Significance
This study enhances the knowledge on the molecular pathogenesis of idiopathic pulmonary fibrosis (IPF). To date, there is no information available on the role of peroxisomes in lung fibrosis. In our study we demonstrate that peroxisomal biogenesis and metabolism is compromised in tissue samples as well as in fibroblasts of IPF patients and in bleomycin-induced fibrosis mouse model. Moreover, RNAi-mediated knockdown of peroxisomal biogenesis leads to a profibrotic response in control and IPF fibroblasts suggesting that the reduction of peroxisomal function in IPF would contribute to the profibrotic phenotype of this devastating disease. Our work opens a new field of research in the area of lung fibrosis and might lead to novel treatment strategies against IPF by modulating the peroxisomal compartment.
Keywords: peroxisome, lung, fibrosis, inflammation, TGF-β1
Abstract
Idiopathic pulmonary fibrosis (IPF) is a devastating disease, and its pathogenic mechanisms remain incompletely understood. Peroxisomes are known to be important in ROS and proinflammatory lipid degradation, and their deficiency induces liver fibrosis. However, altered peroxisome functions in IPF pathogenesis have never been investigated. By comparing peroxisome-related protein and gene expression in lung tissue and isolated lung fibroblasts between human control and IPF patients, we found that IPF lungs exhibited a significant down-regulation of peroxisomal biogenesis and metabolism (e.g., PEX13p and acyl-CoA oxidase 1). Moreover, in vivo the bleomycin-induced down-regulation of peroxisomes was abrogated in transforming growth factor beta (TGF-β) receptor II knockout mice indicating a role for TGF-β signaling in the regulation of peroxisomes. Furthermore, in vitro treatment of IPF fibroblasts with the profibrotic factors TGF-β1 or tumor necrosis factor alpha (TNF-α) was found to down-regulate peroxisomes via the AP-1 signaling pathway. Therefore, the molecular mechanisms by which reduced peroxisomal functions contribute to enhanced fibrosis were further studied. Direct down-regulation of PEX13 by RNAi induced the activation of Smad-dependent TGF-β signaling accompanied by increased ROS production and resulted in the release of cytokines (e.g., IL-6, TGF-β) and excessive production of collagen I and III. In contrast, treatment of fibroblasts with ciprofibrate or WY14643, PPAR-α activators, led to peroxisome proliferation and reduced the TGF-β–induced myofibroblast differentiation and collagen protein in IPF cells. Taken together, our findings suggest that compromised peroxisome activity might play an important role in the molecular pathogenesis of IPF and fibrosis progression, possibly by exacerbating pulmonary inflammation and intensifying the fibrotic response in the patients.
Idiopathic pulmonary fibrosis (IPF) is a chronic, devastating, and lethal fibrotic disorder in human lung. IPF is characterized by a worsening of pulmonary function and persistent alterations of the lung parenchyma as a result of fibrotic foci formation by activated fibroblasts/myofibroblasts and excessive production and deposition of extracellular matrix components (ECM) (1–4). It is well accepted that transforming growth factor beta (TGF-β) signaling plays a critical role in IPF development. Inhibition of TGF-β signaling by blocking its downstream Smad3 gene expression protects against bleomycin-induced fibrosis in animal models (5, 6). In addition, there is increasing evidence that tumor necrosis factor alpha (TNF-α) also plays an important role in initiation and perpetuation of the fibrotic processes, possibly by activating TGF-β signaling pathway (7). However, the mechanisms by which TGF-β and TNF-α promote the fibrotic response in IPF are incompletely known.
Peroxisomes are single membrane bounded ubiquitous organelles, present in all types of cells. Particularly, type II alveolar epithelial cells and club cells (Clara) in the lung have highly abundant peroxisomes (8). These organelles are involved in a variety of metabolic pathways, including degradation of reactive oxygen species (ROS) and bioactive lipid mediators (prostaglandins and leukotriens) and synthesis of antioxidant lipids (polyunsaturated fatty acids, plasmalogens, etc.) (9). Absence or dysfunction of peroxisomes results in increased cellular oxidative stress, leading to severe pathological consequences in many organ systems (10, 11). Lung is one of the organs with highest exposure to various forms of reactive oxygen and nitrogen species (ROS and RNS) due to oxygen and different environmental oxidants in the inspired air, causing oxidation of cellular DNA, proteins and lipids, consequently a direct lung injury (12). Studies have shown that the most severe phenotype of a peroxisome biogenesis disorder (e.g., Zellweger syndrome) is associated with progressive liver fibrosis or cirrhosis, leading to early death of the patients during childhood (11). Moreover, mice with peroxisome dysfunction caused by PEX11β knockout died during their first days of life and exhibit morphological alterations of the lungs (13). In contrast, treatment of rats with an agonist specific for peroxisome proliferator-activated receptor alpha (PPAR-α) significantly ameliorated tubulointerstitial renal fibrosis (14). Despite the fact that peroxisomal metabolism might play an important role in other tissue fibrosis, the role of peroxisomes in lung fibrosis onset and progression seen in IPF patients has never been reported (1, 15).
Herein, using human IPF and control fibroblast cultures as well as a bleomycin-induced mouse lung fibrosis model, we demonstrate that peroxisomal biogenesis and metabolism is compromised in the lung and in fibroblasts of IPF patients, in which a down-regulation of peroxisomal proteins leads to activation and release of profibrotic factors such as TGF-β1 and collagen. In contrast, peroxisome proliferation by treatment with PPAR-α agonist (ciprofibrate, WY14643) significantly reduces the TGF-β1–induced myofibroblast differentiation in IPF fibroblast cultures.
Results
Peroxisome Biogenesis, Lipid Metabolism, and Redox Balance Are Compromised in IPF Patients.
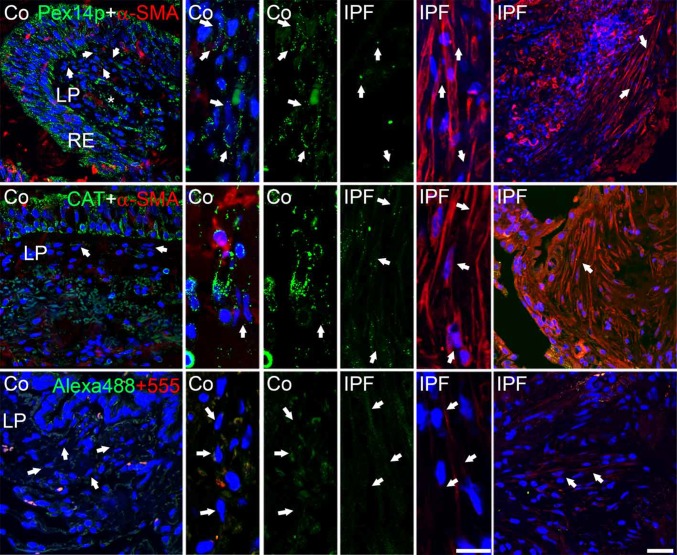
First we analyzed control and patient tissue samples for peroxisomal alterations. Stainings of paraffin-embedded tissue sections of human lung biopsies of controls and IPF patients revealed that peroxisomal markers (PEX14p and cat) in fibroblasts were found to be significantly (Fig. S1E) reduced in IPF lungs (Fig. 1). Next we analyzed whereas lung fibroblasts isolated from these human IPF patients would exhibit similar peroxisomal alterations. IPF fibroblasts retained their profibrotic activity also in cell culture such as the expression of α-SMA (Fig. 2A) and expressed increased mRNA levels of the profibrotic markers TGF-β1, COL1A2, and IL-6 (Fig. 2B) compared with the fibroblasts isolated from controls. The profibrotic phenotype was also confirmed by increased TGF-β signaling in IPF cells via luciferase reporter assay studies using a smad-binding element (SBE)-luciferase reporter plasmid (Fig. 2C).
Fig. 1.
Expression of peroxisomal proteins in human lung biopsies of control and IPF tissues. Immunofluorescence for PEX14p and catalase (CAT) in control and IPF lung tissue. Negative control for the secondary antibody reaction with donkey anti-rabbit Alexa488 and donkey anti-mouse Alexa555. Co, control; IPF, idiopathic pulmonary fibrosis; LP, Lamina propria; RE, respiratory epithelium. Arrow, fibroblasts in the lamina propria. (Scale bar: 10 µm.)
