Keratinocyte stem cell colonies can be identified by analyzing cell motion, an emergent stem cell property.
Abstract
Image-based identification of cultured stem cells and noninvasive evaluation of their proliferative capacity advance cell therapy and stem cell research. Here we demonstrate that human keratinocyte stem cells can be identified in situ by analyzing cell motion during their cultivation. Modeling experiments suggested that the clonal type of cultured human clonogenic keratinocytes can be efficiently determined by analysis of early cell movement. Image analysis experiments demonstrated that keratinocyte stem cells indeed display a unique rotational movement that can be identified as early as the two-cell stage colony. We also demonstrate that α6 integrin is required for both rotational and collective cell motion. Our experiments provide, for the first time, strong evidence that cell motion and epidermal stemness are linked. We conclude that early identification of human keratinocyte stem cells by image analysis of cell movement is a valid parameter for quality control of cultured keratinocytes for transplantation.
Introduction
Ex vivo maintenance and expansion and subsequent transplantation of adult stem cells are indispensable for successful cell therapy of self-renewing tissues, such as the epidermis and cornea epithelium. Adult stem cells maintain their stem cell properties throughout cell culture. After transplantation, they permanently engraft, self-renew, and properly produce functional progenies, which results in long-term therapeutic success (De Luca et al., 2006; Barrandon et al., 2012). Human epidermal keratinocyte stem cells (holoclones; Barrandon and Green, 1987b) can be cultivated under suitable conditions (Rheinwald and Green, 1975), and a single holoclone can generate a progeny large enough to entirely reconstitute the epidermis of an adult human for a lifetime (Rochat et al., 1994; Mathor et al., 1996). This has enabled the autologous transplantation of cultured keratinocytes onto patients with extensive burns (Gallico et al., 1984; Pellegrini et al., 1999; Ronfard et al., 2000) and genetic disorders (Mavilio et al., 2006; De Rosa et al., 2014), and the successful application of human stem cells for regenerative medicine (De Luca et al., 2006; Green, 2008; Barrandon et al., 2012; Fuchs, 2012).
A human keratinocyte culture also contains other clonogenic keratinocytes with restricted growth capabilities (Barrandon and Green, 1987b): progenitor cells (meroclones), which can regenerate an epidermis for a short duration, and transient amplifying cells (paraclones) that cannot regenerate an epidermis at all. Holoclones are eventually converted into meroclones or paraclones during serial cultivation (Barrandon et al., 2012; Rochat et al., 2012), and the loss of holoclones hinders successful transplantation (Rama et al., 2010; Pellegrini et al., 2013). Hence, for regenerative medicine, the determination of number of holoclones in a keratinocyte culture is the best criteria to assess quality (Rama et al., 2010; Barrandon et al., 2012; Rochat et al., 2012; Pellegrini et al., 2013). However, holoclones have been discriminated from meroclones and paraclones by ex post clonal analysis (Barrandon and Green, 1987b) and expression of transcription factor p63 (Pellegrini et al., 2001). A holoclone assay takes 19 d to perform (Barrandon and Green, 1987b; Barrandon et al., 2012), and that has limited its usefulness for regenerative medicine applications.
Expression of cell surface proteins including α1, α2, α4, α6, β1, and β4 integrin subunits, transferin receptor, ATP-binding cassette subfamily G member 2, Delta1, melanoma chondroitin sulfate proteoglycan, and leucine-rich repeats and immunoglobulin-like domains protein 1 (Lrig1) can also be used to identify human keratinocyte stem cells (Jones and Watt, 1993; Jones et al., 1995; Li et al., 1998; Lowell et al., 2000; Legg et al., 2003; Terunuma et al., 2003; Jensen and Watt, 2006; Schlüter et al., 2011). However, none of these cell surface proteins is stem cell specific, and the number of clonogenic keratinocytes in populations enriched by cell sorting based on these cell surface markers is always smaller than that of stem cells simply selected based on size with a Pasteur pipet (up to 28%; Barrandon and Green, 1985). This does not exclude the existence of reliable cell surface markers for human keratinocyte stem cells, but indicates that the cell-sorting procedure might negatively impact the stem cells (discussed in Claudinot et al. [2005] and Barrandon et al. [2012]). Thus, selective cultivation of human keratinocyte stem cells has never been established.
Here, we dissect the motion dynamics of cultured human epidermal keratinocyte stem cells by a combination of motion analysis and physics of multiparticle systems and demonstrate that a keratinocyte stem cell colony can be identified by the analysis of cell motion, an emergent property of the stem cells.
Results
Rotational speed of keratinocytes in the two-cell colony stage is associated with their proliferative capacity
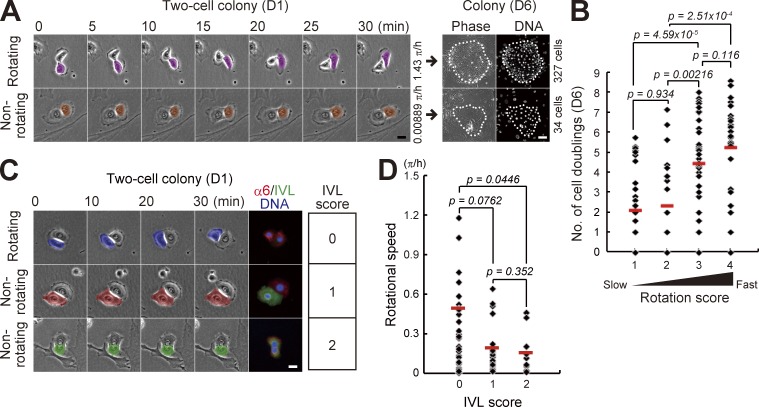
Normal human epidermal keratinocytes formed two-cell colonies by a single cell division of individual cells, which were observed at day 1 after seeding. It has been reported that the number of rotating two-cell colonies of normal human keratinocytes decreases along with passages (Hata et al., 2005); however, the association between the rotational speed and clonal growth remains unknown. We first examined whether the rotational speed of two-cell colonies is associated with proliferative potential in normal human epidermal keratinocytes. Rotational motion of two-cell colonies of keratinocytes was clearly observed 1 d after cells were seeded on one fourth of the normal number of feeder cells (Fig. 1 A and Videos 1 and 2). The rotational speed of each two-cell colony was measured by time-lapse observation. After addition of feeder cells, the colony was further cultivated for 5 d. Each keratinocyte colony was traced, and the number of cells in the colonies was counted (Fig. 1 A). This analysis revealed that rotational speed of two-cell colonies of normal human keratinocytes positively correlated with proliferative capacity (Fig. 1 B and Fig. S1 A). The level of involucrin (IVL) expression is also related to proliferative capacity of keratinocytes (Watt and Green, 1981; Barrandon and Green, 1985). We further found that faster rotating two-cell colonies did not express IVL, whereas the colonies expressing IVL in one or both keratinocytes rarely rotated (Fig. 1, C and D). These data indicate that the keratinocytes with the ability to rotate around the neighbor in the two-cell colony stage also had the ability to proliferate and make a large colony by clonal growth.
Figure 1.
Rotational speed of two-cell colonies of keratinocytes is positively correlated with their proliferative capacity. (A, left) Motion of paired human epidermal keratinocytes. (right) Clonal growth of two-cell colonies was traced and proliferative capacity was evaluated by counting cell number in the colony after 6 d of cultivation with Hoechst 33325. Bars, 20 µm. (B) Correlation between rotational speed in the two-cell colony stage (D1) and proliferative capacity (number of cell doublings) after 6 d of cultivation. Two-cell colonies were categorized in four groups depending on rotational speed: rotation score 1 (<0.125 π/h), rotation score 2 (≥0.125 π/h and <0.25 π/h), rotation score 3 (≥0.25 π/h and <0.5 π/h), and rotation score 4 (≥0.5 π/h). The data shown (n = 126 colonies) were obtained from nine independent experiments. P-value was calculated by Mann-Whitney nonparametric U test. Full set of data is shown in Fig. S1 A. (C) Rotational motion and expression of IVL in two-cell colonies of keratinocytes. IVL expression was categorized in three groups depending on the number of IVL-expressing cells: IVL score 0, no cells; IVL score 1, single cell; and IVL score 2, two cells. Bar, 20 µm. (D) Frequency of two-cell colonies of keratinocytes with each rotation and IVL scores. The data shown (n = 63 colonies) were obtained from triplicate experiments. P-value was calculated by Mann-Whitney nonparametric U test.
