Abstract
Complement activation is common in patients with IgA nephropathy (IgAN) and associated with disease severity. Our recent genome-wide association study of IgAN identified susceptibility loci on 1q32 containing the complement regulatory protein-encoding genes CFH and CFHR1–5, with rs6677604 in CFH as the top single-nucleotide polymorphism and CFHR3–1 deletion (CFHR3–1∆) as the top signal for copy number variation. In this study, to explore the clinical effects of variation in CFH, CFHR3, and CFHR1 on IgAN susceptibility and progression, we enrolled two populations. Group 1 included 1178 subjects with IgAN and available genome-wide association study data. Group 2 included 365 subjects with IgAN and available clinical follow-up data. In group 1, rs6677604 was associated with mesangial C3 deposition by genotype–phenotype correlation analysis. In group 2, we detected a linkage between the rs6677604-A allele and CFHR3–1∆ and found that the rs6677604-A allele was associated with higher serum levels of CFH and lower levels of the complement activation split product C3a. Furthermore, CFH levels were positively associated with circulating C3 levels and negatively associated with mesangial C3 deposition. Moreover, serum levels of the pathogenic galactose-deficient glycoform of IgA1 were also associated with the degree of mesangial C3 deposition in patients with IgAN. Our findings suggest that genetic variants in CFH, CFHR3, and CFHR1 affect complement activation and thereby, predispose patients to develop IgAN.
Keywords: IgA nephropathy, complement activation, complement factor H, complement factor H receptor 3-1Δ deletion
IgA nephropathy (IgAN) is the most common type of primary GN worldwide1–3 and widely considered to be a polygenic disease.4–6 Although the exact pathogenesis is still unclear, a multihit mechanism has been proposed for IgAN, including at least four hits (production of galactose-deficient IgA1 [Gd-IgA1], production of antiglycan antibodies, formation of IgA1-containing immune complexes, and glomerular injury after mesangial deposition).7 In recent years, thanks to rapid advances in genotyping technology, genome-wide association studies (GWASs) have been used to identify common genetic factors that influence health and disease.8 In IgAN, three groups of scientists from England, the United States, and China independently performed GWASs and identified several IgAN susceptibility loci.9–11 In our previous GWAS for IgAN, 1q32 was identified as one of these loci. The genomic region 1q32 contains complement factor H (CFH) and the CFH-related protein genes (CFHR3, CFHR1, CFHR4, CFHR2, and CFHR5). In this region, rs6677604 in CFH was the top single-nucleotide polymorphism (SNP), and a deletion spanning CFHR3 and CFHR1 (CFHR3–1∆) was the top signal in the genome-wide copy number polymorphism analysis.10 The variant rs6677604 is a noncoding SNP located in intron 11 of the CFH gene. Although associated with genetic susceptibility for many complex diseases, such as age-related macular degeneration (AMD)12 and SLE,13 its functional significance is still unclear. The minor allele frequency of rs6677604 varies substantially among individuals from different ethnic groups, ranging from 35% in Africans to 6% in South Asians. In addition, strong linkage disequilibrium (LD) was reported between rs6677604 and CFHR3–1∆, which also hindered the in-depth functional exploration of both variants.
CFH and CFHR genes are arranged in tandem within the regulators of complement activation cluster.14 CFH protein, encoded by gene CFH, is a well known inhibitor of complement activation with a variety of functions, including competition with factor B to hinder the formation of C3 convertase, facilitation of the dissociation of C3 convertase, and support of proteolytic cleavage of C3b by factor I.15,16 Although complement regulatory functions have been attributed to CFH, CFHR3, and CFHR1, their precise biologic roles are not identical. CFHR1 was reported to inhibit C5 convertase and terminal complex formation,17 whereas CFHR3 was found to function as a cofactor for factor I in inactivating C3b.18
In IgAN, the complement system has attracted great attention. In addition to IgA deposition, C3 is the most commonly codeposited molecule, affecting approximately 90% of patients.19 Serologic complement activation has been identified in IgAN.20 Additionally, we recently reported elevated urine CFH levels in patients with IgAN.21 Accumulating evidence from plasma, urine, and renal biopsy samples suggests the involvement of CFH and complement activation in IgAN pathogenesis.
