Abstract
To expand the capabilities of whole-cell biocatalysis, we have engineered Escherichia coli to produce various esters. The alcohol O-acyltransferase (ATF) class of enzyme uses acyl-CoA units for ester formation. The release of free CoA upon esterification with an alcohol provides the free energy to facilitate ester formation. The diversity of CoA molecules found in nature in combination with various alcohol biosynthetic pathways allows for the biosynthesis of a multitude of esters. Small to medium volatile esters have extensive applications in the flavor, fragrance, cosmetic, solvent, paint and coating industries. The present work enables the production of these compounds by designing several ester pathways in E. coli. The engineered pathways generated acetate esters of ethyl, propyl, isobutyl, 2-methyl-1-butyl, 3-methyl-1-butyl and 2-phenylethyl alcohols. In particular, we achieved high-level production of isobutyl acetate from glucose (17.2 g l−1). This strategy was expanded to realize pathways for tetradecyl acetate and several isobutyrate esters.
The natural occurrence of small (C4–C12) volatile esters in many ripening fruits and flowers serves an important role in attracting animals and for protection against pathogens1. Some fungi such as yeast also generate small volatile esters during fermentation (i.e., wine and beer)2. Various foods and beverages contain added flavor esters to enhance their taste, and the fragrance and cosmetic industries add small esters to achieve a particular fruity or floral scent3. In 2012, the global market for flavors and fragrances was $16.6 billion (http://www.ialconsultants.com/). Small esters are also used for solvents, coatings and paints3.
Low-molecular-weight esters are commonly produced by acid-catalyzed esterification synthesis of an organic acid with an alcohol3,4. In water, the reaction thermodynamically favors hydrolysis of the ester at ambient temperatures (ΔG = −5 kcal mol−1)5. To overcome this, conventional water-based esterification relies on high temperatures and acidic conditions to shift the equilibrium toward ester formation3. Enzymatic catalysis using an esterase or lipase also suffers from the same thermodynamic disadvantage as chemical synthesis6,7.
Whole-cell biocatalysts are increasingly attractive as a renewable method for producing chemicals and fuels8. Using an intact microorganism as a catalyst offers several advantages over conventional synthesis, such as high enantioselectivity and regioselectivity8,9. Another advantage of whole-cell biocatalysis is the ability to achieve multipart syntheses, whereby multiple intermediates generated in parallel in the same vessel are combined into a final product.
Another feature of whole-cell biocatalysis is the ability to catalyze reactions under ambient temperatures and in aqueous solutions. This advantage can also be realized in ester synthesis. The ATF10 and wax ester synthases/acyl-CoA:diacylglycerol acyltransferase11 class of enzymes use acyl-CoA units as the acid component for ester formation (Fig. 1a). This reaction takes advantage of the stored energy in the thioester bond of the acyl-CoA molecule, which has a higher energy of hydrolysis than oxygen esters (ΔG = −7.5 kcal mol−1 versus ΔG = −5 kcal mol−1)5. Thus, the release of free CoA upon esterification of the acyl group with an alcohol provides the free energy needed to facilitate ester formation in water at ambient temperatures. Wax esters occur widely in nature, and the biochemistry has been extensively studied11–17. Here, we explore the production of small volatile esters (C4–C11) from E. coli.
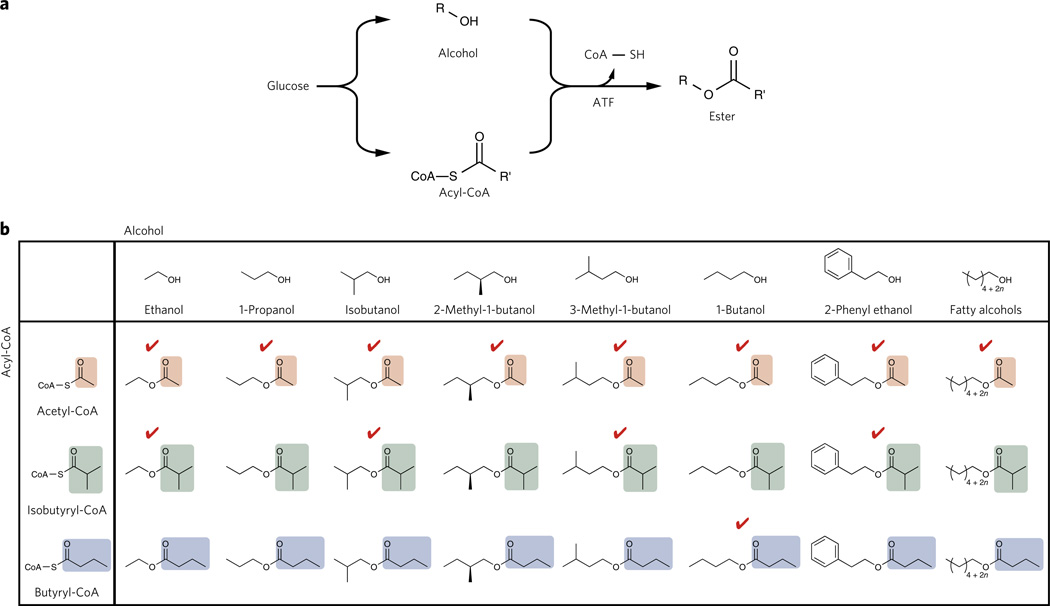
Figure 1. Enzymatic ester synthesis by combining alcohols and acyl-CoAs.
(a) Biological ester synthesis. Various alcohol and acyl-CoA biosynthesis pathways from glucose or other carbon sources have been discovered and/or developed in microorganisms. ATF produces esters by combining alcohols and acyl-CoAs. (b) Matrix of esters by a combination of alcohols and acyl-CoAs. In this study, productions of three types of esters (acetate ester, isobutyrate ester and butyrate ester) were shown. Engineered E. coli has an ability to produce the alcohols and the acyl-CoAs in this figure and, thus, has the potential to produce every ester in this table. Red check mark means the ester was synthesized by an engineered E. coli in this report.
Acetyl-CoA is the most abundant acyl-CoA unit in the cell and, thus, can readily be used to generate a wide range of acetate esters by generating alcohols in the presence of a suitable ATF (Fig. 1b). Efficient C3–C10 alcohol production systems have been developed18–22. Small volatile ester formation by several ATFs (Atf1, Atf2, Eht1 and Eeb1) by Saccharomyces cerevisiae during beer and wine fermentation is well characterized2,10,23–26.
This method can also be applied to production of long-chain alkyl acetate esters (C14–C18), which have applications in cosmetics. In addition, long-chain alkyl acetate esters are similar in size, structure and density to fatty acid ethyl esters or fatty acid methyl esters and therefore may be viable replacements for diesel fuel8. Currently, there are several pathways by which fatty aldehydes can be generated: the LuxCDE system from luminescent bacteria27, fatty acyl reductases from plants and bacteria11,28 and acyl-ACP reductases from cyanobacteria29.
Importantly, this chemistry can be further extended to using larger acyl-CoA units, such as isobutyryl-CoA and butyryl-CoA. In many organisms, branched-chain CoAs are intermediates in the degradation of branched-chain amino acids30. These products are formed by the branched-chain keto acid dehydrogenase complex (KDHC)30. Fatty branched-chain CoAs such as phytanoyl-CoA can be generated from isoprenoid-derived fatty acids by an isoprenoid/acyl-CoA synthetase12,15. The present focus is on generating branched chain-CoA molecules via amino acid biosynthesis pathways for medium chain ester biosynthesis. Given the existence of these metabolic pathways, a matrix of small esters can be theoretically synthesized using whole-cell biocatalysis by combinatorially pairing acyl-CoA pathways with alcohol pathways in the presence of a suitable ATF (Fig. 1b).
