Abstract
Commercial cultivars of garlic, a popular condiment, are sterile, making genetic studies and breeding of this plant challenging. However, recent fertility restoration has enabled advanced physiological and genetic research and hybridization in this important crop. Morphophysiological studies, combined with transcriptome and proteome analyses and quantitative PCR validation, enabled the identification of genes and specific processes involved in gametogenesis in fertile and male-sterile garlic genotypes. Both genotypes exhibit normal meiosis at early stages of anther development, but in the male-sterile plants, tapetal hypertrophy after microspore release leads to pollen degeneration. Transcriptome analysis and global gene-expression profiling showed that >16,000 genes are differentially expressed in the fertile vs. male-sterile developing flowers. Proteome analysis and quantitative comparison of 2D-gel protein maps revealed 36 significantly different protein spots, 9 of which were present only in the male-sterile genotype. Bioinformatic and quantitative PCR validation of 10 candidate genes exhibited significant expression differences between male-sterile and fertile flowers. A comparison of morphophysiological and molecular traits of fertile and male-sterile garlic flowers suggests that respiratory restrictions and/or non-regulated programmed cell death of the tapetum can lead to energy deficiency and consequent pollen abortion. Potential molecular markers for male fertility and sterility in garlic are proposed.
Keywords: tapetum, microsporogenesis, gene expression, protein profiling, mitochondrial dysfunction, energy deficiency
Introduction
The main functions of the plant reproductive system are flowering, fertilization, and seed development. During flower differentiation, the diploid pollen mother cells undergo meiotic divisions to form tetrads of haploid microspores, which further divide mitotically to vegetative and generative cells of the pollen grain. The latter produce the two gametes required for double fertilization (Borg et al., 2009). Pollen formation, germination and fertilization require high amounts of energy from starch, proteins and sugars (Bhandari, 1984), and anther tissues, mainly the tapetum, play an important supportive role in nourishing the developing microspores (Goldberg et al., 1993). Microsporogenesis is controlled by concerted gene expression in both somatic and gametophytic cells (Mascarenhas, 1990; McCormick, 2004), and some of these genes are anther-specific (McCormick, 2004; Borg et al., 2009). Their function and the effect of their products on pollen development and fertility, however, are obscure, even in model plants.
Garlic (Allium sativum L.) represents a sizeable group of useful plants with a large nuclear genome (Arumuganathan and Earle, 1991). The genome of diploid garlic (2n = 2x = 16) is estimated at 15.9 Gbp, 32 times larger than the rice genome. Although full sequencing of the garlic genome remains a challenging task, transcriptome assembly by next generation sequencing may be used for the effective generation of functional genomic data (Kamenetsky et al., 2015).
Classical hybridization, genetic studies, and breeding of this important crop were impossible for hundreds of years, since no commercial garlic varieties produce flowers or seeds. In recent decades, however, seeds have been obtained under experimental conditions from several garlic genotypes (Etoh et al., 1988; Pooler and Simon, 1993, 1994; Jenderek and Hannan, 2000; Kamenetsky et al., 2005). Physiological studies have paved the way for fertility restoration and seed production in bolting genotypes (Kamenetsky et al., 2005; Kamenetsky, 2007; Shemesh et al., 2008), thus facilitating the studies of garlic genetics and breeding (Jenderek and Zewdie, 2005; Rotem et al., 2007, 2011). Bolting and flowering garlic genotypes vary in their reproductive traits, including male fertility and sterility. We have shown that although the temperature has a significant impact on flower development in garlic, in male-sterile plants, fertility cannot be restored by temperature manipulations. We therefore suggested that garlic male sterility is controlled by genetic factors (Shemesh-Mayer et al., 2013, 2015).
In general, the mechanism leading to male sterility in plants is still far from being understood. Male sterility is known to be controlled by genetic and biochemical factors (Hanson, 1991; Mo et al., 1992; Sawhney and Shukla, 1994), or caused by adverse growth conditions, diseases, radiation or chemicals (Islam et al., 2014). Studies in Arabidopsis, rice and maize have revealed a large number of genes encoding male fertility, and mutations in any of them could adversely affect the normal development of anthers, or interfere with microgametogenesis, resulting in infertility. Interruptions may occur during chromosome pairing or meiotic segregation, thus resulting in reduced functionality of the tapetum, abnormal pollen-wall formation, inhibition of filament elongation or accelerated anther dehiscence (Scott et al., 2004; Feng and Dickinson, 2007; Chang et al., 2011; Zhang and Yang, 2014). Comparative analyses between the genetic pathways of pollen development in Arabidopsis, rice (Wilson and Zhang, 2009), and the bulbous species Lilium longiflorum (Mori et al., 2003; Hsu et al., 2007) and Alstroemeria (Igawa et al., 2009) have revealed a great similarity in microgametogenesis among plant species, thus indicating a high degree of conservation in the early regulatory network of pollen production.
In plants, three types of inherited male sterility are known: genetic, cytoplasmic, and cytoplasmic–genetic. Genetic male sterility (GMS) is usually regulated by a single recessive nuclear gene with monogenic Mendelian inheritance. The maternally inherited cytoplasmic male sterility (CMS) is under extranuclear genetic control, often associated with unusual open reading frames (ORFs) in the mitochondrial genome (Hanson, 1991; Schnable and Wise, 1998). CMS is known in more than 150 flowering plant species (Laser and Lersten, 1972), which can have N (normal) or aberrant S (sterile) cytoplasm types. In many instances, fertility can be restored specifically by nuclear-encoded fertility-restoring (Rf) genes. An interaction between CMS and GMS (cytoplasmic–genetic male sterility, CGMS) is quite common in the genus Allium (Budar and Pelletier, 2001). It was identified in bulb onion (Allium cepa L.) more than 70 years ago (Jones and Emsweller, 1937; Jones and Clarke, 1943; Havey, 1993, 1995), and later in the bunching onion (A. fistulosum L.) (Nishimura and Shibano, 1972; Moue and Uehara, 1985) and in chives (A. schoenoprasum L.) (Tatlioglu, 1982). Our living collection of flowering garlic genotypes has enabled us to investigate their reproductive traits and especially fertility/infertility phenomena (Shemesh et al., 2008; Shemesh-Mayer et al., 2013, 2015).
In model plants and in plant species with large and non-sequenced genomes, transcriptome and proteome profiling have revealed marked differences in gene expression during anther and pollen development (Libault et al., 2010; Wei et al., 2010; Kamenetsky et al., 2015). In Arabidopsis male meiocytes, the expression of approximately 20,000 genes has been recorded, with more than 800 preferentially expressed in pollen grains. Homologs of these genes have been found in poplar and in rice (Khurana et al., 2012). A comparison between gene-expression profiles of fertile and male-sterile Arabidopsis plants has revealed differences between genes associated with the MYB and bHLH protein families (Khurana et al., 2012); in Brassica napus, such studies have shown differences related to pollen-wall assembly, binding, catalytic activity, transporter activity, and antioxidant activity (Yan et al., 2013); in chili peppers, differences have been found in ATP synthase, cytochrome oxidase, activated oxygen metabolism and more (Liu et al., 2013).
Proteomic studies have provided information on differential protein profiles between fertile and male-sterile mutants of Arabidopsis, tomato, rice, maize and Brassica (Imin et al., 2001; Mihr et al., 2001; Wen et al., 2003; Sheoran et al., 2009), and between fertile and sterile garlic genotypes (Shemesh-Mayer et al., 2013). The research presented herein focused on transcriptome and proteome analyses and biological processes occurring during microsporogenesis in fertile and male-sterile garlic genotypes. We identified specific genes and proteins involved in bioenergy balance and hypothesized that an interruption in pollen differentiation is associated with a shortage in bioenergy flow.
Materials and methods
Plant material and growth conditions
Following open-pollination, two flowering garlic clones, fertile #87 (F87) and male-sterile #96 (MS96) were raised from two seeds harvested from the same mother plant. The two plants were vegetatively propagated in an insect-proof screenhouse at the ARO, the Volcani Center, Bet Dagan, Israel. In 2012, freshly harvested bulbs were cured and stored in an open shed under ambient conditions, from July to September. After sorting, healthy-looking propagules were stored for 8 weeks at 4°C at a RH of 65–70%. In November, 50 healthy disinfected cloves of each genotype were planted in 20-L containers in a 70:10:20 (v/v) mix of 0.8-mm volcanic tuff particles:perlite:ground coconut peels. Plants were grown under 30% shade in a screenhouse and regularly fertigated with “Shefer” liquid fertilizer (N:P:K = 59:35:94 g L−1, Dshanim, Israel).
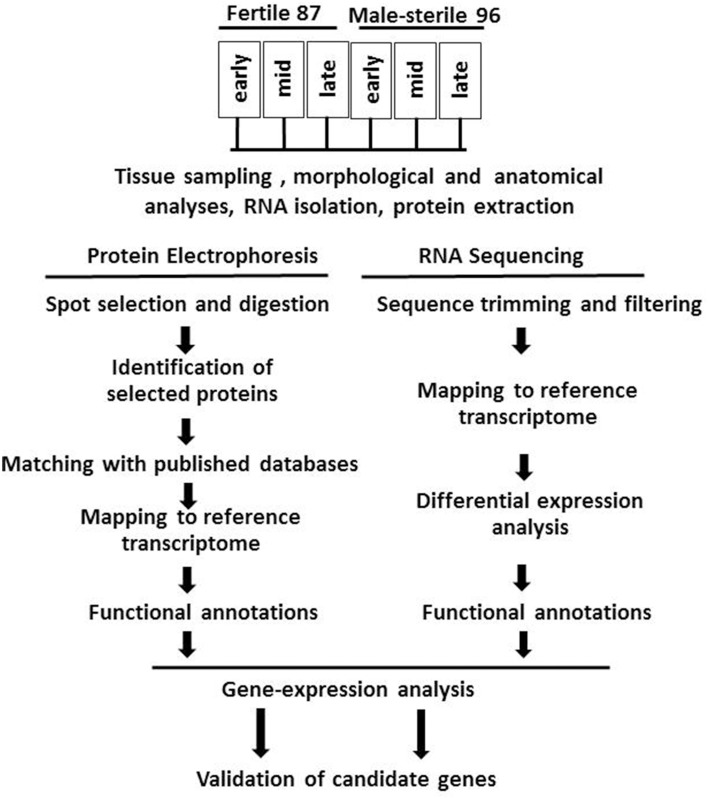
For RNA and proteomic analysis, flowers were collected at the early (green tepal 2.5–3-mm long), mid (green tepal 3–4-mm long) and late (pink tepal 3–4-mm long) stages of development (Shemesh-Mayer et al., 2013). The experimental design is presented in Figure 1.