In many complement-related disorders, such as AMD, atypical hemolytic uremic syndrome, and CFHR5 nephropathy, functional exploration of genetic variants in CFH and CFHRs has helped to reveal underlying mechanisms.22–24 In IgAN, these types of studies have been limited. Considering the genetic association of variants of CFH, CFHR3, and CFHR1 with IgAN, the involvement of complement activation in IgAN pathogenesis, and the regulatory role of CFH and CFHRs in complement activation, we chose in this study to investigate CFH levels and complement activation status in patients with IgAN with different variants of CFH, CFHR3, and CFHR1 to explore the genetic basis for the association of CFH, CFHR3, and CFHR1 with IgAN susceptibility.
Results
The rs6677604 Variant of CFH Was Associated with Intensity of Mesangial C3 Deposits in IgAN
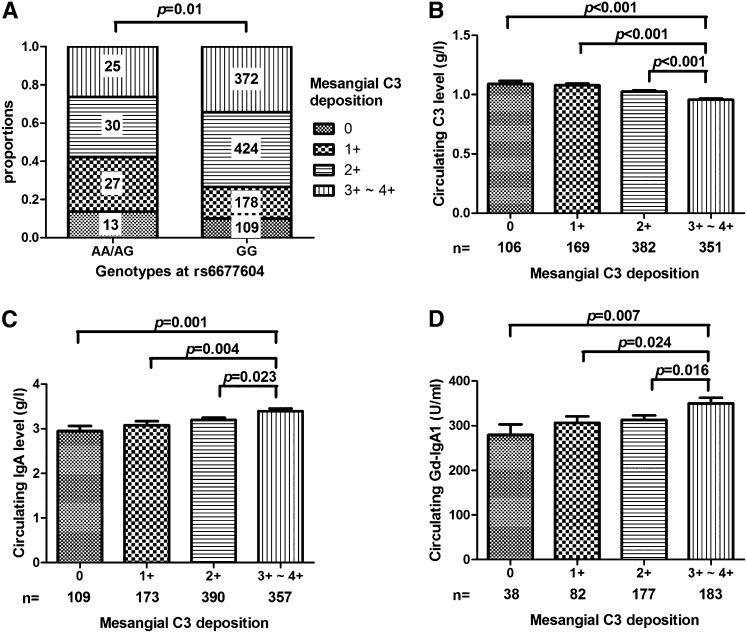
We analyzed an association between rs6677604 and glomerular C3 deposition to evaluate genotype–phenotype correlation in patients with IgAN. Patients with the rs6677604-AA/AG genotype had less intense mesangial C3 deposits, whereas those with the rs6677604-GG genotype had mesangial C3 deposits of greater intensity (0, 1+, 2+, and 3+–4+: 13.7%, 28.4%, 31.6%, and 26.3%, respectively, in the rs6677604-AA/AG genotype versus 10.1%, 16.4%, 39.2%, and 34.3%, respectively, in rs6677604-GG; P=0.01) (Figure 1A).
Figure 1.
Contributors associated with mesangial C3 deposits in IgAN. (A) The rs6677604-A allele was associated with lower intensity of mesangial C3 deposits. Patients with IgAN with the AA/AG genotype of rs6677604 (n=95) had high intensity of mesangial C3 deposits compared with those with the rs6677604-GG genotype (n=1083) (0, 1+, 2+, and 3+–4+: 13.7%, 28.4%, 31.6%, and 26.3%, respectively, in rs6677604-AA/AG versus 10.1%, 16.4%, 39.2%, and 34.3%, respectively, in rs6677604-GG; P=0.01). (B) Lower circulating C3 levels were associated with greater mesangial C3 deposition. Circulating C3 levels decreased significantly from 0 to 3+–4+ mesangial C3 deposition in patients with IgAN (0, 1+, 2+, and 3+–4+: 1.09±0.26, 1.08±0.20, 1.03±0.23, and 0.96±0.21 g/L, respectively; P<0.001; n=106, n=169, n=382, and n=351, respectively). (C and D) Higher circulating IgA and Gd-IgA1 levels were associated with greater mesangial C3 deposition. From 0 to 3+–4+ mesangial C3 deposition, significantly increased levels of circulating IgA (0, 1+, 2+, and 3+–4+: 2.95±1.19, 3.08±1.22, 3.19±1.10, and 3.39±1.23 g/L, respectively; P=0.001; n=109, n=173, n=390, and n=357, respectively) and Gd-IgA1 levels (0, 1+, 2+, and 3+–4+: 279.61±144.68, 306.50±128.64, 313.29±128.89, and 350.15±165.54 units/ml, respectively; P<0.009; n=38, n=82, n=177, and n=183, respectively) were observed in patients with IgAN.