Fig. 2.
Compromised peroxisomal biogenesis and metabolism in IPF fibroblasts. (A) Single immunofluorescence of the fibrotic marker α-SMA in control and IPF fibroblasts. (B) Expression of fibrotic markers at mRNA level TGF-β1, Col1A2 and IL-6 in control and IPF fibroblasts. (C) Luciferase reporter activity of SBE (Smad binding element) in control and IPF fibroblasts. The activity of firefly luciferase was measured in cell lysates and normalized to the activity of renilla. (E.V., empty vector). (D and E) Immunofluorescence staining of peroxisomal proteins Pex13p, ABCD3, ACOX1 and catalase in control and IPF fibroblasts. (F) Expression of PEX13 at mRNA and protein level by Western blotting in both control and IPF fibroblasts. (G) Generation of reactive oxygen species (ROS) detection with dihydroethidine (DHE) in control and IPF fibroblasts. Co, control; IPF, idiopathic pulmonary fibrosis. Data represent ± SD of three independent experiments. P value, unpaired Student t test. (Scale bar: 10 µm.)
Analysis of protein abundance by immunofluorescence revealed that the peroxisomal biogenesis protein PEX13p, the lipid transporter protein ABCD3, the β-oxidation enzyme acyl-CoA oxidase 1 (ACOX1), as well as the antioxidative enzyme catalase were reduced in IPF fibroblasts (Fig. 2 D and E). The down-regulation of the peroxisomal biogenesis protein PEX13p was also observed at the mRNA level between control and IPF fibroblasts by qRT-PCR and confirmed by Western blot analysis (Fig. 2F). Because catalase (Fig. 2E) was down-regulated, we hypothesized that IPF fibroblasts might have an impaired antioxidant response. DHE stainings revealed that IPF fibroblasts exhibited a higher ROS production in comparison with control fibroblasts (Fig. 2G). Interestingly, a series of antioxidant enzymes such as SOD1, heme oxygenase (HO-1), glutathione reductase (GR), and the redox-sensitive transcription factor Nrf2 were decreased in IPF fibroblasts (SI Materials and Methods and Fig. S1 A–D). Moreover, reporter gene analyses showed that Nrf2 binding element (ARE)-driven luciferase activity was significantly decreased in IPF fibroblasts (fivefold reduction) (P < 0.05) (Fig. S1F), whereas the luciferase expression of the AP1 reporter construct was not significantly changed in IPF fibroblasts in basal unstimulated conditions (Fig. S1F). To summarize, these results indicate that in IPF tissues as well as in IPF fibroblasts peroxisomal proteins were significantly down-regulated and IPF fibroblast exhibit an imbalance in the antioxidant response.
PEX13p Knockdown Activates Smad-Dependent TGF-β1 Pathway and Increases COL1 Production.
Alterations in peroxisomal proteins in IPF samples may be a collateral effect due to persistent fibrosis or could also be a significant factor that contributes to the pathogenesis of this devastating condition. To address this question, PEX13p, one of the peroxin proteins involved in peroxisomal biogenesis, was knocked down using a siRNA-mediated approach. The strong knockdown of PEX13 expression in both control and IPF fibroblasts was verified by quantitative RT-PCR and Western blot analysis (Fig. 3 A and B), and disruption of peroxisomal biogenesis, leading in consequence to mistargeting of catalase into the cytoplasm (Fig. S4C). Interestingly, disruption of the peroxisomal biogenesis triggered the production of the profibrotic markers COL1A2 and TGF-β1 at mRNA level and of the COL1 protein in Western blot analysis (Fig. 3 A and B). This disruption was associated with increased collagen and TGF-β1 also in the culture medium (Fig. 3 C and D). Furthermore, increased COL1A2 promoter activity and activation of TGF-β signaling upon PEX13 knockdown was confirmed in COL1A2 and SBE luciferase reporter gene assays, respectively (Fig. 3 E and F). It is noteworthy that also control fibroblasts exhibit an increased fibrotic phenotype after peroxisomal knockdown, even though to a lesser extent in comparison with the transfected IPF fibroblasts (Fig. 3 A–F). Additionally, PEX13 knockdown also led to the intracellular elevation of profibrotic markers such as collagen I, collagen 3A1 (COL3A1), and prolyl 4-hydroxylase beta polypeptide (PDI) as revealed by immunofluorescence studies and also by increased mRNA levels of matrix metalloproteinase 2 (MMP2) which have been implicated in excessive TGF-β1 activation (Figs. S2 and S3 A and E). The increased fibrotic response of TGF-β1 and COL1A2 was also observed with semiquantitative RT-PCR in cells with a PEX13 knockdown (Fig. S3 A–D). To summarize, these results indicate that the down-regulation of peroxisomes in both control and IPF fibroblasts leads to an increased fibrotic phenotype in these cells associated with an increased production of collagen and TGF-β1 as well as an activation of TGF-β signaling.
Fig. 3.
Activation of TGF-β1 Smad dependent pathway in PEX13 siRNA treated control and IPF fibroblasts. (A) qPCR mRNA expression of peroxisome biogenesis PEX13 and fibrotic markers TGF-β1 and Col1A2 in PEX13 siRNA fibroblasts. The expressions of 28S rRNA and of the HPRT1 gene were used as controls for normalization, *P < 0.005. (B) Western blots depicting the abundance of peroxisomal biogenesis genes and collagen I in PEX13 knockdown. The expression of GAPDH was used as control. (C) Collagen Sircol assay, measuring the production of collagen released in medium by control and IPF fibroblasts. Sc (scrambled siRNA control), si-1 (siRNA PEX13-1), si-2 (siRNA PEX13-2), si-3 (siRNA PEX13 1, 2). (D) TGF-β1 release in supernatant measured by TGF-β1 ELISA Assay. (E and F) SBE, COL1A2, luciferase reporter assays in siRNA treated control and IPF fibroblasts. The activity of firefly luciferase was measured in cell lysates and normalized to the activity of renilla. (E.V., empty vector). Data represent ± SD of three independent experiments. P value, unpaired Student t test.
Knockdown of Peroxisomes Leads to Increased ROS and Proinflammatory Cytokine IL-6 in Fibroblasts.
As shown above, IPF fibroblasts exhibit an increased production of ROS (Fig. 2G). Because peroxisomes are able to produce and scavenge ROS and are down-regulated in IPF fibroblasts, we questioned whether they are involved in the cellular ROS production observed. Indeed, the knockdown of PEX13 led to an increase in the production of ROS as measured by dihydroethidine staining both in control as well as IPF fibroblasts (Fig. 4A). However, unlike in the basal conditions of IPF fibroblasts (Fig. S1F), the increased ROS production was also paralleled with an increase in Nrf2 and AP-1 activity (Fig. 4B), and also induced a high antioxidative response indicated with the up-regulation of antioxidative enzymes such as HO-1, GR, and of Nrf2 expression (Fig. S4 A and B).
Fig. 4.
Induction of ROS and cytokine production (IL-6) in PEX13 knockdown control and IPF fibroblasts. (A) Generation of reactive oxygen species (ROS) detection with dihydroethidine (DHE) and quantification in control and IPF siRNA treated fibroblasts, (N-number of cells for quantification). (B) ARE and AP-1 luciferase reporter assays in PEX13 knockdown control and IPF fibroblasts, si vs. Sc (E.V., empty vector). The activity of firefly luciferase was measured in cell lysates and normalized to the activity of renilla. (C) qPCR mRNA expression of cytokines (TNF-α and IL-6) in PEX13 knockdown of control and IPF fibroblasts. (D) Human IL-6 secretory levels measured by Quantikine ELISA in siRNA PEX13 treated control and IPF fibroblasts. Sc (scrambled siRNA control), si-1 (siRNA PEX13-1), si-2 (siRNA PEX13-2). Data represent ± SD of three independent experiments. P value, unpaired Student t test.