Here, we demonstrated the production of several acetate esters using the well-established branched-chain keto acid–based alcohol pathways. In particular, we employed the isobutanol pathway with Atf1 from S. cerevisiae to produce 17.2 g l−1 isobutyl acetate at a yield of 80% of the theoretical maximum. Then we demonstrated the production of the long-chain alkyl ester, tetradecyl acetate, using the luminescence pathway from Vibrio harveyi. Finally, we developed a pathway to generate branched-chain CoA molecules using the KDHC system from Pseudomonas putida and explore the potential to form various isobutyrate esters.
RESULTS
Developing biological routes to acetate esters in E. coli
With acetyl-CoA, a variety of acetate esters can be made in combination with alcohol production pathways. Several alcohols (i.e., ethanol, isopropanol, isobutanol and 1-butanol) have been produced in high yield and titer18–21, allowing, in principle, for similar yields of esters using a suitable ATF (Fig. 1b). As wild-type E. coli is only naturally capable of generating a single alcohol, ethanol, we first developed a minimal platform to generate several alcohols as a means to evaluate the potential of acetate ester production. We used E. coli strain JCL260 (ΔadhE Δfrd ΔldhA Δpta ΔpflB Δfnr)18 as a production host (Supplementary Results, Supplementary Table 1), which lacks all of the major fermentative pathways. This provides increased pyruvate for acetate ester formation.
Next, to generate a pool of alcohols, we took advantage of well-established 2-keto acid–based alcohol biosynthesis18. A promiscuous keto acid decarboxylase (Kdc) can be used to generate the corresponding aldehydes (Fig. 2a)18. These aldehydes are subsequently converted to alcohols by endogenous NAD(P)H-dependent aldehyde reductases/alcohol dehydrogenases (Adhs)31. Thus, kivD from Lactococcus lactis32 was expressed in E. coli to convert pyruvate, 2-ketobutyrate, 2-ketoisovalerate, 2-keto-3-methylvalerate, 2-keto-4-methylvalerate and phenylpyruvate into ethanol, 1-propanol, isobutanol, 2-methyl-1-butanol, 3-methyl-1-butanol and 2-phenylethanol, respectively.
Figure 2. Constructing acetate ester synthesis pathways in E. coli.
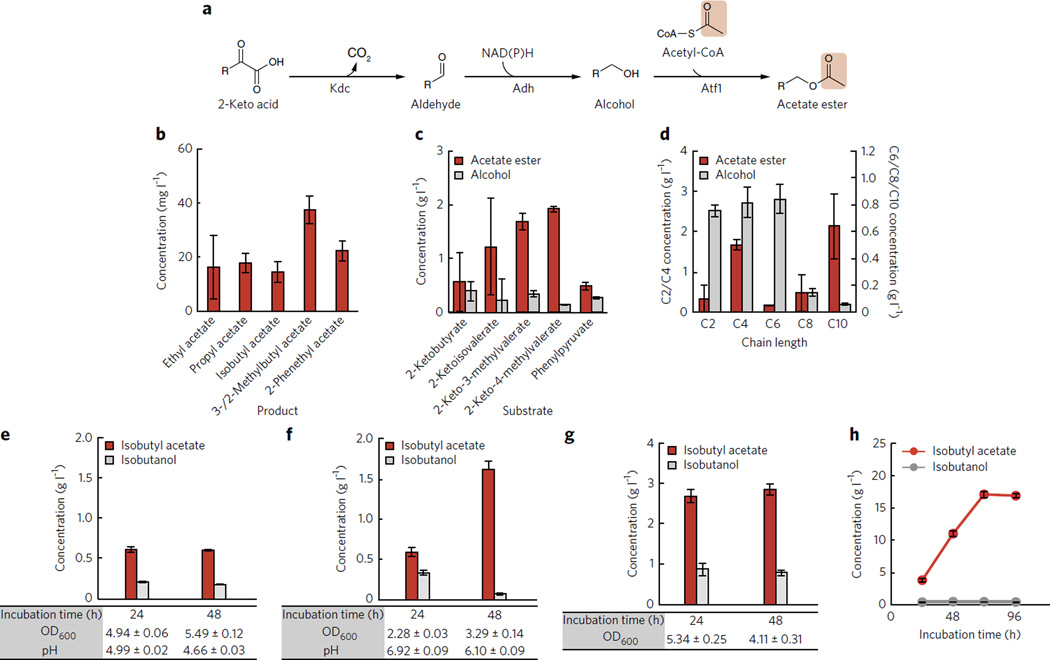
(a) In vivo acetate ester production from 2-keto acid. 2-Keto acid is converted to alcohol, and then acetate ester is synthesized by combining the alcohol and an acetyl-CoA. (b) In vivo production of a mixture of acetate esters from glucose in strain 1 (Table 1). (c) In vivo conversion of individually fed 2-keto acids to acetate esters in strain 1. (d) Production of acetate ester by feeding individual straight-chain alcohols (C2–C10) in strain 2 (Table 1). (e–g) Production of isobutyl acetate in strain 3 (e), strain 4 (f) and strain 5 (g) (Table 1). (h) Production of isobutyl acetate from glucose by strain 5 (Table 1) in 250-ml screw-cap flasks containing a hexadecane layer. Concentrations of isobutyl acetate and isobutanol are in red and gray, respectively in e–h. All of the strains were induced at D600 nm ~0.4 with 1 mM IPTG. Error bars represent s.d. (n = 3).
We introduced kivD and a codon-optimized ATF1 (S. cerevisiae), the minimal set of genes necessary to produce acetate esters in E. coli, into JCL260 (Table 1; strain 1). This strain was incubated for 24 h in M9 minimal medium containing 50 g l−1 glucose and 5 g l−1 yeast extract (hereby referred to as M9P). This strain produced all of the expected esters: ethyl acetate, propyl acetate, isobutyl acetate, 2-methyl-butyl acetate, 3-methyl-butyl acetate and 2-phenylethyl acetate (Fig. 2b). Roughly 15–20 mg l−1 of each ester was detected, totaling ~100 mg l−1. This demonstrated that Atf1 was active in E. coli and had broad substrate specificity for alcohols. No esters were detected from a strain lacking Kdc (Table 1; strain 2), confirming that both genes are required for ester formation.
Table 1.
