Abstract
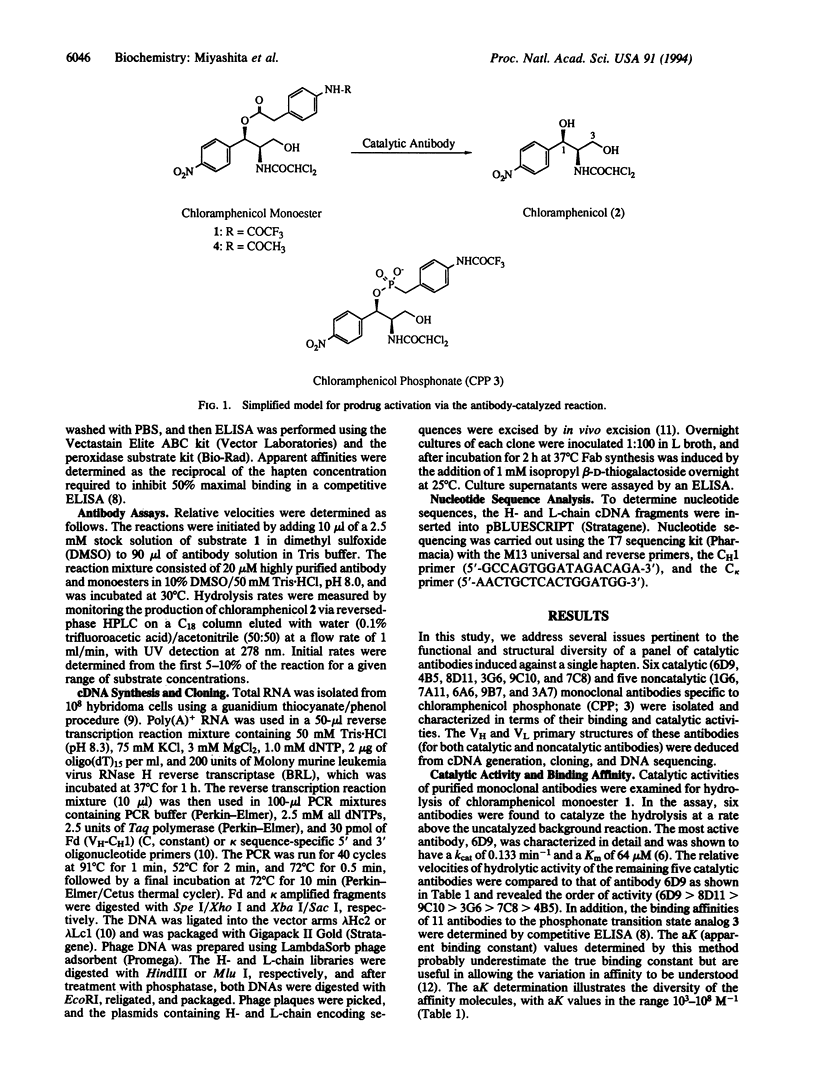
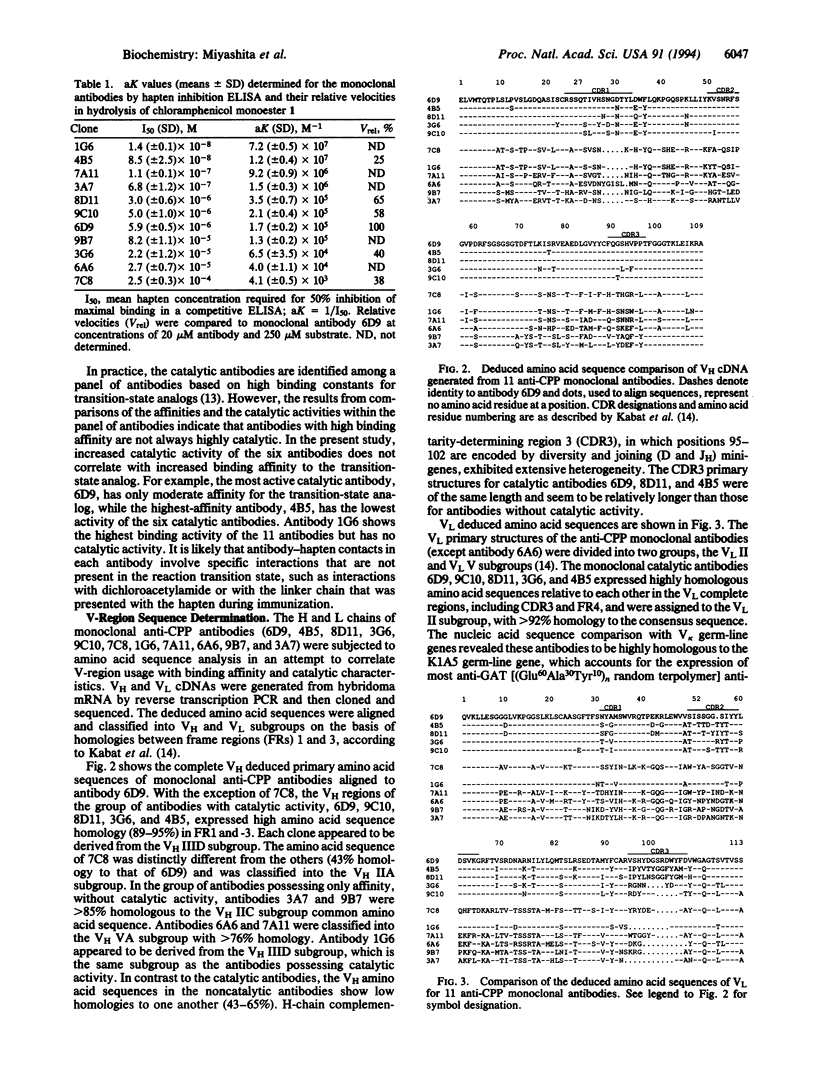
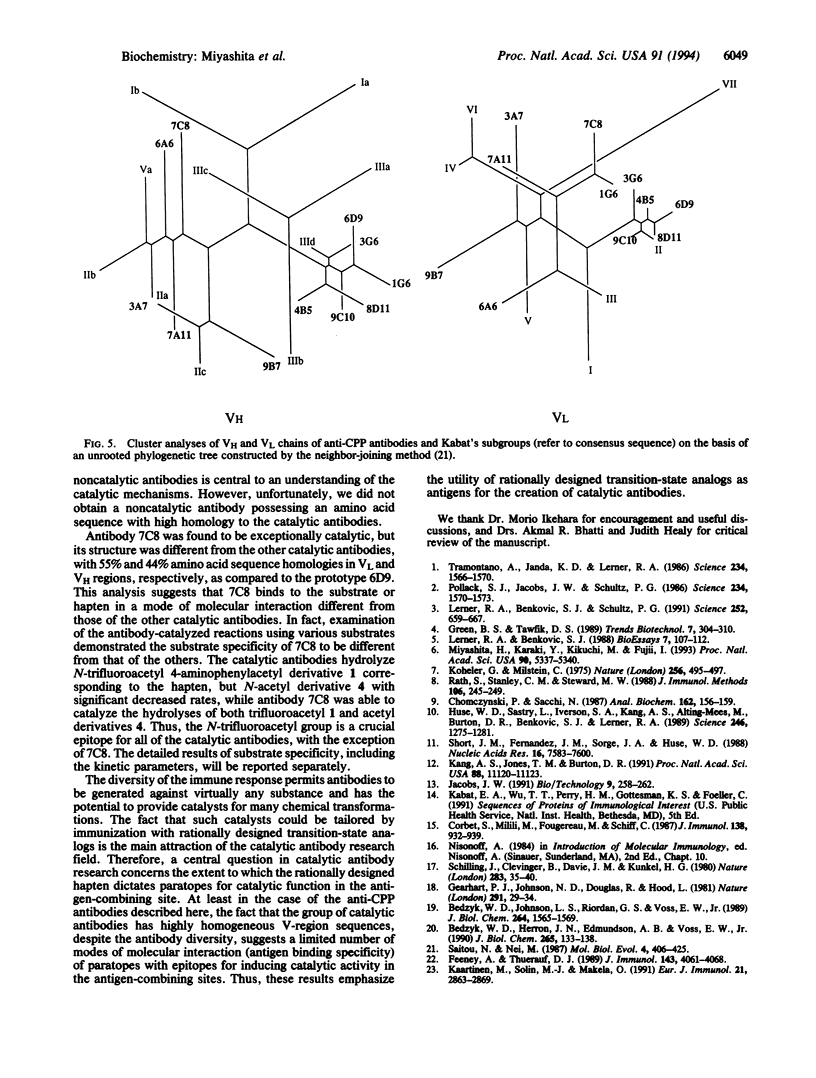
Immunization with a single haptenic transition-state analog generates a few catalytic antibodies among the dozens of antibodies capable of binding the hapten. The diversity of the immune response has raised some fundamental issues, such as How do catalytic and noncatalytic antibodies differ on a structural basis? To address this issue, the variable region primary sequences of 11 antibodies (including 6 catalytic and 5 noncatalytic antibodies) elicited against a single haptenic transition-state analog were deduced from cDNA sequences. Cluster analyses using phylogenetic trees constructed by the neighbor-joining method have revealed that the amino acid sequences of noncatalytic antibodies bear no relationship to one another, while the catalytic antibodies share significant structural identity. Furthermore, no catalytic antibodies possessing amino acid sequences with high homology to those of noncatalytic antibodies were detected. Five catalytic antibodies examined showed 89-95% and 74-84% sequence homologies in the complete light- and heavy-chain variable regions, respectively. Thus, it seems likely that the catalytic antibodies elicited against a single hapten use the canonical set of variable region genes. Interestingly, one catalytic antibody showed only limited sequence similarity to the other catalytic antibodies and was found to exhibit a distinctly different substrate specificity. From the broad range of their binding constants to the hapten, it is unlikely that highly homologous catalytic antibodies are generated as a result of simple high-affinity choices. These results emphasize the utility of rationally designed transition-state analogs for the induction of antibody molecules with catalytic activity.