We also investigated the motion of keratinocytes grown under conditions that were serum free and did not require feeder layers for keratinocyte growth. In these conditions, individual keratinocytes seeded at clonal density divided normally, but generated two daughter cells that immediately detached from each other and did not make two-cell colonies (Fig. S1 B). Therefore, the correlation between rotational speed in the two-cell colonies of keratinocytes and their clonal growth could not be examined. In high density inoculation, however, keratinocytes formed two-cell colonies and some of them exhibited a rotational motion (Videos 3–5), as reported previously (Hata et al., 2005). This indicates that keratinocytes rotate around the neighboring cells when two cells are in contact with each other, even if maintained in serum-free media.
Rotational motion gives rise to collective dynamics, which predicts epidermal stemness
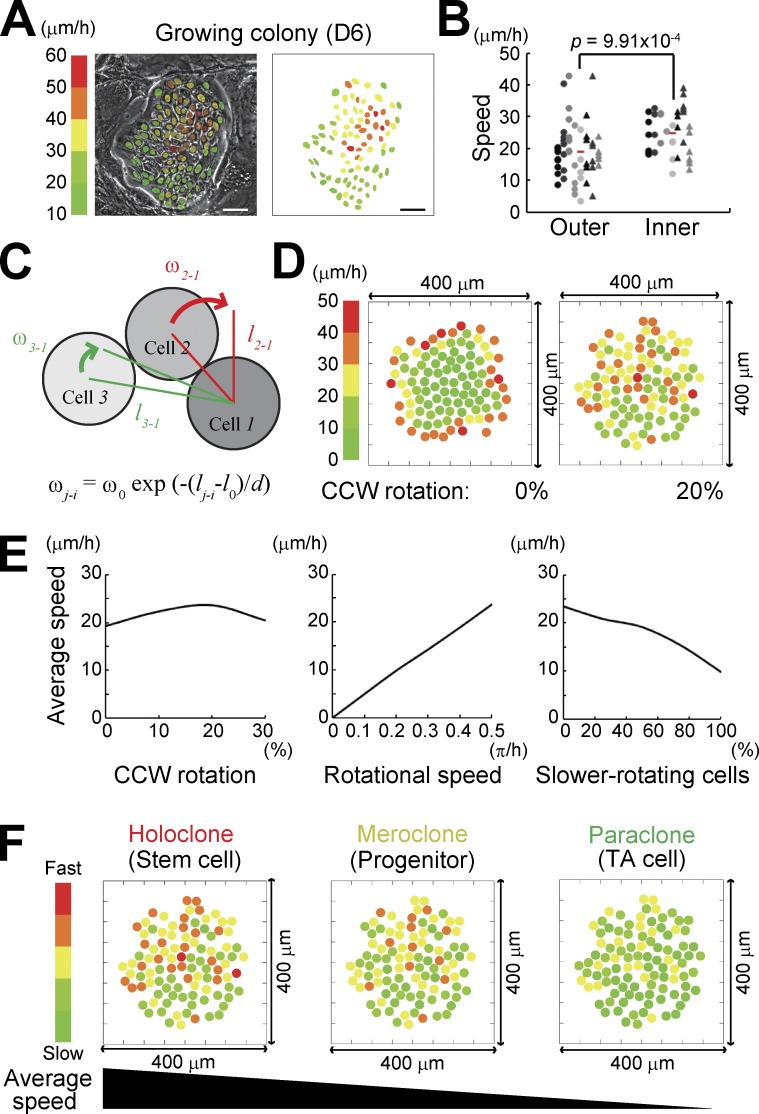
Clonogenic keratinocytes form progressively growing colonies (Barrandon and Green, 1987a,b; Nanba et al., 2013b) that contain more than a hundred cells with differing proliferative capacities (Barrandon and Green, 1987b). In progressive growing colonies, individual keratinocytes spontaneously moved with different locomotion speeds (Fig. 2 A; Fig. S2, A and B; and Video 3); cells located at the inner portion of the colony moved faster than those at the outer portion (Fig. 2 B). We next performed a simulation experiment to analyze whether the collective motion observed in the growing colony could emerge if a hundred cells with a rotational motion accumulated as a colony. In this simulation, all cells were provided with 20 µm of a planer body with 4 µm of variable extension. Hence, the equilibrium distance between the centers of contacting cells was assumed as l0 = 24 µm. As two-cell colonies of proliferative keratinocytes rotated at >0.5 π/h (Fig. 1 B and Fig. S1 A), all cells were provided with the rotational speed ω0 = 0.5 π/h, which means that cells can rotate 0.5 π around the neighboring cells in 1 h when cells are in contact with each other. In this condition, each one of the two cells rotates around the center of two cells at π per hour. We also set an interactive effect of rotation depending on the distance between cells when the number of cells increased as follows. The rotational speed of cell j against cell i, ωj-i, was defined as ωj-i = ω0exp(−(lj-i − l0)/d) (Fig. 2 C). Here, lj-i is the distance between the centers of cell i and cell j and d = l0/2 is a constant that is fixed by comparing the numerical results with observations in culture (Fig. 2, A and B). 99 cells can directly or indirectly interact with and rotate around a cell i (i ≠ j; i, j = 1, 2, 3, … , 100). We made this procedure to all cells; n = 100 in a colony. The positions of all the cells were provided and the locomotion speed of individual cells was calculated.
Figure 2.
In silico reconstitution of keratinocyte stem cell colony dynamics. (A) Distribution of locomotion speed of keratinocytes in a progressively growing colony after 6 d of cultivation. Each color indicates different locomotion speeds that were calculated with 3-h time-lapse observation. (B) Locomotion speed of cells located at the inner and outer portion of progressively growing colonies. Each dot represents the speed of each keratinocyte in the colony. Dots arranged vertically indicate the speed of cells in the same colony. Mean speed of each cell fraction is shown with red bars. Locomotion speed was calculated with 30-min time-lapse observation. 51 outer cells and 35 inner cells in seven progressive growing colonies were analyzed. P-values were calculated by Student’s t test. (C) Two-dimensional modeling of interaction of rotating keratinocytes. Cells were provided with 24 µm (=l0) of a planar diameter and can rotate around the contacting cells at 0.5 π/h (=ω0). The rotational speed of cell j against cell i, ωj-i, was defined as ωj-i = ω0exp(−(lj-i − l0)/d). lj-i is the distance between the centers of cell i and cell j, and d = l0/2 is a constant. i, j = 1, 2, 3, … , 100 and i ≠ j. The input parameters used are described in the Results section. (D) Distribution of locomotion speed of keratinocytes in a progressively growing colony in silico. The speed of each cell was calculated by a simulation experiment based on our modeling (See Materials and methods). (left) All cells constituting the colony rotate clockwise. (right) 20% of cells in the colony rotate CCW. Each color indicates different locomotion speeds. The data shown were obtained from a single representative experiment using a random number generator with three random seeds (triplicate experiments). (E, left) The colony containing 20% of CCW cells showed the highest mean speed of locomotion even if all cells had the same rotational speed (0.5 π/h). (middle) Rotation speed of cells determined cell locomotion speed in the colony. The percentage of CCW rotating cells was fixed at 20%. (right) Variation of locomotion speed of cells was reproduced with a mixture of fast (0.5 π/h) and slow (0.2 π/h) rotating cells. The percentage of CCW rotating cells was fixed at 20%. The data shown were obtained from a single representative experiment using a random number generator with three random seeds (triplicate experiments). (F) Schematic representation of cell locomotion speed in each clonal type. The simulation predicted that a holoclone colony could be identified by measuring cell locomotion speed.
First, when all 100 cells rotated clockwise in the colony in silico, locomotion speed of cells had a gradient along the inner to outer axis of the colony (Fig. 2 D). However, appearance of counterclockwise (CCW) rotating cells generated motion dynamics in the cells located at the inner portion of the colony, and distribution of locomotion speed was almost identical to that of a progressively growing colony in culture (Fig. 2, A, B, and D). The colonies containing 20% of CCW rotating cells showed the maximum mean speed of cell locomotion (Fig. 2 E). Locomotion speed of cells in the colony was completely dependent on rotational speed of cells (Fig. 2 E). Various mixtures of fast (0.5 π/h) and slow (0.2 π/h) rotating cells in the colony gave rise to variations in the mean locomotion speed of cells in the colony (Fig. 2 E). This scenario revealed the possibility of predicting a keratinocyte stem cell colony by measuring cell locomotion speed, if rotational speed of keratinocytes was indeed associated with their proliferative capacity (Fig. 2 F).