List of strains used in this study
| Strain no. |
E. coli strain |
ColE1 plasmids | P15Aplasmids |
|---|---|---|---|
| 1 | JCL260 | PLlacO1: kivD-ATF1 | – |
| 2 | JCL260 | PLlacO1: ATF1 | – |
| 3 | JCL88 | PLlacO1: kivD-ATF1 | PLlacO1: alsS-ilvCD |
| 4 | JCL260 | PLlacO1: kivD-ATF1 | PLlacO1: alsS-ilvCD |
| 5 | JCL260 | PLlacO1: alsS-ilvCD, PLlacO1: kivD-adhA | PLlacO1: ATF1 |
| 6 | AL1050 | ATF1 | PLlacO1: luxCDE |
| 7 | AL1050 | – | PLlacO1: luxCDE |
| 8 | AL704 | PLlacO1: luc | – |
| 9 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | – |
| 10 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | cmR; PLlacO1: mRFP1 |
| 11 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | cmR; PLlacO1: EEB1 |
| 12 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | cmR; PLlacO1: EHT1 |
| 13 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | cmR; PLlacO1: ATF1 |
| 14 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | kanR; PLlacO1: mRFP1 |
| 15 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | kanR; PLlacO1: EEB1 |
| 16 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | kanR; PLlacO1: EHT1 |
| 17 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | kanR; PLlacO1: ATF1 |
| 18 | AL704 | PLlacO1: bkdA1-bkdA2-bkdB-lpdV | kanR; PLlacO1: cat |
| 19 | JCL260 | PLlacO1: EHT1, PLlacO1:bkdA1-bkdA2-bkdB-lpdV | PLlacO1: alsS-ilvCD |
| 20 | JCL260 | PLlacO1: cat, PLlacO1:bkdA1-bkdA2-bkdB-lpdV | PLlacO1: alsS-ilvCD |
| 21 | AL704 | PLlacO1: kivD-mhpF | cmR; PLlacO1: mRFP1 |
| 22 | AL704 | PLlacO1: kivD-mhpF | cmR; PLlacO1: EEB1 |
| 23 | AL704 | PLlacO1: kivD-mhpF | cmR; PLlacO1: EHT1 |
| 24 | AL704 | PLlacO1: kivD-mhpF | cmR; PLlacO1: ATF1 |
To test the ability of this pathway to convert high concentrations of substrates, we fed 3 g l−1 of each 2-keto acid into the culture (Fig. 2c). The amount of acetate ester conversion as well as the remaining alcohol in the medium was measured after 24 h. By introducing 2-keto acids, we observed 0.5–2 g l−1 of each acetate ester (Fig. 2c). This indicated that Atf1 was capable of converting the alcohols to esters efficiently if increased flux to the alcohols is provided. This also showed that the strain produced sufficient acetyl-CoA, and, thus, overexpression of acetyl-CoA–generating enzymes such as pyruvate dehydrogenase complex was not necessary for this level of productivity (~2 g l−1 in 24 h). These experiments also defined the relative rates for different alcohols; branched-chain alcohols underwent >80% conversion to the acetate ester, whereas 1-propanol and 2-phenylethanol only underwent ~50% conversion after 24 h (Fig. 2c)
As straight-chain alcohols of C4–C10 have been previously generated using reverse β-oxidation22, we explored the specificity of Atf1 toward these (C2–C10) using a similar feeding experiment (Fig. 2d). Ethyl acetate formation was relatively low (330 mg l−1). Atf1 is known to produce ethyl acetate but does so to a lesser extent than Atf2 (ref. 26). Butyl acetate was formed at a higher rate than propyl acetate (1.7 g l−1 versus 0.57 mg l−1). For C6–C10 alcohols, it was only possible to add 500–750 mg l−1 into the culture owing to solubility and toxicity. The E. coli strain was not able to grow in the presence of 500 mg l−1 hexanol, and very little hexyl acetate was produced. Octyl acetate formation was also low (140 mg l−1). In contrast, ~600 mg l−1 decyl acetate was formed (>80% conversion), and the culture was able to grow normally. Thus, it appeared that Atf1 was capable of forming acetate esters from a broad range of substrates, and the strain was able to provide ample amounts of acetyl-CoA for the acetate ester formation.
Optimization of isobutyl acetate production from glucose
After demonstrating high flux acetate ester conversion from 2-keto acids by expressing kivD and ATF1 in JCL260, we set out to achieve efficient acetate ester production from a single renewable carbon source. As isobutanol production was demonstrated to approach maximal theoretical yields33 with titers up to 50 g l−1 (ref. 34), we evaluated the performance of isobutyl acetate production from glucose (Fig. 2e–h and Supplementary Fig. 1).
The high productivity of isobutanol production created concerns that acetyl-CoA availability might be limiting for isobutyl acetate production. Thus, we tested two different E. coli strains as production hosts for their ability to produce isobutyl acetate: JCL260 and JCL88 (Supplementary Table 1)18. The difference between the two strains is that JCL88 contains pyruvate formate lyase encoded by pflB, which may provide increased availability of acetyl-CoA. We introduced the isobutanol pathway (AlsS (from Bacillus subtilis), IlvCD (E. coli), Kdc (L. lactis)) and Atf1 (S. cerevisiae) into JCL88 and JCL260 (Table 1; strain 4). We excluded the previously used Adh step as it has been shown that endogenous Adhs efficiently carry out the reduction of isobutyraldehyde to isobutanol31. In M9P supplemented with MOPS buffer (to maintain pH 7), both strains produced 0.6 g l−1 isobutyl acetate after 24 h (Fig. 2e,f). However, at 48 h, strain 4 (JCL260) produced 1.6 g l−1 isobutyl acetate and converted nearly all of the isobutanol to the ester, whereas strain 3 (JCL88) did not produce any additional isobutyl acetate after 24 h. Compared to strain 3, strain 4 (JCL260) exhibited higher titer, higher ester/alcohol ratio and better pH stability. Thus, we used JCL260 as a production host for further analyses.
On the basis of the high ester/alcohol ratio, we suspected that isobutanol production was limiting. To test this, we introduced the entirety of the isobutanol pathway (including adhA (L. lactis))35 onto a high copy plasmid (with a ColE1 origin of replication, ~40 copies per cell) while moving ATF1 to a medium copy plasmid (p15A, ~15 copies) (Table 1; strain 5). In principle, this allows high-level expression of the isobutanol pathway and also introduces adhA (L. lactis) to further increase flux to isobutanol. This strain produced 4.5-fold more isobutyl acetate (2.7 g l−1) in the first 24 h compared to strain 4 (Fig. 2f,g). This production represents 60% of the theoretical maximum yield (0.42 g isobutyl acetate per g glucose). However, from 24–48 h, the strain only marginally increased the titer to 3.0 g l−1, and the cell density dropped 20% (D600 nm 5.34 to 4.11; Fig. 2g). This suggested toxicity of isobutyl acetate. Indeed, toxicity experiments revealed that E. coli was unable to grow in the presence of 3 g l−1 isobutyl acetate, undergoing cell lysis (Supplementary Fig. 2).
To alleviate isobutyl acetate toxicity and further increase titer and yield, we incorporated a hexadecane layer to our production culture to achieve in situ product removal. This two-layer method has been successfully used to extract toxic products such as 3-methyl-1-butanol from cultures36. We chose hexadecane because it does not contain hydrogen-bonding elements. This makes isobutanol and acetate less soluble in the hexadecane layer and isobutyl acetate more soluble, facilitating its removal. Hexadecane is also nontoxic to E. coli, has low miscibility and cannot be degraded by E. coli. Thus, we repeated production with strain 5 (Table 1) and added an equal volume of hexadecane over the culture.
The use of a hexadecane layer enabled strain 5 to produce 3.9 g l−1 isobutyl acetate in 24 h (Fig. 2h), an increase of 44% compared to the same strain without the use of a hexadecane layer (Fig. 2g). Furthermore, 97% of the isobutyl acetate was extracted into the hexadecane (Fig. 2h), indicating that the choice of solvent was functioning as intended to successfully remove product from the culture. In contrast, isobutanol partitioning displayed the opposite behavior; 86% remained in the culture (Supplementary Fig. 1b).
Production continued into 48 h and 72 h, accumulating 11.3 g l−1 and 17.5 g l−1 isobutyl acetate, respectively. Production stopped after 72 h, with the titer remaining stable at 17.2 g l−1 at 96 h. Final glucose consumption was measured at 96 h, resulting in an isobutyl acetate yield of 0.334 g per g glucose (17.2 g l−1 isobutyl acetate produced per 51.6 g l−1 glucose consumed), which is 80% of the theoretical maximum from glucose (Supplementary Table 2). We also observed no degradation of isobutyl acetate by E. coli JCL260 when incubated with isobutyl acetate for 24 h (Supplementary Table 3). We expect that production of other esters using efficient pathways (for example, 1-butanol19,22) will also give high titers and yields of esters such as butyl acetate or butyl butyrate using this platform.
