Gefitinib.42 Because uPAR activates potent pro-migratory cell-signaling factors, such as Rac1 and ERK1/2,27,28,43–46 we used the Dox-repressible EGFRvIII expression model to study the effects of EGFRvIII neutralization on cell migration. Cells were treated with Dox or vehicle and transfected with uPAR-specific or NTC siRNA. Representative images of cells that migrated to the underside of Transwell membranes are shown in Fig. 5A. Neutralization of EGFRvIII expression was associated with a 2.3 ± 0.4-fold increase in cell migration (p<0.05) (Fig. 5B). uPAR gene-silencing blocked the increase in cell migration observed in Dox-treated cells. When uPAR was silenced, Dox-treated and control cells migrated equivalently.
Figure 5.
Effects of EGFRvIII neutralization on migration of U373MG GBM cells. (a) U373MG cells that express EGFRvIII under the control of a Dox repressible promoter were treated with Dox or with vehicle for 4 days. uPAR-specific siRNA (siuPAR) or NTC siRNA were introduced for 12 h. The cells were re-equilibrated in serum-containing medium for 24 h (with or without Dox) and then added to Transwell chambers. Cell migration was allowed to occur in Transwell chambers for 18 h. Serum was present exclusively in the lower chambers. Representative photomicrographs show cells that migrated to the underside surface of the membranes. (b) Cells that migrated to the underside surfaces of the membranes were counted (mean ± SEM, n=3, *, p<0.05). (c) U373MG GBM cells were cultured in the presence of Dox or vehicle (without Dox) for 4 days. These cells and ESC cell lines were transfected with NTC siRNA (black bars) or uPA-specific siRNA (grey bars). After re-equilibration in serum-containing medium for 24 h, cells were added to Transwell chambers. Cells were allowed to migrate for 18 h. The number of migrating cells was standardized against that observed with U373MG cells that were not treated with Dox and transfected with NTC siRNA (mean ± SEM, n=3, *, p<0.05; **, p<0.01). (d) EGFRvIII-expressing U373MG cells were treated with Gefitinib (1 µM) or vehicle (DMSO) for 4 days. The cells were transferred to SFM for 24 h. uPA mRNA expression was determined by qPCR and standardized against the level present in cells treated with vehicle (mean ± SEM; n=3, *, p<0.05). (e) EGFRvIII-expressing U373MG cells were treated with Gefitinib (1 µM) or vehicle (DMSO) for 4 days. Cell migration was allowed to occur in Transwell chambers for 18 h. The number of migrating cells was standardized against that observed with vehicle-treated U373MG cells (mean ± SEM, n=3, *, p<0.05). (f) EGFRvIII-expressing U373MG cells were treated with Gefitinib (1 µM) or with vehicle for 4 days and then transfected with uPA-specific siRNA (grey bar) or NTC siRNA (black bar). The cells were allowed to recover in serum-containing medium for 24 h (with continued treatment with Gefitinib or vehicle) before addition to Transwell chambers. Cell migration occurred for 18 h. The number of migrating cells was standardized against that observed with U373MG cells that were treated with vehicle and transfected with NTC siRNA (mean ± SEM, n=3, *, p<0.05).
Next, we examined ESC cells, in which EGFRvIII deficiency was induced in vivo.42 All three ESC cell lines (ESC1, ESC2, ESC5) demonstrated significantly increased cell migration compared with EGFRvIII-expressing U373MG cells, from which the ESC cells were derived (Fig. 5C). Although uPAR-initiated cell-signaling occurs in the presence and absence of uPA, the full potential of uPAR in cell-signaling requires uPA.23,43,46,47 Thus, we tested whether uPA gene-silencing affects migration of EGFRvIII-expressing and –deficient U373MG cells and ESC cells. As shown in Fig. 5C, uPA gene-silencing completely blocked the increase in cell migration observed when EGFRvIII expression was neutralized by treating U373MG cells with Dox in vitro. uPA gene-silencing also substantially inhibited migration of the ESC cells, effectively neutralizing any advantage in cell migration compared with the parental cells. Supplementary Fig. 2 confirms that uPA gene-silencing was >95% effective in the three ESC cell lines.
To complement our model system in which EGFRvIII gene expression was neutralized genetically, we treated EGFRvIII-expressing U373MG cells with 1.0 µM Gefitinib for 4 days. Control cells were treated with vehicle. At the conclusion of the incubation, uPA mRNA expression was increased 4.9 ± 1.1-fold (p<0.05) in the Gefitinib-treated cells (Fig. 5D). This is the first report demonstrating increased uPA expression induced by Gefitinib in GBM cells. The increase in uPA expression was accompanied by a 1.7-fold increase in cell migration (p<0.05) (Fig. 5E). To assess the role of uPA in migration of Gefitinib-treated U373MG cells, we transfected EGFRvIII-expressing cells with uPA-specific or NTC siRNA. uPA gene-silencing had a modest effect on migration of the control cells; however, in the Gefitinib-treated cells, a more robust effect was observed. uPA gene-silencing inhibited migration of Gefitinib-treated cells by 66 ± 3% (p<0.01). The control and Gefitinib-treated cells migrated equivalently when uPA was not available.