Figure 1.
Flowchart of experimental design, including morphological and anatomical studies of garlic genotypes F87 and MS96, transcriptome and proteome analyses and validation of candidate genes.
Flower phenology, morphology, and anatomy
Flower bud samples were collected in May–June 2013 from spathe break to flower senescence. Morphological studies of 15 flowers per genotype were carried out at each development stage under a stereoscope (Zeiss Stemi 2000-C, Zeiss, Germany). Anatomical studies were performed on flowers fixed in FAA solution (100% acetic acid, 40% formalin, 95% ethanol at 1:2:10 v/v). For plastic embedding, fixed samples were dehydrated in a graded ethanol series and gradually replaced by LR-White resin (Sigma–Aldrich, St. Louis, MO, USA) according to Ruzin (1999). Following polymerization at 60°C for 48–72 h, 2-μm tissue slices obtained by rotary microtome (Leica RM2245) were stained with 0.05% (w/v) toluidine blue and studied under a light microscope (Leica DMLB, Germany).
Transcriptome analysis
For RNA isolation, tissues were frozen in liquid nitrogen upon sampling and then stored at −80°C until required. Total RNA from flower buds was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Extract quality was verified using the Agilent 2100 Bioanalyzer with a minimum RNA integrity number value of 7. In total, 18 libraries were prepared, consisting of two genotypes and three replicates of three developmental stages.
RNA library processing was performed by Roy J. Carver Biotechnology Center, W.M. Keck Center for Comparative and Functional Genomics, Urbana, IL, USA. The RNA-Seq libraries were prepared by applying Illumina's “TruSeq Stranded RNA Sample Prep kit” (www.Illumina.com). The pools were quantified by real-time quantitative PCR (rt-qPCR) and sequenced in two lanes each for 101 cycles from one end of the fragments on a HiSeq2000 using TruSeq SBS sequencing kits version 3. Fastq files were generated with Casava 1.8.2. Reads were 100 nt in length.
The sequencing data are deposited in NCBI Sequence Read Archive (SRA) database as bioprojects PRJNA264944 [SSRR1632269-RR1632277] for fertile garlic genotype F87 and PRJNA264945 for male sterile genotype MS96 [SRR1632278-SRR1632288].
Raw reads were cleaned using FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13.2) including:
trimming reads' end nucleotides with quality scores <30 using fastq_quality_trimmer,
reads consisting of less than 70% base pairs with quality score ≤30 were removed using fastq_quality_filter.
A recently released organ-specific A. sativum transcriptome catalog comprising 240,000 contigs from six vegetative and reproductive organs was employed as a reference (NCBI bioproject PRJNA243415, Kamenetsky et al., 2015). The cleaned reads from each library were aligned separately to the transcriptome catalog using Bowtie aligner (Langmead et al., 2009), and the abundance estimation was calculated, via expectation maximization, using the run _RSEM_align_n_estimate.pl script Perl, following the Trinity protocol (Haas et al., 2013).
Differential expression analysis of the sequence count data for each pair of samples (overall seven comparisons) was performed by Bioconductor DESeq package in the R environment (Anders and Huber, 2010). Differential expression was defined as an over four-fold difference in transcription expression with a false discovery-corrected statistical significance of at most 0.001 (i.e., FDR < 0.001).
The transcripts that were differentially expressed in at least one pairwise sample comparison between genotypes and/or developmental stages were examined using cluster analysis. Following the Trinity protocol (Haas et al., 2013), expression normalization was calculated by TMM (trimmed mean of M-values) normalization and FPKM (fragments per kilobase of transcript per mllion mapped reads) calculation. Based on the average values in the three replications, hierarchical clustering of transcripts and samples was performed and clusters were extracted using R scripts.
Functional classification of the differentially expressed transcripts was performed using the Blast2GO (Conesa et al., 2005), based on gene ontology (GO) terms obtained from the garlic transcriptome catalog (Kamenetsky et al., 2015). The REVIGO web server (Supek et al., 2011) was used for visualization of the GO terms.
Mitochondrial genes were identified by comparing the garlic transcriptome catalog (NCBI bioproject PRJNA243415) with genomic data for mitochondria of Arabidopsis thaliana (NC_001284.2) and Oryza sativa Japonica Group (NC_011033.1), using BLASTN algorithm. Further, the 23 sequences that were most similar to the published mitochondrial gene sequences were compared with the NCBI-nr database (http://www.ncbi.nlm.nih.gov).
Proteome analysis
Upon sampling, fresh flower tissues were dipped in liquid nitrogen, lyophilized and shipped to the Institute of Plant Genetics, Polish Academy of Sciences, Poznan, Poland, where proteins were extracted according to Hurkman and Tanaka (1986). The assay consisted of two biological replicates and three technical repeats. Protein concentration was determined using a 2-D Quant Kit (GE Healthcare, Buckinghamshire, UK), and 2D gel electrophoresis was carried out according to Kosmala et al. (2009) and Perlikowski et al. (2014a). The abundance of each spot was normalized to a relative volume (% volume) and calculated by Image Master 2-D Platinum software (GE Healthcare) as the ratio of the volume of each particular spot to the total volume of all spots present on the electrophoretic gel. Liquid chromatography coupled with mass spectrometry was used for identification of the selected proteins in the Laboratory of Mass Spectrometry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences in Warsaw as described in detail by Perlikowski et al. (2014a).
The obtained peptide masses and fragmentation spectra were matched with NCBI-nr using a Viridiplantae filter (1,032,142 sequences) and the Mascot search engine (Mascot Daemon v. 2.3.0, Mascot Server v. 2.4.0, MatrixScience). The following search parameters were applied: enzyme specificity was set to trypsin, peptide mass tolerance to ± 30 ppm and fragment mass tolerance to ± 0.1 Da. Protein mass remained unrestricted and monoisotopic mass values were used, but one missed cleavage was allowed. Cysteine alkylation by carbamidomethylation was set as fixed, and methionine oxidation as a variable modification. Proteins were characterized by selection for the highest Mascot-assigned protein score (MudPIT-type) and/or for the highest number of peptide sequences. When only “hypothetical” or “predicted” proteins were identified, amino acid sequence was blasted using the BLASTP algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the one with the highest score was selected as its functional homolog (Perlikowski et al., 2014b).
Comparison between transcriptome and proteome
To compare the transcriptome and proteome data, protein annotations were aligned against the garlic transcriptome catalog (NCBI bioproject PRJNA243415) using TBLASTN algorithm. The matched contigs were compared with the sequences deposited in the NCBI-nr protein database using the BLASTX (Altschul et al., 1990) algorithm with an E-value cut-off of 10−5. The expression level (in FPKM) of the matched contigs was compared between fertile and male-sterile genotypes.
Real-time quantitative PCR validation
The expression of 10 selected genes was determined by rt-qPCR using the RNA samples previously employed for the construction of libraries and transcriptome analyses. All rt-qPCR experiments were run in three biological and two technical replications. Garlic homologs of actin (AY821677) and tubulin (AY148156) served as reference genes for data normalization and calculation of relative amounts of mRNA in the studied samples. The sequences are available in the garlic transcriptome catalog (NCBI bioproject PRJNA243415).
Specific probes were designed for the genes: nad7, Cox2, 18S rRNA, ccmC, GPAT2, and SOD using Universal Probe Library software (https://www.roche-applied-science.com). The commercial probe numbers, their sequences, primers and efficiency are detailed in Supplementary Table 1. Differential expression was studied using these probes and the reaction mix of FastStart Universal Probe Master (2X; Roche, Indianapolis, IN, USA). The rt-qPCR (Light-Cycler 480 Real-Time PCR System, Roche) analyses were performed with a 20-μl reaction mixture that included 0.5 μM primers, 5 μl cDNA, 1 μl probe and HPLC-grade H2O, in a multiwell plate. Each plate had a negative control without cDNA template, and a positive control consisting of plasmid as a template for calibration (Pfaffl et al., 2002). For each primer set, PCR efficiency was determined by the standard curve method. The reactions were performed under the following cycling conditions: an initial denaturing step of 10 min at 95°C, following by 35 cycles consisting of 10 s at 95°C and 30 s at 60°C. The candidate genes were quantified using the ΔΔCt method (Pfaffl, 2001), and the n-fold change was calculated with LightCycler 480 software, provided by the manufacturer.
The SYBR Green method (Tyler et al., 2004), used to determine the expression of MS2, MMD1, AP3, and Flavonol synthase, was performed with a 20-μl qPCR reaction mixture consisting of 5 μl template cDNA (1.5 μmol/μl). 10 pmol of each primer and 10 μl of 10X FastStart SYBR Green I Master (Roche). Reactions were performed under the following cycling conditions: an initial denaturing step of 10 min at 95°C, and 35 cycles of 10 s at 95°C, 10 s at 60°C and 10 s at 72°C. The results were analyzed using LightCycler 480 software, provided by the manufacturer.
Both the specific probes and SYBR Green were used for assessment of the reference genes actin and tubulin.
Results
Phenology, morphology, and anatomy of fertile and male-sterile garlic flowers
Plants of both F87 and MS96 genotypes produced dense inflorescences with ca. 300–400 flowers per umbel. However, developmental morphology of the individual flowers varied with genotype. In F87, the filaments of the purple anthers elongated following anthesis; the anthers opened and the pollen dehisced. In contrast, the MS96 anthers turned yellow and degenerated, and no pollen shedding occurred (Figure 2). Female reproductive organs of both genotypes had a similar morphology, appeared to be intact, and when pollinated, both genotypes produced viable seeds.