Production of tetradecyl acetate from glucose in E. coli
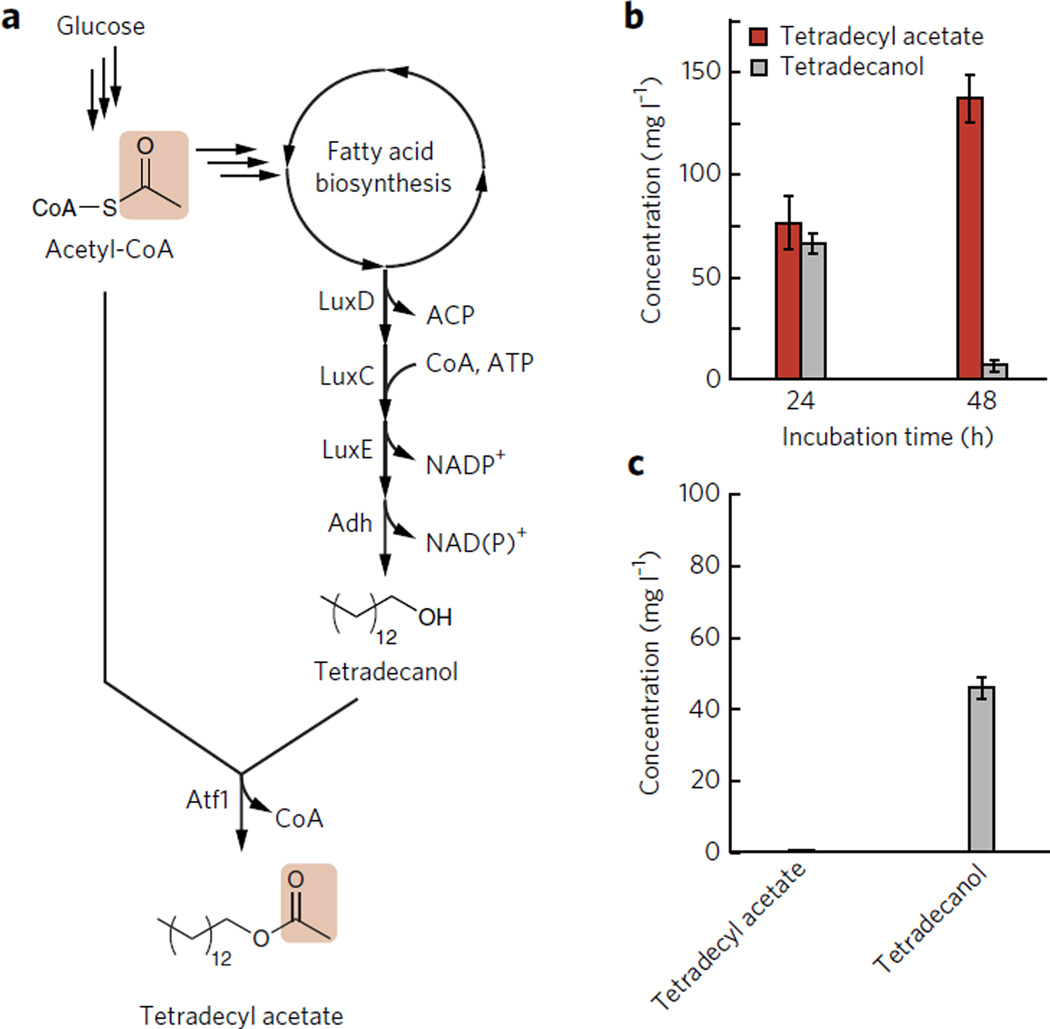
To demonstrate the viability of this method for a wider range of acetate ester products, we next designed a system for production of long-chain (>C10) alkyl acetate esters. We expressed the LuxCDE enzyme system from V. harveyi37,38 along with Atf1 in E. coli strain AL1050 (Table 1; strain 6) (Fig. 3a). The LuxCDE system is known to produce tetradecyl aldehyde specifically, which is converted to tetradecyl alcohol by E. coli38. Strain AL1050 produced 137 mg l−1 tetradecyl acetate after 48 h with 95% tetradecyl alcohol converted to ester (Fig. 3b). With a strain lacking ATF1 (Table 1; strain 7), only tetradecyl alcohol was produced (50 mg l−1) (Fig. 3c).
Figure 3. Tetradecyl acetate production from glucose in E. coli.
(a) Biosynthesis pathway for tetradecyl acetate from glucose. LuxDCEcomplex produces tetradecyl aldehyde from tetradecyl-ACPin the fatty acid biosynthesis cycle. E. coli Adh converts the tetradecyl aldehyde to tetradecanol. (b,c) Consequently, tetradecyl acetate is synthesized by Atf1 from tetradecanol and acetyl-CoA. Production of tetradecyl acetate from glucose in strain 6 (b) and strain 7 (c) (Table 1). All of the strains were induced at D600 nm ~0.4 with 1 mM IPTG. Error bars represent s.d. (n = 3).
Developing branched-chain CoA pathways for higher esters
With a full range of acetate esters produced from native pools of acetyl-CoA, production of more complex esters necessitates first producing higher-chain (either longer-chain or branched) CoA units (Fig. 1b). Production of longer straight-chain CoA units has been achieved by extensive manipulation of E. coli metabolic pathways: reverse β-oxidation was previously used to generate CoA units in lengths from butyryl-CoA up to decanoyl-CoA22. Here, we developed a pathway to generate branched-chain CoA molecules from amino acid biosynthesis.
As Kdc already can generate various aldehydes, these aldehydes can be converted to acyl-CoAs by an acylating aldehyde dehydrogenase such as MhpF (E. coli), which converts acetaldehyde to acetyl-CoA and generates NADH39,40. It can also act on butanal with lower activity22,39. Thus, with Kdc and MhpF, a 2-keto acid can potentially be converted to the corresponding acyl-CoA (Supplementary Fig. 3a).
Alternatively, in many organisms, branched-chain CoAs such as isobutyryl-CoA, isovaleryl-CoA and 3-methylvaleryl-CoA are intermediates in the degradation of l-valine, l-leucine and l-isoleucine, respectively30. These products are formed by the branched-chain KDHC30 (Supplementary Fig. 3a), which is analogous to the pyruvate dehydrogenase complex (encoded by aceEF-lpd) (Supplementary Fig. 4).
We introduced the full KDHC operon of P. putida41,42 into AL704 (JCL260 with ΔyqhD ΔadhP ΔeutG ΔyiaY ΔyjgB ΔfucO)31 (Table 1; strain 9), which exhibits low endogenous aldehyde reductase/alcohol dehydrogenase activity. By feeding 3 g l−1 of various 2-keto acids into the culture, the resulting products were observed and compared to a strain without KDHC (Table 1; strain 8). Strain 9 generated 150–300 mg l−1 isobutanol, 2-methyl-1-butanol and 3-methyl-1-butanol, whereas only ~75 mg l−1 of 1-butanol was produced (Fig. 4a). No alcohols were detected from strain 8. This suggested that the KDHC from P. putida was active in E. coli. This experiment also revealed that these branched-chain CoA products were being converted to aldehydes and alcohols by an unknown enzyme (or enzymes) as adhE was removed from AL704.
Figure 4. Constructing higher chain ester biosynthesis pathway in E. coli.
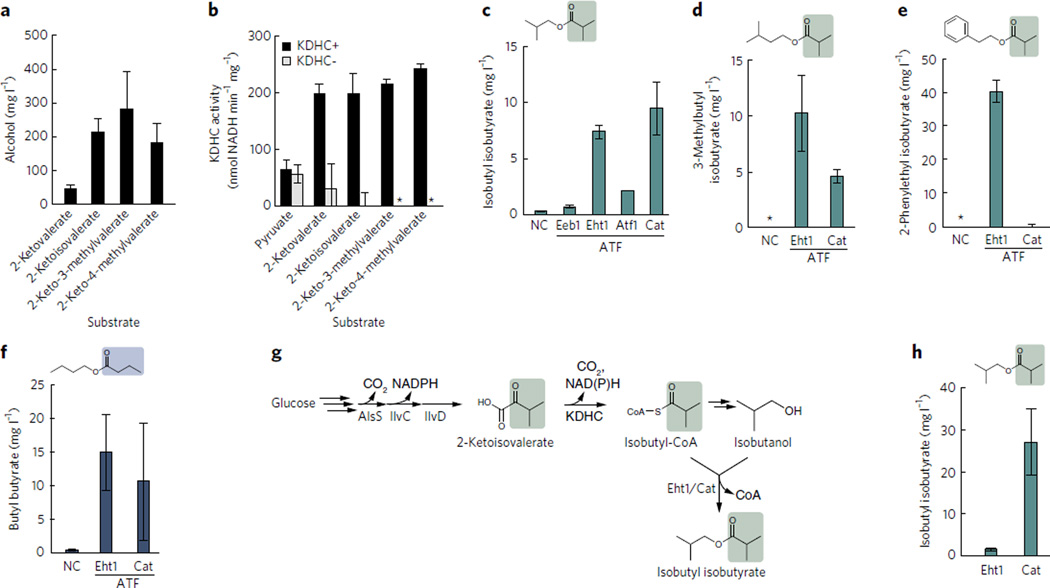
KDHC activity for 2-keto acids was tested in vivo (a) and in vitro (b). For isobutyrate ester production, strains 14 (negative control (NC)), 15 (Eeb1), 16 (Eht1), 17 (Atf1) and 18 (Cat) (Table 1) were grown in M9Pmedia containing 3 g l−1 2-ketoisovalerate and 3 g l−1 isobutanol (c), 3 g l−1 3-methyl-1-butanol (d) or 3 g l−1 2-phenylethanol (e) for 24 h. For butyrate ester, strain 14 (NC), 16 (Eht1), and 18 (Cat) were grown in M9Pmedia with 3 g l−1 2-ketovalerate and 3 g l−1 1-butanol for 24 h (f). To produce isobutyl isobutyrate from glucose, alsS, ilvCD, the KDHCgenes and ATF (EHT1 or cat) were expressed in same cell (g). Strains 19 (Eht1) and 20 (Cat) (Table 1) were grown in M9Pmedium containing 10 g l−1 glucose for 24 h (h). All of the strains were induced at D600 nm ~0.4 with 1 mM IPTG. In c–e, asterisks indicate compound was not detectable. Error bars represent s.d. (n = 3).