Targeting SFKs blocks the increase in cell migration associated with activation of the uPA-uPAR system in GBM cells
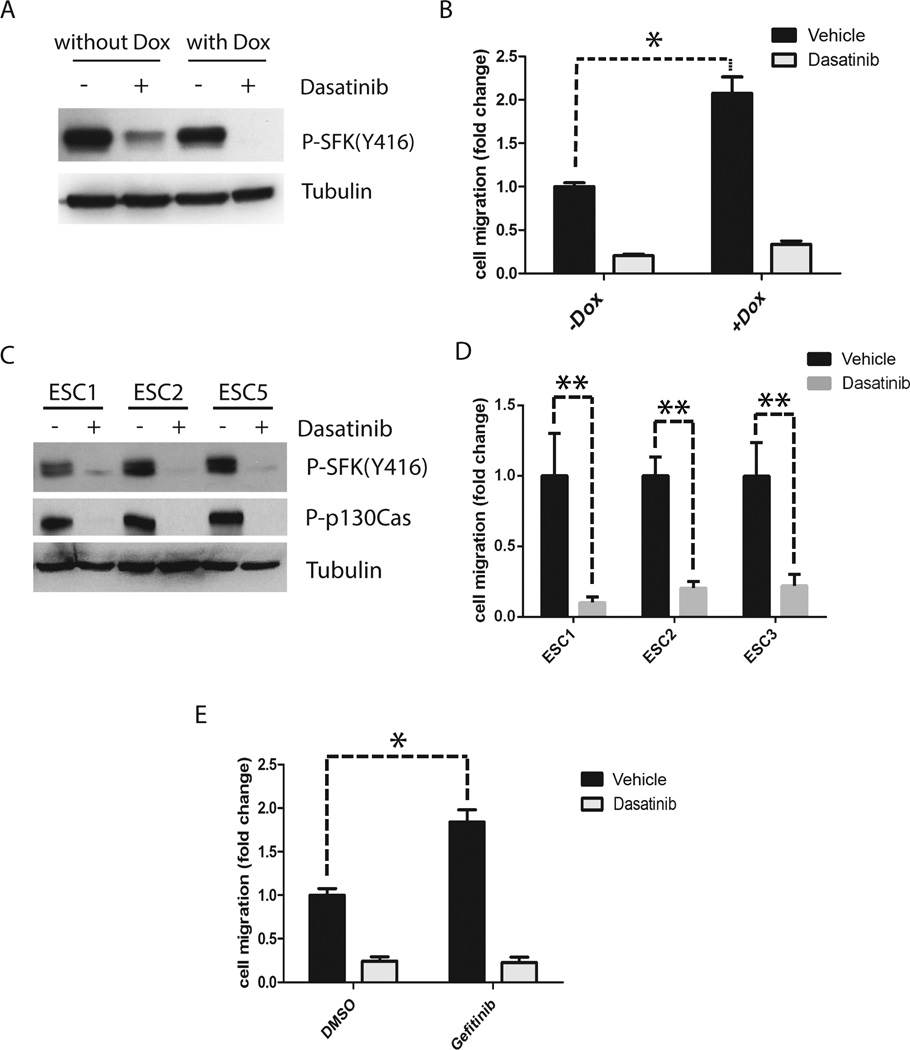
Given the reported role of SFKs as upstream mediators of uPAR-initiated cell-signaling to ERK1/2 and Rac1,25–28 we tested whether the anticancer drug, Dasatinib, which targets SFKs, inhibits migration of cells in which EGFRvIII is neutralized. Fig. 6A shows that treatment with 0.3 µM Dasatinib for 4 h almost entirely blocked activation of SFKs, as determined by measuring phosphorylation of SFK Tyr-416 in U373MG cells that express EGFRvIII (not treated with Dox) and in U373MG cells that were EGFRvIII-deficient (Dox treated). Dasatinib was extremely effective at inhibiting U373MG cell migration, especially when EGFRvIII expression was neutralized and cell migration stimulated by uPA (Fig. 6B). Under these conditions, inhibition of cell migration was greater than 80% complete.
Figure 6.
Dasatinib decreases SFK activation and blocks cell migration promoted by activation of the uPA-uPAR signaling system. (a) U373MG cells that express EGFRvIII under the control of a Dox repressible promoter were treated with Dox or with vehicle for 4 days and then with 0.3 µM Dasatinib (+) or vehicle (DMSO) (−) in SFM for 4 h. Immunoblot analysis was performed to detect phosphorylated Tyr-416 in SFKs and tubulin as a control for load. (b) U373MG cells were treated with Dox or vehicle for 4 days. The cells were then added to Transwells in the presence of 0.3 µM Dasatinib (grey bar) or vehicle (DMSO, black bar). Cells were allowed to migrate for 18 h. Serum (10% FBS) was added only to the lower chamber. The number of migrating cells was determined and standardized against that observed with vehicle-treated U373MG cells (mean ± SEM, n=3, *, p<0.05). (c) ESC cell lines (ESC1, ESC2 and ESC5) were treated with 0.3 µM Dasatinib (+) or vehicle (DMSO) (−) in SFM for 4 h. Immunoblot analysis was performed to detect phosphorylated Tyr-416 in SFKs, phosphop130Cas, and tubulin as a control for load. (d) Escaper cells (three separate cell lines) were treated with Dasatinib (gray bars) or DMSO (black bars) by adding drug to the top and bottom chambers of Transwells. Cell migration was then studied (mean ± SEM, n=3. **, p<0.01). (e) EGFRvIII-expressing U373MG cells were treated with Gefitinib (1 µM) or vehicle (DMSO) for 4 days. These cells were then treated with 0.3 µM Dasatinib (grey bar) or vehicle (black bar) by adding drug to the top and bottom chambers of Transwells. Cells were allowed to migrate for 18 h. The number of migrating cells was standardized to that observed with vehicle-treated U373MG cells (mean ± SEM, n=3, *, p<0.05).
