Abstract
Stepped wedge designs are increasingly commonplace and advantageous for cluster randomized trials (CRTs) when it is both unethical to assign placebo and it is logistically difficult to allocate an intervention simultaneously to many clusters. We study marginal mean models fit with generalized estimating equations (GEE) for assessing treatment effectiveness in stepped wedge CRTs. This approach has advantages over the more commonly used mixed models that (1) the population-average parameters have an important interpretation for public health applications and (2) they avoid untestable assumptions on latent variable distributions and avoid parametric assumptions about error distributions, therefore providing more robust evidence on treatment effects. However, CRTs typically have a small number of clusters, rendering the standard GEE sandwich variance estimator biased and highly variable and hence yielding incorrect inferences. We study the usual asymptotic GEE inferences (i.e., using sandwich variance estimators and asymptotic normality) and four small-sample corrections to GEE for stepped wedge CRTs and for parallel CRTs as a comparison. We show by simulation that the small-sample corrections provide improvement, with one correction appearing to provide at least nominal coverage even with only 10 clusters per group. These results demonstrate the viability of the marginal mean approach for both stepped wedge and parallel CRTs. We also study the comparative performance of the corrected methods for stepped wedge and parallel designs, and describe how the methods can accommodate interval censoring of individual failure times and incorporate semiparametric efficient estimators.
Keywords: Vaccine efficacy trial, Cluster randomization, Generalized Estimating Equations, Marginal mean model, Phase 3, Phase 4, Small-sample variance correction
1 Introduction
Cluster randomized trials (CRTs) randomize groups of individuals to interventions and assess population-level treatment effects. Examples of CRTs include smoking prevention trials where the unit of randomization is school or city1,2 and HIV prevention trials where the unit of randomization is community or workplace.3,4
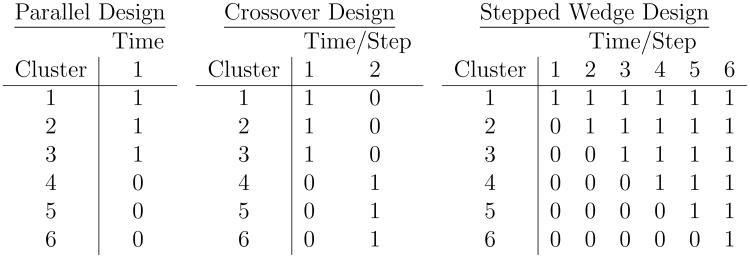
We consider stepped wedge (SW) CRTs and standard parallel CRTs for comparison. The SW design is a one-way crossover CRT design that follows enrolled clusters for at least two “steps” of time intervals. Typically the clusters are crossed over from control to active vaccine/treatment. In our formulation at least one cluster receives vaccine in the first step while the remainder receive control and then are crossed over at randomly assigned steps to receive vaccine. Figure 1 (adapted from Hussey and Hughes5) illustrates the differences between the parallel, crossover, and SW designs.
Figure 1.
Treatment schedules for basic parallel, crossover, and stepped wedge cluster randomized trial designs. A “0” represents control and a “1” represents active treatment/vaccine.
While most CRTs have used parallel designs, recently SW designs have received attention for two main reasons. First, parallel designs assign some communities the control condition for the entire study, which poses an ethical and enrollment challenge if there is prior evidence for intervention/vaccine efficacy. Second, parallel designs implement the intervention simultaneously in all the randomized communities, but this is sometimes logistically impossible. For example, in an HIV prevention trial that evaluates circumcision versus control in 20 versus 20 villages, it may be difficult for medical/surgical teams to deploy immediately in all 20 villages, whilst a traveling team could service four villages at a time in five steps. In addition, there is a large need to develop a scientific framework to guide Phase 4 post-licensure health-care scale-up in developing countries, due to the “formidable gap between innovations in health and their delivery to communities in the developing world”.6 The SW design is a recent tool under development that contributes to this scientific framework as well as to the scientific framework for Phase 3 licensure trials.
SW trials are increasingly being conducted. Since it was introduced in the Gambia Hepatitis B study,7 several other trials have used this design for many different types of outcomes including those involving HIV infection,8–10 waterborn diseases,11 and childhood malnutrition.12 Statistical methods for SW designs, including power and sample size calculations, have been developed by Hussey and Hughes5, Moulton et al.,13 and others.14
Statistical analysis of SW designs must accommodate both within-cluster correlation and time effects (as treatment is stepped up in a one-way crossover manner). Traditional analyses of CRTs handle within-cluster correlation by either a cluster-level or individual-level analysis.15 Cluster-level analysis compares cluster-level summary statistics (e.g., averages) between groups using two-sample methods for independent data. Individual-level analysis uses the individual as the analysis unit and accounts for within-cluster correlation using either marginal mean models (typically fit by GEE with a sandwich variance estimator) or mixed models. The mixed models account for the within-cluster correlation either by including random cluster effects or by conditioning out the random cluster effects.
For handling time effects, cluster-level analyses in SW trials are complicated by the fact that the treatment effect may change over time. Hussey and Hughes5 conducted cluster-level analysis using a linear mixed model that summarized cluster responses with estimated means for each cluster at each time. Hussey and Hughes5 also described individual-level analyses via linear models using both random effects models and marginal mean models. Moulton et al.13,16 regard event times of individuals as right censored and used a Cox regression-type of analysis, where at each observed failure time they compare treatment groups by conditioning on the number of events and the number at risk. To account for within-cluster correlation, either a sandwich or bootstrap estimator of variance is used, with cluster as the unit.13 While methods using sandwich estimators of variance have been shown useful for large cluster trials not requiring bias-correction,17,18 standard sandwich estimators of variance are generally anti-conservative when there are a small number of clusters. Although various methods have been investigated to reduce the bias and under-coverage with small cluster sizes in CRTs,19–24 none have been evaluated in SW designs.