Next we analyzed the effect of the PEX13 knockdown on the production of proinflammatory cytokines such as TNF-α and IL-6, which have been proposed to play an important role in the pathogenesis of fibrosis. At the mRNA level both TNF-α and IL-6 were significantly induced in PEX13 knockdown control and IPF fibroblast in comparison with the respective cells transfected with the negative control siRNA (Fig. 4C). IL-6 was readily detectable and also significantly increased in the culture supernatants of PEX13 knockdown fibroblasts (Fig. 4D). In contrast, by using the same supernatants under similar experimental conditions for a TNF-α ELISA, the concentration of this cytokine was too low for reliable detection. In summary, knockdown of peroxisomes leads to increased ROS production and IL-6 release in both control and IPF fibroblasts.
TGF-β1 Signaling Down-Regulates Peroxisomal Biogenesis Proteins in IPF Fibroblasts and in a Bleomycin-Induced Mouse Model of Fibrosis.
Having considered the pivotal role of TGF-β1 in the pathogenesis of lung fibrosis, we thought to examine the possibility that it might modulate the expression of the PEX13 gene, and that activated TGF-β signaling could account for impaired peroxisome biogenesis and metabolism in IPF. The fibrotic response of TGF-β1 treatment was demonstrated by the up-regulation of COL1A2 and IL-6 mRNAs, which was blocked specifically with the TGF-β1 receptor inhibitor LY364947 (Fig. 5A). Elevated levels of IL-6 in culture supernatants of lung fibroblasts treated with TGF-β1 and inhibition of the same with the TGF-β1 receptor inhibitor LY364947 were confirmed by ELISA (Fig. S5A). Similarly, the activation of the TGF-β1-Smad pathway in these cells was also confirmed by increased SBE luciferase reporter activity, increased TGF-β1 mRNA (Fig. S5 B and D and Fig. 5A) and by increased Smad3 translocation (Fig. S5C). We then analyzed whether the expression of the PEX13 gene would be affected by TGF-β1 stimulation. Interestingly, TGF-β1 treatment indeed resulted in the down-regulation of PEX13 mRNA and protein, suggesting that TGF-β1 inhibits peroxisomal biogenesis (Fig. 5 B and C and Fig. S5D). This effect was reversed when TGF-β signaling was specifically blocked using a TGF-β1 receptor inhibitor (Fig. 5 B and C and Fig. S5D).
Fig. 5.
TGF-β1 signaling suppresses PEX13 peroxisomal biogenesis protein in control/IPF and in a bleomycin-induced mouse model of lung fibrosis. Confluent control and IPF fibroblasts were pretreated with 5 µM LY364947 (TGF-β1 inhibitor) for 1 h, followed by a stimulation with 5 ng/mL TGF-β1, or combined for 24 h. (A and B) RNA expression of TGF-β1, COL1A2, IL-6, and PEX13 was examined by real-time qRT-PCR. The results were normalized with 28S rRNA and HPRT mRNA. (C) Total protein was isolated following 24 h incubation with TGF-β1, LY364947, or combinations and subjected to Western blotting for indicated proteins. Relative density in donor and IPF fibroblasts treated with TGF-β1 and the specific TGF-β1 inhibitor LY364947. GAPDH was used as loading control. (D) Double immunofluorescence of PEX14 and α-SMA in bleomycin-induced mouse model of pulmonary fibrosis. Bleo, Bleomycin; Ctrl, Control; RII-KO, TGF-β receptor II knockout; WT, wild type. (E) TGF-β1–induced reactive oxygen species (ROS) detection with dihydroethidine (DHE) in control and IPF fibroblasts. C, control; LY, LY364947; T, TGF.β1; T+LY, TGF-β-1+LY364947. Data represent ± SD of three independent experiments. P value, unpaired Student t test. (Scale bar: 10 µm.)
To extend these findings to the in vivo situation, TGF-β receptor II knockout mice were used. Because the anti-PEX13p antibody does not work properly for stainings of PFA-fixed paraffin-embedded tissue (16), an antibody against PEX14p, a binding partner of PEX13p in the docking complex of the peroxisomal membrane that was also reduced in IPF lungs (Fig. 1) was used. Indeed, bleomycin treatment in control mice down-regulated peroxisomes (PEX14p) on day 7 after treatment, followed by a recovery on day 14 and 28 compared with day 7, but still at lower protein abundance than in appropriate control animals (Fig. 5D). Strikingly, bleomycin treatment in TGF-β receptor II knockout mice did not induce the down-regulation of peroxisomes as detected by staining with PEX14p, indicating a direct relation for TGF-β–induced signaling in the down-regulation of peroxisomes (Fig. 5D). TGF-β1 treatment also increased ROS production in these fibroblasts (Fig. 5E). In summary, these findings indicate that TGF-β1 signaling down-regulates peroxisomes in fibroblasts and induce the production of ROS.
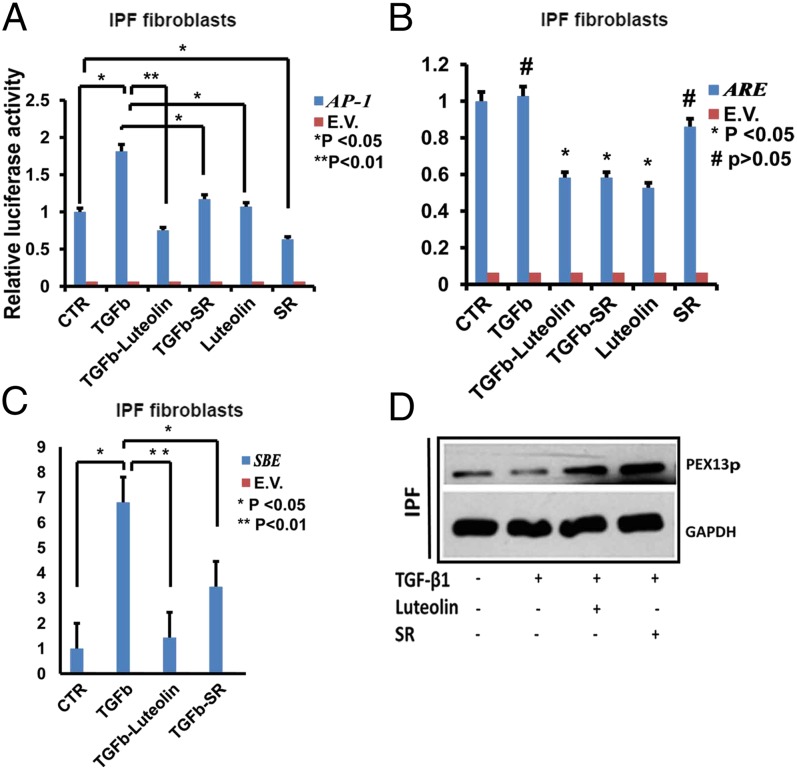
AP-1 Signaling Is Involved in TGF-β1–Mediated Down-Regulation of PEX13 in Human IPF Fibroblasts.
Earlier reports indicate a cross-talk between TGF-β1 signaling and the transcriptional factor AP-1, which as shown was also up-regulated in PEX13 knockdown fibroblasts. Hence, we questioned whether the transcriptional factor AP-1, normally activated during profibrotic and proinflammatory responses, would play a role in the observed down-regulation of PEX13p. For a comparison we also used the luciferase reporter vector (ARE) for the ROS-activated transcriptional factor Nrf2. Indeed, stimulation with TGF-β1 induced the activity of the AP-1 luciferase reporter construct (Fig. 6A), but the activity of the ARE-luciferase construct remained unchanged (Fig. 6B). Furthermore, the AP-1–specific inhibitor SR11302 partially blocked the TGF-β1 stimulated activity of both the AP-1-luciferase construct and also the ARE-luciferase construct (Fig. 6 A and B). This inhibition was expected as the AP-1 binding element shares the consensus sequence of the Nrf2 binding element but not vice versa. The luciferase reporter assays revealed that the inhibitor luteolin used generally as Nrf2 inhibitor is not specific, because it also inhibits the TGF-β1–induced activation of AP-1 (Fig. 6A) in addition to the inhibition of ARE luciferase (Fig. 6B). Interestingly, pretreatment of cells with an AP-1 specific inhibitor SR11302 or the Nrf2/AP-1 inhibitor luteolin blocked the TGF-β1–mediated SBE activation, indicating a role for AP-1 in the TGF-β1–mediated Smad-dependent pathway (Fig. 6C). Moreover, pretreatment with SR11302 and luteolin also reversed the TGF-β1–mediated down-regulation of the PEX13p protein (Fig. 6D). To summarize, the profibrotic factor TGF-β1 might down-regulate PEX13 through the transcriptional factor AP-1.