Next we examined the effects of Dasatinib on migration of ESC cells. Fig. 6C shows that treatment with 0.3 µM Dasatinib for 4 h blocked SFK activation in each of the three ESC cell lines, as determined by measuring SFK phospho-Tyr-416 and phospho-p130Cas. Fig. 6D shows that Dasatinib inhibited migration of the ESC cells by 80–90% (p<0.01). These results suggest that Dasatinib, which is used in human patients, may effectively antagonize the uPAR signaling system and its effects on cell migration.
An equivalent response was observed in Dasatinib treatment experiments when we studied cells that were pre-treated for 4 days with Gefitinib. Cells that were treated with Gefitinib migrated more rapidly (p<0.05), confirming the results presented in Fig. 5E (Fig. 6E). Including Dasatinib in the Transwell chambers inhibited migration of the control cells by about 75% and migration of the Gefitinib-treated cells by close to 90%. Thus, in all of our model systems, targeting SFKs blocked the increase in cell migration associated with EGFRvIII neutralization and activation of the uPAR signaling system.
DISCUSSION
EGFRvIII induces an aggressive phenotype in GBM cells, despite its attenuated activity compared with EGF-ligated wt-EGFR.18,22,37,48–50 Cells that express EGFRvIII may make neighboring cells in the tumor microenvironment more aggressive,48,51 so even a small population of EGFRvIII-positive cells in a GBM may significantly promote tumor progression. Given these properties, it was rational to test the efficacy of EGFR-selective TKIs, such as Gefitinib and Erlotinib, in EGFRvIII-positive GBM. Unfortunately, treated tumors rapidly escape from control, re-establishing aggressive growth and invasion within a short time.52 Failure of EGFR TKIs to induce sustained remission may reflect emergence of novel genomic alterations, activation of signaling systems that replace EGFRvIII, and/or selection of subpopulations of cancer cells that are drug-resistant. One known cause of resistance to EGFR TKIs is mutation in PTEN, which sustains activation of signaling pathways that are downstream of the EGFR.40,48,52 We identified the uPA-uPAR system as one that may allow GBM cells to escape from control by EGFR TKIs.15
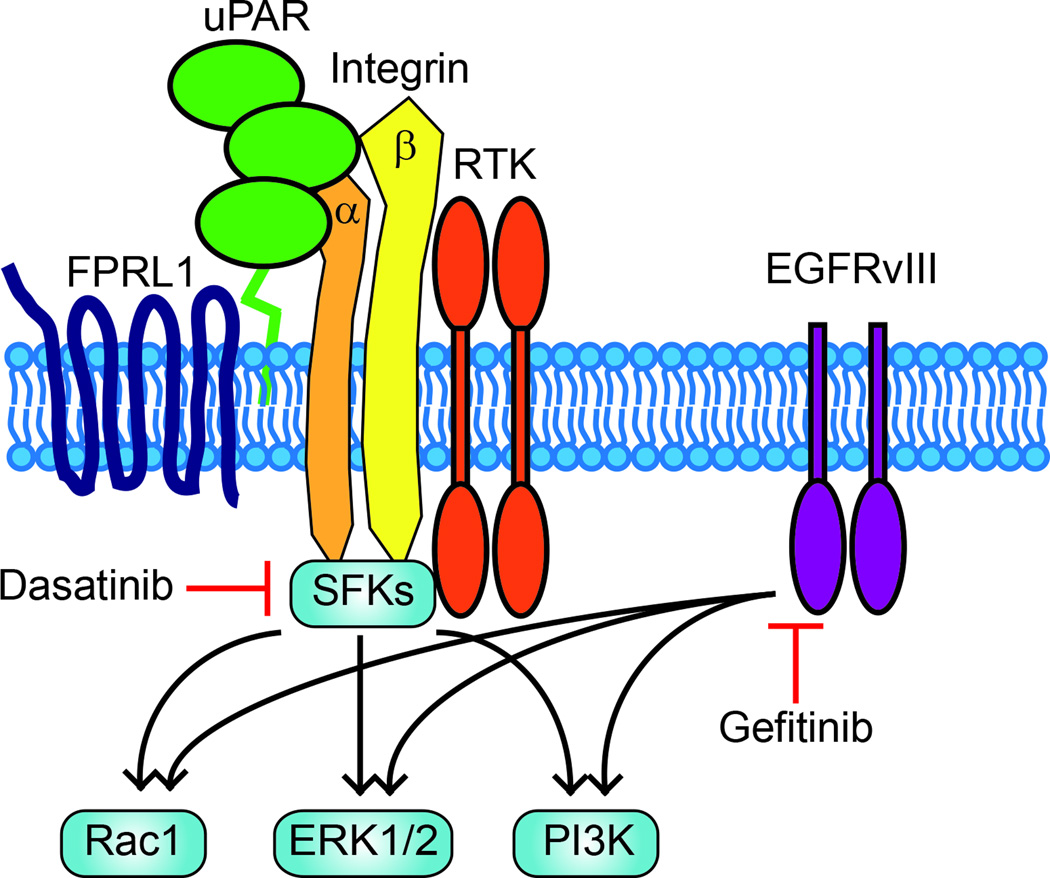
The hypothesis that activation of uPAR-initiated cell-signaling may compensate for loss of EGFR activity in GBM is supported by previous studies that have demonstrated substantial overlap in key downstream targets for uPAR and the EGFR, including ERK1/2, the PI3K-Akt pathway, and Rac1 (Fig. 7).27–29,43–45,47,53–55 These common downstream targets support cell survival; however, the same signaling factors also play key roles in cell migration and in cancer cells, invasion and metastasis.27–29,46,47,54 Fig. 7 shows that uPAR is a member of a multi-protein signaling receptor complex, which may include different RTKs including the EGFR, the G-protein coupled receptor (GPCR) FPRL1, and integrins such as α5β1 and α4β1.23,24,56–59 Because SFKs associate with integrin cytoplasmic tails, the central role of SFKs in uPAR-initiated signaling may reflect a complex in which uPAR is bridged to SFKs through integrins; however, SFKs also are important effectors of RTK signaling and GPCR signaling.5–11,55
Figure 7.