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedzyk W. D., Herron J. N., Edmundson A. B., Voss E. W., Jr Active site structure and antigen binding properties of idiotypically cross-reactive anti-fluorescein monoclonal antibodies. J Biol Chem. 1990 Jan 5;265(1):133–138. [PubMed] [Google Scholar]
- Bedzyk W. D., Johnson L. S., Riordan G. S., Voss E. W., Jr Comparison of variable region primary structures within an anti-fluorescein idiotype family. J Biol Chem. 1989 Jan 25;264(3):1565–1569. [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corbet S., Milili M., Fougereau M., Schiff C. Two V kappa germ-line genes related to the GAT idiotypic network (Ab1 and Ab3/Ab1') account for the major subfamilies of the mouse V kappa-1 variability subgroup. J Immunol. 1987 Feb 1;138(3):932–939. [PubMed] [Google Scholar]
- Feeney A. J., Thuerauf D. J. Sequence and fine specificity analysis of primary 511 anti-phosphorylcholine antibodies. J Immunol. 1989 Dec 15;143(12):4061–4068. [PubMed] [Google Scholar]
- Gearhart P. J., Johnson N. D., Douglas R., Hood L. IgG antibodies to phosphorylcholine exhibit more diversity than their IgM counterparts. Nature. 1981 May 7;291(5810):29–34. doi: 10.1038/291029a0. [DOI] [PubMed] [Google Scholar]
- Huse W. D., Sastry L., Iverson S. A., Kang A. S., Alting-Mees M., Burton D. R., Benkovic S. J., Lerner R. A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989 Dec 8;246(4935):1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- Jacobs J. W. New perspectives on catalytic antibodies. Biotechnology (N Y) 1991 Mar;9(3):258–262. doi: 10.1038/nbt0391-258. [DOI] [PubMed] [Google Scholar]
- Kaartinen M., Solin M. L., Mäkelä O. V genes of oxazolone antibodies in 10 strains of mice. Eur J Immunol. 1991 Nov;21(11):2863–2869. doi: 10.1002/eji.1830211131. [DOI] [PubMed] [Google Scholar]