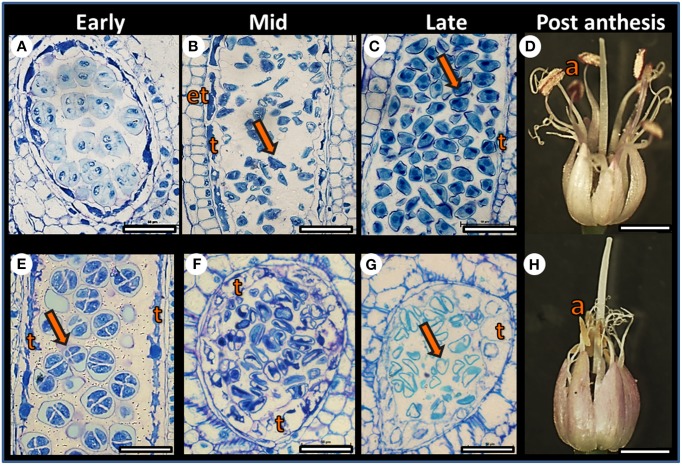
Figure 2.
Comparative developmental anatomy of anthers in the F87 (A–D) and MS96 (E–H) garlic genotypes during microgametogenesis. Comparisons were made between early, mid and late stages of flower development. (A) Cross section of F87 pollen sac at the tetrad stage. Bar = 40 μm. (B) Longitudinal section of F87 pollen sac after microspore (arrow) release from the callose. Endothecium (et), and tapetum (t) are visible. Bar = 60 μm. (C) Longitudinal section of an anther with mature microspores (arrow) that contain vegetative and generative cells. Tapetum (t) is degenerated, and only remains are visible. Bar = 30 μm. (D) Mature F87 flower. Long filaments, dehisced anthers (a) and long style are visible. Bar = 1.5 mm. (E) Longitudinal section of MS96 pollen sac at the tetrad (arrow) stage. Typical tapetum (t) cells are visible. Bar = 30 μm. (F) Cross section of an MS96 anther, with microspores released from the callose. Note hypertrophy of the tapetum (t) cells. Bar = 45 μm. (G) Considerable enlargement of the tapetum (t) cells and degenerated microspores (arrow) in MS96. Bar = 45 μm. (H) Mature MS96 flower. Degenerated yellow anthers (a) are visible and the style is elongated. Bar = 1.5 mm.
Anatomical studies at the early stages of anther development revealed normal meiosis in both genotypes (Figures 2A,E); however, differences between genotypes were obvious post-meiosis. Breakdown of the tetrad callose wall of F87 was followed by microspore release into the locular space of the pollen sac, and nourishment was provided by the tapetum layer (Figure 2B). At the late stage of anther development, mitotic divisions and pollen maturation occurred, and the tapetum disintegrated (Figure 2C). In MS96, hypertrophy of the tapetal cells was already observed when the microspores were released (Figure 2F); this was followed by pollen degeneration, including depletion of pollen content and disintegration of the nuclei (Figure 2G).
Transcriptome analysis of fertile and sterile flowers
Six pools of mRNA samples, representing three floral developmental stages per genotype, served for the construction of high-throughput parallel RNA-Seq libraries. Each of the six cDNA libraries yielded 19–23 million 100-bp one-end reads. Quality trimming and filtration reduced the number to 17–21 million clean reads, ca. 90% of which were mapped to the de-novo assembled transcript catalog of A. sativum (Table 1).
Table 1.
Number of RNA-Seq reads in F87 and MS96 garlic flowers.
| Genotype | Stage of flower development | Reads (in millions) | Reads discarded after cleaning (%)a | Clean reads (in millions)b | Reads mapped to garlic transcriptome catalog (%)c |
|---|---|---|---|---|---|
| MS96 | Early | 22.1 ± 1.2 | 7.11 ± 0.04 | 20.5 ± 1.1 | 89.75 ± 0.29 |
| Mid | 22.7 ± 2.4 | 7.28 ± 0.10 | 21.0 ± 2.2 | 90.42 ± 0.20 | |
| Late | 19.2 ± 1.2 | 7.16 ± 0.05 | 17.8 ± 1.1 | 90.33 ± 0.15 | |
| F87 | Early | 20.5 ± 1.4 | 6.94 ± 0.08 | 19.1 ± 1.3 | 89.94 ± 0.79 |
| Mid | 19.9 ± 2.2 | 7.14 ± 0.08 | 18.5 ± 2.0 | 92.66 ± 0.18 | |
| Late | 19.2 ± 0.4 | 7.10 ± 0.06 | 17.8 ± 0.4 | 91.84 ± 0.55 |
Samples, collected in May–June 2013 at early, mid and late stages of flower development, were used for transcriptome analysis. Presented data are means of three replications ± standard error values.
After trimming and filtration.
Mapping to the transcript catalog of A. sativum (NCBI bioproject PRJNA243415).
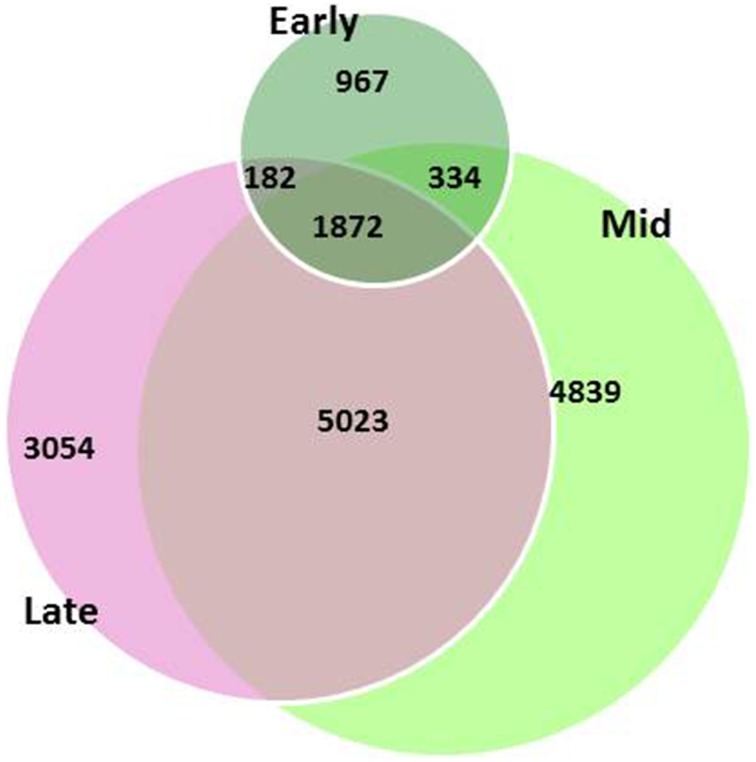
Global gene-expression profiling showed differential patterns between the two genotypes. A total of 16,271 differentially expressed genes (DEGs) were found between the two genotypes throughout the three developmental stages, but only 12% of the DEGs were common in the flowers throughout all developmental stages; 6% of the DEGs were specific to the early stage, 30% to the mid stage, and 19%—to the late stage of flower development (Figure 3).
Figure 3.
Differentially expressed genes (DEGs) between garlic genotypes F87 and MS96 at three stages of flower development. A total of 16,271 DEGs were differentially expressed; 1872 DEGs were common for all three stages, while 967, 4839, 3054 were specific for the early, mid and late stages, respectively.
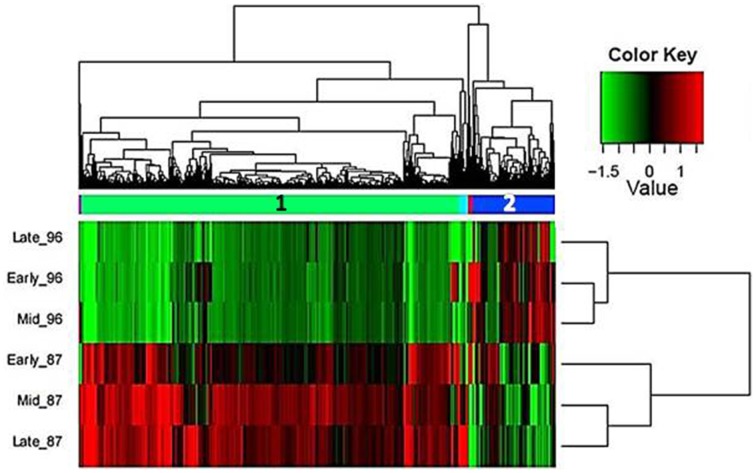
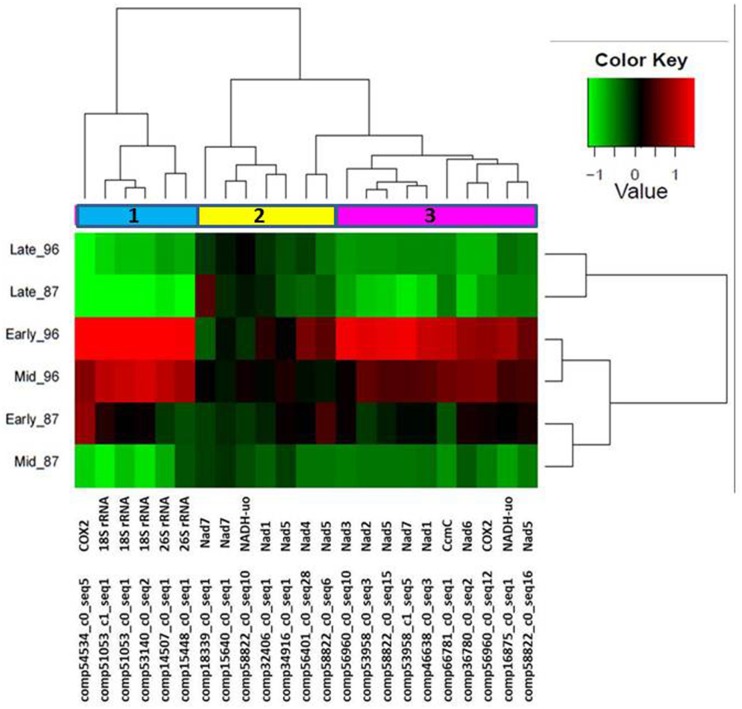
Hierarchical cluster analysis of gene expression revealed clearly differentiated patterns between the two investigated genotypes, with two large and three small gene clusters (Figure 4). Interestingly, in the male-sterile genotype, genes expressed in early and mid stages were clustered together, whereas in the fertile genotype, higher proximity was found between the mid and late stages of flower development, which were distinct from the early stage (Figure 4).