Fig. 6.
AP-1 signaling is activated in TGF-β1–mediated down-regulation of PEX13 in human IPF fibroblasts. Confluent IPF fibroblasts were pretreated 1 h before with luteolin 25 µM or SR11302 (SR) 10 µM. Then, cells were challenged with 5 ng/mL TGF-β1 for 24 h as indicated. (A–C) AP1, ARE, and SBE luciferase reporter assays in IPF fibroblasts. The activity of firefly luciferase was measured in cell lysates and normalized to the activity of renilla. (E.V., empty vector). (D) Protein analysis of PEX13p in IPF fibroblasts treated with TGF-β1, luteolin or SR11302. GAPDH was used as loading control. Data represent ± SD of three independent experiments. P value, unpaired Student t test.
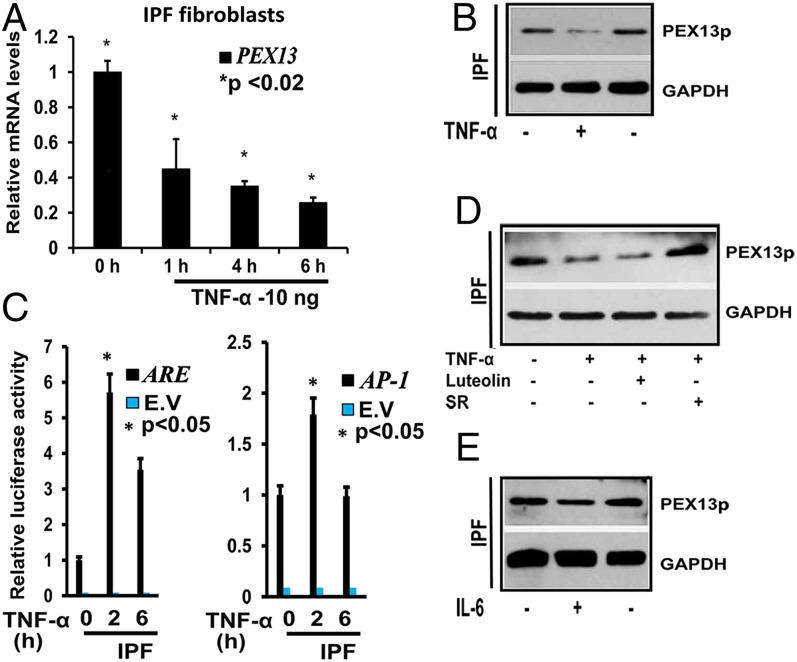
Proinflammatory Cytokines TNF-α and IL-6 also Suppress the Peroxisome Biogenesis Protein PEX13p in Human IPF Fibroblasts.
In a complex condition such as IPF, factors other than TGF-β1 may also contribute to the down-regulation of peroxisomal genes in vivo. Macrophage-mediated TNF-α production might play an important paracrine role in this process. To determine whether TNF-α affects peroxisome biogenesis, IPF fibroblasts were treated with 10 ng/mL of TNF-α for the indicated time points (Fig. 7 A and C). TNF-α induced a significant down-regulation of the PEX13 mRNA as early as 1 h (Fig. 7A), as well as the protein abundance of PEX13p after 10 h (Fig. 7B). Similar to TGF-β1, TNF-α also induced the activity of the AP-1 luciferase construct and also increased the luciferase activity of the ARE-luciferase construct (Fig. 7C). Interestingly, the AP-1 inhibitor SR11302 reversed the TNF-α–mediated down-regulation of PEX13p (Fig. 7D). Finally, treatment with the proinflammatory cytokine IL-6 also induced the down-regulation of PEX13p (Fig. 7E). In summary, proinflammatory cytokines (TNF-α and IL-6) down-regulate PEX13p in IPF fibroblasts. Moreover, TNF-α–mediated down-regulation of PEX13p is at least partially mediated through AP-1 signaling.
Fig. 7.
TNF-α suppresses peroxisome biogenesis by induction of AP1 in human IPF fibroblasts. (A) IPF fibroblasts were treated with 10 ng/mL TNF-α for the indicated times, and the expression of PEX13 mRNA was determined by using qRT-PCR. (B) IPF fibroblasts were treated with 10 ng/mL TNF-α for for 6 h, and cells were lysed for Western blot analysis. As loading control GAPDH was used. (C) ARE and AP-1 dual luciferase reporter assays of IPF fibroblasts treated with 10 ng/mL TNF-α for indicated times. The activity of firefly luciferase was measured in cell lysates and normalized to the activity of renilla luciferase. (D) IPF fibroblasts were treated with 10 ng/mL TNF-α for 6 h, cells were pretreated 1 h before with ARE inhibitor Luteolin and AP-1 inhibitor SR11302 (SR), PEX13 abundance was analyzed with Western blotting. GAPDH was used as loading control. (E) IPF fibroblasts were treated with 20 ng/mL IL-6 cytokine and PEX13 abundance was analyzed with Western blotting. GAPDH was used as loading control. Data represent the results of at least three experiments performed in triplicates (E.V., empty vector). (Mean ± SEM, relative units, n = 3). P value, unpaired Student t test.
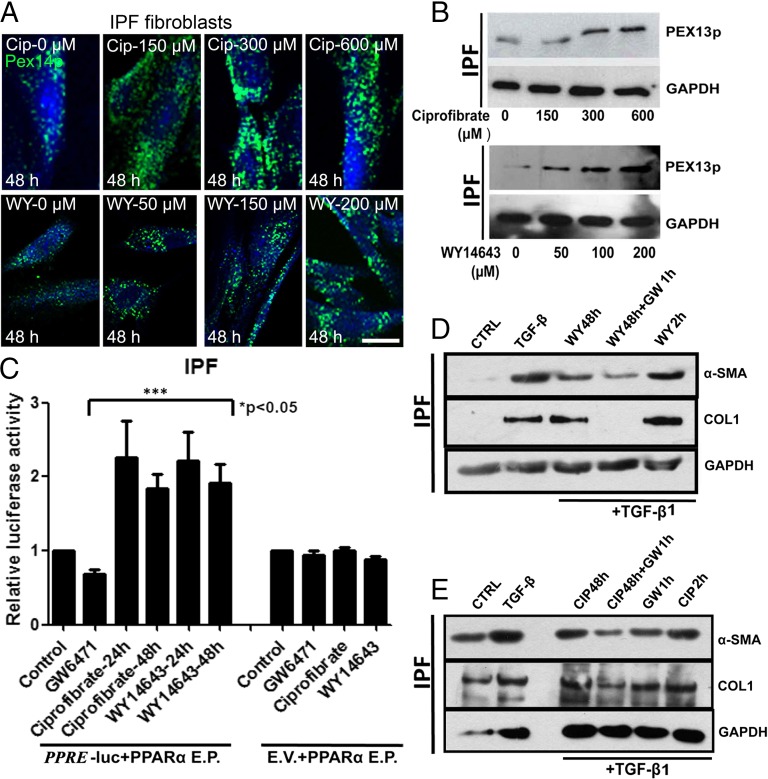
PPAR-α Agonists Proliferate Peroxisomes and Block the TGF-β1–Induced Profibrotic Response in IPF Fibroblasts.