- Kang A. S., Jones T. M., Burton D. R. Antibody redesign by chain shuffling from random combinatorial immunoglobulin libraries. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11120–11123. doi: 10.1073/pnas.88.24.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Benkovic S. J. Principles of antibody catalysis. Bioessays. 1988 Oct;9(4):107–112. doi: 10.1002/bies.950090402. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Benkovic S. J., Schultz P. G. At the crossroads of chemistry and immunology: catalytic antibodies. Science. 1991 May 3;252(5006):659–667. doi: 10.1126/science.2024118. [DOI] [PubMed] [Google Scholar]
- Miyashita H., Karaki Y., Kikuchi M., Fujii I. Prodrug activation via catalytic antibodies. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5337–5340. doi: 10.1073/pnas.90.11.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack S. J., Jacobs J. W., Schultz P. G. Selective chemical catalysis by an antibody. Science. 1986 Dec 19;234(4783):1570–1573. doi: 10.1126/science.3787262. [DOI] [PubMed] [Google Scholar]
- Rath S., Stanley C. M., Steward M. W. An inhibition enzyme immunoassay for estimating relative antibody affinity and affinity heterogeneity. J Immunol Methods. 1988 Feb 10;106(2):245–249. doi: 10.1016/0022-1759(88)90204-9. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schilling J., Clevinger B., Davie J. M., Hood L. Amino acid sequence of homogeneous antibodies to dextran and DNA rearrangements in heavy chain V-region gene segments. Nature. 1980 Jan 3;283(5742):35–40. doi: 10.1038/283035a0. [DOI] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontano A., Janda K. D., Lerner R. A. Catalytic antibodies. Science. 1986 Dec 19;234(4783):1566–1570. doi: 10.1126/science.3787261. [DOI] [PubMed] [Google Scholar]