Whereas the predominant approach to analyzing CRTs has been to use individual-level outcomes and random effects models, e.g.,5,13 we focus on the marginal mean modeling approach that has target parameter “total vaccine efficacy” (V ET).25 This parameter combines direct and indirect vaccine efficacies in comparing outcomes for individuals in vaccinated populations versus those for individuals in unvaccinated populations, and is defined mathematically in Section 2.3.25 We focus on the V ET parameter (and its time-varying version) because in Phase 3 and 4 trials it is of particular interest to use population-average estimands of group-level summary statistics that are most relevant for guiding public health policy decisions. The population-average approach also has advantages in the weaker and more testable assumptions it requires compared to mixed model approaches. In particular, mixed models require untestable assumptions about latent variable distributions, require correctly specified error distributions for consistent estimation, and standard error estimates are not robust to model mis-specifications.26 In contrast, marginal mean models do not make assumptions about latent variables and do not require correctly specified error distributions for consistent estimation, and, consequently, results from the latter models may be interpreted as carrying a greater weight of evidence.26 A potential pitfall of the marginal mean models is insufficient numbers of clusters to allow unbiased and stable standard error estimation, however, which has limited its use in practice.27 The significance of this work is that we show that small cluster size methods for marginal mean models can be used for correct inference about total vaccine efficacy overall and over time, thereby justifying the restored use of the highly interpretable population-average target parameter that is appropriate for stepped wedge and parallel CRTs.
The article is organized as follows. Section 2 describes a marginal mean model for the cluster-step outcomes for the SW and parallel designs, and defines the intervention/vaccine efficacy estimands of interest in terms of parameters in the model. This model allows interval censored time to events. Section 3 summarizes five approaches to estimation and testing of the estimands, using standard sandwich SEs and four small-sample methods that better protect the type I error and improve the accuracy of confidence intervals19–22 with additional details in the Web Supplementary Materials. Section 4 provides a simulation study to evaluate the five methods and compare their operating characteristics in SW and parallel designs. Section 5 provides discussion. R code implementing the developed methods for CRTs is provided at the last author's website.
2 Marginal mean model
2.1 Notation
We consider a prospective cohort CRT. Subjects testing HIV negative are enrolled during a fixed accrual period. Following the accrual period subjects are followed through J additional fixed calendar time intervals, each of duration M months, and are tested for HIV infection once within each calendar interval. Upon enrollment within the initial/first calendar interval, a subject's HIV testing date within the second calendar interval is scheduled, and evenly-spaced HIV testing dates within the subsequent calendar intervals 3, …, J are scheduled.
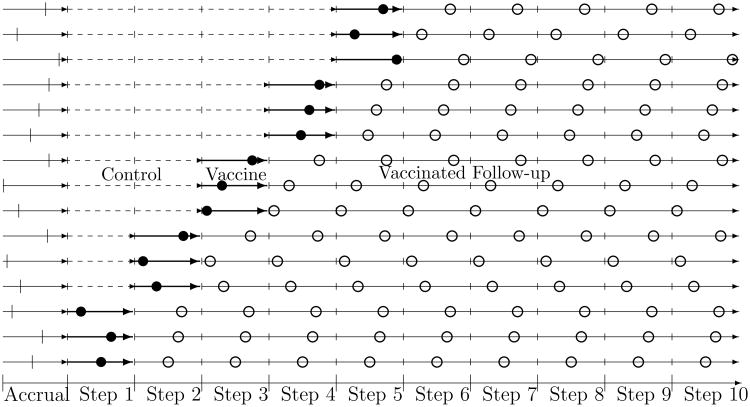
As an example used throughout, we consider a 72-month study with 12-month accrual period and ten subsequent 6-month calendar intervals, such that J = 10 and M = 6 months. Corresponding to the J calendar intervals, each subject is followed through J “steps,” with Step 1 defined as the interval between enrollment and his/her scheduled HIV test in calendar interval 2, Step 2 defined as the interval between the scheduled HIV tests in calendar intervals 2 and 3, and so on. Thus the steps are defined based on study time and the HIV testing schedule, conforming to the usual approach for HIV prevention trial design. For the SW design, the ith cluster starts the intervention (crosses over from the control condition) at a randomly assigned calendar interval . Subjects in cluster i start vaccination a day or two after their HIV test (if it is negative) within calendar interval . The two-group parallel design is the same except the ith cluster is assigned either to or to the control condition throughout the follow-up period of the study. Figure 2 describes the follow-up and HIV testing schedule for the SW design.
Figure 2.
A sample of three participants from each of five clusters are shown, each randomized to start vaccine in different steps. The dashed lines represent the non-vaccinated follow-up while the solid lines represent the vaccinated follow-up. The hash marks during the accrual period represent participants' dates of entry into the study. The open circles denote the times at which participants are tested for HIV infection and the filled circles denote the times at which the participants are tested for HIV infection and vaccinated if HIV negative.
Let Yijk be the indicator of whether individual k in cluster i is HIV infected during step j for i = 1, …, I and j = 1, …, J. Let X1ij be the treatment indicator (1=vaccine; 0=control), and X2ij be a vector of other cluster-step level covariates that may be useful for bias-correction and/or improving precision. We study marginal mean models of cluster-step level responses Yij, with Yij an estimate of E[Yij], the incidence of new infections in cluster i during step j. (Recall that our approach analyzes cluster-level summary statistics, not the individual outcomes Yijk.) In Section 3.1 we discuss different possible estimators of the cluster-step incidences E[Yij].
2.2 Generalized linear model
We model the marginal mean μij = E[Yij] with a generalized linear model (glm)
| (1) |
where g(·) is a link function, β2 adjusts for calendar/secular trends, and , where is the step (j) at which cluster i is randomized to start vaccination, and a+ = a if a > 0 and 0 otherwise. While β2 measures how the marginal mean varies across steps defined based on an individual's HIV testing schedule, it approximately measures how the marginal mean varies over calendar time, because the accrual period is a small fraction of the total study follow-up period. The term sij is the number of steps since cluster i started vaccination, and ranges from 0 to .
In model (1) with β3 = 0, β1 represents the treatment effect averaged over all clusters (i.e., the expected change in the response in the population when all clusters change from the control condition to the intervention). If the treatment effect changes with time, then the interaction coefficient β3 is nonzero. We use the coding sij for time effects because this allows assessment of how the treatment effect varies with time since vaccination. We focus on the identity link g(y) = y, although the method can also be implemented straightforwardly by modeling Yij as a count variable (the number of new infections in cluster i during step j) and using a log link, as was done in the first author's PhD dissertation.28 These approaches lead to different types of treatment efficacy estimands, as described in Section 2.3.