A model showing overlap in downstream targets for EGFRvIII and uPAR-initiated cell-signaling. uPAR signals as a component of a multiprotein receptor complex, which probably includes integrins, the GPCR, FPRL1, and RTKs. Gefitinib targets EGFRvIII signaling and may activate uPAR as a compensatory, pro-survival response. SFKs, which are targeted by Dasatinib, serve as key upstream signaling factors in the uPAR cell-signaling system. PI3K and ERK1/2 are well described pro-survival factors. PI3K, ERK1/2, and Rac1 have been implicated in cancer cell migration.
In this study, we demonstrated that uPAR is a major activator of SFKs in EGFRvIII-expressing GBM cells. The effects of uPAR on SFK activation provide an explanation for the previously reported role of uPAR in Tyr-845 phosphorylation in EGFRvIII.15 uPAR also promoted phosphorylation of the SFK substrate, p130Cas. In a human GBM propagated as a xenograft, the extent of phosphorylation of Tyr-845 in EGFRvIII correlated with uPAR immunopositivity at the single cell level. SFK activation is a known determinant of GBM cell aggressiveness.1–4 Cooperation between EGFRvIII and uPAR may explain the cancer-promoting activity of EGFRvIII in GBM cells, despite its attenuated enzymatic activity.22,37
In diverse forms of neoplasia, uPAR-initiated cell-signaling is known to prevent apoptosis and anoikis and release cancer cells from states of dormancy.56,61–63 Thus, it is not surprising that activation of uPAR-initiated cell-signaling compensates for neutralization of EGFRvIII in GBM cells.15 Other compensatory cell-signaling systems have been described, which also may promote GBM survival and growth following EGFR TKI treatment.52 In our model systems, when EGFRvIII was neutralized, uPAR remained an important activator of SFKs; however, the decrease in SFK activation induced by uPAR gene-silencing was incomplete, suggesting that, in these cells, SFKs may be activated by proximal receptors other than EGFRvIII and in addition to uPAR.
uPAR promotes cell migration by two major mechanisms.23,57 By binding and promoting activation of uPA as a protease at the cancer cell surface, uPAR assembles a cell-surface protease system, which may aid in the degradation of tissue boundaries for the migrating cancer cell.22,57–59,64 uPAR also promotes cell migration by its effects on pro-migratory cell-signaling factors, such as Rac1 and ERK1/2.27–29,43–45,47,53–55 Because uPA was expressed at increased levels in GBM cells when EGFRvIII was genetically neutralized or when the cells were treated with Gefitinib, we explored the effects of these treatments on GBM cell migration. The Transwell cell migration method applied in this study does not incorporate tissue boundaries and thus, is more sensitive to changes in cell-signaling, as opposed to effects on cell-surface protease activity. In GBM cells in which EGFRvIII expression was blocked in vitro, in ESC cells in which EGFRvIII expression was blocked in vivo, and in cells treated with Gefitinib, increased cell migration was observed and the increase was entirely attributed to the uPA-uPAR signaling system. Silencing of uPA or uPAR blocked the increase in cell migration associated with EGFRvIII inactivation. These results provide an example of an important principle in cancer biology, relevant to uPAR. The cell-signaling systems activated downstream of uPAR to promote cell survival are overlapping with those that promote cancer cell migration.23,61 The increase in uPA expression and activation of uPAR signaling probably represents a cellular response to the stress imposed by loss of constitutive EGFRvIII signaling. uPAR-signaling provides resistance to the negative consequences of EGFRvIII neutralization and allows the cells to survive. Increased cell migration is a potentially important side-effect of activation of this pro-survival signaling system.
Parker et al.65 recently reported that Gefitinib selectively inhibits migration of GBM cells in which the EGFR gene is amplified. Although these investigators did not specifically study EGFRvIII, the more important difference between the study reported by Parker et al.65 and this investigation probably involves the methods applied. The tissue slice approach used by Parker et al.,65 although elegant, did not provide a sufficiently long exposure of the cells to Gefitinib to allow activation of compensatory signaling pathways, like the uPA-uPAR system. Understanding the mechanisms by which uPA expression is up-regulated in cells in which EGFRvIII is neutralized is an important topic for future investigation.
GBM is typically lethal due to local invasion as opposed to metastasis.48 Although the capacity for cancer cell migration correlates with the capacity for invasion in vivo, further work will be necessary to understand how the uPAR cell-signaling system affects GBM invasion in vivo, in new tumors and in treated malignancies. A number of strategies for specifically targeting uPAR in cancer are currently under development.66 If uPAR-initiated cell-signaling allows tumor cells to escape from control by EGFR TKIs in patients with GBM and promotes more rapid tumor spreading through the CNS, our results suggest that the responsible signaling pathways may be antagonized by Dasatinib. In our model systems, Dasatinib blocked migration of GBM cells by as much as 90%, after EGFRvIII expression was inhibited or its activity antagonized. The possibility that targeting SFKs may represent an alternative to direct targeting of uPAR is supported by our model in which SFKs function as proximal signaling factors, linking receptors in the uPAR cell-signaling system to key downstream signaling factors (Fig.7).