To confirm the activity of KDHC, cell lysates from strain 8 and 9 were prepared, and activity was tested in vitro with pyruvate, 2-ketovalerate, 2-ketoisovalerate, 3-methyl-ketovalerate or 4-methylketovalerate as substrate. Activity was measured by detecting the reduction of NAD+ in the presence of CoA. Activity of KDHC with each of the 2-keto acid substrates was similar (~225 nmol NADH per min per mg protein) except for pyruvate (67 nmol NADH per min per mg protein) (Fig. 4b). The negative control strain showed nearly identical activity for pyruvate as the other strains, indicating that activity toward pyruvate was coming from the endogenous pyruvate dehydrogenase complex. A small amount of activity (30 nmol NADH per min per mg protein) with 2-ketovalerate was observed from the negative control, perhaps owing to slight promiscuity of the pyruvate dehydrogenase complex (Fig. 4b).
Characterizing ATFs for isobutyrate ester biosynthesis
Having now designed a method for production of branched-chain CoAs, we could use it to develop a method for production of isobutyrate esters. We characterized three ATFs (Eeb1, Eht1 and Atf1) for their ability to produce isobutyl isobutyrate from isobutyryl-CoA and isobutanol in E. coli. We fed 3 g l−1 2-ketoisovalerate and 3 g l−1 isobutanol into the cultures of the strains expressing the KDHC genes and either EEB1, EHT1, ATF1 or mRFP1 (negative control) (Table 1; strains 11, 12, 13 and 10, respectively). We also tested whether Kdc-MhpF could replace KDHC for production of isobutyryl-CoA by measuring isobutyl isobutyrate production from strains expressing Kdc-MhpF and one of the same ATFs (Table 1; strains 21, 22, 23 and 24, respectively). Of the strains harboring Kdc-MhpF, only the strain with Eht1 showed a small amount (~0.5 mg l−1) of isobutyl isobutyrate (Supplementary Fig. 3b). In contrast, all of the strains expressing the KDHC pathway formed 1–5 mg l−1 isobutyl isobutyrate (Supplementary Fig. 3b), including the negative control. The strains with EHT1 and ATF1 showed the highest production of isobutyl isobutyrate. However, the Atf1 strain also produced >1 g l−1 isobutyl acetate, indicating a high preference of Atf1 for acetyl-CoA (Supplementary Fig. 3c). This high preference for acetyl-CoA makes Atf1 a poor choice for higher-chain CoA ester formation (Supplementary Fig. 3c). The Eht1 strain produced only a few mg l−1 of isobutyl acetate, suggesting low activity of Eht1 toward acetyl-CoA (Supplementary Fig. 3c).
The isobutyl isobutyrate formation was unexpectedly detected with the negative control (Supplementary Fig. 3b). Coincidentally, the plasmids carrying the ATF and mRFP1 genes contained the chloramphenicol resistance gene (cat) encoding chloramphenicol acetyltransferase, which is the same class of enzyme as the ATFs and was also recently shown to have activity toward some aromatic alcohols43. To determine whether Cat was involved in isobutyl isobutyrate formation, we replaced cat with a kanamycin resistance (kanR) gene (Table 1; strains 15, 16, 17 and 14). We also placed cat under the PLlacO1 promoter (Table 1; strain 18) to investigate the formation of ester from a strain with increased cat expression. As expected, the new negative control strain did not produce isobutyl isobutyrate, whereas the strain with cat under control of PLlacO1 (strain 18) showed the highest isobutyl isobutyrate formation (9.5 mg l−1) (Fig. 4c). The Eht1 strain (strain 16) produced 7.4 mg l−1 isobutyl isobutyrate in this condition. Thus, we found that Cat was best among the ATFs tested for isobutyl isobutyrate formation.
To test the alcohol substrate specificity of Cat and Eht1 with respect to isobutyl-CoA as a co-substrate, we added 2-ketoisovalerate and either 3-methyl-1-butanol or 2-phenethanol to cultures and measured the isobutyrate esters produced. Although Cat was the best ATF for isobutyrate ester production with isobutanol (Fig. 4c), Eht1 was the best ATF for isobutyrate ester production with the larger substrates 3-methyl-1-butanol and 2-phenethanol (Fig. 4d,e). Only a small amount of ethyl isobutyrate was detected from strain 16, which contains Eht1 (0.1 mg l−1) (Supplementary Fig. 3d). Cat only produced low levels of (4.6 mg l−1) 3-methylbutyl isobutyrate and did not produce any detectable 2-phenylethyl isobutyrate from the larger co-substrate isobutyryl-CoA. In contrast, 3-methyl-1-butanol and 2-phenylethanol were good substrates for Eht1, allowing production of 10 mg l−1 3-methylbutyl isobutyrate and 40 mg l−1 2-phenethyl isobutyrate, respectively (Fig. 4d,e). Cat and Eht1 both preferred acetyl-CoA over isobutyryl-CoA as a co-substrate, producing 100–400 mg l−1 3-methyl-1-butyl acetate (Supplementary Fig. 5a) and 2-phenylethyl acetate (Supplementary Fig. 5b). Although Eht1 is known as ethanol hexanoyl transferase23, according to the present results, Eht1 preferred larger alcohols as substrates. These varying titers (5–40 mg l−1) suggested that the activity of these ATFs with large substrates was limiting, rather than the supply of isobutyryl-CoA from KDHC. An ATF with higher activity toward C4–C6 CoAs could improve the titers of these pathways.
Butyl butyrate is valuable as a solvent and flavoring44 and has been successfully produced from engineered Clostridium acetobutylicum45. As the KDHC used in this study was able to accept 2-ketovalerate as a substrate (Fig. 4b), it suggests an alternative means to effectively generate butyryl-CoA besides reverse β-oxidation. This would allow application of our biochemical strategy to produce butyl butyrate, using KDHC as a source of the required straight-chain CoA, butyryl-CoA. Thus, we fed 3 g l−1 each of 1-butanol and 2-ketovalerate into cultures of E. coli expressing the KDHC genes and either EHT1 or cat. After 24 h, the strains with EHT1 or cat produced 14.9 mg l−1 and 10.6 mg l−1 butyl butyrate, respectively (Fig. 4f). Butyl butyrate was not detected from strains without an ATF.