Circulating C3 Levels Were Associated with Mesangial C3 Deposition
Among 1008 patients with IgAN in group 1 with data available on circulating C3 levels, 233 patients had levels below the lower limit of our laboratory reference range (<85 mg/dl), implying the possibility of systemic complement activation in these patients with IgAN. In this study, a negative correlation was found between circulating C3 levels and mesangial C3 deposition. Patients with 3+–4+ mesangial C3 deposition presented with significantly lower circulating C3 levels than those with lower grades of C3 deposition (0, 1+, 2+, and 3+–4+: 1.09±0.26, 1.08±0.20, 1.03±0.23, and 0.96±0.21 g/L, respectively; P<0.001) (Figure 1B).
Both Circulating IgA and Gd-IgA1 Levels Were Associated with Intensity of Mesangial C3 Deposits
On the basis of our understanding of IgAN pathogenesis, circulating Gd-IgA1 molecules are the initial pathogenic factor in IgAN. Here, we compared circulating IgA and Gd-IgA1 levels among patients with IgAN with different degrees of mesangial C3 deposition. We found that patients who showed greater intensity of mesangial C3 deposits had higher levels of circulating IgA (0, 1+, 2+, and 3+–4+: 2.95±1.19, 3.08±1.22, 3.19±1.10, and 3.39±1.23 g/L, respectively; P=0.001) (Figure 1C) and Gd-IgA1 (0, 1+, 2+, and 3+–4+: 279.61±144.68, 306.50±128.64, 313.29±128.89, and 350.15±165.54 units/ml, respectively; P<0.01) levels (Figure 1D), indicating a positive correlation between the intensity of mesangial C3 deposits and the levels of circulating IgA and Gd-IgA1.
Minor Allele A of rs6677604 Tagged CFHR3–1∆
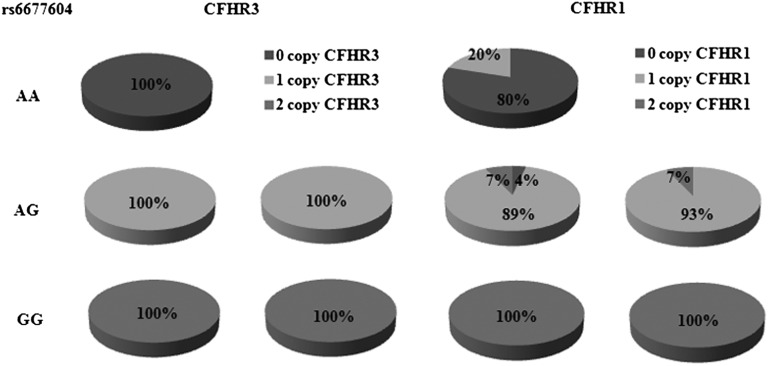
Strong LD between the CFHR3–1∆ and the rs6677604-A allele has been observed in SLE and AMD.13,25 Here, we detected copy number variations (CNVs) in the CFHR3–1 region in a subgroup of group 2 that included 83 patients with IgAN and 88 healthy controls by multiplex ligation-dependent probe amplification (MLPA). Because we were aiming to establish the genetic linkage between rs6677604 and the CFHR3–1∆, more individuals with minor genotypes of rs6677604 (AA/AG) were analyzed compared with their proportion in the general population.