MATERIALS AND METHODS
Cell lines
U373MG cells, which express EGFRvIII under the control of a Dox-repressible promoter and U373MG cells that express wt-EGFR, are previously described.42 Escaper tumor cell lines (ESC1, ESC2, ESC5), which were derived from U373MG cell xenografts that re-established growth following loss of EGFRvIII in vivo, also are previously described.42 These cells were maintained in DMEM supplemented with 10% tetracycline-approved fetal bovine serum (FBS) (Clontech), Dox (1 µg/mL), puromycin (1 µg/mL), and Geneticin (200 µg/mL). U87MG cells that express EGFRvIII or over-express wt-EGFR are previously described.37
Antibodies and reagents
Antibody that detects phospho-Tyr-416 in c-Src and cross-reacts with the equivalent epitope in other SFKs was from Cell Signaling Technology. Antibodies specific for the phosphorylated form of p130Cas (phopho-Tyr-410) and total ERK1/2 also were from Cell Signaling Technology. Antibody that detects phospho-Tyr-845 in the EGFR was from Invitrogen. Antibody that detects uPA was from American Diagnostica and antibody that detects total EGFR was from Millipore. Human uPAR-specific antibody was from R&D Systems. Horseradish peroxide-conjugated anti-rabbit IgG and anti-mouse IgG were from GE Healthcare. Quantitative PCR (qPCR) reagents, including primers and probes for uPA, uPAR, and hypoxanthine phosphoribosyltransferase 1 (HPRT-1) were from Applied Biosystems. EGF was from R&D Systems. Dasatinib was from Eton Bioscience. Gefitinib was from LC Laboratories
Real-Time qPCR
Total RNA was isolated using the RNeasy Kit (Qiagen). cDNA was synthesized with the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed on a System 7300 instrument (Applied Biosystems) with a one-step program: 95°C for 10 min, 95°C for 30 s, and 60°C for 1 min for 40 cycles. HPRT-1 gene expression was measured as a normalizer. Results were analyzed by the relative quantity method. Experiments were performed in triplicate with internal duplicate determinations.
Immunoblot analysis
Cell extracts were prepared in RIPA buffer [20 mM sodium phosphate, 150 mM NaCl, pH 7.4, 1% Nonidet P-40, 0.1% SDS, and 0.5% deoxycholic acid] containing complete protease inhibitor mixture (Roche). Protein concentrations were determined by bicinchoninic acid assay (Sigma-Aldrich). Equal amounts of cell extract were subjected to SDS-PAGE, electro-transferred to PVDF membranes, and probed with primary antibodies.
siRNA Transfection
uPAR-specific siRNA (5′-GCCGUUACCUCGAAUGCAU-3′) and uPA-specific siRNA (5′CAUGUUACUGACC-AGCAAC-3′) are previously described.15,67 Non-targeting control (NTC) siRNA was from Dharmacon. siRNAs (25 nM) were introduced into cells by incubation with Lipofectamine 2000 (Invitrogen) in SFM for 4 h. Cultures were allowed to recover in serum-containing medium for 12 h. The extent of gene-silencing was determined by qPCR and immunoblot analysis.
Cell Migration Assays
Cell migration was studied using 6.5-mm Transwell chambers with 8 µm pores (Costar, Corning, NY). Cells (1.5×105) that were transfected with siRNA targeting uPAR or uPA or with NTC siRNA were added to the top chamber and allowed to migrate at 37°C. When specified, Dasatinib was added to both chambers. The bottom chamber contained 10% FBS. After 18 h, the upper surface of each membrane was cleaned with a cotton swab. The membranes were then stained with Diff-Quick (Dade-Behring, Deerfield, IL). The number of cells on the bottom surface of each membrane was counted (cells/field). Four fields from each membrane were examined. Each condition was studied at least in triplicate.
SH2 domain pulldown assays
A construct encoding a glutathione-S-transferase (GST) fusion protein with the SH2 domain of c-Src (GST-SH2) was kindly provided by Dr. J.T. Parsons (University of Virginia) and expressed in Escherichia coli. Bacterial cells that express GST-SH2 were suspended in 20 mM sodium phosphate, 150 mM NaCl, pH 7.4, with 0.1% Triton X-100 and protease inhibitor cocktail and lysed by sonication. GST-SH2 was purified by glutathione-Sepharose affinity chromatography.
To assess SH2 docking sites in the EGFR, an equivalent number of different GBM cells was extracted in RIPA buffer. The extracts were subjected to centrifugation at 14,000×g for 10 min at 4°C. The supernatants were incubated with G ST-SH2 coupled to glutathinone-Sepharose for 3 h at 4°C. The Sepharose beads were was hed three times with RIPA buffer and resuspended in SDS sample buffer for SDS-PAGE. EGFR that associated with GST-SH2 was determined by immunoblot analysis. In control experiments, EGFR failed to associate with glutathinone-Sepharose that was not loaded with GST-SH2.
Quantum dot immunofluorescence (IF) microscopy
An EGFRvIII-expressing human GBM (GBM39) was propagated as a xenograft40 and kindly provided by C. David James (Department of Neurological Surgery, University of California San Francisco). Harvested tumor tissue was formalin-fixed, paraffin-embedded, and cut into 4 µm sections for mounting on positively-charged slides. Antigen retrieval was performed using protease 2 (Ventana). Sections were immunostained with primary antibodies targeting phospho-Tyr-845 (1:150; Abcam) and human uPAR (1:75; Dako) for 1 h at 37°C using the Ventana Discovery Ultra Platform. Q-dot-linked fluorescent secondary antibodies (1:150; Invitrogen) were added for 1 h. The slides were rinsed and cover-slipped with Prolong Gold and DAPI (Invitrogen). Slides were visualized on a Zeiss Axio Imager2 using Cambridge Research Instruments Nuance Multispectral Imaging System software to capture images and visualize individual fluorophore spectra free from auto-fluorescence noise. In control experiments, phospho-epitope labeling was validated using protein phosphatase treatment, which eliminated signal.