As suggested by the studies above, reduced peroxisome biogenesis is associated with an increased profibrotic response as shown by the activation of TGF-β1 signaling and collagen production. This reduction raises the possibility that increasing peroxisomal biogenesis may be beneficial as a treatment strategy in IPF. To evaluate this we used two structurally distinct PPAR-α agonists (ciprofibrate and WY14643), classical peroxisome proliferators, and investigated the relationship between peroxisome proliferation and TGF-β1–induced myofibroblast differentiation (as shown by α-SMA) and up-regulation of collagen I protein. Treatment with either ciprofibrate or WY14643 for 48 h resulted in proliferation of peroxisomes as detected by PEX14p stainings (Fig. 8A). The peroxisomal biogenesis protein PEX13p was also induced after treatment of IPF cells with ciprofibrate or WY14643 (Fig. 8B). PPAR-α has been shown to exert multiple effects on cellular targets that are independent of peroxisome proliferation, wherefore it is important to distinguish the peroxisome-dependent antifibrotic effects of PPAR-α agonists. To do this, the experimental setup (mentioned in detail in SI Materials and Methods) contained two different controls: (i) IPF cells, which were pretreated with PPAR-α agonists for 48 h, after which the medium was replaced with the PPAR-α antagonist to block endogenous PPAR-α activation. Ideally, these cells then contain proliferated peroxisomes but further PPAR-α activation is blocked. (ii) IPF cells pretreated with PPAR-α agonists only for 2 h before TGF-β1 stimulation and hence will exhibit an activation of PPAR-α but no peroxisome proliferation due to the insufficiently short time period of drug treatment. The concentration of PPAR-α agonist and antagonist that was used for this approach activated and inhibited the PPAR-response-element (PPRE)-luciferase reporter constructs, respectively (Fig. 8C). Interestingly, IPF cells pretreated with ciprofibrate or WY14643 for 48 h showed a significant reduction in the TGF-β1–induced myofibroblast differentiation represented by the abundance of the α-SMA protein. The strongest reduction was observed in the cells pretreated with PPAR-α agonist for 48 h followed by pretreatment with PPAR-α antagonist for 1 h before the addition of TGF-β1 (Fig. 8 C and D), whereas pretreatment with PPAR-α agonist for 2 h before TGF-β1 stimulation did not block the TGF-β1–induced α-SMA protein. Similarly, TGF-β1–induced COL1 protein was also blocked by the addition of PPAR-α agonists treated for 48 h followed by antagonist, but not in IPF cells pretreated with PPAR-α agonists alone for 2 h or 48 h (Fig. 8 C and D). In summary, these findings suggest that IPF cells containing proliferated peroxisomes block the TGF-β1–induced up-regulation of myofibroblast differentiation and COL1 protein and endogenous PPAR activity might interfere with this mechanism.
Fig. 8.
Peroxisome proliferation by PPAR-α agonists ciprofibrate and WY14643 blocks the TGF-β1–induced profibrotic response in IPF fibroblasts. (A) Staining of IPF fibroblasts treated with ciprofibrate or WY14643 for 48h at the indicated concentrations with the peroxisomal marker PEX14p. (B) Western blot analysis of PEX13p in IPF cells treated with ciprofibrate or WY14643 for 48 h. (C) IPF cells cotransfected with PPAR-α expression vector (PPAR-α E.P.) and PPRE-lucfierase-reporter vector (PPRE)/empty vector (E.V) were treated with ciprofibrate (200 µM) or WY14643 (100 µM) or GW6471 (10 µM) for the indicated times after which the firefly luciferase activity was measured in cell lysates and normalized to the activity of renilla luciferase. (D and E) Western blot analysis of IPF cells pretreated with WY14643 (100 µM) or ciprofibrate (200 µm) and/or GW6471 (10 µM) for the indicated times followed by treatment with TGF-β1 (5 ng/mL) for another 24 h. Note: After 48 h treatment of ciprofibrate/WY14643. the medium was replaced with fresh serum free medium before simulation with TGF-β1, whereas the medium was not replaced before the addition of TGF-β1 in cells pretreated with ciprofibrate/WY14643 for 2 h or GW6471 for 1 h. CIP, ciprofibrate; CTRL, control; GW, GW6471; WY, WY14643. Data are a representative of at least three reproducible experiments. Statistical analysis for luciferase assays was performed by ANOVA. (Scale bar: 10 µm.)
Discussion
The findings presented here provide compelling evidence that peroxisomes are protective organelles against the development of pulmonary fibrosis. Importantly, we show that, in lung tissue samples of IPF patients as well as in IPF fibroblast cultures, peroxisomal proteins are down-regulated. Moreover, siRNA-mediated down-regulation of the peroxisomal biogenesis protein PEX13p elicits a profibrotic response, characterized by the activation of TGF-β signaling and increased collagen production. Furthermore, treatment with PPAR-α agonists increased the peroxisomal abundance in IPF fibroblasts and decreased the TGF-β1–induced myofibroblast differentiation.
Peroxisomes are present in different pulmonary cell types and exhibit strong heterogeneity in their abundance and enzyme composition (8). It is well known that the lung is one of the organs mostly exposed to the various forms of reactive oxygen species due to its high oxygen environment (12). In this respect, the protective role of peroxisomes in pulmonary fibrosis is closely associated with their functions in diminishing ROS species thus preventing excessive ROS production and inflammatory reactions (1, 8, 15). Accordingly, our findings also indicate that dysfunctional peroxisomes lead to increased cellular ROS production in IPF cells. This notion is in line with previous studies showing that the deficiency of peroxisomal proteins leads to increased ROS production and oxidative stress (10, 17).
The observation that IPF samples have reduced peroxisomal proteins is of particular interest because one of the clinical features of Zellweger syndrome, a peroxisomal disorder with complete absence or reduced number of peroxisomes, is the development of hepatic fibrosis (11). A continuous trigger would be necessary to induce this consistent down-regulation of peroxisomes in in vitro IPF fibroblast cultures because peroxisomal biogenesis will complement for this down-regulation over time. Based on our findings that the SBE-luciferase activity was higher in IPF cells in their basal state and also contained increased concentrations of TGF-β1 in the cell culture medium (Figs. 2C and 3D), we propose that the persistent activation of TGF-β signaling in these cells might be responsible for the observed down-regulation of peroxisomal proteins. This notion is also supported by the in vivo findings in the bleomycin-induced TGF-β receptor II knockout mouse model studies in which loss of TGF-β signaling prevented the bleomycin-induced down-regulation of peroxisomes (Fig. 5D). To our knowledge, this is the first study to show a direct role for TGF-β signaling in the regulation of peroxisomal genes.
In contrast, down-regulation of the peroxisomal genes by the proinflammatory cytokine TNF-α has been shown previously in the liver (18). In this study we extend our knowledge on the molecular mechanisms leading to the TNF-α–mediated down-regulation of peroxisomes by showing that TNF-α mediates this effect through activation of AP-1 (Fig. 7 C and D). It is known that TNF-α−/− mice develop less liver fibrosis in comparison with littermate controls, exhibit reduced levels of α-SMA, a marker for activated myofibroblasts, and reduced TGF-β1 mRNA (19). Consistent with our findings are the existing evidences from the literature that TNF-α is crucial in initiation and progression of the fibrotic processes via AP-1 in Swiss 3T3 fibroblasts (7, 20). Similarly, the interplay between Smad-dependent TGF-β signaling and AP-1 that we observed in our study has also been reported in numerous studies and are contradicting. In one study, the transcriptional factor AP-1 was reported to be essential for ROS-mediated TGF-β1 activation and TGF-β1–induced IL-6 production (21), which is in line with our finding that AP-1 signaling activates Smad-dependent SBE activation (Fig. 6C). In contrast to this observation, Verrecchia and colleagues reported that the Jun family of AP-1 factors act as inhibitors of Smad-dependent signaling (22). The AP-1 family of transcriptional factors is a broad class of transcriptional factors that can form hetero and homo dimers and are shown to be both profibrotic and antifibrotic based on the specific factors activated in different conditions (23, 24). Further studies to specifically identify the AP-1 factors activated in our experimental conditions are required to understand this observed profibrotic nature in our experimental conditions.
In the proposed mechanism TGF-β1–mediated down-regulation of peroxisomes would lead to a synergistic effect on the proinflammatory and profibrotic responses and induce a vicious cycle. In agreement with this notion are the findings that PEX13 knockdown in fibroblast induce activation of TGF-β1 signaling, increased ROS, collagen and IL-6 production. Several studies have reported that ROS and release of proinflammatory cytokines are the main triggers of TGF-β1 signaling pathway, shown also during the peak of inflammation on day 7 using the bleomycin-induced lung fibrosis mouse model (1, 25). Interestingly, in our bleomycin model, the strongest down-regulation of peroxisomes is also observed during day 7 (Fig. 5D), which is consistent with our in vitro findings that proinflammatory cytokines TNF-α and IL-6 down-regulate peroxisomes in IPF fibroblasts (Fig. 7 D and E). IL-6 is known to mediate many inflammatory processes in the lung and has been implicated in the pathogenesis of a variety of respiratory disorders and a possible association between IL-6 and development of fibrosis (26, 27). In addition, IL-6 plays an important role in development of bleomycin-induced lung inflammation and subsequent fibrotic changes through the activation of TGF−β1 (28).