Keratinocyte stem cell colonies can be identified by measuring cell locomotion speed
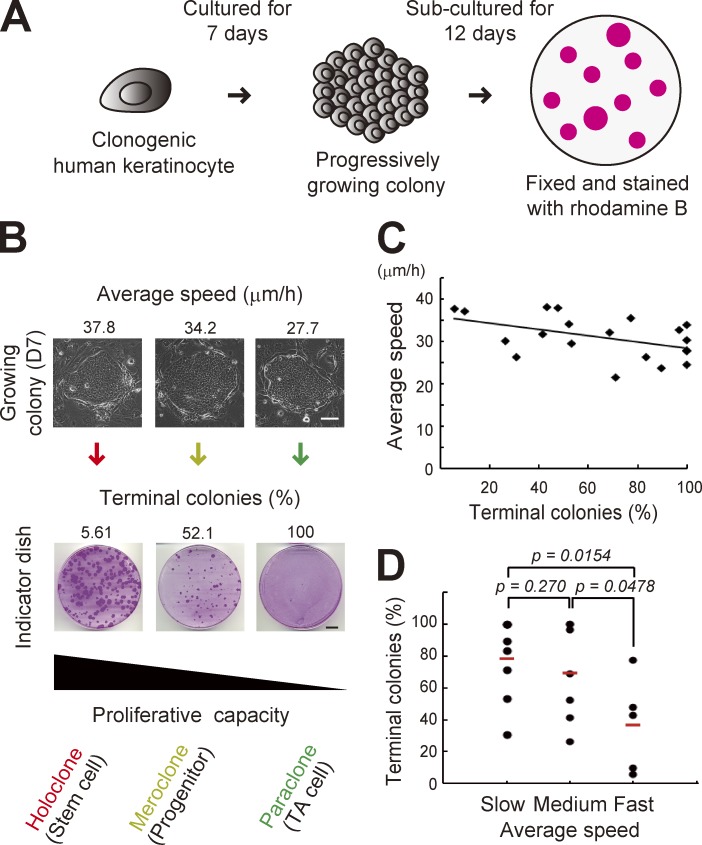
To validate this prediction, we measured cell locomotion speed in each growing colony by time-lapse observation in 7 d of culture, and then assessed proliferative capacity by clonal analysis (Barrandon and Green, 1987b; Fig. 3, A and B). The mean speed of cell locomotion in the colony at day 7 was ∼25–40 µm/h (Fig. 3, B and C), but that in the colony at day 6 and in silico prediction was ∼15–25 µm/h (Fig. 2, B and D), which might be accounted for by the increase of the number of cells. This combination analysis revealed that the ratio of terminal colonies after passages negatively correlated with the mean speed of keratinocyte locomotion in the colony before passages. It also showed that colonies consisting of higher locomotive cells contained keratinocytes with higher growth potential (Fig. 3, C and D), as predicted. Analysis of experiments done in the same day showed stronger correlation between mean speed and ratio of terminal colonies (Fig. S2 C), probably because of the same passage numbers and culture conditions of keratinocytes and 3T3 feeder cells in the experiment. This result indicates that a keratinocyte colony retaining the greatest growth capacity, and which was derived from a holoclone, showed the highest locomotive phenotype among progressively growing colonies of keratinocytes. Simulation experiments revealed that rotational motion of keratinocytes generated collective motion of cells and that rotational speed was correlated with the mean speed of cell locomotion in the colony (Fig. 2). Therefore, the result of this combination analysis confirmed that the rotational speed of keratinocytes was correlated with their proliferative capacity, as shown in Fig. 1, and also strongly suggests that keratinocytes with the highest rotational speed in a culture were keratinocyte stem cells.
Figure 3.
Keratinocyte stem cell colonies can be identified by measuring cell locomotion speed. (A) Clonal analysis of human clonogenic keratinocytes (Barrandon and Green, 1987b). A progressive growing colony derived from a single keratinocyte is subcultured into a 10-cm cell culture dish (indicator dish) so its growing capacity can be evaluated. (B, top) Progressive growing colonies showed variations in the mean cell locomotion speed in the colony after 7 d of cultivation. The mean cell locomotion speed in the colony was obtained by measuring locomotion speed of randomly selected 15–20 cells in each colony. Bar, 100 µm. (bottom) Each growing colony in the top panel was trypsinized and passaged into an indicator dish arranged in the same column. The culture was maintained for 12 d, and then fixed and stained with rhodamine B. Proliferative capacity of growing colonies was assessed by determining the ratio of terminal colonies in the indicator dish. Bars, 10 mm. (C) Correlation of mean speed of cells in each colony and its proliferative capacity (Pearson’s r = −0.477, P = 0.0335, and n = 20 colonies). (D) Growing colonies were categorized in three groups depending on the mean speed of cells: slow (≤30 µm/h), medium (>30 µm/h and ≤35 µm/h), and fast (>35 µm/h). P-values were calculated by Student’s t test.
The expression level of α6 integrin is associated with locomotion speed and proliferative capacity
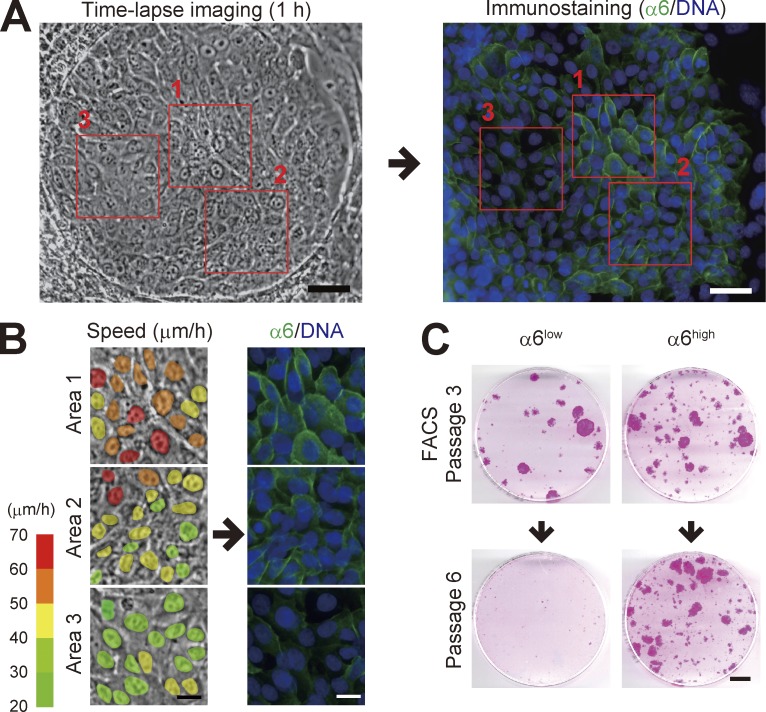
In a progressively growing colony, a variation in integrin α6 subunit expression in individual keratinocytes was observed (Fig. 4 A); keratinocytes with higher α6 integrin expression moved faster than those with lower expression (Fig. 4 B). The level of α6 integrin expression in human keratinocytes is correlated with their proliferative capacity (Li et al., 1998) and clonal types (Fortunel et al., 2010). We next fractionated keratinocytes in progressively growing colonies into α6 integrin high- and low-expressing populations (α6high and α6low). The culture of keratinocytes on a feeder layer of 3T3 cells also contains terminal colonies that have an irregular shape and is mostly composed of large and flattened basal cells (Nanba et al., 2013b). Cellular fractions mainly containing keratinocytes derived from such colonies were identified and discriminated from keratinocytes comprising growing colonies by IVL expression and flow cytometric signatures of each subfraction (Fig. S3). We then found that α6high but not α6low keratinocytes in the progressively growing colony maintained proliferative capacity through serial cultivation (Fig. 4 C). Flow cytometric analysis also revealed that α6high keratinocyte population contained the smallest cells (Fig. S3 D), which possess higher clonogenic ability (Barrandon and Green, 1985). Collectively, these data indicate that keratinocytes with higher expression of α6 integrin in growing colonies possessed both higher locomotive and proliferative capacities.
Figure 4.
Expression of α6 integrin is associated with both locomotive and proliferative capacities. (A) Serial images of a progressive growing colony were collected for 60 min, and then the colony was immediately fixed and stained with an antibody against α6 integrin (17D11). Bars, 50 µm. (B) Magnification of the area outlined by the red boxes in A. The colors in the left column indicate the locomotion speed (µm/h) of each cell in the colony. Bars, 20 µm. (C) Colony-forming efficiency of α6high and α6low keratinocyte population after serial cultivation. For more detail, see Fig. S3 F. Bar, 10 mm.