All individuals with the rs6677604-GG genotype (41 patients with IgAN and 28 healthy controls) carried two copies of CFHR3 and CFHR1. Approximately 90% of individuals with the rs6677604-AG genotype were heterozygous for CFHR3–1∆ (39 of 42 patients with IgAN and 49 of 55 healthy controls). Of the remaining nine patients, seven patients carried heterozygous CFHR3∆ alone (three patients with IgAN and four healthy controls), and two patients were heterozygous CFHR3∆/homozygous CFHR1∆ (two healthy controls). Finally, among five patients with the rs6677604-AA genotype (all were healthy controls), four (80%) patients were homozygous for the CFHR3–1∆, whereas one patient was heterozygous for CFHR1∆ alone. Thus, our results indicated that the rs6677604-A allele completely tagged CFHR3∆ and highly tagged CFHR1∆ (Figure 2).
Figure 2.
Minor allele A of rs6677604 highly tagged CFHR3–1 deletion (CFHR3–1∆). Pie charts show the proportion of different copy numbers of CFHR3 and CFHR1 in patients with IgAN and healthy controls with rs6677604-AA, -AG, and -GG genotypes. In our population, 100% of individuals (41 of 41 patients with IgAN and 28 of 28 healthy controls) with rs6677604-GG had two copies of CFHR3 and CFHR1. All individuals (42 of 42 patients with IgAN and 55 of 55 healthy controls) with rs6677604-AG had one copy of CFHR3, whereas all individuals (5 of 5 healthy controls) with rs6677604-AA had no copies of CFHR3. In patients with IgAN with rs6677604-AG, 93% (39 of 42) had one copy of CFHR1, and the remaining 7% (3 of 42) had two copies of CFHR1. In healthy controls with rs6677604-AG, 89% (49 of 55) had one copy of CFHR1, 7% (4 of 55) had two copies of CFHR1, and 4% (2 of 55) had no copies of CFHR1. Of five controls with rs6677604-AA, four (80%) had no copies of CFHR1, and one (20%) had one copy of CFHR1.
The rs6677604-A Allele Was Associated with Higher CFH and Lower C3a Levels
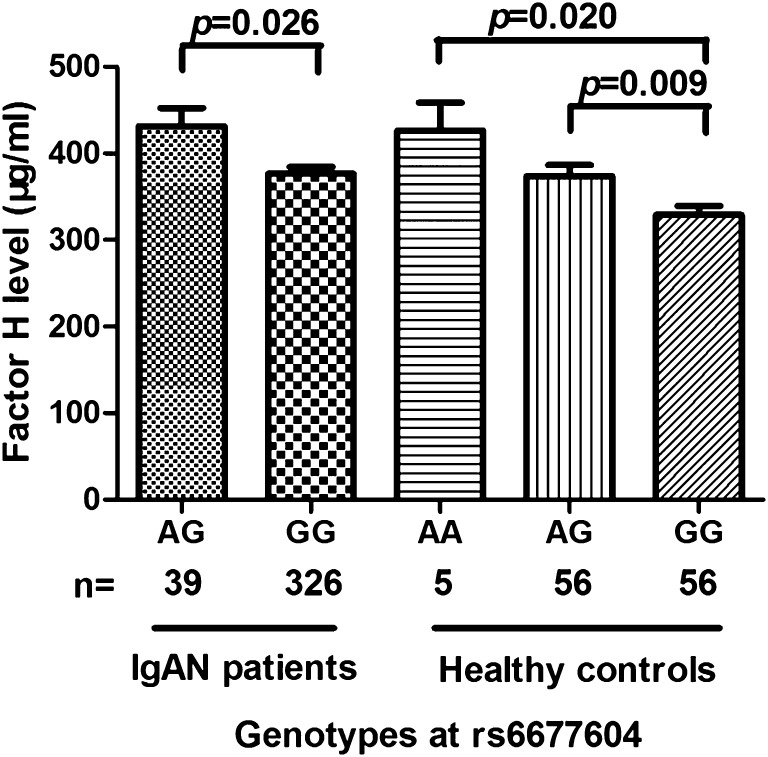
To explore the underlying mechanism of the relationship between rs6677604 and mesangial C3 deposition, we detected circulating CFH levels in 365 patients with IgAN and 117 healthy controls in group 2. Individuals with the rs6677604-AG genotype presented with higher circulating CFH levels compared with those with the rs6677604-GG genotype in patients with IgAN and healthy controls (Figure 3). Healthy individuals with the rs6677604-AA genotype had even higher CFH levels than those with the rs6677604-AG genotype (patients with IgAN: AG versus GG: 431.29±131.71 versus 376.27±147.06 µg/ml; P=0.03; healthy controls: AA versus AG versus GG: 426.26±71.73 versus 373.37±97.71 versus 328.81±80.03 µg/ml; P<0.01) (Figure 3).