Exploring isobutyl isobutyrate from glucose in E. coli
Up to this point we have only described strains capable of isobutyrate ester production from added alcohols and keto acids. Thus, the next step is to simplify the system by constructing a strain able to generate complex esters using only simple sugars. As the isobutanol pathway23 is capable of generating high flux to 2-ketoisovalerate, it is ideal for generating branched chain-CoA in the form of isobutyryl-CoA. This, in turn, would allow a biological route to produce the symmetric ester isobutyl isobutyrate (Fig. 4g). Isobutyl isobutyrate has a water solubility of just 520 mg l−1. This would allow for bilayer formation at high titers, which would simplify product removal during or after production and limit the potential toxicity of the ester to E. coli.
We introduced the first half of the isobutanol pathway (AlsS and IlvCD) on a medium copy plasmid, with the expected rate-limiting steps (KDHC and either Cat or Eht1) on a high copy plasmid (Table 1; strains 19 and 20, respectively). Our characterization of KDHC indicated that isobutyryl-CoA was endogenously converted to isobutyraldehyde and isobutanol (Fig. 4a). Thus, an AdhE-type enzyme, which would convert isobutyryl-CoA to isobutyraldehyde and then isobutanol, was excluded from the pathway.
Production was carried out using 5-ml cultures in screw-cap tubes. After 24 h, strain 20 (Cat) produced 27.1 mg l−1 isobutyl isobutyrate, which is much higher than the 1.4 mg l−1 produced by strain 19 (Eht1) (Fig. 4h). More than 1 g l−1 of isobutanol remained in the supernatant (data not shown), suggesting that the ATFs characterized in this work, though functional, had weak activity toward branched-chain CoAs. We observed no degradation of isobutyl isobutyrate by E. coli after 24 h (Supplementary Table 4). This strategy would represent a good starting point for production of a range of larger esters from simple substrates using whole-cell catalysts.
DISCUSSION
Thioester bonds enable thermodynamically favorable whole-cell synthesis of esters. The diversity of CoA thioesters found in biological systems in combination with well-engineered alcohol biosynthetic pathways allows for the biosynthesis of a wide variety of esters (Fig. 1b). The biochemistry of ATFs complements the well-established pathways to produce alcohols from C2 up to C18. Although microbial production of wax esters has already been explored11–17, this study bridges the gap with the biosynthetic production of short and medium chain esters. These small to medium volatile esters have extensive applications in the flavor, fragrance, cosmetic, solvent, paint and coating industries3.
Achieving high titer and yield is paramount to commercialization of whole-cell biological production. To accomplish this, careful consideration of pathway construction, enzyme mechanisms and enzymatic activity are needed. Irreversible reactions have a large role in the success of high-flux biological pathways19,20. In the isobutanol pathway, AlsS and Kdc achieve irreversibility via CO2 loss, and both enzymes are also highly active18. Acetyl-CoA formation also involves the irreversible decarboxylation of pyruvate. The final step of esterification capitalizes on the stored energy of the thioester bond to provide the driving force necessary to favor ester formation in water at ambient temperatures. Tuning the pathway at the genetic level by changing copy numbers of various genes successfully eliminated a bottleneck in isobutanol formation. These engineering steps enabled the high titer and yield of isobutyl acetate achieved in this work (Fig. 2h).
Despite the successful acetate esters production, further optimization is required for medium-chain esters. Although C6–C10 CoA formation has been previously demonstrated, industrially relevant productivity has yet to be achieved for this range of molecules22. We introduced KDHC to generate branched-chain CoAs, which generated similar product titers, as previously demonstrated by alternate methods22 (Fig. 4a). This pathway requires further improvement to achieve higher branched-chain CoA flux. Similarly, the collection of ATFs that were tested proved inadequate to achieve high titers of isobutyl isobutyrate. Characterization of additional ATFs or engineering of existing ones is needed to realize high titers of larger esters such as isobutyl isobutyrate. Several wax ester synthases have been characterized16 that may better accept the bulkier isobutyl groups, yet few medium chain alcohol acyltransferases have been discovered. Although the ATFs tested in this study are only weakly active toward higher chain CoA molecules, these higher ester pathways are now possible with adequate flux of alcohol and CoA molecules.
ONLINE METHODS
Reagents
All enzymes were purchased from New England Biolabs. All synthetic oligonucleotides were ordered from Integrated DNA Technologies. DNA sequencing services were done by Davis Sequencing. All chemicals for gas chromatography (GC) standards except for ethanol (VWR) and tetradecyl acetate (Ark Pharm, Inc.) were purchased from Sigma Aldrich. 2-Keto acids (pyruvate (≥99%), 2-ketobutyrate (≥95%), 2-ketovalerate (≥98%), 2-ketoisovalerate (≥95%), 2-keto-3-methylvalerate (≥98%), 2-keto-4-methylvalerate (≥98%) and phenylpyruvate (≥98%)) were purchased from Sigma-Aldrich.
Plasmid construction, cloning and transformations
All plasmids and oligonucleotides are listed in Supplementary Tables 1 and 5, respectively. The target gene(s) and vector fragments were amplified with the pairs of primers from the templates listed in Supplementary Table 6. The resulting fragments were combined by sequence and ligation-independent cloning (SLIC)46. Vector PCR product was treated with DpnI for 1 h at 37 °C. A 10 µl reaction containing 1× NEB Buffer 2, 100–1000 ng of vector and insert fragments, and 0.75 U of T4 DNA Polymerase (NEB) was incubated at room temperature for 10 min, then placed on ice to stop the reaction. 2.5 µl of the solution was used for transformation of E. coli. Plasmids were verified by colony PCR, by digestion with restriction enzymes, and by sequencing.
Cell culturing
Overnight cultures were grown in 5 ml Luria Broth (LB) (Fisher BioReagents) containing appropriate antibiotics. Antibiotic concentrations were as follows: kanamycin (50 µg/ml) (IBI Scientific), chloramphenicol (40 µg/ml) (Fisher BioReagents), ampicillin 250 (µg/ml) (Fisher BioReagents), tetracycline (20 µg/ml) (Fisher BioReagents). Production was carried out with M9 medium (33.7 mM Na2HPO4, 22 mM KH2PO4, 8.55 mM NaCl, 9.35 mM NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2) (BD Bacto), 5 g l−1 yeast extract (BD Bacto), 50 g l−1 or 10 g l−1 glucose (Fisher BioReagents), and 1,000-fold dilution of A5 trace metal mix (2.86 g H3BO3 (Fisher Chemical), 1.81 g MnCl2·4H2O (MP Biomedicals), 0.222 g ZnSO4·7H2O (Sigma-Aldrich), 0.39 g Na2MoO4·2H2O (Alfa Aesar), 0.079 g CuSO4·5H2O (Sigma-Aldrich), 49.4 mg Co(NO3)2·6H2O (Sigma-Aldrich) per liter water). In this work, this media is referred to as M9P. 50 g l−1 glucose was used for C2–C10 acetate ester experiments and 10 g l−1 glucose was used for tetradecyl acetate, isobutyrate and butyrate ester experiments. Optical densities (D) were measured at 600 nm with a Synergy H1 Hybrid Plate Reader (BioTek Instruments, Inc.).
Substrate feeding experiments
Overnight cultures were inoculated 1% in 5 ml M9P in 15 ml screw-cap culture tubes. Cells were grown to a D600 nm of ~0.4 at 37 °C in a rotary shaker (250 r.p.m.), followed by adding 1 mM isopropyl-β-d-thio-galactoside (IPTG) (Promega). The cultures were incubated for 1 h after induction at 30 °C. Then metabolites of interest were added to the cultures. Production was performed at 30 °C in a rotary shaker (250 r.p.m.) for 24 h. Screw-cap tubes were tightly sealed to prevent evaporation of products. 1.5 ml of culture was taken for analysis every 24 h. The 1.5 ml of the cultures were centrifuged at 17,000g for 3 min, and then 1 ml of the supernatants were transferred to 2-ml GC vials for GC analysis. Excretion of small products such as alcohols and acids by E. coli is well known8,9,18.