α6 integrin is required for rotational and collective motion
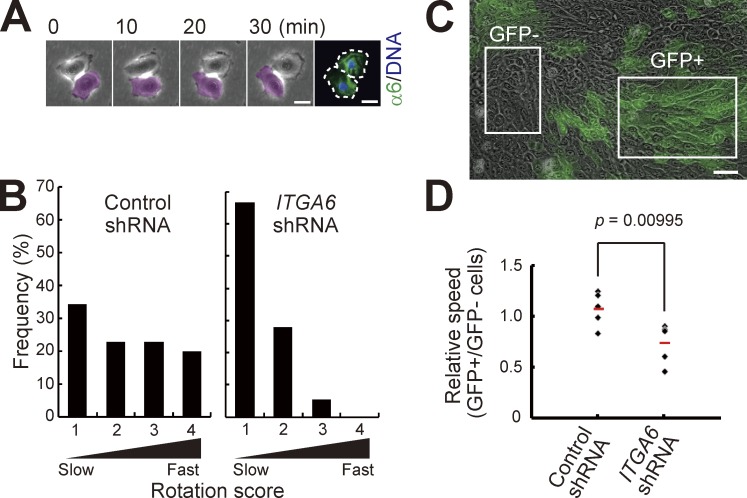
α6 integrin was also expressed in rotating two-cell colonies of normal human epidermal keratinocytes (Fig. 5 A). We next examined whether α6 integrin is involved in the rotational motion of two-cell colonies of keratinocytes by using a doxycyclin-inducible α6 integrin gene (ITGA6)–specific knockdown system (Matsushita et al., 2013; Nanba et al., 2013b). shRNA-expressing cells were easily traced as GFP-positive (GFP+) cells, and a lentivirus carrying shRNA targeted to ITGA6 efficiently decreased α6 expression in human epidermal keratinocytes (Fig. S4, A–D). Rotational speed was significantly decreased in ITGA6, but not control shRNA-transduced two-cell colonies (Fig. 5 B). We further found that locomotion speed of GFP+ cell populations in progressive growing colonies was significantly slower than that of GFP-negative (GFP−) cell populations in colonies infected with a lentivirus carrying shRNA targeted to ITGA6, but not the control shRNA (Fig. 5, C and D). These results indicate that α6 integrin was required for rotational and collective motion of human keratinocytes. Altogether, we revealed that keratinocytes expressing higher amounts of α6 integrin, which is required for rotational motion of keratinocytes, also possessed higher proliferative capacity, which corroborates that the rotational speed of keratinocytes was correlated with their proliferative capacity, as shown in Fig. 1.
Figure 5.
α6 integrin is required for rotational and collective motion. (A) Rotational motion of a two-cell colony of human epidermal keratinocytes and distribution of α6 integrin. Serial images of a two-cell stage colony were collected for 60 min, and then the colony was immediately fixed and stained with an antibody against α6 integrin (17D11). Bars, 20 µm. (B) Distribution of rotational speed of two-cell colonies of keratinocytes transduced with control and ITGA6 shRNA. Rotation score is described in Fig. 1. The data shown (n = 35 and 36 colonies of control shRNA and ITGA6 shRNA-transfected keratinocytes, respectively) were obtained from triplicate experiments. (C) Mixture of ITGA6 shRNA-transduced and nontransduced keratinocytes in a large progressively growing colony. Bar, 50 µm. (D) Effect of α6 integrin knockdown on keratinocyte locomotion in the colony. The locomotion speed of cells was measured after transduction of control and ITGA6 shRNA. shRNA-transduced cells were identified as GFP-positive (GFP+) cells. The values were obtained from analyzing five colonies in each condition. P-values were calculated by Student’s t test.
α6β4 integrin is required for collective motion
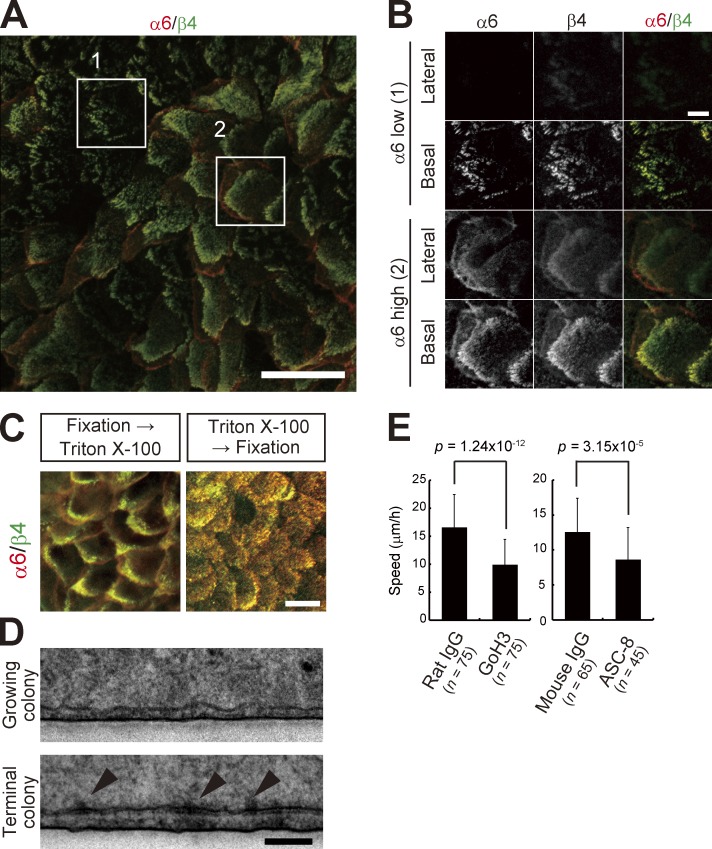
α6 integrin forms a heterodimer with β4 subunit and functions as a surface receptor for laminin-332 (Margadant et al., 2008). We confirmed the polarized expression of α6β4 integrin (Marchisio et al., 1991) at the basal surface of keratinocytes (Fig. 6, A and B). α6β4 integrin can be linked with cytokeratins via cytoplasmic proteins, and cytoskeleton-linked α6β4 integrin is detergent insoluble and can be detected even if cells are treated with detergent before fixation (Rabinovitz et al., 1999). In cultured human epidermal keratinocytes, α6β4 integrin was also detectable when treated with 0.5% Triton X-100 before fixation (Fig. 6 C). Basal, but not lateral, distribution of α6β4 integrin was more evident by Triton X-100 pretreatment (Fig. 6 C). However, electron microscopy demonstrated that hemidesmosomes, which appear as electron-dense cytoplasmic plaques at the basal surface of cells, were only detected in the terminal colony but hardly observed in the progressively growing colony (Fig. 6 D). We further examined whether α6β4 integrin is involved in keratinocyte locomotion. Because of the limited access of inhibitory antibodies to the basal surface of cells in the colony, we investigated keratinocyte movements in two-dimensional cell aggregates, which were formed after high density keratinocyte inoculation (Fig. S5 and Materials and methods). Locomotion speed of keratinocytes incubated with anti-α6 (GoH3) and -β4 integrin (ASC-8) antibodies in two-dimensional aggregates significantly decreased, whereas it was not affected by normal rat and mouse IgG (Fig. 6 E). These data indicate that α6β4 integrin was not organized into anchoring complexes of hemidesmosomes in growing colonies, but was associated with cytokeratins and involved in collective motion of keratinocytes by mediating functional adhesion to extracellular substrates in the progressively growing colony.
Figure 6.
α6β4 integrin is involved in collective motion. (A) A maximum intensity projection image of immunofluorescence against α6 and β4 integrin in a growing colony by confocal microscopy. Bar, 50 µm. (B) Magnification of the area outlined by the white boxes in A. Optical sections show α6β4 integrin on basal and lateral surfaces. Bar, 10 µm. (C) Detergent-insoluble α6β4 integrin on basal surface. α6β4 integrin can be linked with cytokeratins via cytoplasmic proteins, and cytoskeleton-linked α6β4 integrin is detergent insoluble and can be detected even if cells are treated with detergent before fixation (Rabinovitz et al., 1999). In keratinocytes, α6β4 integrin was also detectable when treated with 0.5% Triton X-100 before fixation. Bar, 20 µm. (D) Electron micrographs of the growing and terminal colonies of human epidermal keratinocytes. Arrowheads indicate well-defined electron-dense structures of hemidemosomes in the basal surface. Bar, 500 nm. (E) Effect of inhibitory antibodies against α6 (GoH3) and β4 integrin (ASC-8) on keratinocyte locomotion in keratinocyte aggregates. Two-dimensional keratinocyte aggregates were formed after high density keratinocyte inoculation with antibodies. P-values were calculated by Student’s t test.