While standard glm modeling fit by GEE can be used for inference on β ≡ (β0, β1, β2, β3, β4)′, the fact that the number of clusters is small implies that the typical sandwich variance estimators for β̂ tend to underestimate the variance, which leads to anti-conservative inference.29,30 Accordingly, we will implement and evaluate four small-sample corrected SE methods for fitting model 1 for the CRT application with a small number of clusters.
2.3 Treatment efficacy/vaccine efficacy estimands
Vaccine efficacy, V E, is a measure of the form 1 − RR (one minus some measure of relative risk, see Halloran et al.25 Chapter 2). We use the term treatment efficacy, T E, as a more general term to denote any parameter that measures a treatment effect (e.g., a difference, a ratio, or 1 − RR). We focus on the glm with identity link and Yij the estimated incidence of cluster i in step j. From model (1), in the case that β3 = 0, the population-average parameter of interest is the total treatment efficacy T ET ≡ β1, which measures a combination of the direct and indirect effects of the intervention, as an additive difference (T ET is interpreted as the mean difference in cluster-step incidences for treated versus control clusters). In the general case where β3 may be nonzero, for a fixed s ∈ {1, …, J − 1}, define T ETs = β1 + β3 * s, which is the treatment/vaccine effect s steps (i.e., sM months) after vaccination. The parameters T ETs measure how the treatment efficacy changes with time since vaccination.
While we do not evaluate it here, the glm with log link and Yij the estimated mean number of infections is also of interest. In this model the number of person-years at risk PYij is included as a fixed covariate (entered as an offset term). If β3 = 0, the population-average total treatment efficacy parameter of interest is T ET = 1 − exp(β1), the multiplicative reduction in the mean cluster-step incidences for treated versus control clusters. If the intervention is vaccine, then T ET equals V ET as defined by Halloran et al.31 The parameter T ETs = 1 − exp(β1 + β3 * s) is the treatment/vaccine effect s steps after treatment/vaccination.
3 Estimation and testing
In this section we first describe approaches to estimating the HIV infection incidence for a given cluster-step. Second we summarize the standard sandwich SE and small-sample corrections.
3.1 Estimation of cluster-step incidences
Estimates of the cluster-step incidences, E[Yij], are used as the summary statistic responses in the glm. As such the estimation of the E[Yij] and of the parameters in the glm are completely separate steps, and the goal in the first step is to choose an estimator that performs well in bias and variance. In the following, the beginning and end of step j for a subject is the time of the scheduled HIV test in calendar interval j − 1 and in calendar interval j, respectively. Let Nij be the number of subjects at-risk (HIV uninfected) and under follow-up at the beginning of step j, for j = 1, …, J − 1. If all subjects in cluster-step (i,j) have an HIV test at the beginning and end of the step, then Nij is known (based on HIV negative results at the beginning of the step) and E[Yij] may be estimated by p̂ij, the fraction of the Nij subjects with a positive test result at the end of the step. If some subjects are missing HIV test results at either edge of the step and if the missingness is completely at random, then the above estimator computed in subjects with complete testing data is consistent. However, if a missing at random (MAR) mechanism is more tenable and hence the complete-case estimator is not consistent, a method designed for MAR data is superior. (Here MAR means that whether HIV testing results are missing depends only on collected information from the subject, which could include individual- and cluster-level data.)
One MAR method would model the incidence of infections parametrically, and estimate E[Yij] by maximum likelihood. Because consistent estimation for this approach would rely on a correct parametric model, an alternative approach that would provide consistent estimation under a mis-specified parametric model would be augmented inverse probability weighting (AIPW)32. This method, while advantageous as a doubly robust method, could perform poorly if there is no reasonable model predicting whether a subject's HIV test results are observed, or if some of these estimated probabilities are outliers near zero33. The collaborative targeted maximum likelihood (collaborative tMLE) method is another option for a doubly robust method. It has been shown in simulations to often perform well in settings with outlying weights near zero, which is due to its targeting of the mean-variance tradeoff on the parameter of interest and to the fact that it is a substitution estimator that is guaranteed to fall in the parameter space34. Augmented GEE35 is another approach, which, like the above approaches, can increase precision for estimating E[Yij] by incorporating individual-level covariates that predict whether individuals are infected during the step.
A limitation of the methods described above is that HIV tests have a time-lag between the date of HIV acquisition and the time at which the test would yield a positive result (for antibody-based tests about 3 weeks and for antigen-based tests such as HIV-specific PCR about 1 week). Therefore, the most accurate way to assess HIV infection is to consider the infection time as interval censored, where there is a known window period during which each diagnosed infection is known to occur (for a typical HIV prevention trial the window would be 1 week before the last negative PCR test to 1 week before the first PCR positive test). To explicitly handle the interval censoring, a nonparametric maximum likelihood estimator of the distribution of all subjects may be used to obtain a consistent estimate of E[Yij] for each i, j.36
3.2 Standard sandwich variance and corrections for handling a small number of clusters
We consider GEE for estimation and inference in model (1). This approach, introduced by Liang and Zeger,37 estimates β as the solution to an estimating equation (see Web Appendix A). GEE analysis typically uses the sandwich estimator of the variance matrix of β, which is consistent and asymptotically normal even when the working correlation is mis-specified; however, the estimator is biased (see Theorem 5.4 of Ziegler).29 This bias tends to underestimate the variance which leads to under-coverage of confidence intervals and inflated type I errors, particularly for the Wald test in small sample settings.
Numerous studies have been conducted and several solutions proposed to correct this bias and undercoverage (e.g., see Lu et al.38, the papers cited below, and the reviews of Ziegler29 and Dahmen and Ziegler30). One solution is to abandon the use of the sandwich estimator and resort to either a Jackknife or bootstrap estimator of the variance. Other approaches use the sandwich estimator with adjustments to the Wald test, and less commonly to the score test39. We focus on the more common Wald tests in this paper.