Supplementary Material
Supplementary Figure 1 (a) U373MG were treated with Dox or vehicle for 4 days and then transfected with NTC siRNA (black bars) or uPAR-specific siRNA (grey bars). uPAR mRNA levels were determined by qPCR and standardized against the levels present in vehicle-treated cells transfected with NTC siRNA. (b) ESC1, ESC2 and ESC5 cells were transfected with NTC siRNA (black bars) or uPAR-specific siRNA (grey bars). uPAR mRNA levels were determined by qPCR and standardized against the levels present in ESC1 cells treated with NTC siRNA.
Supplementary Figure 2 U373MG, ESC1, ESC2 and ESC5 cells were transfected with NTC siRNA (black bars) or uPA-specific siRNA (grey bars). uPA mRNA levels were determined by qPCR and standardized against the levels present in cells treated with NTC siRNA.
ACKOWLEDGEMENTS
This work was supported by NIH R01 CA169096 (to S.L.G.), R01 NS080939 (to F.B.F), and the Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society (to W.K.C and F.B.F.). W.K.C. is a Fellow of the National Foundation for Cancer Research. The authors would like to thank Aran Merati and Nancy Du for their technical assistance with some of the experiments.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Stettner MR, Wang W, Nabors LB, Bharara S, Flynn DC, Grammer JR, et al. Lyn kinase activity is the predominant cellular SRC kinase activity in glioblastoma tumor cells. Cancer Res. 2005;65(13):5535–5543. doi: 10.1158/0008-5472.CAN-04-3688. [DOI] [PubMed] [Google Scholar]
- 2.Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27(1):77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, Ahkavan D, et al. Fyn and Src are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69(17):6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahluwalia MS, de Groot J, Liu WM, Gladson CL. Targeting Src in glioblastoma tumors and brain metastases: rationale and preclinical studies. Cancer Lett. 2010;298(2):139–149. doi: 10.1016/j.canlet.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4(6):470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 6.Alper O, Bowden ET. Novel insights into c-Src. Curr Pharm Des. 2005;11(9):1119–1130. doi: 10.2174/1381612053507576. [DOI] [PubMed] [Google Scholar]
- 7.Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18(9):2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenfeld DP, Schwartzberg PL, Venegas A, Tse R, Sheetz MP. Selective regulation of integrin-cytoskeleton interactions by the tyrosine kinase Src. Nat Cell Biol. 1999;1(4):200–206. doi: 10.1038/12021. [DOI] [PubMed] [Google Scholar]
- 9.Ma YC, Huang J, Ali S, Lowry W, Huang XY. Src tyrosine kinase is a novel direct effector of G proteins. Cell. 2000;102(5):635–646. doi: 10.1016/s0092-8674(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 10.Kubota Y, Tanaka T, Kitanaka A, Ohnishi H, Okutani Y, Waki M, et al. Src transduces erythropoietin-induced differentiation signals through phosphatidylinositol 3-kinase. EMBO J. 2001;20(20):5666–5677. doi: 10.1093/emboj/20.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servitja JM, Marinissen MJ, Sodhi A, Bustelo XR, Gutkind JS. Rac1 function is required for Src-induced transformation. Evidence of a role for Tiam1 and Vav2 in Rac activation by Src. J Biol Chem. 2003;278(36):34339–34346. doi: 10.1074/jbc.M302960200. [DOI] [PubMed] [Google Scholar]
- 12.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci U S A. 1999;96(4):1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. J Biol Chem. 2003;278(3):1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 14.Jo M, Thomas K, Takimoto S, Gaultier A, Hsieh E, Lester R, et al. Urokinase receptor primes cells to proliferate in response to epidermal growth factor. Oncogene. 2007;26(18):2585–2594. doi: 10.1038/sj.onc.1210066. [DOI] [PubMed] [Google Scholar]
- 15.Hu J, Jo M, Cavenee WK, Furnari F, VandenBerg SR, Gonias SL. Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc Natl Acad Sci U S A. 2011;108(38):15984–15989. doi: 10.1073/pnas.1113416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chumbalkar V, Latha K, Hwang Y, Maywald R, Hawley L, Sawaya R, et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of DeltaEGFR. J Proteome Res. 2011;10(3):1343–1352. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24(16):7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313(5998):144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 19.Schlegel J, Stumm G, Brandle K, Merdes A, Mechtersheimer G, Hynes NE, et al. Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J Neurooncol. 1994;22(3):201–207. doi: 10.1007/BF01052920. [DOI] [PubMed] [Google Scholar]
- 20.Schlegel J, Merdes A, Stumm G, Albert FK, Forsting M, Hynes N, et al. Amplification of the epidermal-growth-factor-receptor gene correlates with different growth behaviour in human glioblastoma. Int J Cancer. 1994;56(1):72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- 21.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272(5):2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 23.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3(12):932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Aguirre Ghiso J, Estrada Y, Ossowski L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell. 2002;1(5):445–457. doi: 10.1016/s1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 25.Bohuslav J, Horejsi V, Hansmann C, Stockl J, Weidle UH, Majdic O, et al. Urokinase plasminogen activator receptor, beta 2-integrins, and Src-kinases within a single receptor complex of human monocytes. J Exp Med. 1995;181(4):1381–1390. doi: 10.1084/jem.181.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degryse B, Resnati M, Rabbani SA, Villa A, Fazioli F, Blasi F. Src-dependence and pertussis-toxin sensitivity of urokinase receptor-dependent chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. Blood. 1999;94(2):649–662. [PubMed] [Google Scholar]