Figure 4.
Hierarchical cluster analysis of gene-expression patterns at the three developmental stages of garlic genotypes F87 and MS96 shows the relative expression levels of each gene (column) in each sample (row). Two large (1, 2) and three small gene clusters were differentially expressed in one or more samples. The expression values (FPKM; average of three replications) were log2-transformed and then median-centered by transcript.
Annotation and GO-based functional analysis were performed separately for each of the five DEG clusters. Clusters 1 and 2 had the most substantial differences between the two genotypes (Figure 5).
Figure 5.
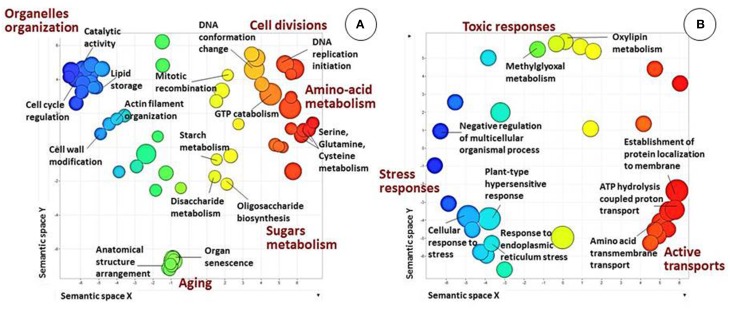
Biological processes in clusters 1 and 2 (see Figure 4), as revealed by analysis of GO term distribution using Blast2GO and REVIGO algorithms. GO terms are represented by circles and are plotted according to semantic similarities to other GO terms (adjoining circles are most closely related). Circle size is proportional to the abundance of the GO term in the cluster, while color indicates semantic similarities. Only GO terms with higher than 1% frequency in the cluster are shown. (A) Cluster 1. Main patterns are related to the general development of reproductive tissues, metabolism, microsporogenesis and cell-division processes and specific related fertility processes. (B) Cluster 2. Main patterns are related to energy-consuming activities and/or response to stress.
Bioinformatics analysis revealed that the largest cluster (cluster 1, highlighted in green in Figure 4) consists of 6729 genes, 1194 of which were annotated. At all three developmental stages, most of these genes were highly expressed in genotype F87 compared to genotype MS96. Biological processes, as assessed by Blast2GO, were divided into two main groups (Figure 5A). The first group was related to the general development of reproductive tissues, including, inter alia, regulation of meristem development, floral organ development, regulation of cellular-component organization, polysaccharide-catabolism processes, protein polymerization and anthocyanin metabolism. The second group consisted of specific microsporogenesis and cell-division processes, including cell-wall thickening, lipid storage, chromosome organization, DNA replication and packaging, chromatin assembly, mitotic cell cycle, cytoskeleton organization, and dehiscence. Numerous processes specifically associated with fertility (by Blast2GO) were abundant in this cluster, i.e., starch and glycerol-3-phosphate metabolism, hexose biosynthesis, fatty-acyl-CoA transport, naringenin-chalcone synthase activity, Ras and Rab GTPase activity, and more.
Bioinformatics analysis of cluster 2 (highlighted in blue in Figure 4) revealed 1449 genes, 367 of them annotated. In contrast to cluster 1, most of these genes were expressed more strongly in MS96 than in F87. Molecular functions of the annotated genes included the energy-consuming activities of transmembrane transporters, e.g., carboxylic acid and organic acid transmembrane transporters, and ATP synthase (data not shown). Regulation of cellular responses to stress and responses to endoplasmic reticulum stress was abundant in this cluster. Biological processes, assessed by Blast2GO, included, among others, stress responses, energy-coupled proton transmembrane transport, ATP proton transport, and protein targeting and localization to the membrane (Figure 5B).
We hypothesized that garlic male sterility might be associated with mitochondrial activity. Therefore, we further selected 23 annotated transcripts with high similarity to the published sequences of mitochondrial genes (Supplemenary Table 2). Hierarchical cluster analysis of the expression patterns of these genes revealed three gene clusters (Figure 6). Most of the identified mitochondrial genes were abundant in the early stage of flower development, and gradually decreased in later stages in MS96 (Figure 6).
Figure 6.
Hierarchical cluster analysis of the expression patterns of 23 genes with high similarity to the published sequences of plant mitochondrial genes at three flower-development stages of garlic genotypes F87 and MS96. Three clusters were identified. Note enhanced representation in the early stage of MS96. The relative expression levels of each gene (column) in each sample (row) are shown. Average of three replications' expression values (FPKM) were log2-transformed and then median-centered by transcript.
Proteome analysis and transcriptome–proteome matching
For each developmental stage of both genotypes, 2D-gel protein maps were produced. Each map generated ca. 800 protein spots (data not shown). The level of map similarity was very high, but a quantitative comparison revealed 36 protein spots with significant differences, i.e., presence/absence in the genotype or at a specific developmental stage. “Difference” was defined as an at least two-fold change in the normalized volume of a particular spot. The 19 protein spots with the most significant differences in accumulation pattern between genotypes or developmental stages were excised and the proteins identified by mass spectrometry. Out of these, nine proteins were identified in genotype MS96, but not in F87. The predicted protein sequences of these nine proteins were mapped to the garlic transcriptome. Surprisingly, some of these sequences matched a number of transcripts with a high percentage of identity (Table 2).
Table 2.
Differentially accumulated proteins and matching transcripts.
| Identified protein | Accession in NCBI-nr | Reference species | Matched transcripts in garlic transcriptome catalog | Identity (%) | E-value | |
|---|---|---|---|---|---|---|
| 1 | 26S protease regulatory subunit 6A | XP_002271397 | Vitis vinifera | comp56296_c2_seq6 | 97 | 0.0 |
| homolog A | comp56296_c2_seq7 | 97 | 0.0 | |||
| 2 | Flavonol synthase | AAO63023 | Allium cepa | comp47853_c1_seq1 | 94 | 0.0 |
| 3 | ADP-ribosylation factor 1 | P51821 | Hyacinthus orientalis | comp14342_c0_seq1 | 99 | 8e-079 |
| comp14342_c0_seq2 | 99 | 8e-079 | ||||
| comp14638_c0_seq1 | 98 | 3e-052 | ||||
| comp47637_c0_seq1 | 99 | 3e-079 | ||||
| comp47637_c0_seq2 | 99 | 3e-079 | ||||
| comp47637_c0_seq4 | 99 | 1e-078 | ||||
| 4 | Biotin carboxyl carrier protein subunit of Het-ACCase (BCCP2) | XP_002526099 | Ricinus communis | comp53901_c0_seq11 | 66 | 3e-52 |
| 5 | Putative Cu/Zn superoxide dismutase | BAG16516 | Capsicum chinense | comp46488_c0_seq1 | 82 | 9e-73 |
| 6 | Cytosolic ascorbate peroxidase | AAY21068 | Capsicum annuum | comp31384_c0_seq1 | 85 | 1e-124 |
| comp42393_c0_seq1 | 81 | 1e-119 | ||||
| comp58999_c0_seq1 | 91 | 1e-084 | ||||
| 7 | Class-1 LMW heat shock protein | AAM28293 | Ananas comosus | comp56702_c0_seq11 | 73 | 7e-063 |
| comp56702_c0_seq9 | 73 | 7e-063 | ||||
| comp56702_c0_seq26 | 73 | 7e-063 | ||||
| comp56702_c0_seq20 | 73 | 1e-062 | ||||
| comp56702_c0_seq17 | 73 | 1e-062 | ||||
| comp56702_c0_seq4 | 73 | 1e-062 | ||||
| comp56702_c0_seq19 | 73 | 1e-062 | ||||
| comp56702_c0_seq6 | 73 | 1e-062 | ||||
| comp56702_c0_seq10 | 73 | 1e-062 | ||||
| 8 | Copper/zinc superoxide dismutase, partial | ADB28989 | Allium sativum | comp46488_c0_seq1 | 98 | 2e-084 |
| 9 | Calmodulin | CAA78058 | Arabidopsis thaliana | comp39196_c0_seq1 | 98 | 3e-074 |
| comp42519_c0_seq1 | 91 | 3e-067 | ||||
| comp42519_c0_seq3 | 90 | 8e-059 |
Nine proteins were identified only in genotype MS96. The proteins were identified by mass spectrometry analysis, followed by matching with NCBI-nr. The predicted protein sequence was mapped to the garlic transcriptome (NCBI bioproject PRJNA243415) using TBLASTN.
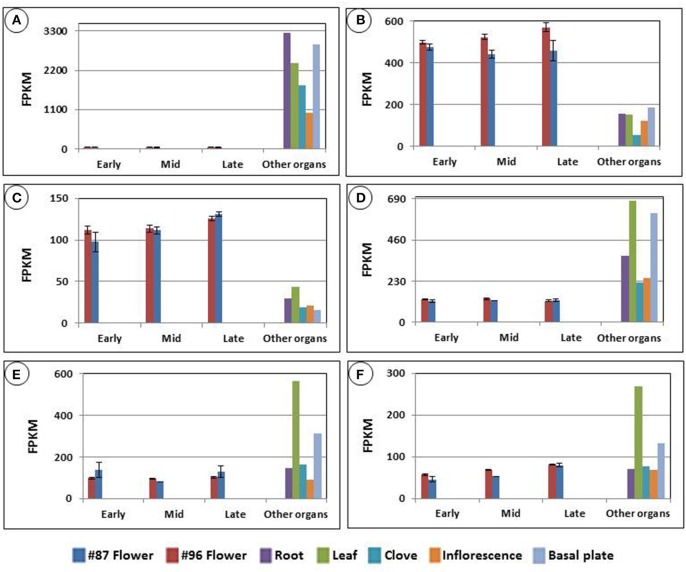
We examined the expression patterns of the transcripts that matched the nine proteins. The previously assembled organ-specific garlic transcript catalog enabled us to compare expression of these transcripts in the developing flowers with those in roots, leaves, cloves, basal plates and inflorescences. For instance, Figure 7 displays the expression of six transcripts mapped to ADP-ribosylation factor 1. Although most transcripts were mapped with high identity to the protein, their expression in garlic tissues differed dramatically. Thus, one of the transcripts was not detected in flowers (Figure 7A), whereas two others were expressed mainly in the reproductive tissues (Figures 7B,C). It should be noted that although at the protein level, the predicted ADP-ribosylation factor 1 was identified exclusively in MS96, transcript accumulation was found in both genotypes, and only one transcript showed significant expression differences between the two genotypes (Figure 7B).