Discussion
The proliferative capacity of human epidermal keratinocyte stem cells has been well studied. In contrast, their locomotive capacity has not been well investigated because stem cells are assumed to be anchored in their niche. Our present work has four major findings concerning the motion of human keratinocyte stem cells. First, keratinocytes with higher proliferative capacity show a rotational motion in two-cell stage colonies. Second, the rotational motion of keratinocyte stem cells triggers collective motion of the cells within the colony. Third, collective motion analysis identifies keratinocyte stem cell colonies. Fourth, α6 integrin is involved in rotational and collective motion of keratinocytes.
The mechanism of rotational motion of the two-cell colony of keratinocytes and how it is associated with growth capacity is still unclear. We have previously reported that single keratinocytes can create helical tunnels in fibrin matrix (Ronfard and Barrandon, 2001), which might be related with the rotational motion in two-dimensional culture. Mammary epithelial cells also exhibit rotational motion (Kino-oka et al., 2004), which is required for sphere formation in three-dimensional culture (Tanner et al., 2012). In mammalian epidermis, keratinocyte stem cells are located in the basal layer of the interfollicular epidermis and epidermal appendages (Rochat et al., 1994; Jones et al., 2007; Blanpain and Fuchs, 2009). Lineage tracing experiments have clearly demonstrated the clonal growth of single keratinocyte stem/progenitor cells in the interfollicular epidermis (Clayton et al., 2007; Mascré et al., 2012), but the motion of keratinocyte stem/progenitor cells during homeostasis is still unknown. Importantly, after wounding, keratinocyte stem cells mainly contribute to reepithelialization and long-term regeneration of the epidermis, whereas progenitor cells make a limited contribution (Mascré et al., 2012). Keratinocyte stem cells located in the bulge region of hair follicles also participate in the regeneration of interfollicular epidermis after wounding (Taylor et al., 2000; Oshima et al., 2001; Claudinot et al., 2005; Ito et al., 2005). These studies have suggested that the locomotive capacity of keratinocyte stem cells is higher than that of progenitor cells, resulting in a substantial contribution to epidermal wound healing. This notion has been proven, in part, by our study with cultured human keratinocytes. We have demonstrated here that cultured keratinocyte stem cells lost hemidesmosomal structures, even though they highly expressed α6β4 integrin. This phenotype is also observed in keratinocytes at the leading edge of migrating epidermis after wounding (Krawczyk and Wilgram, 1973; Cavani et al., 1993). Although directed migration is indispensable for wound closure, collective motion generated by keratinocyte stem cells might also be involved in epidermal wound healing.
Image-based identification of cultured stem cells and evaluation of their potency are required for quality assurance of stem cell culture in regenerative medicine (Ker et al., 2011) and also for high-throughput screening in stem cell research (Erdmann et al., 2012). Recent studies have established noninvasive and morphology-based evaluation methods to identify mesenchymal stem/progenitor cell potency (Kino-oka et al., 2009; Seiler et al., 2012; Matsuoka et al., 2013). Maintenance and self-renewal of keratinocyte stem cells in culture are definitely associated with successful transplantation for epidermal regeneration. For regenerative medicine, the determination of number of holoclones in a keratinocyte culture is the best criteria to assess quality (Rama et al., 2010; Barrandon et al., 2012; Rochat et al., 2012; Pellegrini et al., 2013). However, a holoclone assay takes 19 d to perform (Barrandon and Green, 1987b; Barrandon et al., 2012), and that has limited its usefulness for regenerative medicine applications. Image-based noninvasive identification of cultured human keratinocyte stem cells enables us to monitor and validate their successful cultivation in situ, which is essential for engraftment of transplanted cells and the long-term maintenance of the regenerated epidermis and cornea epithelium in the patients (Gallico et al., 1984; Pellegrini et al., 1997, 1999; Ronfard et al., 2000; Rama et al., 2001; Rama et al., 2010). This technology is also available for selection of normal and genetically modified keratinocyte stem cells in cultured keratinocytes derived from the patient’s skin (Mavilio et al., 2006) or differentiated from embryonic stem cells and patient-specific induced pluripotent stem cells for therapy (Green et al., 2003; Metallo et al., 2008; Guenou et al., 2009; Itoh et al., 2011).
Our findings are the basis of a noninvasive method that relies on the analysis of cell motion to estimate the growth capacity of keratinocytes in culture and monitor and validate the quality of these cells for transplantation. In practice, the combination of time-lapse observation of keratinocyte cultures and an image processing technology that can recognize and trace individual cells within the densely packed colonies can achieve in situ identification of keratinocyte stem cells through the automatic measurement of cell locomotion speed. This enables us to monitor whether the culture contains enough keratinocyte stem cells with higher locomotive capacity and eliminate the cultures that are not suitable for transplantation before making confluent keratinocyte sheets. Selective cultivation of keratinocyte stem cells can also be achieved in combination with automated aspiration of stem cell colonies from the feeder cell layer (Schneider et al., 2008). Advances in our simulation based on physics of multiparticle systems, by including the cell division and the growth of a colony, will also contribute to profound understanding of collective dynamics of keratinocyte stem cells during keratinocyte sheet formation. Although further investigations and technological developments are still required, phase-contrast time-lapse imaging with an image processing algorithm will definitely improve the clinical outcome of cell therapy and the efficiency of cell manufacturing for regenerative medicine.
Materials and methods
Cell culture
Normal human epidermal keratinocytes were isolated from neonatal skin. Frozen keratinocytes were thawed and cultivated at clonal density on a feeder layer of irradiated or mitomycin C–treated 3T3-J2 cells, at 37°C and 10% CO2 in a 3:1 mixture of the DMEM and Ham’s F12 medium supplemented with 10% FCS, 5 µg/ml insulin, 0.4 µg/ml hydrocortisone, 10−10 M cholera toxin, and 2 × 10−9 M triiodothyronine, as described previously (Rheinwald and Green, 1975; Rochat et al., 1994). Human epidermal keratinocytes were also cultivated with CnT-PR medium (CELLnTEC), EpiLife medium containing supplement S7 (Life Technologies), and MCDB153 medium containing bovine pituitary extract (Hashimoto et al., 1994). Keratinocytes were used between passages 2 and 7. The medium was changed every 4 d. Cultures were fixed with 3.7% buffered formaldehyde and stained with 1% rhodamine B, and keratinocyte colonies were counted under a binocular microscope (SMZ645; Nikon). Clonal analysis was performed as described previously (Barrandon and Green, 1987b). In brief, human keratinocytes were cultivated at clonal density on a feeder layer of irradiated or mitomycin C–treated 3T3-J2 cells. Well-isolated colonies were then individually trypsinized in a cloning ring and subcultured into a 10-cm cell culture dish so clonal types could be evaluated.
Time-lapse imaging and image processing
Human epidermal keratinocytes were seeded at clonal density in a 35-mm cell culture dish (Corning) with irradiated or mitomycin C–treated 3T3-J2 cells and grown for 6 or 7 d without EGF. For time-lapse imaging, cells were maintained at 37°C and 10% CO2 in a chamber mounted on a microscope (Axiovert 200M; Carl Zeiss). Images were obtained at 5-min intervals, for 30, 60, and 180 min, depending on experimental conditions, with a monochrome camera (AxioCamHR3; Carl Zeiss) using Enhanced Contrast Plan Neofluar 5×/0.16 and 10×/0.30 and LD A-Plan 20×/0.30 objective lenses. Time-lapse images were acquired with AxioVision (Carl Zeiss) and analyzed with MetaMorph (Molecular Devices) or Volocity (PerkinElmer) on the graphic tablet (Wacom). The angle of contact surface between cells in a two-cell colony was given by the top right angle of contact surface against the perpendicular axis. The rotational speed of a two-cell colony was assessed as the amount of change in the angle of the colony.