ROS interferes with many cellular functions and results in activation of the master regulator of the cellular response to oxidative stress NRF2 and the antioxidant machinery (29). Although we were able to identify the activation of Nrf2 in our PEX13 knockdown fibroblasts based on the ARE-luciferase activity, luteolin which is commonly used in studies as an Nrf2 inhibitor was also found to inhibit the transcriptional factor AP-1. Hence at present we cannot conclude that the observed effect of luteolin on SBE activation is dependent on its ability as an Nrf2 inhibitor.
Finally, we also show that pretreatment of IPF cells with PPAR-α agonists for longer time points reduced the TGF-β1–induced collagen and myofibroblast differentiation, provided that the endogenous PPAR-α activation during TGF-β1 stimulation is blocked. This result is particularly interesting because several independent studies that reported antifibrotic effects of PPAR-α agonists have not considered peroxisome proliferation by these agonists into context. Their observation was largely based on the vast amount of accumulating evidence in the literature describing its broad antifibrotic and antiinflammatory properties of PPARs. In particular are the studies which report that PPAR−α agonists (i) inhibit cardiac fibrosis by inhibiting the proliferation of cardiac fibroblasts (30), (ii) reduce the lung injury induced by bleomycin (31), and (iii) inhibit TGF-β–induced transcription of β5 integrin in vascular smooth muscle cells (32). PPAR-α as a transcription factor mediates the peroxisome proliferation in rodent liver. A functional PPRE is found about 8.4 kb downstream of the PEX11α promoter (33). Moreover, PEX11α is one of the PEX genes responsible for peroxisome proliferation (33, 34). Lack of specific and potent peroxisome proliferators that are independent of PPARs is one of the main technical limitations in distinguishing the beneficial effects of peroxisome proliferation. Although the findings presented here suggest that peroxisome proliferation rather than the endogenous PPAR-α activation mediates the antifibrotic effects observed, we cannot rule out the possibility of other molecular targets being altered during this 48 h pretreatment with PPAR-α agonists. Similarly, it should also be taken into consideration that the PPAR-α antagonist, GW6471 used in this study might also interfere in the activation/regulation of other PPAR family members leading to secondary effects which could result in the inhibitory effect observed on TGF-β1–induced α-SMA and collagen. Future studies have to be carried out to confirm the specificity of this PPAR-α antagonist and to get more insights into the complex interactions of distinct PPARs on the PPREs of dependent genes, e.g., genes for peroxisomal proteins. Our study also highlights the necessity to design and synthesize new drugs with selective peroxisome proliferation activity that is independent of PPARs to resolve the technical difficulties in studying the beneficial effects of peroxisome proliferation in disease models.
Taken together, activation of TGF-β signaling during lung injury and subsequent induction of proinflammatory mediators such as TNF-α, ROS and IL-6 in IPF, leads to the down-regulation of peroxisomes specifically PEX13 via AP-1 transcription factor, thus enabling the persistence of a fibrotic phenotype, which in turn generates more ROS and elevates secretion of proinflammatory cytokines (e.g., IL-6) (Fig. 9). Moreover, the activation of TGF-β signaling (Smad-dependent pathway) by peroxisome down-regulation promotes an increased extracellular matrix production and generation of a fibrotic phenotype (Fig. 9). In summary, this study identifies a functionally relevant and potentially possible target for future development of new viable therapeutic approaches, and significantly extends the role of this organelle in the maintenance of normal cellular function by scavenging ROS, metabolizing lipid mediators, and by protecting against inflammatory processes leading eventually to exacerbations in patients with pulmonary fibrosis.
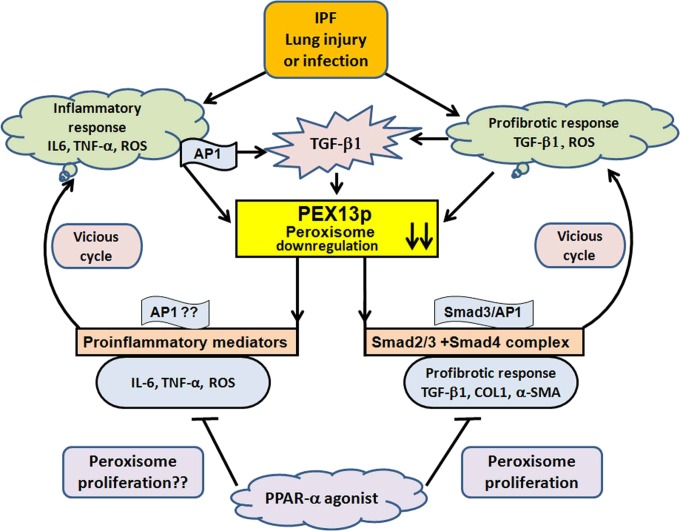
Fig. 9.
Mechanism: Schematic illustration of TGF-β1 effects on peroxisome function, described as proposed model in this study. In idiopathic pulmonary fibrosis, lung injury leads to the production of proinflammatory mediators such as TNF-α, IL-6, and ROS and the activation of profibrogenic TGF-β and AP-1 signaling. This damage leads to down-regulation of peroxisomes specifically PEX13p, which in turn generates more ROS, elevates secretion of cytokines such as IL-6 and promotes the activation of TGF-β1 and AP-1 signaling in a vicious cycle thus enabling the persistence of fibrotic phenotype and inflammatory exacerbation phases in IPF patients. In addition, this cycle also leads to increased production of collagen. In contrast, treatment with PPAR-α agonists induce the proliferation of peroxisomes and inhibit the profibrogenic factors such as α-SMA and collagen.
Materials and Methods
Detailed descriptions of cell culture, animal model, reagents, enzymatic treatments, confocal fluorescence microscopy, immunofluorescence, Western blotting, qRT-PCR and RT-PCR, and ELISAs are provided in SI Materials and Methods.
Cell Culture, Cell Isolation, and Tissue Sections.
Lung tissue and fibroblasts were obtained from 10 IPF patients with typical IPF characteristics (mean age 49 ± 13 y; four females and six males) and 10 control subjects (organ donors, 56 ± 10 y, five females and five males) from the Giessen DZL-biobank at Universities of Giessen and Marburg Lung Center. The study protocol was approved by the Ethics Committee of the Justus-Liebig-University School of Medicine (AZ 31/93) in accordance with the national law and with the “Good Clinical Practice/International Conference on Harmonisation.” Informed consent was obtained in written form from each subject for the study protocol. Control and IPF fibroblasts were cultured in Dulbecco's Modified Eagle's Medium (DMEM) low-glucose media supplemented with 2 mM l-glutamine, 10 U of penicillin/mL, 100 μg of streptomycin/mL, and 10% FBS and maintained at 37 °C with 5% CO2. Paraffin embedding sections and isolation of cells are described in detail in SI Materials and Methods.
Animal Model.
C57BL/6J mice were housed under standard conditions in the central animal facility of the University of Southern California. Floxed TGF-β receptor II (TβRII) mice were provided Harold Moses (Vanderbilt University, Nashville, TN) (35, 36). C57BL/6J and TGF-β receptor II (TβRII) mice were used in experimental model in accordance with the National Institutes of Health (NIH) guidelines for animal care as approved by the USC Institutional Animal Care and Use Committee.
Bleomycin-Induced Pulmonary Fibrosis.
C57BL/6J control and TGF-β receptor II (TβRII) female mice 8-wk-old, anesthetized with pentobarbital sodium (30-40 µg/g ip) were administered with 4 U/kg bleomycin (BLM) (Sigma) diluted in 120 μL of saline, or saline alone by intratracheal instillation using an intratracheal aerosolizer (MicroSprayer Aerosolizer, Model IA, Penn-Century) (37) on day 0 (6). The mouse lungs were then harvested 7, 14, and 28 d after BLM treatment. The harvesting and further processing of paraffin embedded sections was done as described in SI Materials and Methods.