Figure 7.
Six transcripts mapped to the predicted protein ADP-ribosylation factor 1, accumulated in garlic genotype MS96. The protein was identified by mass spectrometry analysis, followed by matching with NCBI-nr. Expression was calculated using TMM normalization and FPKM calculations. The comparison was made with transcript expression in roots, leaves, cloves, basal plates and inflorescences, using the organ-specific garlic transcriptome catalog (NCBI bioproject PRJNA243415). Transcript-expression levels of (A) 14342_c0_seq1, (B) 14342_c0_seq2, (C) 14638_c0_seq1, (D) 47637_c0_seq4, (E) 47637_c0_seq1, (F) 47637_c0_seq2.
Data mining for genes involved in male fertility and sterility and validation of candidate genes
Transcriptome- and proteome-assisted mining for the candidate genes associated with microsporogenesis was performed using the following approaches (Supplementary Image 1):
Bioinformatics tools were employed to select the number of the DEGs associated with male fertility. The total number of DEGs in fertile vs. male-sterile genotypes was 16,271 (Figure 3). Based on anatomical studies, differences between anthers of the two genotypes were already observed at the mid stage of development (Figure 2), and we therefore focused only on transcripts expressed in the early and mid-stages of development, thus reducing the DEG number to ca. 5500. Furthermore, only annotated DEGs were selected, reducing the list to ca. 1800 DEGs; designating the DEGs expressed in the reference transcriptome catalog only in flowers reduced the number to ca. 350.
Revision of the reduced list by GO annotation and manual search for the keywords “male sterility,” “tapetum,” and “pollen” led to selection of the homologs of three genes: MALE STERILITY 2 (MS2) which is associated with pollen development, MALE MEIOCYTE DEATH 1-LIKE (MMD1) which is associated with anther wall tapetum morphogenesis, and GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 2 (GPAT2) which is involved in pollen sperm cell differentiation.
In addition, a literature search for fertility-related genes led to focusing on homologs of the gene APETALA 3 (AP3) which is known to be associated with morphogenetic processes in the developing anthers and pollen. The sequences of AP3 were retrieved from NCBI-nr and compared with the garlic transcript catalog using TBLASTN, with an E-value cut-off of 10−5, and similarity 92% (Table 3).
Four annotated genes: NADH dehydrogenase subunit 7 (nad7), CYTOCHROME OXIDASE SUBUNIT 2 (COX2), cytochrome C maturation C (ccmC), and 18S rRNA were retrieved from the list of DEGs with high similarity to the published sequences of the mitochondrial genes. These DEGs were chosen according to their expression pattern at the transcriptome level, mainly if the expression was higher in MS96 in the early and mid-developmental stages.
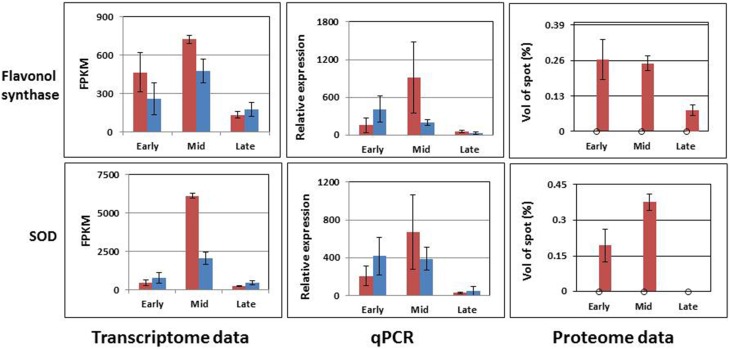
Finally, two proteins from the proteomic data, flavanol synthase and copper/zinc superoxide dismutase (SOD), and their transcripts were chosen from the list of nine proteins that were found only in the MS96 protein analysis. They were chosen according to a literature survey suggesting their involvement in fertility.
Table 3.
Transcript sequences of the 10 candidate genes for validation, retrieved from NCBI-nr databases.
| Transcript number | Candidate gene | Sequence length (bp) | Accession in NCBI-nr | Reference species | Identity (%) | E-value |
|---|---|---|---|---|---|---|
| comp35626_c0_seq1 | APETALA 3 (AP3) | 1062 | JX661502.1 | Allium cepa | 92 | 0.0 |
| comp48389_c0_seq2 | MALE STERILITY 2 (MS2) | 965 | XM_004973057.1 | Setaria italica | 57 | 1e-101 |
| comp57254_c0_seq52 | MALE MEIOCYTE DEATH 1-LIKE (MMD1) | 700 | XM_004288545 | Fragaria vesca | 55 | 6e-013 |
| comp10285_c0_seq1 | GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 2 (GPAT2) | 1237 | XM_003633174 | Vitis vinifera | 53 | 5e-081 |
| comp53958_c1_seq5 | NADH dehydrogenase subunit 7 (nad7) | 1530 | DQ381459.1 | Beta vulgaris | 97 | 0.0 |
| comp66781_c0_seq1 | Cytochrome C maturation C (ccmC) | 590 | KF798326.1 | Amborella trichopoda | 93 | 0.0 |
| comp56960_c0_seq12 | CYTOCHROME OXIDASE SUBUNIT 2 (COX2) | 930 | GU253307.1 | Allium cepa | 98 | 0.0 |
| comp53140_c0_seq2 | 18S rRNA | 950 | KC465231 | Dendrobium officinale | 99 | 0.0 |
| comp47853_c1_seq1 | Flavanol synthase | 1215 | AY221247.1 | Allium cepa | 92 | 0.0 |
| comp46488_c0_seq1 | Copper/zinc superoxide dismutase (SOD) | 876 | GU290312.2 | Allium sativum | 98 | 0.0 |
Altogether, 10 candidate genes were retrieved for further validation (Supplementary Image 1, Table 3). Bioinformatics analysis using organ-specific garlic transcript catalog, showed that most of these genes are expressed mainly in flower tissues, and less so in roots, basal plate, leaves and cloves (data not shown). However, two mitochondrial genes (COX2, 18S rRNA) were expressed not only in flowers, but also in the other organs.
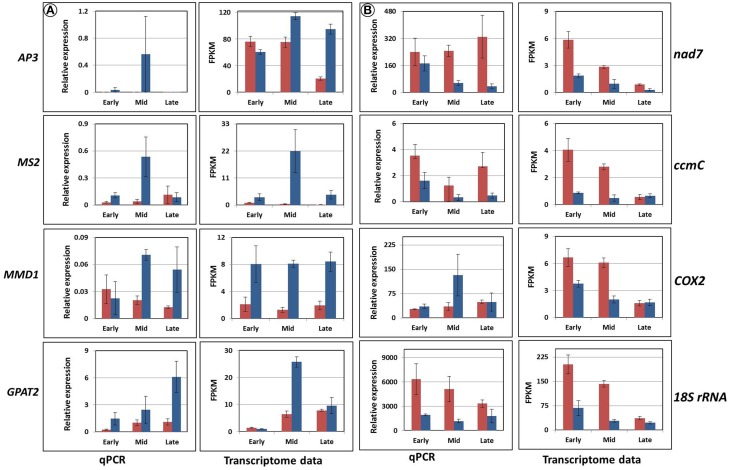
The expression analysis revealed three distinct groups of genes of interest (Figures 8, 9). The first one, abundant in F87, consisted of homologs of AP3, MS2, MMD1, and GPAT2, with putative functions in flower and pollen development (Figure 8A). The second group, abundant mainly in MS96, consisted of homologs of the mitochondrial genes nad7, ccmC, COX2, and 18S rRNA (Figure 8B).
Figure 8.
Validation of the expression of eight candidate genes by rt-qPCR and transcriptome analyses. Red columns represent gene expression in MS96, blue columns in F87. (A) Homologs of the genes AP3, MS2, MMD1, and GPAT2, expressed mainly in F87. (B) Homologs of the genes nad7, ccmC, COX2, and 18S rRNA, expressed mainly in MS96.
Figure 9.
Accumulation patterns of two predicted proteins and their matched transcripts, as estimated by bioinformatics tools and rt-qPCR. Red columns represent the expression in MS96, blue columns in F87.
The third group contained two sequences that matched predicted proteins. Although the proteins were detected only in samples from MS96, they were expressed in both genotypes at the transcript level. Moreover, the expression patterns varied between the transcriptome and rt-qPCR data (Figure 9).
Discussion
The question of fertility and male sterility is of special importance for both genetics and breeding activities in many crops. In garlic, which for years was propagated only vegetatively, this problem is of special scientific interest. The availability of a large collection of flowering garlic genotypes (Kamenetsky et al., 2005; Shemesh et al., 2008) has allowed for in-depth physiological, anatomical, morphological and genetic studies of flowering and seed production. In the present report, we combined the information obtained from morphophysiological studies with analyses of transcriptome and proteome profiles, to identify differences in global gene expression and to pinpoint the specific processes and genes involved in gametogenesis and male sterility in garlic.
Microsporogenesis and tapetal development in fertile and male-sterile plants
Our histological observations suggested that in young flower buds, microsporogenesis is intact in both fertile and male-sterile genotypes. Later, in the mid and late stages of flower development, differences in pollen differentiation became obvious. Differential expression analysis supported these morphophysiological observations: a relatively low number of genes were differentially expressed during the early stage of anther development, but this number increased significantly in the later stages of development (Figure 3).
Tapetal hypertrophy in the anthers of the male-sterile garlic genotype is associated with pollen deterioration and anther withering prior to first mitosis. In anthers of the fertile plant, no tapetal hypertrophy occurs, pedicels elongate and anthers shed viable pollen (Shemesh-Mayer et al., 2015; Figure 2). Abnormal tapetal development has been reported as one of possible reasons for male sterility in bulb onion (Monosmith, 1928; Saini and Davis, 1969; Holford et al., 1991), Arabidopsis (Chaudhury et al., 1994), rice (Nishiyama, 1976), alfalfa (Childers, 1952), and tomato (Rick, 1948), among others. In maize, sunflower and petunia, premature tapetal degeneration is one of the first predictive signs of CMS (Schnable and Wise, 1998). Similarly, in garlic, tapetal hypertrophy, caused by either genetic or abiotic factors, may lead to malnutrition of the released microspores, with the consequent development of empty, non-viable pollen grains (Shemesh-Mayer et al., 2015).