Figure 3.
Comparison of CFH levels according to rs6677604 genotype in patients with IgAN and healthy controls. Individuals with rs6677604-A alleles showed significantly higher plasma CFH levels than those with rs6677604-G alleles in both patients with IgAN (AG versus GG: 431.29±131.71 versus 376.27±147.06 µg/ml; P=0.03; n=39 and n=326, respectively) and healthy controls (AA versus AG versus GG: 426.26±71.73 versus 373.37±97.71 versus 328.81±80.03 µg/ml; P<0.01; n=5, n=56, and n=56, respectively). P values for each comparison between two groups are shown.
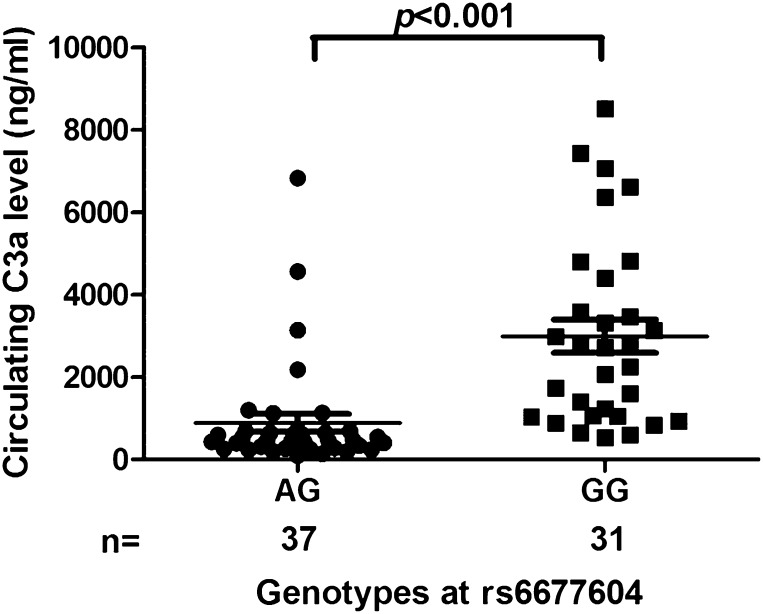
Because CFH is a complement regulatory protein, we then detected levels of the complement activation split product C3a in 68 patients with IgAN. Circulating C3a was significantly decreased in patients with the rs6677604-AG genotype compared with patients with the rs6677604-GG genotype (C3a: 464.0 [275.3–687.6] versus 2715.1 [1044.8–4405.3] ng/ml; P<0.001) (Figure 4), suggesting a greater degree of systemic complement activation in patients with IgAN with the rs6677604-GG genotype.
Figure 4.
Comparison of C3 activation split product C3a serum levels according to the rs6677604 genotype in patients with IgAN. Patients with IgAN with the rs6677604-AG genotype showed significantly lower C3a levels than those without the genotype (464.0 [275.3–687.6] versus 2715.1 [1044.8–4405.3] ng/ml; P<0.001; n=37 and n=31, respectively). Each symbol corresponds to a value from one individual.