PEX 13 siRNA Transfection.
Control and IPF fibroblasts were transfected twice with 15 nM PEX 13 siRNA (Ambion, catalog no. AM16708), (Ambion, catalog no. AM16773), or silencer select negative control siRNA (Ambion, catalog no. 4390843), with Interferin reagent (Peqlab, catalog no. 13-409-10), processed after 72 h for harvesting and immunofluorescence as described in SI Materials and Methods.
IL-6 Treatment.
IPF fibroblasts were seeded as described at a density of 8 × 104 cells per well in 12-well plates. After 24 h, they were challenged with 20 ng/mL human IL-6 (Biomol, catalog no. 50435), for 6 h duration. At the end of incubation period, the cells were processed for protein analysis with Western blotting.
Treatment of Fibroblast Cultures with Cytokines and Drugs.
Treatment of cells with rhTGF-β1 5 ng/mL, 24 h (R&D catalog no. 240-B); LY364947 5 µM, 24 h (Tocris catalog no. 2718); rhTNF-α 10 ng/mL, 0 h, 1 h, 4 h, and 6 h duration (Biomol catalog no. 50435); interleukin 6, human recombinant (rHuIL-6) (Biomol, catalog no. 50436) for 6 h; SR11302 (Tocris, catalog no. 2476), inhibitor of activator protein-1 (AP-1) transcription factor activity, for indicated times and concentrations; Luteolin (Sigma, L9283), an Nrf2 inhibitor for indicated concentration and duration; ciprofibrate, a PPAR-α agonist (Sigma-Aldrich Chemie), for 48 h, with the indicated concentrations: 0 µM, 150 µM, 300 µM, 600 µM; WY14643, a selective PPAR-α agonist (Tocris, catalog no. 1312) for 48 h for indicated concentrations: 0 µM, 50 µM, 100 µM, 200 µM; GW6471, a PPAR-α–specific antagonist (Tocris catalog no. 4618), for 24 h with the indicated concentration 10 µM, were performed in cell culture for the indicated times and conditions as described in SI Materials and Methods.
Immunofluorescence.
Control and IPF fibroblasts were plated on poly-l-lysine coated coverslips in 24-well plates for 24 h. Thereafter, cells were treated with the compounds mentioned above and described in detail in SI Materials and Methods for different time points. After treatment, they were subjected to an indirect immunofluorescence staining protocol as described (38).
Western Blot Analysis.
Total cell lysates of control and IPF fibroblasts were separated by SDS/PAGE, transferred to PVDF membranes, and incubated with antibodies as described in SI Materials and Methods (8).
Quantitative RT-PCR.
Control and IPF cells were grown in basal conditions as well as treated with the compounds described above and the cells were harvested after the respective time-points. Total RNA was isolated using the RNeasy kit (Qiagen), and cDNA was synthesized by reverse transcription with SuperScript II as described by the manufacturer (Applied Biosystems). The relative expression, fold change of a defined gene was calculated using the ddCT method. All primer pairs and incubation conditions are given in SI Materials and Methods.
Luciferase Reporter Gene Assay and Plasmid Constructs.
Luciferase reporter gene assays were done for ARE, AP1, PPRE reporter, SBE elements, and the COL1A2 promoter. Plasmids were obtained from B. Vogelstein (SBE), E. Jung (COL1A2), W. E. Fahl (p-ARE), and C. A. Hauser (AP-1). PPRE reporter plasmid was from Qiagen Cignal PPAR Reporter (luc) kit. PPAR-α expression plasmid pSG5 PPAR alpha was a gift from B. Spiegelman (Addgene plasmid no. 22751). Empty control vectors pGL2-basic and pGL3-basic were obtained from Promega. Transfection of plasmid DNA into cells was done with TransIT-LT1 (Mirus Bio) as described (39). See SI Materials and Methods.
Measurement of Reactive Oxygen Species.
Reactive oxygen species (ROS) production was detected with dihydroethidine (DHE) final concentration of 5 μM, incubated for 20 min. DHE is oxidized by superoxide to its fluorescent product, ethidine. Ethidine remains intracellularly after it is oxidized, thus allowing quantitative estimations of the intracellular ROS level (40). See SI Materials and Methods.
Cytokine ELISAs and Sircol Collagen Assay.
Cytokine ELISAs and the collagen assay were used according to the manufacturer’s instructions as described in detail in SI Materials and Methods. Collagen Sircol Assay (Biocolor; catalog no. S1000), human TGF-β1 immunoassay (R&D, catalog no. DB100B) and human IL-6 Quantikine ELISA Kit (R&D, catalog no. D6050).
Statistics.
All values are expressed as means ± SEM where n = 3 or 4. An unpaired Student’s t test or ANOVA were used to assess the difference between two groups. Image J was used for quantification of RT-PCR expression. Differences were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We thank Srinu Tumpara, Omelyan Trompak, Ranjithkumar Rajendran, Petra Hahn-Kohlberger, Andrea Textor, Elke Richter, Bianca Pfeiffer, and Gabriele Thiele for excellent technical assistance. We also thank Dr. Bert Vogelstein (The Ludwig Center and the Howard Hughes Medical Institute at Johns Hopkins Kimmel Cancer Center) for providing the luciferase reporter gene construct SBE (Smad binding element), Dr. Eunsum Jung (BioSpectrum LifeScience Institute) for the COL1A2 luciferase construct, Dr. William E. Fahl (University of Wisconsin, Madison) for p-ARE luciferase plasmid, and Dr. C. A. Hauser (The Burnham Institute) for providing pAP-1 luciferase construct. Further, we also thank Denis I. Crane (Griffith University), Paul P. Van Veldhoven (Catholic University) and Alfred Völkl (Ruprecht-Karls-University), for providing us with some antibodies.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415111112/-/DCSupplemental.
This article is a PNAS Direct Submission. D.A.S. is a guest editor invited by the Editorial Board.