Gene expression of the fertile and male-sterile genotypes exhibits opposite patterns
In the fertile genotype, vital pollen development is associated with the activity of genes involved in general development of the reproductive tissues, e.g., regulation of meristem structural organization, floral organ development, regulation of cellular-component organization and sugar metabolism (Figure 5). In addition, the processes of cell division associated with microsporogenesis were abundant. Similarly, in Brassica napus, maize and petunia, fertility was associated with genes involved in pollen-wall assembly, energy transfer and pollen function and germination (Yan et al., 2013).
Pollen grains are rich in triglycerides, starch and other storage compounds for energy supply during germination, and also contain large amounts of the enzymes involved in glycolysis and mitochondrial respiration (Bhandari, 1984; Dorion et al., 1996; Kerim et al., 2003; Miernyk et al., 2011). In maize, the level of hexoses is much higher in the anthers of male-fertile than male-sterile plants (Datta et al., 2002). Similarly, in fertile garlic flowers, GO analysis has shown an abundance of proteins participating in processes associated with polysaccharides, which are required for energy supply (Figure 5).
Expression analysis of four fertility-related genes (homologs of AP3, MMD1, MS2, and GPAT2) confirmed their higher expression in the fertile vs. sterile genotype (Figure 8). The AP3 gene is a MADS-box transcription factor that controls flower differentiation (Honma and Goto, 2001), and a homolog of this gene might be involved in normal garlic florogenesis. MMD1, one of the homologs of MS1, is involved in the formation of pollen exine and cytosolic components, as well as tapetum development (Ito et al., 2007), and is also required for male meiosis (Yang et al., 2003). In Arabidopsis and rice, mutations in these genes result in lack of normal tapetal programmed cell death (PCD), lack of tapetal DNA fragmentation, delayed tapetal degeneration, abnormal pollen-wall formation and microspore abortion (Ito and Shinozaki, 2002; Li et al., 2011). Similar phenomena were observed in the male-sterile genotype of garlic, with low expression of MMD1 homolog (Figure 8). We therefore suggest that MMD1 plays a role in controlling normal pollen formation. MS2 is strongly related to fertility and encodes a predicted fatty acyl-CoA reductase, involved in fatty-acyl-CoA transport, which is required for exine formation. MS2 protein specifically accumulates in the tapetum. In Arabidopsis ms2 mutants, microspores collapse shortly after their release from the tetrads, and pollen-wall formation is not detected (Aarts et al., 1997; de Azevedo Souza et al., 2009). In fertile garlic, the MS2 homolog showed markedly higher expression, mostly at the mid stage of flower development, following microspore release from callose (Figures 2, 8). We therefore propose that this gene might play an essential role in pollen formation, and can be considered a potential marker for fertility in garlic. Members of the GPAT family are involved in pollen development and tapetum viability in Arabidopsis (Zheng et al., 2003). Similarly, abundance of the GPAT2 homolog in the fertile genotype indicates its involvement in normal functioning of the garlic tapetum (Figure 8).
Male-sterility in garlic is associated with respiration restriction (the energy deficiency model)
According to the GO analysis, the flowers of male sterile garlic have a higher abundance of genes associated with energy flow, respiration and mitochondrial functions. In general, in higher plants, CMS is associated with alterations in the mitochondrial genome that affect the functions of anthers, pollen or male gametes. The genes that determine CMS have chimeric open-reading-frames (ORFs) of unknown origin and function (Woodson and Chory, 2008; Shaya et al., 2012; Islam et al., 2014). Many of these genes consist of portions of essential mitochondrial genes involved in respiration pathways, including NADH-dehydrogenase subunit genes, cytochrome oxidase, and ATP synthase. Such fused genes may disrupt the mitochondrial function by impairing its membrane integrity and interfering with the expression of intact mitochondrial genes, with a consequent reduction in respiration and energy supply (Chen and Liu, 2014). This phenomenon has been described as the “Energy Deficiency Model” (Woodson and Chory, 2008; Chen and Liu, 2014). In male-sterile garlic, a relatively high expression of mitochondrial genes is already noted at the meiotic stage, prior to the appearance of the anatomical evidence of sterility (Figure 6). Later, when tapetum heterotrophy and pollen abortion are evident in the male-sterile anthers (Figure 2, Shemesh-Mayer et al., 2015), mitochondrial gene expression decreases (Figure 6). We therefore propose that in garlic, similar to other plant species (Woodson and Chory, 2008; Shaya et al., 2012; Chen and Liu, 2014), the chimeric ORFs might be fused to mitochondrial genes, impairing their respiratory system. Furthermore, the internal feedback from low energy in the cell urges the mitochondria to overcome the shortage, thus resulting in a high expression of mitochondrial genes.
This hypothesis is supported by higher expression of three specific mitochondrial genes in male-sterile as compared to fertile flowers (Figure 8). The association between these genes and CMS has been also reported in sterile phenotypes of Petunia and Nicotiana sylvestris (NADH-dehydrogenase subunit gene; Rasmussen and Hanson, 1989; Pla et al., 1995), in Brassica napus (ccmC, a participant in the respiratory complex; Leino et al., 2005; Kim et al., 2009b) and in maize (18S rRNA; Wen and Chase, 1999). Although COX2 is associated with CMS in sugar beet (Senda et al., 1991), our analysis of the garlic COX2 homolog gave controversial results in the transcriptome analysis vs. qPCR validation, probably due to low sensitivity of the primers.
Oxidative stress might induce male sterility in garlic (the aberrant PCD model)
The “Aberrant PCD Model” suggests that the increased production of reactive oxygen species (ROS) in mitochondria stimulates unregulated PCD in the tapetum, resulting in pollen abortion (Chen and Liu, 2014; Touzet and Meyer, 2014). Mitochondrial ROS, generated by oxidative stress, are abundant in the anthers of the CMS lines of sunflower, cotton and rice (Jiang et al., 2007; Touzet and Meyer, 2014). In male-sterile garlic, this model might explain the results of the GO analysis, which indicated processes of oxidative stress and negative regulation of PCD, suggesting premature tapetal PCD and consequent microspore degeneration (Figure 5B).
Correlation of mRNA and protein accumulation
Numerous reports have suggested that RNA transcript accumulation is not always conveyed to the final product—protein. For example, a negative correlation between mRNA and protein accumulation patterns was found in Arabidopsis in response to cold treatment (Nakaminami et al., 2014) and in Japanese apricot buds following gibberellin treatment (Zhuang et al., 2013). Therefore, the changes in protein might not always be associated with the changes in function.
In garlic, proteome analysis generated 36 proteins whose levels varied significantly between the fertile and sterile flowers; 9 of them presented exclusively in the latter. Within this latter group, a few genes are known to be involved in pollen development. Thus, misregulation of ADP ribosylation factor contributes to the defects in tapetal cell division and callose secretion (e.g., in maize, Wang et al., 2012); SOD acts as an antioxidant and is involved in sporogenous cell division (e.g., in cotton, Jiang et al., 2007). Our results indicate that at the mid stage of male-sterile flower development, when hypertrophy of the tapetum first becomes evident, the transcript and protein levels of SOD are high (Figure 9). It is possible that the stress from ROS production at this developmental stage serves as a signal to increase SOD activity due to a feedback mechanism, as has been described in cotton (Jiang et al., 2007). Later, a very low level of SOD expression was registered, when massive pollen abortion occurs (Figures 2, 9). This decrease might result from high ROS level and oxidative damage to the developing pollen.
It is suggested that in garlic, separate transcriptome or proteome analyses are not sufficient to understand the underlying genetic and molecular mechanisms. For a comprehensive interpretation of developmental processes, including mechanisms of male sterility, future research and integration of physiological and anatomical studies with transcriptome, proteome and metabolome analyses are required.
Future research and practical use of garlic fertility and male sterility
A comparison of morphophysiological and molecular traits of fertile and male-sterile garlic flowers suggests that the most vulnerable stage of pollen development is the post-meiotic, uninuclear stage (Shemesh-Mayer et al., 2015; Figure 2). During this stage, respiratory restrictions caused by a disruption in mitochondrial functions can lead to energy deficiency and consequent pollen abortion. This process can also be linked to oxidative stress, which negatively affects tapetum development and pollen nutrition. Similar to onion, bunching onion and chives, garlic male sterility might result from the interaction between CMS and GMS (CGMS). This notion warrants further investigation. Transcriptome and proteome analyses suggest that garlic homologs of MS2, MMD1, and GPAT2 can be further developed as possible molecular markers for fertility, and nad7, ccmC, 18S rRNA as possible markers for male sterility in garlic.
In order to verify CMS gene candidates, functional data analysis is required. One of the main technical barriers to such analysis is the lack of a successful method to transform plant mitochondrial genomes. As an alternative strategy, recombinant constructs of the CMS candidate genes (the chimeric ORFs fused with a mitochondrial targeting signal sequence) from bean, rice, sugar beet, pepper and other plants were produced and transferred into the nuclei of tobacco plants. This procedure caused male sterility in tobacco, thus verifying the CMS functions of the fused genes (Chen and Liu, 2014). The second option can be use of the transformation system in garlic. However, garlic is a bulbous perennial plant with a complicated flower biology, and its transformation system is not available yet. Therefore, unlike in model plants systems, the application of functional data analysis in garlic is not feasible at this point. Future research is needed to define garlic CMS mitochondrial genes and to provide their functional analysis in model plants or in garlic.
Male sterility is known to be a practical and cost-effective mechanism for commercial F1 hybrid seed production in onion (Havey, 2004). Male-sterile plants have a selective advantage: as a result of reallocation of the energy saved from pollen production, they produce more or better seeds than the corresponding hermaphroditic plants (Touzet and Meyer, 2014). Male-sterile garlic clones are characterized by enhanced seed setting compared to fertile plants, and therefore can be used in the commercial production of hybrid seeds. Searching for specific chimeric ORFs fused to mitochondrial genes in garlic will facilitate further identification of molecular markers related to male sterility, as has been done in onion (Kim et al., 2009a), and will enable the future use of male sterility in the commercial hybridization of garlic.