There are three main ways of adjusting the Wald tests for small samples. One solution implements some form of bias correction on the variance, with approaches developed by Fay and Graubard,19 Mancl and DeRouen,20 Kauermann and Carroll,21 and Morel et al.22 Theorem 5.21 of Ziegler29 shows that the Mancl and DeRouen,20 and modified Fay and Graubard29 (mFG) estimators are less biased than the usual sandwich variance estimator. This proof extends to the Kauermann and Carroll21 (KC) estimator since the KC variance-covariance estimator is identical to that of mFG (proved in Web Appendix B). While the mFG and KC procedures are identical analytically, their implementation requires numerical inversion of different matrices. Typically this matrix has lower dimension for the mFG approach, such that the mFG method may be preferred to provide more numerically stable inference. Although we ran simulations with KC, the results are so close to those with mFG that the KC results are not presented. Based on the estimator formulas the Mancl and DeRouen20 (MD) variance estimator will tend to be larger than those of mFG and KC,38 which is confirmed in the simulations. The Fay and Graubard variance estimator (see Web Appendix A) is similar to the mFG approach. The adjusted estimator of Morel et al.22 (MBN) is additive and always positive such that the type I error rate of Wald tests is always smaller than Wald tests based on the uncorrected sandwich estimator. In summary, all three bias-corrected SE estimators (MD, mFG/KC, MBN) are guaranteed to confer better Type I error control of Wald tests in GEE than the standard sandwich estimator, and the simulation study provides insight into the comparative performance. Web Appendices A and B provide additional details on the four corrected SE methods that we study.
A second way of adjusting the Wald test is to assume that the working variance is correctly specified and there is a common correlation structure, and then to reduce the variability of the sandwich estimator by smoothing. For example, Pan (2001)40 smoothed by replacing individual Pearson residuals with their mean, Gosho et al. (2014)41 added a simple bias correction to Pan's method, and Wang and Long (2011)42 used regularization for smoothing. Since these approaches are less robust, we do not pursue these in this paper.
A third adjustment takes into account the variability of the sandwich estimator and bases inferences on a t-distribution (or F-distribution) instead of a normal (or chi-square) distribution, an idea that parallels using a t-test instead of a Z-test.43 Combinations of the first and third solutions have been proposed19,21,43–45. Because (1) these combination approaches are often very similar, (2) the most recent work of Fan et al. (2012)45 does not show substantial improvement over the combination approach of Fay et al. (2001)19, and (3) the software is readily available in R to implement Fay et al. (2001), we focus on the latter combination approach in this article. In particular, in addition to studying three (mFG, MD, MBN) of the corrected-SE methods mentioned above using a reference normal distribution, we also study the Fay and Graubard19 δ5 (FG d5) approach (method=d5 in the saws R function). This approach uses the FG bias-corrected SE estimator and a reference t-distribution with degrees of freedom estimated by a weighted average of covariance estimates of the terms of the estimating equation (d̃H in the paper). In models with more than one parameter, the FG d5 approach in general has different degrees of freedom for different parameters, which addresses the different amounts of variability in the variance estimator for each parameter.
Other small sample adjustments not explored in this paper are an adjustment to the mean parameters (as opposed to adjustments to the variance estimators noted above), see Paul and Zhang46. In addition to the F-statistic adjustments mentioned above, McCaffrey and Bell (2006)44 studied saddlepoint approximations for calculating p-values with bias corrected sandwich estimators.
4 Simulation study
4.1 Objectives of the simulation study
We address several scientific questions via a simulation study for a glm with identity link g(y) = y and hence a mean difference treatment/vaccine efficacy parameter T E. Negative values of T E indicate treatment/vaccine efficacy. For SW and parallel CRTs, we evaluate the following properties of inference based on GEE assuming asymptotic normality with the standard SE estimator and GEE with a small sample correction:
Standard errors and coverage probabilities of Wald-based confidence intervals for the regression parameters in the glm.
Size of tests for and for for s = 0, …, J − 1.
Power of tests for and at various alternatives.
We study standard sandwich SE and MD-, MBN-, and mFG-corrected SEs; results for KC-corrected SEs are not reported because the results were almost identical to those for mFG. We also study the FG d5 method.
We study three cluster sizes, 10, 20 and 50. The first choice was based on the need to have at least 10 clusters to allow adequately stable inference. The second and third numbers were chosen to represent a typical number and a number that should be representative of asymptotic results and is near the maximum of what is typically used in stepped wedge CRTs. A literature review of all published stepped wedge CRTs with at least 50 individuals per cluster on average showed that the mean and median number of clusters was 33 and 12, respectively, with interquartile range 7–29 (Supplementary Table 1 in Web Appendix C).
4.2 Simulation of vaccine trials
We study the following three scenarios for the true vaccine efficacy parameters:
Scenario 1:T ETs = T ET = 0 for all s = 0, …, J − 1 (complete null)
Scenario 2:T ETs = T ET for all s = 0, …, J − 1 and T ET < 0 (beneficial efficacy that is time-constant)
Scenario 3:T ETs decreasing in time s = 0, …, J − 1 (efficacy increases over the steps) CRTs are simulated in R version 2.14.0 by generating the number of infections for each cluster step from the linear marginal mean model in (1) without any extra covariates (X2ij):
| (2) |
where X1ij is defined as in Section 2.2 for cluster i and step j = 1, …, J. For parallel CRTs, for all i, such that sij = j − 1 for all i, j, whereas for SW CRTs, as described in Section 2.2.
The parameter β2 specifies how much the background incidence changes over time, and for simplicity we set β2 = 0. We choose β0 such that the mean HIV incidence over a step in non-vaccinated cluster-steps is either 0.04 (Scenario 1) or 0.05 (Scenarios 2–3). Based on the fact that the additive difference vaccine efficacy parameters equal
in terms of model (2), we choose β1 and β3 to create the three scenarios: (1) T ETs = 0 for all s = 0, …, J − 1; (2) T ETs = −0.015 for all s = 0, …, J − 1; and (3) T ET0 = −0.008, T ET(J−1) = −0.03, and T ETs decreases linearly with s = 1, …, J − 2. Scenario 2 represents an antibody-based vaccine that protects only through a reduction in HIV acquisition, with protection established quickly without waning, whereas scenario 3 represents an antibody/T-cell based combination vaccine that protects through indirect effects (on infectiousness and disease progression) that take time to accrue. Setting T ET0 near zero for scenario 3 is reasonable for a vaccine with no effect to reduce susceptibility, T ES = 0, given the time needed for indirect efficacy to accrue.