References
- 1.Cui Y, et al. Oxidative stress contributes to the induction and persistence of TGF-β1 induced pulmonary fibrosis. Int J Biochem Cell Biol. 2011;43(8):1122–1133. doi: 10.1016/j.biocel.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: Clinical relevance of pathologic classification. Am J Respir Crit Care Med. 1998;157(4 Pt 1):1301–1315. doi: 10.1164/ajrccm.157.4.9707039. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn C, McDonald JA. The roles of the myofibroblast in idiopathic pulmonary fibrosis. Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol. 1991;138(5):1257–1265. [PMC free article] [PubMed] [Google Scholar]
- 4.Pardo A, Selman M. Molecular mechanisms of pulmonary fibrosis. Front Biosci. 2002;7:d1743–1761. doi: 10.2741/pardo. [DOI] [PubMed] [Google Scholar]
- 5.Warburton D, Shi W, Xu B. TGF-β-Smad3 signaling in emphysema and pulmonary fibrosis: An epigenetic aberration of normal development? Am J Physiol Lung Cell Mol Physiol. 2013;304(2):L83–L85. doi: 10.1152/ajplung.00258.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, et al. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol. 2002;282(3):L585–L593. doi: 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan DE, Ferris M, Nguyen H, Abboud E, Brody AR. TNF-alpha induces TGF-beta1 expression in lung fibroblasts at the transcriptional level via AP-1 activation. J Cell Mol Med. 2009;13(8B):1866–1876. doi: 10.1111/j.1582-4934.2008.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karnati S, Baumgart-Vogt E. Peroxisomes in mouse and human lung: Their involvement in pulmonary lipid metabolism. Histochem Cell Biol. 2008;130(4):719–740. doi: 10.1007/s00418-008-0462-3. [DOI] [PubMed] [Google Scholar]
- 9.Karnati S, Baumgart-Vogt E. Peroxisomes in airway epithelia and future prospects of these organelles for pulmonary cell biology. Histochem Cell Biol. 2009;131(4):447–454. doi: 10.1007/s00418-009-0566-4. [DOI] [PubMed] [Google Scholar]
- 10.Ahlemeyer B, Gottwald M, Baumgart-Vogt E. Deletion of a single allele of the Pex11β gene is sufficient to cause oxidative stress, delayed differentiation and neuronal death in mouse brain. Dis Model Mech. 2012;5(1):125–140. doi: 10.1242/dmm.007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinberg SJ, et al. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763(12):1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Rahman I, Swarska E, Henry M, Stolk J, MacNee W. Is there any relationship between plasma antioxidant capacity and lung function in smokers and in patients with chronic obstructive pulmonary disease? Thorax. 2000;55(3):189–193. doi: 10.1136/thorax.55.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol Cell Biol. 2002;22(12):4358–4365. doi: 10.1128/MCB.22.12.4358-4365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boor P, et al. The peroxisome proliferator-activated receptor-α agonist, BAY PP1, attenuates renal fibrosis in rats. Kidney Int. 2011;80(11):1182–1197. doi: 10.1038/ki.2011.254. [DOI] [PubMed] [Google Scholar]
- 15.Rahman I, MacNee W. Regulation of redox glutathione levels and gene transcription in lung inflammation: Therapeutic approaches. Free Radic Biol Med. 2000;28(9):1405–1420. doi: 10.1016/s0891-5849(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 16.Grant P, et al. The biogenesis protein PEX14 is an optimal marker for the identification and localization of peroxisomes in different cell types, tissues, and species in morphological studies. Histochem Cell Biol. 2013;140(4):423–442. doi: 10.1007/s00418-013-1133-6. [DOI] [PubMed] [Google Scholar]
- 17.Baarine M, et al. Evidence of oxidative stress in very long chain fatty acid—treated oligodendrocytes and potentialization of ROS production using RNA interference-directed knockdown of ABCD1 and ACOX1 peroxisomal proteins. Neuroscience. 2012;213:1–18. doi: 10.1016/j.neuroscience.2012.03.058. [DOI] [PubMed] [Google Scholar]
- 18.Beier K, Völkl A, Fahimi HD. TNF-alpha downregulates the peroxisome proliferator activated receptor-alpha and the mRNAs encoding peroxisomal proteins in rat liver. FEBS Lett. 1997;412(2):385–387. doi: 10.1016/s0014-5793(97)00805-3. [DOI] [PubMed] [Google Scholar]
- 19.Gäbele E, et al. TNFalpha is required for cholestasis-induced liver fibrosis in the mouse. Biochem Biophys Res Commun. 2009;378(3):348–353. doi: 10.1016/j.bbrc.2008.10.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verjee LS, et al. Unraveling the signaling pathways promoting fibrosis in Dupuytren’s disease reveals TNF as a therapeutic target. Proc Natl Acad Sci USA. 2013;110(10):E928–E937. doi: 10.1073/pnas.1301100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junn E, et al. Requirement of hydrogen peroxide generation in TGF-beta 1 signal transduction in human lung fibroblast cells: Involvement of hydrogen peroxide and Ca2+ in TGF-beta 1-induced IL-6 expression. J Immunol. 2000;165(4):2190–2197. doi: 10.4049/jimmunol.165.4.2190. [DOI] [PubMed] [Google Scholar]
- 22.Verrecchia F, et al. Smad3/AP-1 interactions control transcriptional responses to TGF-beta in a promoter-specific manner. Oncogene. 2001;20(26):3332–3340. doi: 10.1038/sj.onc.1204448. [DOI] [PubMed] [Google Scholar]
- 23.Rajasekaran S, Vaz M, Reddy SP. Fra-1/AP-1 transcription factor negatively regulates pulmonary fibrosis in vivo. PLoS ONE. 2012;7(7):e41611. doi: 10.1371/journal.pone.0041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, et al. Fra-2 mediates oxygen-sensitive induction of transforming growth factor beta in cardiac fibroblasts. Cardiovasc Res. 2010;87(4):647–655. doi: 10.1093/cvr/cvq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, et al. Antiflammin-1 attenuates bleomycin-induced pulmonary fibrosis in mice. Respir Res. 2013;14:101. doi: 10.1186/1465-9921-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantelidis P, Fanning GC, Wells AU, Welsh KI, Du Bois RM. Analysis of tumor necrosis factor-alpha, lymphotoxin-alpha, tumor necrosis factor receptor II, and interleukin-6 polymorphisms in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2001;163(6):1432–1436. doi: 10.1164/ajrccm.163.6.2006064. [DOI] [PubMed] [Google Scholar]
- 27.Qiu Z, Fujimura M, Kurashima K, Nakao S, Mukaida N. Enhanced airway inflammation and decreased subepithelial fibrosis in interleukin 6-deficient mice following chronic exposure to aerosolized antigen. Clin Exp Allergy. 2004;34(8):1321–1328. doi: 10.1111/j.1365-2222.2004.02013.x. [DOI] [PubMed] [Google Scholar]
- 28.Saito F, et al. Role of interleukin-6 in bleomycin-induced lung inflammatory changes in mice. Am J Respir Cell Mol Biol. 2008;38(5):566–571. doi: 10.1165/rcmb.2007-0299OC. [DOI] [PubMed] [Google Scholar]
- 29.Fourtounis J, et al. Gene expression profiling following NRF2 and KEAP1 siRNA knockdown in human lung fibroblasts identifies CCL11/Eotaxin-1 as a novel NRF2 regulated gene. Respir Res. 2012;13:92. doi: 10.1186/1465-9921-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata T, et al. Stimulation of peroxisome-proliferator-activated receptor alpha (PPAR alpha) attenuates cardiac fibrosis and endothelin-1 production in pressure-overloaded rat hearts. Clin Sci (Lond) 2002;103(Suppl 48):284S–288S. doi: 10.1042/CS103S284S. [DOI] [PubMed] [Google Scholar]
- 31.Genovese T, et al. Role of endogenous and exogenous ligands for the peroxisome proliferator-activated receptor alpha in the development of bleomycin-induced lung injury. Shock. 2005;24(6):547–555. doi: 10.1097/01.shk.0000190825.28783.a4. [DOI] [PubMed] [Google Scholar]
- 32.Kintscher U, et al. PPARalpha inhibits TGF-beta-induced beta5 integrin transcription in vascular smooth muscle cells by interacting with Smad4. Circ Res. 2002;91(11):e35–e44. doi: 10.1161/01.res.0000046017.96083.34. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu M, Takeshita A, Tsukamoto T, Gonzalez FJ, Osumi T. Tissue-selective, bidirectional regulation of PEX11 alpha and perilipin genes through a common peroxisome proliferator response element. Mol Cell Biol. 2004;24(3):1313–1323. doi: 10.1128/MCB.24.3.1313-1323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colasante C, Chen J, Ahlemeyer B, Baumgart-Vogt E. Peroxisomes in cardiomyocytes and the peroxisome / peroxisome proliferator-activated receptor-loop. Thromb Haemost. 2015;113(3):452–463. doi: 10.1160/TH14-06-0497. [DOI] [PubMed] [Google Scholar]
- 35.Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32(2):73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 36.Sauer B. Manipulation of transgenes by site-specific recombination: Use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 37.Bivas-Benita M, Zwier R, Junginger HE, Borchard G. Non-invasive pulmonary aerosol delivery in mice by the endotracheal route. Eur J Pharm Biopharm. 2005;61(3):214–218. doi: 10.1016/j.ejpb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Nenicu A, et al. Peroxisomes in human and mouse testis: Differential expression of peroxisomal proteins in germ cells and distinct somatic cell types of the testis. Biol Reprod. 2007;77(6):1060–1072. doi: 10.1095/biolreprod.107.061242. [DOI] [PubMed] [Google Scholar]
- 39.Vijayan V, Baumgart-Vogt E, Naidu S, Qian G, Immenschuh S. Bruton’s tyrosine kinase is required for TLR-dependent heme oxygenase-1 gene activation via Nrf2 in macrophages. J Immunol. 2011;187(2):817–827. doi: 10.4049/jimmunol.1003631. [DOI] [PubMed] [Google Scholar]
- 40.Ahlemeyer B, Neubert I, Kovacs WJ, Baumgart-Vogt E. Differential expression of peroxisomal matrix and membrane proteins during postnatal development of mouse brain. J Comp Neurol. 2007;505(1):1–17. doi: 10.1002/cne.21448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.