Author contributions
All authors contributed to the experiment and the manuscript. ESM, AS, AK, and RK designed the experiment and drafted the manuscript. ADF carried out the bioinformatics analysis. ESM, TBM, NR, and DP conducted the experimental work. HDR and AS conceived the study, and contributed to the writing of the manuscript. This research is a part of the graduate studies of ESM and TBM. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was partly financed by “Classeed Ltd.” We thank the two reviewers for their helpful comments and suggestions.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00271/abstract
References
- Aarts M. G., Hodge R., Kalantidis K., Florack D., Wilson Z. A., Mulligan B. J., et al. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12, 615–623. 10.1046/j.1365-313X.1997.d01-8.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K., Earle E. D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218 10.1007/BF02672069 [DOI] [Google Scholar]
- Bhandari N. N. (1984). The microsporangium, in Embryology of Angiosperms, ed Johri B. M. (New York, NY: Springer; ), 53–121. [Google Scholar]
- Borg M., Brownfield L., Twell D. (2009). Male gametophyte development: a molecular perspective. J. Exp. Bot. 60, 1465–1478. 10.1093/jxb/ern355 [DOI] [PubMed] [Google Scholar]
- Budar F., Pelletier G. (2001). Male sterility in plants: occurrence, determinism, significance and use. C. R. Acad. Sci. III 324, 543–550. 10.1016/S0764-4469(01)01324-5 [DOI] [PubMed] [Google Scholar]
- Chang F., Wang Y., Wang S., Ma H. (2011). Molecular control of microsporogenesis in Arabidopsis. Curr. Opin. Plant Biol. 14, 66–73. 10.1016/j.pbi.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Chaudhury A. M., Craig S., Dennis E. S., Lavithis M., Taylor P. E., Singh M. B., et al. (1994). Genetic control of male fertility in Arabidopsis thaliana: structural analysis of premeiotic developmental mutants. Sex. Plant Reprod. 7, 17–28. 10.1007/BF00241884 [DOI] [PubMed] [Google Scholar]
- Chen L., Liu Y. G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. 10.1146/annurev-arplant-050213-040119 [DOI] [PubMed] [Google Scholar]
- Childers W. R. (1952). Male sterility in Medicago sativa L. Sci. Agric. 32, 351–364. [Google Scholar]
- Conesa A. S., Götz J. M., Garcia-Gomez J., Talon T. M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Datta R., Chamusco K. C., Chourey P. S. (2002). Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 130, 1645–1656. 10.1104/pp.006908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S. S., Koch S., Kienow L., Schneider K., McKim S. M., et al. (2009). A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21, 507–525. 10.1105/tpc.108.062513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorion S., Lalonde S., Saini H. S. (1996). Induction of male sterility in wheat by meiotic-stage water deficit is preceded by a decline in invertase activity and changes in carbohydrate metabolism in anthers. Plant Physiol. 111, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etoh T., Noma Y., Nishitarumizu Y., Wakamoto T. (1988). Seed productivity and germinability of various garlic clones collected in Soviet Central Asia. Mem. Fac. Agric. Kagoshima Univ. 24, 129–139. [Google Scholar]
- Feng X., Dickinson H. G. (2007). Packaging the male germline in plants. Trends Genet. 23, 503–510. 10.1016/j.tig.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Beals T. P., Sanders P. M. (1993). Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229. 10.1105/tpc.5.10.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. R. (1991). Plant mitochondrial mutations and male sterility. Annu. Rev. Genet. 25, 461–486. 10.1146/annurev.ge.25.120191.002333 [DOI] [PubMed] [Google Scholar]
- Havey M. J. (1993). A putative donor of S-cytoplasm and its distribution among open-pollinated populations of onion. Theor. Appl. Genet. 86, 128–134. 10.1007/BF00223817 [DOI] [PubMed] [Google Scholar]
- Havey M. J. (1995). Identification of cytoplasms using the polymerase chain reaction to aid in the extraction of maintainer lines from open-pollinated populations of onion. Theor. Appl. Genet. 90, 263–268. 10.1007/BF00222212 [DOI] [PubMed] [Google Scholar]
- Havey M. J. (2004). The use of cytoplasmic male sterility for hybrid seed production, in Molecular Biology and Biotechnology of Plant Organelles, eds Daniell H., Chase C. (Dordrecht: Springer; ), 623–634. [Google Scholar]
- Holford P., Croft J., Newbury H. J. (1991). Structural studies of microsporogenesis in fertile and male sterile onions (Allium cepa L.) containing the cms-S cytoplasm. Theor. Appl. Genet. 82, 745–755. 10.1007/BF00227320 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. 10.1038/35054083 [DOI] [PubMed] [Google Scholar]
- Hsu Y. F., Wang C. S., Raja R. (2007). Gene expression pattern at desiccation in the anther of Lilium longiflorum. Planta 226, 311–322. 10.1007/s00425-007-0483-5 [DOI] [PubMed] [Google Scholar]
- Hurkman W. J., Tanaka C. K. (1986). Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81, 802–806. 10.1104/pp.81.3.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Hoshino Y., Yanagawa Y. (2009). Isolation and characterization of the plant glsA promoter from Alstroemeria. Plant Biol. 11, 878–885. 10.1111/j.1438-8677.2008.00177.x [DOI] [PubMed] [Google Scholar]
- Imin N., Kerim T., Weinman J. J., Rolfe B. G. (2001). Characterisation of rice anther proteins expressed at the young microspore stage. Proteomics 1, 1149–1161. [DOI] [PubMed] [Google Scholar]
- Islam M., Studer B., Møller I. M., Asp T. (2014). Genetics and biology of cytoplasmic male sterility and its applications in forage and turf grass breeding. Plant Breed. 133, 299–312 10.1111/pbr.12155 [DOI] [Google Scholar]
- Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19, 3549–3562. 10.1105/tpc.107.054536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Shinozaki K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292. 10.1093/pcp/pcf154 [DOI] [PubMed] [Google Scholar]
- Jenderek M. M., Hannan R. M. (2000). Seed producing ability of garlic (Allium sativum L.) clones from two public US collections, in Proceedings of the Third International Symposium on Edible Alliaceae, ed Randle W. M. (Athens, GA: University of Georgia; ), 73–75. [Google Scholar]
- Jenderek M. M., Zewdie Y. (2005). Within- and between-family variability for important bulb and plant traits among sexually derived progenies of garlic. Hortscience 40, 1234–1236. [Google Scholar]
- Jiang P., Zhang X., Zhu Y., Zhu W., Xie H., Wang X. (2007). Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep. 26, 1627–1634. 10.1007/s00299-007-0351-6 [DOI] [PubMed] [Google Scholar]
- Jones H. A., Clarke A. E. (1943). Inheritance of male sterility in the onion and the production of hybrid seed. Proc. Am. Soc. Hortic. Sci. 43, 189–194. [Google Scholar]
- Jones H. A., Emsweller S. L. (1937). A mail sterile onion. Proc. Am. Soc. Hortic. Sci. 34, 582–585. [Google Scholar]
- Kamenetsky R. (2007). Garlic: botany and horticulture. Hortic. Rev. 33, 123–172 10.1002/9780470168011.ch2 [DOI] [Google Scholar]
- Kamenetsky R., Faigenboim A., Shemesh-Mayer E., Ben Michael T., Gershberg C., Kimhi S., et al. (2015). Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genomics 16:12. 10.1186/s12864-015-1212-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetsky R., London Shafir I., Khassanov F., Kik C., van Heusden A. W., Vrielink-van Ginkel M., et al. (2005). Diversity in fertility potential and organo-sulphur compounds among garlics from Central Asia. Biodivers. Conserv. 14, 281–295 10.1007/s10531-004-5050-9 [DOI] [Google Scholar]
- Kerim T., Imin N., Weinman J. J., Rolfe B. G. (2003). Proteome analysis of male gametophyte development in rice anthers. Proteomics 3, 738–751. 10.1002/pmic.200300424 [DOI] [PubMed] [Google Scholar]
- Khurana R., Kapoor S., Tyagi A. K. (2012). Anthology of anther/pollen-specific promoters and transcription factors. Crit. Rev. Plant Sci. 31, 359–390 10.1080/07352689.2012.664986 [DOI] [Google Scholar]
- Kim S., Lee E. T., Cho D. Y., Han T., Bang H., Patil B. S., et al. (2009a). Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing among three cytoplasm types in onion (Allium cepa L.). Theor. Appl. Genet. 118, 433–441. 10.1007/s00122-008-0909-x [DOI] [PubMed] [Google Scholar]
- Kim S. R., Yang J. I., Moon S., Ryu C. H., An K., Kim K. M., et al. (2009b). Rice OGR1 encodes a pentatricopeptide repeat–DYW protein and is essential for RNA editing in mitochondria. Plant J. 59, 738–749. 10.1111/j.1365-313X.2009.03909.x [DOI] [PubMed] [Google Scholar]
- Kosmala A., Bocian A., Rapacz M., Jurczyk B., Zwierzykowski Z. (2009). Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J. Exp. Bot. 60, 3595–3609. 10.1093/jxb/erp205 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser K. D., Lersten N. R. (1972). Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 38, 425–454 10.1007/BF02860010 [DOI] [Google Scholar]
- Leino M., Landgren M., Glimelius K. (2005). Alloplasmic effects on mitochondrial transcriptional activity and RNA turnover result in accumulated transcripts of Arabidopsis orfs in cytoplasmic male-sterile Brassica napus. Plant J. 42, 469–480. 10.1111/j.1365-313X.2005.02389.x [DOI] [PubMed] [Google Scholar]
- Li H., Yuan Z., Vizcay-Barrena G., Yang C., Liang W., Zong J., et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630. 10.1104/pp.111.175760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Joshi T., Takahashi K., Langley R. J., Franklin L. D., et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. 10.1111/j.1365-313X.2010.04222.x [DOI] [PubMed] [Google Scholar]