Vaccine trials are simulated to satisfy model (2). For each cluster i, the vector of estimated infection incidences (Yi1, …, YiJ)′ is generated from a multivariate normal distribution with E[Yij] = θij, Var[Yij] = 0.0005, and an AR-1 correlation structure (with linear correlation ρ = 0.8) to account for the anticipated within-cluster correlation of the Yij's over the steps. In addition, the case of independent Yij, j=1, …, J, for each i is considered to study sensitivity of inference to the within-cluster correlation structure (results presented in Web Appendix D).
To simulate SW and parallel trials via model (2) that follow each of the three scenarios above, simple math determines maps between values of θij and values of the true regression parameters β0, β1, and β3. In particular:
Scenario 1:θij = 0.04 for all i,j; β0 = 0.04, β1 = β3 = 0.
Scenario 2:θij = 0.05 for all i,j with X1ij = 0; θij = β0 + β1 = 0.035 for all i,j with X1ij = 1; β0 = 0.05, β1 = −0.015, β3 = 0
Scenario 3:θij = 0.05 for all i,j with X1ij = 0; θi1 = β0 + β1 = 0.042 with X1i1 = 1; β0 = 0.05, β1 = −0.008. For the parallel design, θij = 0.05 − 0.008 + β3(j − 1) for all i with X1ij = 1; β3 = (0.02 − β0 − β1)/(J − 1) = −0.022/(J − 1). For the SW design, θij = 0.05 − 0.008 + β3(j − 1) for all i,j with sij = j − 1 and X1ij = 1; β3 = (0.02 − β0 − β1)/(J − 1) = −0.022/(J − 1).
In practice, lacking evidence of the interaction effect in scenarios 1–2 (i.e., analysis consistent with β3 = 0) would result in fitting a reduced form of model (2) excluding the interaction term. Consequently, we report operating characteristics for β1 in scenarios 1–2 based on this reduced model.
This simulation approach supposes that complete HIV testing results are available; we focus on this relatively simple setting to focus attention on the performance of estimation and inference in the marginal mean model. Given the separation of the steps to estimate E[Yij] and to estimate the marginal mean model parameters, we can infer that the marginal mean inferences would perform similarly well (or better) if any of the AIPW, collaborative tMLE, or interval censoring approaches for estimating the E[Yij] were used, as long as they perform well for estimating the E[Yij]. For HIV prevention trial data sets the interval-censoring approach is appealing given that real data sets tend to have interval-censored infection times; however future work would be needed to conjoin this approach with the AIPW or collaborative tMLE methodology. The simulations use 5000 iterations.
4.3 Simulation results
The simulated data are analyzed with the standard sandwich approach and the four small sample corrections, using both the assumed correlation structure matching and mismatching the true correlation structure (as AR-1 for both the truth and the methods or as AR-1 for the truth and exchangeable for the methods), yielding very similar results. We report results for the mismatched case, as in practice some degree of mis-specification is expected. For all methods, we evaluate finite-sample bias of the standard errors of β̂1 and β̂3 in model (2) (or the reduced model for β1 in scenarios 1–2), as well as coverage probabilities of Wald-based 95% confidence intervals for β1 and β3. (We also explored a null reference t-distribution with J-p degrees of freedom with p the number of coefficients in the linear predictor, but it yielded overly conservative Wald tests.) In addition, we investigate power of the Wald tests to reject and to reject .
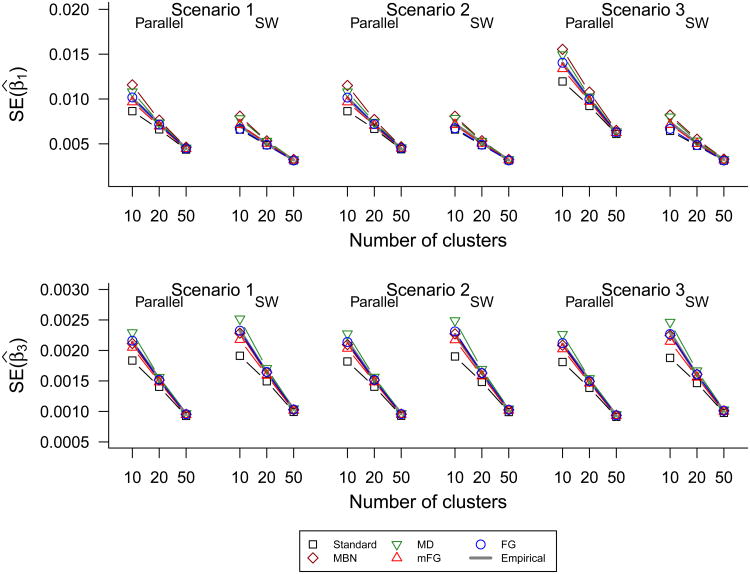
Figure 3 and Web Figure 1 show that all standard error estimators accurately reflect the true variability in coefficient estimation for the setting with a large number of clusters (I=50), as expected. Also as expected, with a small number of clusters the uncorrected GEE standard error estimator is too small whereas all of the corrected estimators are more accurate. The mFG and FG d5 estimators appear to be the most accurate, closely tracking the sample standard errors calculated across the simulation runs, although in Web Figure 1 FG d5 appears to slightly overestimate the SEs. MBN, MD and FG tend to be close or slightly conservative.
Figure 3.
Median standard-GEE, bias-corrected and empirical standard error (SE) estimates of β̂1 and β̂3 using the stepped wedge and parallel designs in simulation scenarios 1–3 with cluster-step incidences satisfying the AR-1 correlation structure. The empirical SE estimate is computed as the sample standard deviation of the β̂ estimates. In scenarios 1–2, a reduced form of model (2) excluding the interaction term is considered for inference about β1.