Positive Correlation between Circulating CFH and C3 Levels
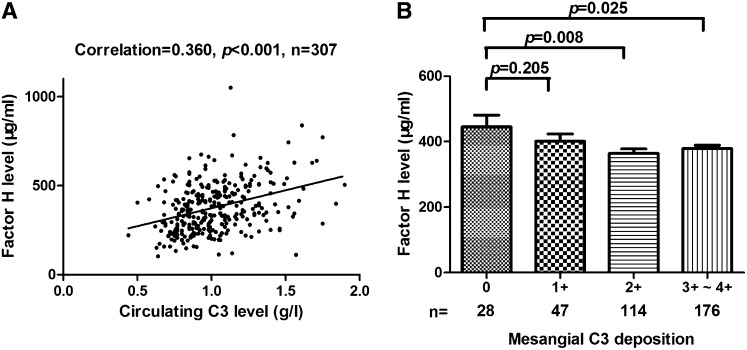
To explore the regulation of systemic complement activation in IgAN, we further analyzed the correlation between circulating CFH and C3 levels. In patients with IgAN from group 2, we found that circulating CFH levels were positively correlated with circulating C3 levels (correlation coefficient=0.360; P<0.001) (Figure 5A), suggesting the involvement of CFH in the regulation of systemic complement activation in IgAN.
Figure 5.
Correlation of CFH levels with circulating C3 and mesangial C3 deposition in patients with IgAN. (A) The circulating CFH level was positively correlated with the circulating C3 level (correlation coefficient=0.360; P<0.001). (B) Lower CFH levels were associated with greater mesangial C3 deposition. Circulating CFH levels decreased significantly from 0 to 3+–4+ mesangial C3 deposition in patients with IgAN (0, 1+, 2+, and 3+–4+: 445.30±186.41, 401.25±150.82, 363.97±147.14, and 378.78±135.09 µg/ml, respectively; 0 versus 1+, P=0.21; 0 versus 2+, P=0.008; 0 versus 3+–4+, P=0.03; n=28, n=47, n=114, and n=176, respectively).
Circulating CFH Was Associated with Mesangial C3 Deposition
As in group 1, patients with IgAN in group 2 with greater mesangial C3 deposition presented with lower circulating C3 levels (0, 1+, 2+, and 3+–4+: 1.11±0.28, 1.12±0.27, 1.04±0.23, and 0.97±0.23 g/L, respectively; P=0.001). Moreover, patients with no C3 deposition had significantly higher circulating CFH levels than those with 2+ and 3+–4+ C3 deposition (0 versus 2+: 445.30±186.41 versus 363.97±147.14 µg/ml; P=0.008; 0 versus 3+–4+: 445.30±186.41 versus 378.78±135.09 µg/ml; P=0.03) (Figure 5B).
Mesangial C3 Deposition but Not rs6677604 Genotype Was Associated with Oxford M, S, and T Grades
To explore the clinical implications of rs6677604 and mesangial C3 deposition in IgAN, we compared clinical and histologic manifestations at biopsy among patients with IgAN with different rs6677604 genotypes and different grades of mesangial C3 deposition. We found that histologic changes were severe in patients with greater mesangial C3 deposition, with a higher proportion classified as M1, S1, and T1/T2 grades, although there was no significant difference in clinical characteristics (Table 1). For patients with IgAN with rs6677604-AG and rs6677604-GG genotypes, no associations with clinical and pathologic findings were observed.
Table 1.
A cross-sectional comparison of baseline patient characteristics according to the rs6677604 genotype and mesangial C3 deposition
| Characteristics | Total | rs6677604 | Mesangial C3 Deposition | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AA | AG | P Value | 0 | 1+ | 2+ | 3+–4+ | P Value | ||
| Patient numbers | 365 | 39 | 326 | 28 | 47 | 114 | 176 | ||
| Age (yr) | 34.09±11.98 | 35.10±11.98 | 33.97±11.99 | 0.58 | 37.32±15.88 | 34.45±14.3 | 32.45±11.13 | 34.54±11.06 | 0.22 |
| Sex (men/women) | 194/171 (53.2%/46.8%) | 22/17 (56.4%/43.6%) | 172/154 (52.8%/47.2%) | 0.67 | 16/12 (57.1%/42.9%) | 24/23 (51.1%/48.9%) | 64/50 (56.1%/43.9%) | 90/86 (51.1%/48.9%) | 0.81 |
| eGFR (ml/min) | 85.87±36.57 | 90.63±31.08 | 84.93±32.52 | 0.30 | 91.75±50.50 | 88.52±28.60 | 87.48±26.47 | 82.50±33.15 | 0.34 |