Production of esters from glucose
For mixed acetate ester (Fig. 2b) and isobutyl isobutyrate (Fig. 4h) production from glucose, overnight cultures were inoculated 1% in 5 ml M9P in 15-ml screw-cap tubes. Cells were grown to a D600 nm of ~0.4 at 37 °C on a rotary shaker (250 r.p.m.), followed by adding 1 mM IPTG. The cultures were allowed to produce at 30 °C on a rotary shaker (250 r.p.m.) for 24 h. 1.5 ml of culture was taken for analysis after 24 h.
For the isobutyl acetate (Fig. 2h and Supplementary Fig. 1) and tetradecyl acetate (Fig. 3c) production experiments, overnight cultures were inoculated 1% in 25 ml M9P in 250 ml baffled flasks (Fig. 2h and Supplementary Fig. 1) or screw-cap flasks (Fig. 3c). Cells were grown to a D600 of ~0.4 at 37 °C on a rotary shaker (250 r.p.m.), followed by adding 1 mM IPTG. Then, 20 ml of culture was transferred to a 250-ml screw-cap flask. For hexadecane layer–assisted isobutyl acetate production (Fig. 2h and Supplementary Fig. 1), 20 ml hexadecane (Sigma-Aldrich) was added to each 250-ml screw-cap flask. The strong agitation at which the flask was shaking caused the hexadecane layer to become an emulsion with micelles of culture, but the majority of the culture layer underneath is discernible. The cultures were allowed to produce at 30 °C on a rotary shaker (250 r.p.m.) for 24–96 h. 1 ml of culture was taken for analysis every 24 h. For hexadecane layer–assisted isobutyl acetate production, 1 ml of the hexadecane layer was also taken for analysis every 24 h.
GC sample preparation
The 1–1.5 ml of the cultures were centrifuged at 17,000g for 3 min, and then 0.5–1 ml of the supernatants were transferred to 2-ml GC vials for GC analysis. For hexadecane layer–assisted isobutyl acetate production, the centrifugation of the hexadecane samples separates the aqueous and hexadecane layer into two clear layers, and then 0.5–1 ml of the culture fraction and hexadecane fraction were transferred to a 2-ml GC vial for GC-FID analysis. The same volume of ethyl acetate (Sigma-Aldrich) as culture was added into the tubes to extract tetradecyl acetate and isobutyl isobutyrate after production at room temperature. These samples were mixed for 1 min and settled for 30 min on ice. The each sample was centrifuged with 20,000g for 1 min. Then, 1 ml of supernatant was taken for GC analysis.
GC analysis
Concentrations of all of the products, except glucose, were analyzed by GC equipped with a flame ionization detector (FID). The GC system is a GC-2010 with an AOC-20 S auto sampler and AOC-20i Auto Injector (Shimadzu). The column used was a DB-Wax capillary column (30 m length, 0.32-mm diameter, 0.50-µm film thickness) (Agilent Technologies). The GC oven temperature was initially held at 40 °C for 3 min and then was increased at a rate of 45 °C min until 235 °C and held for 4 min. The injector temperature was held at 225 °C, and the FID detector was held at 330 °C. The injection volume was 0.5 µl, injected at a 15:1 split ratio. Helium was used as the carrier gas. 1-pentanol was used as an internal standard. In the case of tetradecyl acetate and butyl butyrate, 1-dodecene was used for internal standard. Retention times from samples were compared with external standards. Standard curves were prepared by diluting pure ester or alcohol into water at concentrations of 0.01 g l−1, 0.1 g l−1 and 1 g l−1. 100 mg l−1 of 1-pentanol was added to all of the samples and external standards as an internal standard.
Although GC-FID was used to quantify all of the products in the experiments presented in this study, GS-MS was used to verify the products we produced. 1–2.5 ml GC-grade hexane was used to extract from 5 ml cell culture. The mixture was mixed for 1 min, and then 1.4 ml of the hexane layer was transferred to a 1.5-ml tube. The hexane extract was centrifuged for 3 min at 17,000g, and then 0.5–1 ml was filtered with a 0.45-µm filter into a GC vial. Owing to the volatility of ethyl acetate and propyl acetate, pentane (Sigma-Aldrich) was used for extraction instead of hexane. The GC system is a GC-2010 with an AOC-20i S auto sampler and AOC-20i Auto Injector (Shimadzu). The column used was a SHR5XLB column (30 m length, 0.25-mm diameter, 0.25-µm film thickness) (Shimadzu). The GC oven temperature was held at 40 °C for 4 min and then was increased at a rate of 45 °C per min until 300 °C and held for 3 min. The injector temperature was held at 250 °C. The injection volume was 5 µl, injected at a 10:1 split ratio. Hydrogen was used as the carrier gas. The MS is a GCMS-QP2010S (Shimadzu). The ion source temperature was 200 °C, and the interface temperature was 250 °C. The solvent cut time was 2 min for hexane and 1 min for pentane. Detector voltage was −0.1 kV. The start m/z was 50, and the end m/z was 500. Mass spectra and retention times from samples were compared with external standards.
Glucose analysis by HPLC
To determine isobutyl acetate yield from glucose, the final glucose concentration was measured after 96 h (Supplementary Table 4) by centrifuging the entire culture and hexadecane layer for 10 min at 3,000g. Then, 1 ml of cell supernatant was used for HPLC-RID analysis to obtain a final glucose concentration. Glucose consumption was measured using a 20A HPLC (Shimadzu) equipped with a differential refractive detector (RID) 10A and an Aminex fast acid analysis column (Biorad). 5 mM H2SO4 served as the mobile phase at a flow rate of 0.6 ml/min at 65 °C for 12.5 min. The standard curve for glucose was done by measuring 0.1 g l−1, 1 g l−1 and 10 g l−1 glucose by HPLC-RID.
KDHC activity assay
Cells were grown to an D600 nm of ~0.4 in 5 ml LB medium at 37 °C, followed by adding 1 mM IPTG. Protein expression was performed at 30 °C for 2 h. Then 1.8 ml of cells were centrifuged at 16,000 g for 10 min, resuspended in 300 µl BugBuster Protein Extraction Reagent (Novagen) and incubated at room temperature for 20 min for cell lysis. Then samples were centrifuged for 20 min, 16,000g, at 2 °C. The soluble fractions were transferred to chilled 1.5-ml tubes, and the insoluble fractions were suspended in 200 µl BugBuster, forming a slurry, and were kept on ice. KDHC activities were measured by following the conversion of 2-keto acids to acyl-CoA with NAD+ at 340 nm at 30 °C using a Synergy H1 Hybrid Plate Reader (BioTek Instruments, Inc.). The assay mixture contained 25 mM 2-keto acid (pyruvate, 2-ketovalerate, 2-ketoisovalerate, 2-keto-3-methylvalerate and 2-keto-4-methylvalerate), 50 mM MOPS buffer (pH 7.0), 0.2 mM Tris-Cl (pH 7.00), 0.2 mM NADH (Sigma-Aldrich), 0.2 mM CoA (Sigma-Aldrich) and 12.5 mM potassium phosphate buffer (pH 7.5). One unit of activity is defined as the reduction of 1 µmol of NAD+ per minute per mg protein. Protein concentrations were measured using 5× Advanced Protein Assay Reagent (Cytoskeleton Inc.). Bovine Serum Albumin (BSA) was used to prepare a standard curve. No activity was observed in the soluble fraction of the cell lysates of either strain. However, we resuspended the insoluble fraction with the lysis buffer and upon testing, substantial activity of KDHC was observed (Fig. 4b).
Supplementary Material
Acknowledgments
This work was supported by University of California–Davis startup fund and the Hellman fellowship to S.A. G.M.R. is supported by a US National Institutes of Health Biotechnology Training Grant Fellowship (T32-GM008799) and a Sloan Fellowship. Y.T. is supported by Japan Society for the Promotion of Science postdoctoral fellowship for research abroad. We would like to thank R. Luu and R.E. Parales (University of California–Davis) for graciously providing genomic DNA from P. putida g7 and M. Kato and S.-J. Lin (University of California–Davis) for providing genomic DNA from S. cerevisiae BY4742. We also thank M.D. Toney and C.A. Rabinovitch-Deere (University of California–Davis) for critical reading of the manuscript. Finally, we thank S. Desai for technical assistance with HPLC analysis and for constructing pAL603.