Immunofluorescence microscopy
Keratinocytes were seeded at clonal density in a 35-mm cell culture dish with irradiated or mitomycin C–treated 3T3-J2 cells and grown for 6 d. The cells were fixed in 4% PFA in PBS at 4°C for 10 min, and subsequently treated with 0.5% Triton X-100 in PBS for 10 min. In the detergent-insoluble experiment, the cells were treated with 0.5% Triton X-100 in PBS for 10 min before fixation. After the treatment with 1% BSA, the cells were incubated with primary antibodies for 1 h at RT or overnight at 4°C, washed in PBS, and subsequently incubated with FITC- and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at RT. After a wash with PBS, cells were incubated with Hoechst 33258 (Molecular Probes), mounted with a fluorescent mounting medium (Dako), and examined with a microscope (Axiovert 200M; Carl Zeiss) with a Plan Apochromat 20×/0.8 objective lens (Carl Zeiss). Confocal imaging was performed with an A1 confocal microscope (Nikon) with Plan Apochromat 20×/0.75 and 40×/0.95 lenses (Nikon), and images were obtained and analyzed with NIS-Elements (Nikon). Primary antibodies used for immunofluorescence microscopy were as follows: a mouse monoclonal antibody against α6 integrin (clone 17D11; gift from Y. Hirako, Nagoya University, Nagoya, Japan), a rat monoclonal antibody against α6 integrin (clone GoH3; BD), a mouse monoclonal antibody against β4 integrin (clone 3E1; EMD Millipore), and a mouse monoclonal antibody against IVL (clone SY5; Abcam). The mouse monoclonal antibody against α6 integrin (clone 17D11) was produced by using the extracellular domain of bovine α6 integrin as the antigen.
Flow cytometry and cell sorting
Trypsinized and resuspended keratinocytes were incubated with a FITC-conjugated rat monoclonal antibody against α6 integrin (GoH3; BD) for 1 h on ice. After three washes with HBSS (+), the cells were resuspended in culture medium, sorted using the FACSAria cell sorter (BD), and collected into culture medium. For IVL experiments, trypsinized and resuspended keratinocytes were incubated with mouse monoclonal antibody against IVL (SY5; Abcam) for 1 h on ice. Keratinocytes were then incubated with phycoerythrin-conjugated goat polyclonal antibody against mouse IgG (Santa Cruz Biotechnology, Inc.) for 1 h on ice. After three washes with HBSS (+), the cells were then resuspended in HBSS (+) and analyzed by using the FACSCan flow cytometer (BD).
Plasmid construction and preparation of lentivirus vectors
A doxycyclin-inducible gene-specific knockdown system using a lentivirus was described previously (Matsushita et al., 2013; Nanba et al., 2013b). The HIV-based self-inactivating lentiviral expression vector plasmid CS-CA-MCS was provided by the Institute of Physical and Chemical Research BioResource Center, Japan. Three plasmids, pTet-On-Advanced vector, pTRE-Tight vector (Takara Bio Inc.), and pcDNA 6.2-GW/EmGFP-miR vector (Invitrogen), were obtained, and their components were used to construct a transgene cassette consisting of two expression units for inducible gene knockdown. The cDNA fragment of reverse Tet-controlled transactivator was recovered from the pTet-On-Advanced vector and ligated into the pIRES vector (Takara Bio Inc.), upstream of the internal ribosome entry site (IRES) sequence. The blasticidin-resisitance gene (Bsd) was ligated into this plasmid, downstream of the IRES sequence, resulting in the generation of prtTA-IRES-Bsd, containing one transgene unit. In contrast, the DraI–PvuII fragment of the pcDNA 6.2-GW/EmGFP-miR vector containing EmGFP cDNA and pre-miRNA sequence was recovered and inserted into the pTRE-Tight vector, downstream of the Tet-responsive promoter. This resulted in the vector pTRE-EmGFP-miR, which contained another transgene unit. Finally, the fragment of the rtTA-IRES-Bsd was inserted into the lentivirus expression vector plasmid CS-CA-MCS, directly downstream of the CAG promoter sequence, and then another unit, the TRE-EmGFP-miR fragment, was inserted into the resulting plasmid between the Bsd and WPRE sequence. The TRE-EmGFP-miR unit was arranged in the opposite direction as the CAG-driven rtTA-IRES-Bsd unit, yielding a new single lentivirus expression plasmid for inducible gene silencing, CS-CA-rtTA-IRBsd-TRE-EmGFP-miR (Fig. S4 A). To construct the lentivirus vector for inducible silencing of the human integrin α6 (ITGA6) gene expression, two synthetic oligonucleotides designed by BLOCK-iT miR RNAi technologies were obtained (Life Technologies). The sequences were 5′-TGCTGTAAAGTGTCTGGAAACGTTGCGTTTTGGCCACTGACTGACGCAACGTTCAGACACTTTA-3′ for Hmi408869_top_ITGA6 and 5′-CCTGTAAAGTGTCTGAACGTTGCGTCAGTCAGTGGCCAAAACGCAACGTTTCCAGACACTTTAC-3′ for Hmi408869_bot_ITGA6. The double-stranded DNA was inserted between the 5′-miR and 3′-miR flanking sequences of the vector plasmid. The shRNA-negative control sequence presented by Life Technologies was used to construct a negative control vector. The sequences used as negative controls were 5′-TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′ for the top strand and 5′-CCTGAAATGTACTGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCGCAGTACATTTC-3′ for the bottom strand. Using these ITGA6-inducible knockdown plasmids, lentiviral particles were prepared by standard transfection procedures and used to introduce the transgene into 2 × 105 cells. After transduction of the transgenes at a multiplicity of infection of 1 as previous described (Nanba et al., 2013a), the cells were treated with 100 nM doxycyline to induce silencing of the ITGA6 gene and subsequently analyzed.
Electron microscopy
A keratinocyte culture containing growing and terminal colonies was fixed in 0.1 M cacodylate buffer, pH 7.3, containing 2.5% glutaraldehyde and 2% PFA, and was then immersed in 1% osmium tetraoxide in the same buffer. Dehydration in a gradient series of ethanol was followed by embedding in PolyBed 812 (Polysciences). Ultrathin sections were cut on a Reichert ultramicrotome (Leica), stained with uranyl acetate and lead citrate, and observed with a electron microscope (1200EX; JEOL).
Two-dimensional keratinocyte aggregation assay
Resuspended keratinocytes were incubated with 20 µg/ml of an inhibitory antibody against α6 (clone GoH3; BD) or β4 integrin (clone ASC-8; EMD Millipore) for 1 h on ice. Cells were then washed with HBSS (+) (Invitrogen) twice, and 5.0 × 104 cells were seeded into a 35-mm cell culture dish (Corning) on irradiated or mitomycin C–treated 3T3-J2 cells. Keratinocytes formed two-dimensional cell aggregates during overnight culture. The aggregates were maintained at 37°C and 10% CO2 in a chamber mounted on a microscope (Axiovert 200M; Carl Zeiss), and then serial images of the aggregates were obtained at 5-min intervals for 30 min. Locomotion speed was measured by analyzing time-lapse images with Volocity (PerkinElmer) on the graphic tablet.
Computer simulation
We defined the rotational motion of a target cell as the rotation of surrounding cells around the target cell. The rotational speed of surrounding cells was set to decrease in function of the distance l from the target cell. All cells in a colony were provided with the ability to rotate at π/2 per hour per cell for the two-cell colony. This arrangement of rotation for each cell results in each one of the two cells rotating at π/h around the center of two cells. We set the functional form of the rotational speed in an exponential form as ω = ω0exp(−(l − l0)/d), where l0 is the equilibrium distance between contacting cells and ω0 and d are parameters to be fixed by comparing with observation. The ω0 has to be smaller, as the number of sticking cells increases with the total number N of cells in a colony. To avoid the overlap of cells, we introduced a process where overlapping cells repel each other and go back to the equilibrium distance by the same amount for the overlapping cells. We made this procedure to all cells in a colony with a time step of 1 min. To avoid the outward motion of cells away from a colony, we introduced a boundary with a circle of radius R from the center of the colony. When a cell tried to move out from the cell boundary, the cell was pushed back. With this set-up, we made a Fortran program to simulate the movement of cells in a colony with various numbers of cells. For the simulation, we took all cells as spheres with the radius of rcell = 10 µm, and also provided the variable extension of e = 2 µm. Hence, the equilibrium distance of contacting cells was assumed as l0 = 24 µm. For the presentation of numerical results, we took ω0 = π/2/h for n = 2 and gradually decrease to π/5/h for n = 100 with d = l0/2, to take into account the effect of sticking of cells controlled by the distance parameter d. We defined the radius of the boundary of a colony as R2 = l02(N/2). All cells were distributed randomly in the colony by using the random number generator. When we introduced cells with opposite or reduced rotational speed by some fraction, we also used the random number generator to confer these parameters to the cells. The Fortran program is provided in the online supplemental material.