- 27.Nguyen DH, Webb DJ, Catling AD, Song Q, Dhakephalkar A, Weber MJ, et al. Urokinase-type plasminogen activator stimulates the Ras/Extracellular signal-regulated kinase (ERK) signaling pathway and MCF-7 cell migration by a mechanism that requires focal adhesion kinase, Src, and Shc. Rapid dissociation of GRB2/Sos-Shc complex is associated with the transient phosphorylation of ERK in urokinase-treated cells. J Biol Chem. 2000;275(25):19382–19388. doi: 10.1074/jbc.M909575199. [DOI] [PubMed] [Google Scholar]
- 28.Smith HW, Marra P, Marshall CJ. uPAR promotes formation of the p130Cas-Crk complex to activate Rac through DOCK180. J Cell Biol. 2008;182(4):777–790. doi: 10.1083/jcb.200712050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Z, Liu Y, Johnson JJ, Stack MS. Urinary-type plasminogen activator receptor (uPAR) modulates oral cancer cell behavior with alteration in p130cas. Molecular and Cellular Biochemistry. 2011;357(1–2):151–161. doi: 10.1007/s11010-011-0885-3. [DOI] [PubMed] [Google Scholar]
- 30.Osherov N, Levitzki A. Epidermal-growth-factor-dependent activation of the src-family kinases. European Journal of Biochemistry / FEBS. 1994;225(3):1047–1053. doi: 10.1111/j.1432-1033.1994.1047b.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilde A, Beattie EC, Lem L, Riethof DA, Liu SH, Mobley WC, et al. EGF receptor signaling stimulates Src kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell. 1999;96(5):677–687. doi: 10.1016/s0092-8674(00)80578-4. [DOI] [PubMed] [Google Scholar]
- 32.Cooper JA, MacAuley A. Potential positive and negative autoregulation of p60c-src by intermolecular autophosphorylation. Proc Natl Acad Sci U S A. 1988;85(12):4232–4236. doi: 10.1073/pnas.85.12.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rich JN, Reardon DA, Peery T, Dowell JM, Quinn JA, Penne KL, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 34.Franceschi E, Cavallo G, Lonardi S, Magrini E, Tosoni A, Grosso D, et al. Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO) Br J Cancer. 2007;96(7):1047–1051. doi: 10.1038/sj.bjc.6603669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu-Emerson C, Norden AD, Drappatz J, Quant EC, Beroukhim R, Ciampa AS, et al. Retrospective study of dasatinib for recurrent glioblastoma after bevacizumab failure. J Neurooncol. 2011;104(1):287–291. doi: 10.1007/s11060-010-0489-x. [DOI] [PubMed] [Google Scholar]
- 36.Huveldt D, Lewis-Tuffin LJ, Carlson BL, Schroeder MA, Rodriguez F, Giannini C, et al. Targeting Src family kinases inhibits bevacizumab-induced glioma cell invasion. PLoS One. 2013;8(2):e56505. doi: 10.1371/journal.pone.0056505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91(16):7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato K. Cellular Functions Regulated by Phosphorylation of EGFR on Tyr845. International Journal of Molecular Sciences. 2013;14(6):10761–10790. doi: 10.3390/ijms140610761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piwnica-Worms H, Saunders KB, Roberts TM, Smith AE, Cheng SH. Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c-src. Cell. 1987;49(1):75–82. doi: 10.1016/0092-8674(87)90757-4. [DOI] [PubMed] [Google Scholar]
- 40.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6(3):1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 41.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 42.Mukasa A, Wykosky J, Ligon KL, Chin L, Cavenee WK, Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci U S A. 2010;107(6):2616–2621. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen D, Catling A, Webb D, Sankovic M, Walker L, Somlyo A, et al. Myosin light chain kinase functions downstream of Ras/ERK to promote migration of urokinase-type plasminogen activator-stimulated cells in an integrin-selective manner. J Cell Biol. 1999;146(1):149–164. doi: 10.1083/jcb.146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjoller L, Hall A. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J Cell Biol. 2001;152(6):1145–1157. doi: 10.1083/jcb.152.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Z, Thomas KS, Webb DJ, Moravec R, Salicioni AM, Mars WM, et al. Regulation of Rac1 activation by the low density lipoprotein receptor-related protein. J Cell Biol. 2002;159(6):1061–1070. doi: 10.1083/jcb.200207070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jo M, Takimoto S, Montel V, Gonias S. The urokinase receptor promotes cancer metastasis independently of urokinase-type plasminogen activator in mice. Amer J Pathol. 2009;175(1):190–200. doi: 10.2353/ajpath.2009.081053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eastman BM, Jo M, Webb DL, Takimoto S, Gonias SL. A transformation in the mechanism by which the urokinase receptor signals provides a selection advantage for estrogen receptor-expressing breast cancer cells in the absence of estrogen. Cellular Signalling. 2012;24(9):1847–1855. doi: 10.1016/j.cellsig.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 49.Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56(21):5079–5086. [PubMed] [Google Scholar]