Footnotes
Author contributions
G.M.R., Y.T. and S.A. designed research; G.M.R. and Y.T. performed the experiments; G.M.R., Y.T. and S.A. analyzed data; and G.M.R., Y.T. and S.A. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
References
- 1.Beekwilder J, et al. Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 2004;135:1865–1878. doi: 10.1104/pp.104.042580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verstrepen KJ, et al. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl. Environ. Microbiol. 2003;69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki T, Maegawa Y, Ohshima T, Mashima K. Kirk-Othmer Encyclopedia of Chemical Technology. Vol. 10. New Jersey: John Wiley & Sons; 2012. pp. 497–516. [Google Scholar]
- 4.Liu YJ, Lotero E, Goodwin JG. Effect of water on sulfuric acid catalyzed esterification. J. Mol. Catal. Chem. 2006;245:132–140. [Google Scholar]
- 5.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 5th edn. New York: Sara Tenney; 2008. [Google Scholar]
- 6.Stergiou PY, et al. Advances in lipase-catalyzed esterification reactions. Biotechnol. Adv. 2013;31:1846–1859. doi: 10.1016/j.biotechadv.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Dhake KP, Thakare DD, Bhanage BM. Lipase: a potential biocatalyst for the synthesis of valuable flavour and fragrance ester compounds. Flavour Fragrance J. 2013;28:71–83. [Google Scholar]
- 8.Rabinovitch-Deere CA, Oliver JW, Rodriguez GM, Atsumi S. Synthetic biology and metabolic engineering approaches to produce biofuels. Chem. Rev. 2013;113:4611–4632. doi: 10.1021/cr300361t. [DOI] [PubMed] [Google Scholar]
- 9.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 10.Fujii T, et al. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 1994;60:2786–2792. doi: 10.1128/aem.60.8.2786-2792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowland O, Domergue F. Plant fatty acyl reductases: enzymes generating fatty alcohols for protective layers with potential for industrial applications. Plant Sci. 2012;193–194:28–38. doi: 10.1016/j.plantsci.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Rontani JF, Bonin PC, Volkman JK. Production of wax esters during aerobic growth of marine bacteria on isoprenoid compounds. Appl. Environ. Microbiol. 1999;65:221–230. doi: 10.1128/aem.65.1.221-230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenneman EM, Ohlert JM, Palani NP, Barney BM. Fatty alcohols for wax esters in Marinobacter aquaeolei VT8: two optional routes in the wax biosynthesis pathway. Appl. Environ. Microbiol. 2013;79:7055–7062. doi: 10.1128/AEM.02420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiser S, Somerville C. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 1997;179:2969–2975. doi: 10.1128/jb.179.9.2969-2975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtzapple E, Schmidt-Dannert C. Biosynthesis of isoprenoid wax ester in Marinobacter hydrocarbonoclasticus DSM 8798: identification and characterization of isoprenoid coenzyme A synthetase and wax ester synthases. J. Bacteriol. 2007;189:3804–3812. doi: 10.1128/JB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barney BM, Wahlen BD, Garner E, Wei J, Seefeldt LC. Differences in substrate specificities of five bacterial wax ester synthases. Appl. Environ. Microbiol. 2012;78:5734–5745. doi: 10.1128/AEM.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stöveken T, Kalscheuer R, Malkus U, Reichelt R, Steinbuchel A. The wax ester synthase/acyl coenzyme A:diacylglycerol acyltransferase from Acinetobacter sp. strain ADP1: characterization of a novel type of acyltransferase. J. Bacteriol. 2005;187:1369–1376. doi: 10.1128/JB.187.4.1369-1376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 19.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011;77:2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond-Watts BB, Bellerose RJ, Chang MC. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 21.Inokuma K, Liao JC, Okamoto M, Hanai T. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 2010;110:696–701. doi: 10.1016/j.jbiosc.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 23.Saerens SM, et al. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 2006;281:4446–4456. doi: 10.1074/jbc.M512028200. [DOI] [PubMed] [Google Scholar]
- 24.Lilly M, et al. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast. 2006;23:641–659. doi: 10.1002/yea.1382. [DOI] [PubMed] [Google Scholar]
- 25.Mason AB, Dufour JP. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast. 2000;16:1287–1298. doi: 10.1002/1097-0061(200010)16:14<1287::AID-YEA613>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 26.Fujii T, Yoshimoto H, Tamai Y. Acetate ester production by Saccharomyces cerevisiae lacking the ATF1 gene encoding the alcohol acetyltransferase. J. Ferment. Bioeng. 1996;81:538–542. [Google Scholar]
- 27.Howard TP, et al. Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110:7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin F, Das D, Lin XN, Marsh EN. Aldehyde-forming fatty acyl-CoA reductase from cyanobacteria: expression, purification and characterization of the recombinant enzyme. FEBS J. 2013;280:4773–4781. doi: 10.1111/febs.12443. [DOI] [PubMed] [Google Scholar]
- 29.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 30.Mooney BP, Miernyk JA, Randall DD. The complex fate of α-ketoacids. Annu. Rev. Plant Biol. 2002;53:357–375. doi: 10.1146/annurev.arplant.53.100301.135251. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez GM, Atsumi S. Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb. Cell Fact. 2012;11:90. doi: 10.1186/1475-2859-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Plaza M, Fernandez de Palencia P, Pelaez C, Requena T. Biochemical and molecular characterization of α-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett. 2004;238:367–374. doi: 10.1016/j.femsle.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 33.Bastian S, et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 2011;13:345–352. doi: 10.1016/j.ymben.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Baez A, Cho KM, Liao JC. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 2011;90:1681–1690. doi: 10.1007/s00253-011-3173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atsumi S, et al. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010;85:651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connor MR, Cann AF, Liao JC. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl. Microbiol. Biotechnol. 2010;86:1155–1164. doi: 10.1007/s00253-009-2401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belas R, et al. Bacterial bioluminescence: isolation and expression of the luciferase genes from Vibrio harveyi. Science. 1982;218:791–793. doi: 10.1126/science.10636771. [DOI] [PubMed] [Google Scholar]
- 38.Meighen EA. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 39.Ferrández A, Garcia JL, Diaz E. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 1997;179:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SJ, Ko JH, Kang HY, Lee Y. Coupled expression of MhpE aldolase and MhpF dehydrogenase in Escherichia coli. Biochem. Biophys. Res. Commun. 2006;346:1009–1015. doi: 10.1016/j.bbrc.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Hester K, Luo J, Burns G, Braswell EH, Sokatch JR. Purification of active E1α2β2 of Pseudomonas putida branched-chain-oxoacid dehydrogenase. Eur. J. Biochem. 1995;233:828–836. doi: 10.1111/j.1432-1033.1995.828_3.x. [DOI] [PubMed] [Google Scholar]
- 42.Hester KL, Luo J, Sokatch JR. Purification of Pseudonomas Putida branched-chain keto acid dehydrogenase E1 component. Methods Enzymol. 2000;324:129–138. doi: 10.1016/s0076-6879(00)24226-2. [DOI] [PubMed] [Google Scholar]
- 43.Alonso-Gutierrez J, et al. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Park YC, Shaffer CE, Bennett GN. Microbial formation of esters. Appl. Microbiol. Biotechnol. 2009;85:13–25. doi: 10.1007/s00253-009-2170-x. [DOI] [PubMed] [Google Scholar]
- 45.van den Berg C, Heeres AS, Van der Wielen LA, Straathof AJ. Simultaneous Clostridial fermentation, lipase-catalyzed esterification, and ester extraction to enrich diesel with butyl butyrate. Biotechnol. Bioeng. 2013;110:137–142. doi: 10.1002/bit.24618. [DOI] [PubMed] [Google Scholar]
- 46.Machado HB, Dekishima Y, Luo H, Lan EI, Liao JC. A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng. 2012;14:504–511. doi: 10.1016/j.ymben.2012.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.