Online supplemental material
Fig. S1 shows motion dynamics of keratinocytes seeded at clonal density in various cell culture media. Fig. S2 shows collective motion dynamics of keratinocytes in a progressively growing colony. Fig. S3 shows fractionation of cultured keratinocytes based on α6 integrin and IVL expression. Fig. S4 shows characterization and validation of functional shRNA targeted to ITGA6. Fig. S5 shows that α6β4 integrin is involved in collective motion of keratinocytes. Videos 1 and 2 show the dynamics of two-cell colonies, as shown in Fig. 1 A. Videos 3–5 show motion dynamics of keratinocytes at high density culture in serum-free media CnT-PR, EpiLife, and MCDB153, respectively. Video 6 shows collective motion dynamics of keratinocytes in a progressively growing colony, as shown in Fig. S2. The supplemental program file contains the Fortran program that we used for the simulation experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409024/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201409024.dv.
Supplementary Material
Acknowledgments
We thank Professor H. Green and Japan Tissue Engineering Co., Ltd. for providing 3T3-J2 cells; Drs. F. Gorostidi, Y. Hirako, A. Rochat, Y. Shirakata, D. Tsuruta, and Mr. M. Inoie for valuable discussion and providing materials; Dr. K. Kameda and Mr. M. Shudo for flow cytometry and electron microscopy; and Mrs. S. Matsushita for lentivirus production. We are also grateful to Dr. A.W. Amici for critical reading of the manuscript.
This work was supported by the Japan Society for the promotion of Science KAKENHI grants 22791080 and 24791159, Research Fund for Promotion of Industry-Academia Joint Research and Development from Ehime University to D. Nanba, and the European Community through the seventh framework program (EuroSyStem and OptiStem) to Y. Barrandon.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCW
- counterclockwise
- IVL
- involucrin
References
- Barrandon Y., and Green H.. 1985. Cell size as a determinant of the clone-forming ability of human keratinocytes. Proc. Natl. Acad. Sci. USA. 82:5390–5394 10.1073/pnas.82.16.5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y., and Green H.. 1987a. Cell migration is essential for sustained growth of keratinocyte colonies: the roles of transforming growth factor-α and epidermal growth factor. Cell. 50:1131–1137 10.1016/0092-8674(87)90179-6 [DOI] [PubMed] [Google Scholar]
- Barrandon Y., and Green H.. 1987b. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA. 84:2302–2306 10.1073/pnas.84.8.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y., Grasset N., Zaffalon A., Gorostidi F., Claudinot S., Droz-Georget S.L., Nanba D., and Rochat A.. 2012. Capturing epidermal stemness for regenerative medicine. Semin. Cell Dev. Biol. 23:937–944 10.1016/j.semcdb.2012.09.011 [DOI] [PubMed] [Google Scholar]
- Blanpain C., and Fuchs E.. 2009. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10:207–217 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavani A., Zambruno G., Marconi A., Manca V., Marchetti M., and Giannetti A.. 1993. Distinctive integrin expression in the newly forming epidermis during wound healing in humans. J. Invest. Dermatol. 101:600–604 10.1111/1523-1747.ep12366057 [DOI] [PubMed] [Google Scholar]
- Claudinot S., Nicolas M., Oshima H., Rochat A., and Barrandon Y.. 2005. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc. Natl. Acad. Sci. USA. 102:14677–14682 10.1073/pnas.0507250102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E., Doupé D.P., Klein A.M., Winton D.J., Simons B.D., and Jones P.H.. 2007. A single type of progenitor cell maintains normal epidermis. Nature. 446:185–189 10.1038/nature05574 [DOI] [PubMed] [Google Scholar]
- De Luca M., Pellegrini G., and Green H.. 2006. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen. Med. 1:45–57 10.2217/17460751.1.1.45 [DOI] [PubMed] [Google Scholar]
- De Rosa L., Carulli S., Cocchiarella F., Quaglino D., Enzo E., Franchini E., Giannetti A., De Santis G., Recchia A., Pellegrini G., and De Luca M.. 2014. Long-term stability and safety of transgenic cultured epidermal stem cells in gene therapy of junctional epidermolysis bullosa. Stem Cell Reports. 2:1–8 10.1016/j.stemcr.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann G., Volz C., and Boutros M.. 2012. Systematic approaches to dissect biological processes in stem cells by image-based screening. Biotechnol. J. 7:768–778 10.1002/biot.201200117 [DOI] [PubMed] [Google Scholar]
- Fortunel N.O., Cadio E., Vaigot P., Chadli L., Moratille S., Bouet S., Roméo P.H., and Martin M.T.. 2010. Exploration of the functional hierarchy of the basal layer of human epidermis at the single-cell level using parallel clonal microcultures of keratinocytes. Exp. Dermatol. 19:387–392 10.1111/j.1600-0625.2009.01046.x [DOI] [PubMed] [Google Scholar]
- Fuchs E.2012. The impact of cell culture on stem cell research. Cell Stem Cell. 10:640–641 10.1016/j.stem.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallico G.G. III, O’Connor N.E., Compton C.C., Kehinde O., and Green H.. 1984. Permanent coverage of large burn wounds with autologous cultured human epithelium. N. Engl. J. Med. 311:448–451 10.1056/NEJM198408163110706 [DOI] [PubMed] [Google Scholar]
- Green H.2008. The birth of therapy with cultured cells. BioEssays. 30:897–903 10.1002/bies.20797 [DOI] [PubMed] [Google Scholar]
- Green H., Easley K., and Iuchi S.. 2003. Marker succession during the development of keratinocytes from cultured human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 100:15625–15630 10.1073/pnas.0307226100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenou H., Nissan X., Larcher F., Feteira J., Lemaitre G., Saidani M., Del Rio M., Barrault C.C., Bernard F.X., Peschanski M., et al. 2009. Human embryonic stem-cell derivatives for full reconstruction of the pluristratified epidermis: a preclinical study. Lancet. 374:1745–1753 10.1016/S0140-6736(09)61496-3 [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Higashiyama S., Asada H., Hashimura E., Kobayashi T., Sudo K., Nakagawa T., Damm D., Yoshikawa K., and Taniguchi N.. 1994. Heparin-binding epidermal growth factor-like growth factor is an autocrine growth factor for human keratinocytes. J. Biol. Chem. 269:20060–20066. [PubMed] [Google Scholar]
- Hata N., Agatahama Y., Kino-oka M., and Taya M.. 2005. Relations between individual cellular motions and proliferative potentials in successive cultures of human keratinocytes. Cytotechnology. 47:127–131 10.1007/s10616-005-3749-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Liu Y., Yang Z., Nguyen J., Liang F., Morris R.J., and Cotsarelis G.. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11:1351–1354 10.1038/nm1328 [DOI] [PubMed] [Google Scholar]
- Itoh M., Kiuru M., Cairo M.S., and Christiano A.M.. 2011. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 108:8797–8802 10.1073/pnas.1100332108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.B., and Watt F.M.. 2006. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. USA. 103:11958–11963 10.1073/pnas.0601886103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.H., and Watt F.M.. 1993. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 73:713–724 10.1016/0092-8674(93)90251-K [DOI] [PubMed] [Google Scholar]
- Jones P.H., Harper S., and Watt F.M.. 1995. Stem cell patterning and fate in human epidermis. Cell. 80:83–93 10.1016/0092-8674(95)90453-0 [DOI] [PubMed] [Google Scholar]
- Jones P.H., Simons B.D., and Watt F.M.. 2007. Sic transit gloria: farewell to the epidermal transit amplifying cell? Cell Stem Cell. 1:371–381 10.1016/j.stem.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Ker D.F., Weiss L.E., Junkers S.N., Chen M., Yin Z., Sandbothe M.F., Huh S.I., Eom S., Bise R., Osuna-Highley E., et al. 2011. An engineered approach to stem cell culture: automating the decision process for real-time adaptive subculture of stem cells. PLoS ONE. 6:e27672 10.1371/journal.pone.0027672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino-oka M., Agatahama Y., Hata N., and Taya M.. 2004. Evaluation of growth potential of human epithelial cells by motion analysis of pairwise rotation under glucose-limited condition. Biochem. Eng. J. 19:109–117 10.1016/j.bej.2003.11.004 [DOI] [Google Scholar]