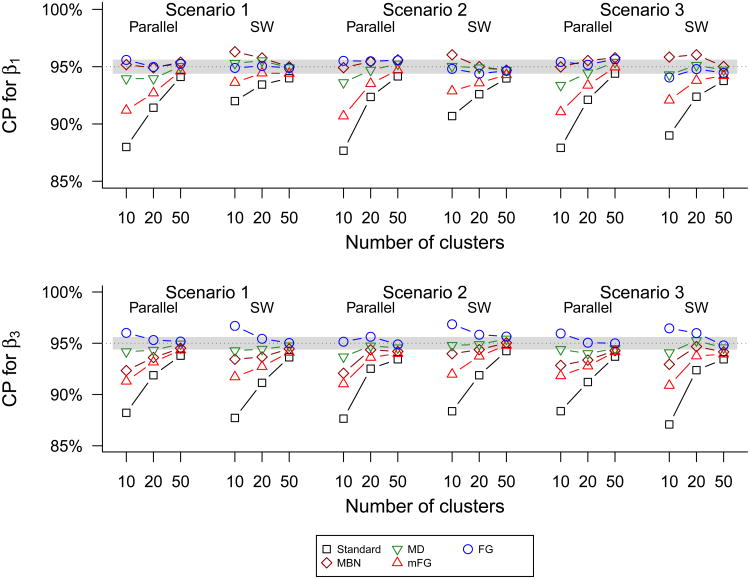
Figure 4 and Web Figure 2 show results on coverage probabilities. Recall that MBN, MD and mFG use the normal reference distribution, while FG d5 uses different t-distributions for β1 and β3. For MBN and MD, the combination of the conservative SE and the anti-conservative use of the normal distribution can lead to close to nominal coverage in many cases. However, considering AR-1 within-cluster correlation, the MBN method has slightly low coverage probabilities for β3. In contrast to MBN and MD, the coverage of mFG tends to be anti-conservative because although its SE is close to the empirical SE, the use of the normal distribution leads to under-coverage. The best method in terms of guaranteeing coverage is the FG d5 method, giving at least nominal coverage even with only 10 clusters per group. The coverage for FG d5 can be overly large for some cases with 10 clusters.
Figure 4.
Coverage probabilities (CP) of 95% standard-GEE and small sample corrected Wald confidence intervals for β1 and β3 using the stepped wedge and parallel designs in simulation scenarios 1–3 with cluster-step incidences satisfying the AR-1 correlation structure. The horizontal band represents ±2 × Monte Carlo standard error. In scenarios 1–2, a reduced form of model (2) excluding the interaction term is considered for inference about β1
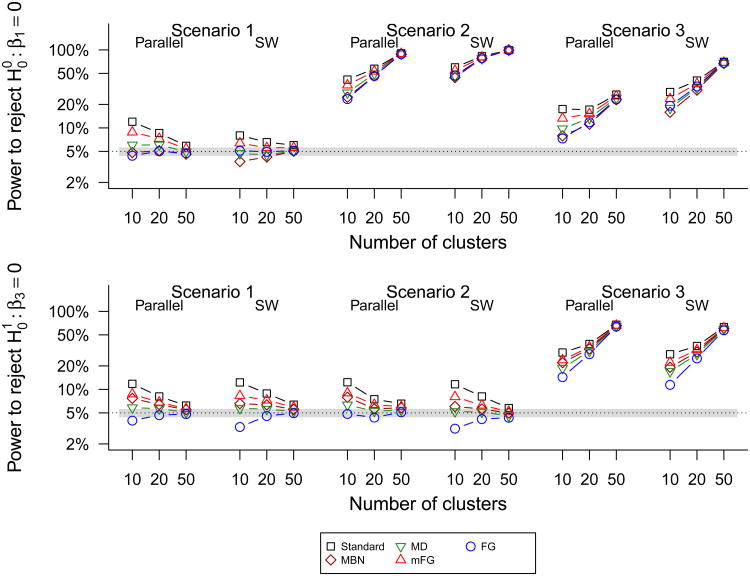
Figure 5 and Web Figure 3 show that all of the corrected-GEE Wald tests have closer to nominal size than the standard Wald test. The apparent gain in power of some methods over others appears to be due primarily to the anti-conservative sizes, since the order of the sizes and the powers appears consistent across scenarios (with Standard having the highest size and power, and mFG having the next highest). Differences in power between the methods become negligible for larger numbers of clusters.
Figure 5.
Size and power of standard-GEE and small sample corrected Wald tests to reject the null hypotheses and at 5% significance level using the stepped wedge and parallel designs in simulation scenarios 1–3 with cluster-step incidences satisfying the AR-1 correlation structure. For readability of the estimated sizes, size and power are plotted on the log scale. The horizontal band represents ±2 × Monte Carlo standard error.
We observe that the underlying within-cluster correlation structure impacts relative efficiency and power comparing the SW versus parallel design. For example, independence of the Yij's leads to superiority of the parallel design in terms of efficiency and power for β1 in scenario 2, whereas an AR-1 correlation leads to superiority of the SW design in the same setting. Overall, the small sample corrected methods have size closer to the nominal level, with the FG d5 being conservative or close to nominal, and MD or MBN being anti-conservative or close to nominal. Thus, the choice between FG d5 and either MD of MBN depends on the importance of guaranteeing coverage versus maximizing power. The Standard or mFG methods are not recommended unless the cluster size is large.
5 Discussion
This article considers the use of generalized linear models for inference on population-average parameters measuring total treatment efficacy over time in stepped wedge and parallel cluster randomized trials (CRTs). We focused on overcoming the challenge that CRTs typically have a small number of clusters, yet in this context the standard sandwich variance estimator for GEE is anti-conservative. We found that for this setting small-sample corrected GEE inferences allow GEE to provide Wald hypothesis testing procedures with closer to nominal size and confidence intervals with more correct coverage probabilities. In particular, our analytical and empirical evaluation suggests that FG d5 has close to nominal or conservative coverage, MD and MBN have close to nominal or anti-conservative coverage, and mFG/KC has generally anti-conservative coverage. Therefore, for settings where anti-conservative inference is strongly unacceptable, we recommend the FG d5 method, and, for settings where a slight inflation of the type I error rate is tolerable and maximizing power is at a premium, we recommend the MD or MBN method. Overall the spread between these three different methods is useful because it allows choices based on the context-dependent relative utility of maintaining type I error versus maximizing power.