- Liu C., Ma N., Wang P. Y., Fu N., Shen H. L. (2013). Transcriptome sequencing and de novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.). PLoS ONE 8:e65209. 10.1371/journal.pone.0065209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J. P. (1990). Gene activity during pollen development. Annu. Rev. Plant Biol. 41, 317–338 10.1146/annurev.pp.41.060190.001533 [DOI] [Google Scholar]
- McCormick S. (2004). Control of male gametophyte development. Plant Cell 16(Suppl. 1), S142–S153. 10.1105/tpc.016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J. A., Pret'ová A., Olmedilla A., Klubicová K., Obert B., Hajduch M. (2011). Using proteomics to study sexual reproduction in angiosperms. Sex. Plant Reprod. 24, 9–22. 10.1007/s00497-010-0149-5 [DOI] [PubMed] [Google Scholar]
- Mihr C., Baumgärtner M., Dieterich J. H., Schmitz U. K., Braun H. P. (2001). Proteomic approach for investigation of cytoplasmic male sterility (CMS) in Brassica. J. Plant Physiol. 158, 787–794 10.1078/0176-1617-00292 [DOI] [Google Scholar]
- Mo Y., Nagel C., Taylor L. P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. U.S.A. 89, 7213–7217. 10.1073/pnas.89.15.7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosmith H. R. (1928). Male sterility in Allium cepa. Ph.D. thesis, University of California, Berkeley, CA. [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2003). Identification of higher plant GlsA, a putative morphogenesis factor of gametic cells. Biochem. Biophys. Res. Commun. 306, 564–569. 10.1016/S0006-291X(03)01005-2 [DOI] [PubMed] [Google Scholar]
- Moue T., Uehara T. (1985). Inheritance of cytoplasmic male sterility in Allium fistulosum L. (Welsh onion). J. Jpn. Soc. Hortic. Sci. 53, 432–437 10.2503/jjshs.53.432 [DOI] [Google Scholar]
- Nakaminami K., Matsui A., Nakagami H., Minami A., Nomura Y., Tanaka M., et al. (2014). Analysis of differential expression patterns of mRNA and protein during cold-and de-acclimation in Arabidopsis. Mol. Cell. Proteomics 13, 3602–3611. 10.1074/mcp.M114.039081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama I. (1976). Male sterility caused by cooling treatment at the young microspore stage in rice plants. XII. Classification of tapetal hypertrophy on the basis of ultrastructure. Proc. Crop Sci. Soc. Jpn. 45, 254–262 10.1626/jcs.45.254 [DOI] [Google Scholar]
- Nishimura Y., Shibano M. (1972). Male sterility in Allium fistulosum L. Cytological and anatomical studies. Abstr. J. Jpn. Soc. Hort. Sci. Spring Meet. 180–181. [Google Scholar]
- Perlikowski D., Kosmala A., Rapacz M., Kościelniak J., Pawłowicz I., Zwierzykowski Z. (2014a). Influence of short-term drought conditions and subsequent re-watering on the physiology and proteome of Lolium multiflorum/Festuca arundinacea introgression forms with contrasting levels of tolerance to long-term drought. Plant Biol. 16, 385–394. 10.1111/plb.12074 [DOI] [PubMed] [Google Scholar]
- Perlikowski D., Wiśniewska H., Góral T., Kwiatek M., Majka M., Kosmala A. (2014b). Identification of kernel proteins associated with the resistance to Fusarium head blight in winter wheat (Triticum aestivum L.). PLoS ONE 9:e110822. 10.1371/journal.pone.0110822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla M., Mathieu C., De Paepe R., Chétrit P., Vedel F. (1995). Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol. Gen. Genet. 248, 79–88. 10.1007/BF02456616 [DOI] [PubMed] [Google Scholar]
- Pooler M. R., Simon P. W. (1993). Garlic flowering in response to clone, photoperiod, growth temperatures and cold storage. HortScience 28, 1085–1086. [Google Scholar]
- Pooler M. R., Simon P. W. (1994). True seed production in garlic. Sex. Plant Reprod. 7, 282–286 10.1007/BF00227710 [DOI] [Google Scholar]
- Rasmussen J., Hanson M. R. (1989). A NADH dehydrogenase subunit gene is co-transcribed with the abnormal Petunia mitochondrial gene associated with cytoplasmic male sterility. Mol. Gen. Genet. 215, 332–336. 10.1007/BF00339738 [DOI] [PubMed] [Google Scholar]
- Rick C. M. (1948). Genetics and development of nine male-sterile tomato mutants. Hilgardia 18, 599–633 10.3733/hilg.v18n17p599 [DOI] [Google Scholar]
- Rotem N., David Schwartz R., Peretz Y., Sela I., Rabinowitch H. D., Flaishman M., et al. (2011). Flower development in garlic: the ups and downs of gaLFY expression. Planta 233, 1063–1072. 10.1007/s00425-011-1361-8 [DOI] [PubMed] [Google Scholar]
- Rotem N., Shemesh E., Peretz Y., Akad F., Edelbaum O., Rabinowitch H. D., et al. (2007). Reproductive development and phenotypic differences in garlic are associated with expression and splicing of LEAFY homologue gaLFY. J. Exp. Bot. 58, 1133–1141. 10.1093/jxb/erl272 [DOI] [PubMed] [Google Scholar]
- Ruzin S. E. (1999). Plant Microtechnique and Microscopy. Oxford: Oxford University Press. [Google Scholar]
- Saini S. S., Davis G. N. (1969). Male Sterility in Allium cepa and some species hybrids. Econ. Bot. 23, 37–49 10.1007/BF02862970 [DOI] [Google Scholar]
- Sawhney V. P., Shukla A. (1994). Male sterility in flowering plants: are plant growth substances involved? Am. J. Bot. 8, 1640–1647. 10.2307/24453439351246 [DOI] [Google Scholar]
- Schnable P. S., Wise R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180 10.1016/S1360-1385(98)01235-7 [DOI] [Google Scholar]
- Scott R. J., Spielman M., Dickinson H. G. (2004). Stamen structure and function. Plant Cell 16(Suppl. 1), S46–S60. 10.1105/tpc.017012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda M., Harada T., Mikami T., Sugiura M., Kinoshita T. (1991). Genomic organization and sequence analysis of the cytochrome oxidase subunit II gene from normal and male-sterile mitochondria in sugar beet. Curr. Genet. 19, 175–181. 10.1007/BF00336484 [DOI] [PubMed] [Google Scholar]
- Shaya F., Gaiduk S., Keren I., Shevtsov S., Zemah H., Belausov E., et al. (2012). Expression of mitochondrial gene fragments within the tapetum induce male sterility by limiting the biogenesis of the respiratory machinery in transgenic tobacco. J. Integr. Plant Biol. 54, 115–130. 10.1111/j.1744-7909.2012.01099.x [DOI] [PubMed] [Google Scholar]
- Shemesh E., Scholten O., Rabinowitch H. D., Kamenetsky R. (2008). Unlocking variability: inherent variation and developmental traits of garlic plants originated from sexual reproduction. Planta 227, 1013–1024. 10.1007/s00425-007-0675-z [DOI] [PubMed] [Google Scholar]
- Shemesh-Mayer E., Ben-Michael T., Kimhi S., Forer I., Rabinowitch H. D., Kamenetsky R. (2015). Effects of different temperature regimes on flower development, microsporogenesis and fertility in bolting garlic (Allium sativum). Funct. Plant Biol. 10.1071/FP14262 [DOI] [PubMed] [Google Scholar]
- Shemesh-Mayer E., Winiarczyk K., Błaszczyk L., Kosmala A., Rabinowitch H. D., Kamenetsky R. (2013). Male gametogenesis and sterility in garlic (Allium sativum L.): barriers on the way to fertilization and seed production. Planta 237, 103–120. 10.1007/s00425-012-1748-1 [DOI] [PubMed] [Google Scholar]
- Sheoran I. S., Ross A. R., Olson D. J., Sawhney V. K. (2009). Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato (Solanum lycopersicum): a proteomic analysis. J. Proteomics 71, 624–636. 10.1016/j.jprot.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatlioglu T. (1982). Cytoplasmic male sterility in chives (Allium schoenoprasum L.). Z. Pflanzenzuecht. 89, 251–262. [DOI] [PubMed] [Google Scholar]
- Touzet P., Meyer E. H. (2014). Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 19, 166–171. 10.1016/j.mito.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S. G., Hu J., Dill A., Alonso J. M., Ecker J. R., et al. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019. 10.1104/pp.104.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Adams C. M., Fernandes J. F., Egger R. L., Walbot V. (2012). A low molecular weight proteome comparison of fertile and male sterile 8 anthers of Zea mays. Plant Biotechnol. J. 10, 925–935. 10.1111/j.1467-7652.2012.00721.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L. Q., Xu W. Y., Deng Z. Y., Su Z., Xue Y., Wang T. (2010). Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11:338. 10.1186/1471-2164-11-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ruesch K. L., Ortega V. M., Kamps T. L., Gabay-Laughnan S., Chase C. D. (2003). A nuclear restorer-of-fertility mutation disrupts accumulation of mitochondrial ATP synthase subunit α in developing pollen of S male-sterile maize. Genetics 165, 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L. Y., Chase C. D. (1999). Mitochondrial gene expression in developing male gametophytes of male-fertile and S male-sterile maize. Sex. Plant Reprod. 11, 323–330 10.1007/s004970050159 [DOI] [Google Scholar]
- Wilson Z. A., Zhang D. B. (2009). From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60, 1479–1492. 10.1093/jxb/erp095 [DOI] [PubMed] [Google Scholar]
- Woodson J. D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Dong C., Yu J., Liu W., Jiang C., Liu J., et al. (2013). Transcriptome profile analysis of young floral buds of fertile and sterile plants from the self-pollinated offspring of the hybrid between novel restorer line NR1 and Nsa CMS line in Brassica napus. BMC Genomics 14:26. 10.1186/1471-2164-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Makaroff C. A., Ma H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15, 1281–1295. 10.1105/tpc.010447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yang L. (2014). Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55. 10.1016/j.pbi.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Xia Q., Dauk M., Shen W., Selvaraj G., Zou J. (2003). Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15, 1872–1887. 10.1105/tpc.012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang W., Gao Z., Wang L., Zhong W., Ni Z., Zhang Z. (2013). Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 64, 4953–4966. 10.1093/jxb/ert284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.