- 50.Besnard-Guerin C, Newsham I, Winqvist R, Cavenee WK. A common region of loss of heterozygosity in Wilms' tumor and embryonal rhabdomyosarcoma distal to the D11S988 locus on chromosome 11p15.5. Hum Genet. 1996;97(2):163–170. doi: 10.1007/BF02265259. [DOI] [PubMed] [Google Scholar]
- 51.Inda M, Bonavia R, Mukasa A, Narita Y, Sah D, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wykosky J, Mukasa A, Furnari F, Cavenee WK. Escape from targeted inhibition: the dark side of kinase inhibitor therapy. Cell Cycle. 2010;9(9):1661–1662. doi: 10.4161/cc.9.9.11592. [DOI] [PubMed] [Google Scholar]
- 53.Jo M, Thomas K, Marozkina N, Amin T, Silva C, Parsons S, et al. Dynamic assembly of the urokinase-type plasminogen activator signaling receptor complex determines the mitogenic activity of urokinase-type plasminogen activator. J Biol Chem. 2005;280(17):17449–17457. doi: 10.1074/jbc.M413141200. [DOI] [PubMed] [Google Scholar]
- 54.Chandrasekar N, Mohanam S, Gujrati M, Olivero WC, Dinh DH, Rao JS. Downregulation of uPA inhibits migration and PI3k/Akt signaling in glioblastoma cells. Oncogene. 2003;22:392–400. doi: 10.1038/sj.onc.1206164. [DOI] [PubMed] [Google Scholar]
- 55.Zhang S, Yu D. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends in Pharmacological Sciences. 2012;33:122–128. doi: 10.1016/j.tips.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguirre Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol. 1999;147(1):89–104. doi: 10.1083/jcb.147.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carriero MV, Franco P, Votta G, Longanesi-Cattani I, Vento MT, Masucci MT, et al. Regulation of cell migration and invasion by specific modules of uPA: mechanistic insights and specific inhibitors. Current Drug Targets. 2011;12(12):1761–1771. doi: 10.2174/138945011797635777. [DOI] [PubMed] [Google Scholar]
- 58.Montuori N, Cosimato V, Rinaldi L, Rea VE, Alfano D, Ragno P. uPAR regulates pericellular proteolysis through a mechanism involving integrins and fMLF-receptors. Thromb Haemost. 2013;109(2):309–318. doi: 10.1160/TH12-08-0546. [DOI] [PubMed] [Google Scholar]
- 59.Carriero MV, Del Vecchio S, Capozzoli M, Franco P, Fontana L, Zannetti A, et al. Urokinase receptor interacts with alpha(v)beta5 vitronectin receptor, promoting urokinase-dependent cell migration in breast cancer. Cancer Res. 1999;59(20):5307–5314. [PubMed] [Google Scholar]
- 60.Yu W, Kim J, Ossowski L. Reduction in surface urokinase receptor forces malignant cells into a protracted state of dormancy. J Cell Biol. 1997;137(3):767–777. doi: 10.1083/jcb.137.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma Z, Webb D, Jo M, Gonias S. Endogenously produced urokinase-type plasminogen activator is a major determinant of the basal level of activated ERK/MAP kinase and prevents apoptosis in MDA-MB-231 breast cancer cells. J Cell Sci. 2001;114(Pt 18):3387–3396. doi: 10.1242/jcs.114.18.3387. [DOI] [PubMed] [Google Scholar]
- 62.Alfano D, Franco P, Vocca I, Gambi N, Pisa V, Mancini A, et al. The urokinase plasminogen activator and its receptor: role in cell growth and apoptosis. Thromb Haemost. 2005;93(2):205–211. doi: 10.1160/TH04-09-0592. [DOI] [PubMed] [Google Scholar]
- 63.Alfano D, Iaccarino I, Stoppelli M. Urokinase signaling through its receptor protects against anoikis by increasing BCL-xL expression levels. J Biol Chem. 2006;281(26):17758–17767. doi: 10.1074/jbc.M601812200. [DOI] [PubMed] [Google Scholar]
- 64.Ellis V, Behrendt N, Dano K. Cellular receptor for urokinase-type plasminogen activator: function in cell-surface proteolysis. Methods in Enzymology. 1993;223:223–233. doi: 10.1016/0076-6879(93)23048-r. [DOI] [PubMed] [Google Scholar]
- 65.Parker JJ, Dionne KR, Massarwa R, Klaassen M, Foreman NK, Niswander L, Canoll P, Kleinschmidt-Demasters BK, Waziri A. Gefitinib selectively inhibits tumor cell migration in EGFR-amplified human glioblastoma. Neuro Oncol. 2013;15:1048–1057. doi: 10.1093/neuonc/not053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazar AP, Ahn RW, O'Halloran TV. Development of novel therapeutics targeting the urokinase plasminogen activator receptor (uPAR) and their translation toward the clinic. Curr Pharm Des. 2011;17:1970–1978. doi: 10.2174/138161211796718152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo M, Lester R, Montel V, Eastman B, Takimoto S, Gonias S. Reversibility of epithelial-mesenchymal transition (EMT) induced in breast cancer cells by activation of urokinase receptor-dependent cell signaling. J Biol Chem. 2009;284:22825–22833. doi: 10.1074/jbc.M109.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 (a) U373MG were treated with Dox or vehicle for 4 days and then transfected with NTC siRNA (black bars) or uPAR-specific siRNA (grey bars). uPAR mRNA levels were determined by qPCR and standardized against the levels present in vehicle-treated cells transfected with NTC siRNA. (b) ESC1, ESC2 and ESC5 cells were transfected with NTC siRNA (black bars) or uPAR-specific siRNA (grey bars). uPAR mRNA levels were determined by qPCR and standardized against the levels present in ESC1 cells treated with NTC siRNA.
Supplementary Figure 2 U373MG, ESC1, ESC2 and ESC5 cells were transfected with NTC siRNA (black bars) or uPA-specific siRNA (grey bars). uPA mRNA levels were determined by qPCR and standardized against the levels present in cells treated with NTC siRNA.