A contribution of this work lies in the fact that most stepped wedge design analyses have used mixed models, yet marginal mean models may be preferred because they avoid the two drawbacks of mixed models that they rely on untestable assumptions about latent variable distributions and they require correctly specified error distributions for consistent estimation. Moreover, we discussed ways in which the approach can be combined modularly with methods to estimate the cluster-step disease incidences, for example allowing use of semiparametric efficient methods and allowing use of the methods that accommodate interval censoring of disease times. While GEE methods with finite-sample corrections have been evaluated for CRTs for parallel designs,23,38,47 we compared the performance of the four small sample corrected methods for stepped wedge versus parallel designs. We found that the level of correlation of cluster-step disease incidences across steps affects their relative power, where greater correlation implies greater relative power of the stepped wedge design, likely due to accounting for within-cluster information as well as between-cluster information.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Numbers R01AI029168 (Allan deCamp, Michal Juraska, Peter Gilbert) and R37AI054165 (Michal Juraska, Peter Gilbert). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary Materials Web Appendices and Figures referenced in Sections 3.2 and 4.3 are available with this paper online.
References
- 1.Peterson AV, Jr, Kealey KA, Mann SL, Marek PM, Sarason IG. Hutchinson Smoking Prevention Project: Long-term randomized trial in school-based tobacco use prevention results on smoking. Journal of the National Cancer Institute. 2000;92(24):1979–1991. doi: 10.1093/jnci/92.24.1979. [DOI] [PubMed] [Google Scholar]
- 2.The COMMIT Research Group. Community intervention trial for smoking cessation (COMMIT): I. Cohort results from a four-year community intervention. American Journal of Public Health. 1995;85(2):183–192. doi: 10.2105/ajph.85.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes RJ, Changalucha J, Ross DA, Gavyole A, Todd J, Obasi AIN, et al. The MEMA kwa Vijana Project: Design of a community randomised trial of an innovative adolescent sexual health intervention in rural Tanzania. Contemporary Clinical Trials. 2005;26:430–442. doi: 10.1016/j.cct.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Corbett EL, Dauya E, Matambo R, Cheung Y, Makamure B, Bassett MT, et al. Uptake of workplace HIV counselling and testing: A cluster-randomised trial in Zimbabwe. PLoS Medicine. 2006;3(7):1005–1012. doi: 10.1371/journal.pmed.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussey MA, Hughes JP. Design and analysis of stepped wedge cluster randomized trials. Contemporary Clinical Trials. 2007;28(2):182–191. doi: 10.1016/j.cct.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Madon T, Hofman KJ, Kupfer L, Glass RI. PUBLIC HEALTH: Implementation Science. Science. 2007 Dec;318:1728–1729. doi: 10.1126/science.1150009. [DOI] [PubMed] [Google Scholar]
- 7.The Gambia Hepatitis Study Group. The Gambia Hepatitis Intervention Study. Cancer Research. 1987 Nov 1;47:5782–5787. [PubMed] [Google Scholar]
- 8.Fairley CK, Levy RW, Rayner CR, Allardice K, Costello K, Thomas C, et al. Randomized trial of an adherence programme for clients with HIV. International Journal of STD and AIDS. 2003;14:805–809. doi: 10.1258/095646203322556129. [DOI] [PubMed] [Google Scholar]
- 9.Levy RW, Rayner CR, Fairley CK, Kong D, Mijch A, Costello K, et al. Multidisciplinary HIV Adherence Intervention: A randomized study. AIDS Patient Care and STDs. 2004;18(12):728–735. doi: 10.1089/apc.2004.18.728. [DOI] [PubMed] [Google Scholar]
- 10.Grant AD, Charalambous S, Fielding KL, Day JH, Corbett EL, Chaisson RE, et al. Effect of routine Isoniazid preventive therapy on tuberculosis incidence among HIV-infected men in South Africa. Journal of the American Medical Association. 2005;293:2719–2725. doi: 10.1001/jama.293.22.2719. [DOI] [PubMed] [Google Scholar]
- 11.Bailey IW, Archer L. The impact of the introduction of treated water on aspects of community health in a rural community in Kwazulu-Natal, South Africa. Water Science and Technology. 2004;50(1):105–110. [PubMed] [Google Scholar]
- 12.Ciliberto MA, Sandige H, Ndekha MJ, Ashorn P, Briend A, Ciliberto HM, et al. Comparison of home-based therapy with ready-to-use therapeutic food with standard therapy in the treatment of malnourished Malawian children: a controlled, clinical effectiveness trial. American Journal of Clinical Nutrition. 2005;81:864–870. doi: 10.1093/ajcn/81.4.864. [DOI] [PubMed] [Google Scholar]
- 13.Moulton LH, O'Brien KL, Reida R, Weatherholtza R, Santoshama M, Siberb GR. Evaluation of the indirect effects of a pneumococcal vaccine in a community-randomized study. Journal of Biopharmaceutical Statistics. 2006;16(4):453–462. doi: 10.1080/10543400600719343. [DOI] [PubMed] [Google Scholar]
- 14.Brown CA, Lilford RJ. The stepped wedge trial design: a systematic review. BMC Medical Research Methodology. 2006;6:54–59. doi: 10.1186/1471-2288-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes RJ, Moulton LH. Cluster Randomized Trials. CRC Press; New York: 2009. [Google Scholar]
- 16.Moulton LH, Golub JE, Durovni B, Cavalcante SC, Pacheco AG, Saraceni V, et al. Statistical design of THRio: a phased implementation clinic-randomized study of a tuberculosis preventive therapy intervention. Clinical Trials. 2007;5:190–199. doi: 10.1177/1740774507076937. [DOI] [PubMed] [Google Scholar]
- 17.Preisser JS, Young ML, Zaccaro DJ, Wolfson M. An integrated population-averaged approach to the design, analysis and sample size determination of cluster-unit trials. Statistics in Medicine. 2003;22(8):1235–1254. doi: 10.1002/sim.1379. [DOI] [PubMed] [Google Scholar]
- 18.Young ML, Preisser JS, Qaqish BF, Wolfson M. Comparison of subject-specific and population averaged models for count data from cluster-unit intervention trials. Statistical Methods in Medical Research. 2011;16(2):167–184. doi: 10.1177/0962280206071931. [DOI] [PubMed] [Google Scholar]