- Kino-oka M., Maeda Y., Sato Y., Maruyama N., Takezawa Y., Khoshfetrat A.B., Sugawara K., and Taya M.. 2009. Morphological evaluation of chondrogenic potency in passaged cell populations. J. Biosci. Bioeng. 107:544–551 10.1016/j.jbiosc.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Krawczyk W.S., and Wilgram G.F.. 1973. Hemidesmosome and desmosome morphogenesis during epidermal wound healing. J. Ultrastruct. Res. 45:93–101 10.1016/S0022-5320(73)90035-X [DOI] [PubMed] [Google Scholar]
- Legg J., Jensen U.B., Broad S., Leigh I., and Watt F.M.. 2003. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development. 130:6049–6063 10.1242/dev.00837 [DOI] [PubMed] [Google Scholar]
- Li A., Simmons P.J., and Kaur P.. 1998. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc. Natl. Acad. Sci. USA. 95:3902–3907 10.1073/pnas.95.7.3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S., Jones P., Le Roux I., Dunne J., and Watt F.M.. 2000. Stimulation of human epidermal differentiation by Delta–Notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10:491–500 10.1016/S0960-9822(00)00451-6 [DOI] [PubMed] [Google Scholar]
- Marchisio P.C., Bondanza S., Cremona O., Cancedda R., and De Luca M.. 1991. Polarized expression of integrin receptors (α6β4, α2β1, α3β1, and αvβ5) and their relationship with the cytoskeleton and basement membrane matrix in cultured human keratinocytes. J. Cell Biol. 112:761–773 10.1083/jcb.112.4.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant C., Frijns E., Wilhelmsen K., and Sonnenberg A.. 2008. Regulation of hemidesmosome disassembly by growth factor receptors. Curr. Opin. Cell Biol. 20:589–596 10.1016/j.ceb.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Mascré G., Dekoninck S., Drogat B., Youssef K.K., Broheé S., Sotiropoulou P.A., Simons B.D., and Blanpain C.. 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 489:257–262 10.1038/nature11393 [DOI] [PubMed] [Google Scholar]
- Mathor M.B., Ferrari G., Dellambra E., Cilli M., Mavilio F., Cancedda R., and De Luca M.. 1996. Clonal analysis of stably transduced human epidermal stem cells in culture. Proc. Natl. Acad. Sci. USA. 93:10371–10376 10.1073/pnas.93.19.10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka F., Takeuchi I., Agata H., Kagami H., Shiono H., Kiyota Y., Honda H., and Kato R.. 2013. Morphology-based prediction of osteogenic differentiation potential of human mesenchymal stem cells. PLoS ONE. 8:e55082 10.1371/journal.pone.0055082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita N., Matsushita S., Hirakawa S., and Higashiyama S.. 2013. Doxycycline-dependent inducible and reversible RNA interference mediated by a single lentivirus vector. Biosci. Biotechnol. Biochem. 77:776–781 10.1271/bbb.120917 [DOI] [PubMed] [Google Scholar]
- Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C., et al. 2006. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 12:1397–1402 10.1038/nm1504 [DOI] [PubMed] [Google Scholar]
- Metallo C.M., Ji L., de Pablo J.J., and Palecek S.P.. 2008. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 26:372–380 10.1634/stemcells.2007-0501 [DOI] [PubMed] [Google Scholar]
- Nanba D., Matsushita N., Toki F., and Higashiyama S.. 2013a. Efficient expansion of human keratinocyte stem/progenitor cells carrying a transgene with lentiviral vector. Stem Cell Res Ther. 4:127 10.1186/scrt338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D., Toki F., Matsushita N., Matsushita S., Higashiyama S., and Barrandon Y.. 2013b. Actin filament dynamics impacts keratinocyte stem cell maintenance. EMBO Mol. Med. 5:640–653 10.1002/emmm.201201839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima H., Rochat A., Kedzia C., Kobayashi K., and Barrandon Y.. 2001. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 104:233–245 10.1016/S0092-8674(01)00208-2 [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Traverso C.E., Franzi A.T., Zingirian M., Cancedda R., and De Luca M.. 1997. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 349:990–993 10.1016/S0140-6736(96)11188-0 [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Ranno R., Stracuzzi G., Bondanza S., Guerra L., Zambruno G., Micali G., and De Luca M.. 1999. The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin. Transplantation. 68:868–879 10.1097/00007890-199909270-00021 [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., and De Luca M.. 2001. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA. 98:3156–3161 10.1073/pnas.061032098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G., Rama P., Matuska S., Lambiase A., Bonini S., Pocobelli A., Colabelli R.G., Spadea L., Fasciani R., Balestrazzi E., et al. 2013. Biological parameters determining the clinical outcome of autologous cultures of limbal stem cells. Regen. Med. 8:553–567 10.2217/rme.13.43 [DOI] [PubMed] [Google Scholar]
- Rabinovitz I., Toker A., and Mercurio A.M.. 1999. Protein kinase C–dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146:1147–1160 10.1083/jcb.146.5.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rama P., Bonini S., Lambiase A., Golisano O., Paterna P., De Luca M., and Pellegrini G.. 2001. Autologous fibrin-cultured limbal stem cells permanently restore the corneal surface of patients with total limbal stem cell deficiency. Transplantation. 72:1478–1485 10.1097/00007890-200111150-00002 [DOI] [PubMed] [Google Scholar]
- Rama P., Matuska S., Paganoni G., Spinelli A., De Luca M., and Pellegrini G.. 2010. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 363:147–155 10.1056/NEJMoa0905955 [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G., and Green H.. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 6:331–343 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- Rochat A., Kobayashi K., and Barrandon Y.. 1994. Location of stem cells of human hair follicles by clonal analysis. Cell. 76:1063–1073 10.1016/0092-8674(94)90383-2 [DOI] [PubMed] [Google Scholar]
- Rochat A., Grasset N., Gorostidi F., Lathion S., and Barrandon Y.. 2012. Regeneration of epidermis from adult human keratinocyte stem cells. Handbook of Stem Cells. Vol. 2 Atala A., and Lanza R., editors. Academic Press, London. 767–780. [Google Scholar]
- Ronfard V., and Barrandon Y.. 2001. Migration of keratinocytes through tunnels of digested fibrin. Proc. Natl. Acad. Sci. USA. 98:4504–4509 10.1073/pnas.071631698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfard V., Rives J.M., Neveux Y., Carsin H., and Barrandon Y.. 2000. Long-term regeneration of human epidermis on third degree burns transplanted with autologous cultured epithelium grown on a fibrin matrix. Transplantation. 70:1588–1598 10.1097/00007890-200012150-00009 [DOI] [PubMed] [Google Scholar]
- Schlüter H., Paquet-Fifield S., Gangatirkar P., Li J., and Kaur P.. 2011. Functional characterization of quiescent keratinocyte stem cells and their progeny reveals a hierarchical organization in human skin epidermis. Stem Cells. 29:1256–1268 10.1002/stem.675 [DOI] [PubMed] [Google Scholar]
- Schneider A., Spitkovsky D., Riess P., Molcanyi M., Kamisetti N., Maegele M., Hescheler J., and Schaefer U.. 2008. “The good into the pot, the bad into the crop!”—a new technology to free stem cells from feeder cells. PLoS ONE. 3:e3788 10.1371/journal.pone.0003788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C., Gazdhar A., Reyes M., Benneker L.M., Geiser T., Siebenrock K.A., and Gantenbein-Ritter B.. 2012. Time-lapse microscopy and classification of 2D human mesenchymal stem cells based on cell shape picks up myogenic from osteogenic and adipogenic differentiation. J. Tissue Eng. Regen. Med. 8:737–746. [DOI] [PubMed] [Google Scholar]
- Tanner K., Mori H., Mroue R., Bruni-Cardoso A., and Bissell M.J.. 2012. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc. Natl. Acad. Sci. USA. 109:1973–1978 10.1073/pnas.1119578109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G., Lehrer M.S., Jensen P.J., Sun T.T., and Lavker R.M.. 2000. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 102:451–461 10.1016/S0092-8674(00)00050-7 [DOI] [PubMed] [Google Scholar]
- Terunuma A., Jackson K.L., Kapoor V., Telford W.G., and Vogel J.C.. 2003. Side population keratinocytes resembling bone marrow side population stem cells are distinct from label-retaining keratinocyte stem cells. J. Invest. Dermatol. 121:1095–1103 10.1046/j.1523-1747.2003.12531.x [DOI] [PubMed] [Google Scholar]
- Watt F.M., and Green H.. 1981. Involucrin synthesis is correlated with cell size in human epidermal cultures. J. Cell Biol. 90:738–742 10.1083/jcb.90.3.738 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.