- 19.Fay MP, Graubard BI. Small-sample adjustments for Wald-type tests using sandwich estimators. Biometrics. 2001 Dec;57:1198–1206. doi: 10.1111/j.0006-341x.2001.01198.x. [DOI] [PubMed] [Google Scholar]
- 20.Mancl LA, DeRouen TA. A covariance estimator for GEE with improved small-sample properties. Biometrics. 2001 Mar;57(1):126–134. doi: 10.1111/j.0006-341x.2001.00126.x. [DOI] [PubMed] [Google Scholar]
- 21.Kauermann G, Carroll RJ. A note on the efficiency of sandwich covariance matrix estimation. Journal of the American Statistical Association. 2001 Dec;96(456):1387–1396. [Google Scholar]
- 22.Morel JG, Bokossa MC, Neerchal NK. Small sample correction for the variance of GEE estimators. Biometrical Journal. 2003;45(4):395–409. [Google Scholar]
- 23.Braun TM. A mixed model-based variance estimator for marginal model analyses of cluster randomized trials. Biometrical Journal. 2007;49(3):394–405. doi: 10.1002/bimj.200510280. [DOI] [PubMed] [Google Scholar]
- 24.Westgate PM, Braun TM. Improving small-sample inference in group randomized trials with binary outcomes. Statistics in Medicine. 2011;30(3):201–210. doi: 10.1002/sim.4101. [DOI] [PubMed] [Google Scholar]
- 25.Halloran ME, Longini IM, Jr, Struchiner CJ. Design and Analysis of Vaccine Studies. Springer; 2010. [Google Scholar]
- 26.Hubbard A, Ahern J, Fleischer N, van der Laan M, Lippman S, Jewell N, et al. To GEE or not to GEE: Comparing population average and mixed models for estimating the associations between neighborhood risk factors and health. Epidemiology. 2010;21:467–474. doi: 10.1097/EDE.0b013e3181caeb90. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z, Diehr P, Peterson A, McLerran D. Selected statistical issues in group randomized trials. Annual Review of Public Health. 2001;22:167–187. doi: 10.1146/annurev.publhealth.22.1.167. [DOI] [PubMed] [Google Scholar]
- 28.Scott J. Unpublished PhD dissertation. University of Washington; Seattle (WA): 2008. Stepped Wedge Cluster Randomized Trials. [Google Scholar]
- 29.Ziegler A. Generalized Estimating Equations. New York: Springer; 2011. [Google Scholar]
- 30.Dahmen G, Ziegler A. Generalized Estimating Equations in Controlled Clinical Trials: Hypotheses Testing. Biometrical Journal. 2004;46(2):214–232. [Google Scholar]
- 31.Halloran ME, Struchiner CJ, Longini IM., Jr Study designs for evaluating different efficacy and effectiveness aspects of vaccines. American Journal of Epidemiology. 1997 Nov 15;146(10):789–803. doi: 10.1093/oxfordjournals.aje.a009196. [DOI] [PubMed] [Google Scholar]
- 32.Robins JM, Rotnitzky A, Zhao LP. Estimation of regression-coefficients when some regressors are not always observed. Journal of the American Statistical Association. 1994;89:846–866. [Google Scholar]
- 33.Kang J, Schafer J. A comparison of alternative strategies for estimating a population mean from incomplete data. Statistical Science. 2007;22:523–539. doi: 10.1214/07-STS227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Laan M, Gruber S. Collaborative double robust targeted maximum likelihood estimation. International Journal of Biostatistics. 2010;6 doi: 10.2202/1557-4679.1181. Article 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens AJ, Tchetgen EJT, De Gruttola V. Augmented GEE for improving efficiency and validity of estimation in cluster randomized trials by leveraging cluster-and individual-level covariates. Statistics in Medicine. 2012;31(10):915. doi: 10.1002/sim.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fay MP, Shaw PA. Exact and asymptotic weighted logrank tests for interval censored data: The interval R package. Journal of Statistical Software. 2010;36(2) doi: 10.18637/jss.v036.i02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang K, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 38.Lu B, Preisser JS, Qaqish BF, Suchindran C, Bangdiwala SI, Wolfson M. A comparison of two bias-corrected covariance estimators for generalized estimating equations. Biometrics. 2007 Sep;63(3):935–941. doi: 10.1111/j.1541-0420.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- 39.Guo X, Pan W, Connett JE, Hannan PJ, French SA. Small-sample performance of the robust score test and its modifications in generalized estimating equations. Statistics in Medicine. 2005;24(22):3479–3495. doi: 10.1002/sim.2161. [DOI] [PubMed] [Google Scholar]
- 40.Pan W. On the robust variance estimator in generalised estimating equations. Biometrika. 2001 Sep;88(3):901–906. [Google Scholar]
- 41.Gosho M, Sato Y, Takeuchi H. Robust covariance estimator for small-sample adjustment in the generalized estimating equations: A simulation study. Science Journal of Applied Mathematics and Statistics. 2014;2(1):20–25. [Google Scholar]
- 42.Wang M, Long Q. Modified robust variance estimator for generalized estimating equations with improved small-sample performance. Statistics in Medicine. 2011;30(11):1278–1291. doi: 10.1002/sim.4150. [DOI] [PubMed] [Google Scholar]
- 43.Pan W, Wall MM. Small-sample adjustments in using the sandwich variance estimator in generalized estimating equations. Statistics in Medicine. 2002;21(10):1429–1441. doi: 10.1002/sim.1142. [DOI] [PubMed] [Google Scholar]
- 44.McCaffrey DF, Bell RM. Improved hypothesis testing for coefficients in generalized estimating equations with small samples of clusters. Statistics in Medicine. 2006;25(23):4081–4098. doi: 10.1002/sim.2502. [DOI] [PubMed] [Google Scholar]
- 45.Fan C, Zhang D, Zhang CH. A comparison of bias-corrected covariance estimators for generalized estimating equations. Journal of Biopharmaceutical Statistics. 2012;23(5):1172–1187. doi: 10.1080/10543406.2013.813521. [DOI] [PubMed] [Google Scholar]
- 46.Paul S, Zhang X. Small sample GEE estimation of regression parameters for longitudinal data. Statistics in Medicine. 2014 doi: 10.1002/sim.6198. doi:10.1002sim.6198. [DOI] [PubMed] [Google Scholar]
- 47.Teerenstra S, Lu B, Preisser JS, van Achterberg T, Borm GF. Sample size considerations for GEE analyses of three-level cluster randomized trials. Biometrics. 2010;66(4):1230–1237. doi: 10.1111/j.1541-0420.2009.01374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.