| Proteinuria (g/d) | 1.40 (0.80, 2.69) | 1.51 (0.58, 2.59) | 1.40 (0.78, 2.71) | 0.55 | 1.38 (0.79, 4.91) | 1.82 (0.75, 3.43) | 1.42 (0.77, 2.45) | 1.40 (0.81, 2.52) | 0.70 |
| SBP (mmHg) | 124.2±14.3 | 121.3±14.0 | 124.6±14.3 | 0.17 | 124.5±11.4 | 126.5±21.0 | 122.8±12.8 | 124.5±13.5 | 0.50 |
| DBP (mmHg) | 79.2±11.0 | 79.6±12.1 | 79.1±10.9 | 0.78 | 78.8±9.4 | 79.2±14.4 | 77.6±10.5 | 80.2±10.5 | 0.26 |
| Oxford classification | |||||||||
| M (M0/M1) | 106/257 (29.2%/70.8%) | 11/28 (28.2%/71.8%) | 95/229 (29.3%/70.7%) | 0.89 | 13/15 (46.4%/53.6%) | 23/24 (48.9%/51.1%) | 40/74 (35.1%/64.9%) | 30/144 (17.2%/82.8%) | <0.001a |
| E (E0/E1) | 140/223 (38.6%/61.4%) | 15/24 (38.5%/61.5%) | 125/199 (38.6%/61.4%) | 0.99 | 10/18 (35.7%/64.3%) | 19/28 (40.4%/59.6%) | 38/76 (33.3%/66.7%) | 73/101 (42.0%/58.0%) | 0.45a |
| S (S0/S1) | 98/265 (27.0%/73.0%) | 13/26 (33.3%/66.7%) | 85/239 (26.2%/73.8%) | 0.84 | 13/15 (46.4%/53.6%) | 15/32 (31.9%/68.1%) | 29/85 (25.4%/74.6%) | 41/133 (23.6%/76.4%) | 0.02a |
| T (T0/T1/T2) | 278/46/39 (76.6%/12.7%/10.7%) | 34/4/1 (87.2%/10.3%/2.6%) | 244/42/38 (75.3%/13.0%/11.7%) | 0.17 | 26/2/0 (92.9%/7.1%/0.0%) | 39/4/4 (83.0%/8.5%/8.5%) | 85/20/9 (74.6%/17.5%/7.9%) | 128/20/26 (73.6%/11.5%/14.9%) | <0.01 |
P value for the linear-by-linear association chi-squared test.
Discussion
A previous IgAN GWAS identified variants in CFH, CFHR3, and CFHR1 as IgAN-susceptible genetic polymorphisms.10 In this study, we found that variants in CFH, CFHR3, and CFHR1 were associated with circulating CFH levels and also affected systemic complement activation in IgAN. Our findings have provided functional annotation for the GWAS locus and revealed a possible genetic mechanism for IgAN susceptibility caused by variants of CFH, CFHR3, and CFHR1.
CFH, CFHR3, and CFHR1 are regulators of the complement system, which plays a key role in immune surveillance and homeostasis.26 In many systemic autoimmune diseases, activation of the complement system is involved in pathogenesis.27 In recent years, it has become more apparent that IgAN is an autoimmune disease, in which IgA1-containing immune complexes initiate glomerular injury.28 As in other autoimmune diseases, complement activation has also been reported in IgAN.19 However, our understanding is limited regarding the precise mechanisms of complement activation in IgAN pathogenesis.
Our previous GWAS of IgAN identified 1q32 as an IgAN susceptibility locus, with rs6677604 in CFH as the top signal. However, in another GWAS in Europeans, 1q32 was not identified,11 which might be because of the relatively small population and low statistical power. Moreover, in a recent IgAN GWAS, in which the southern Chinese were recruited as the discovery population, 1q32 was also not identified.9 In our previous GWAS, the signal at rs6677604 was weaker in the southern Chinese compared with in the northern Chinese10; this discrepancy might be caused by differences in genetic structure between the southern and northern Chinese. In a recent GWAS replication study, the association of 1q32 with IgAN susceptibility was confirmed in eight new independent cohorts.29 On the basis of this genetic association, we further explored the clinical significance of rs6677604 in IgAN. Our finding that the rs6677604 locus was associated with mesangial C3 deposition in IgAN implied a functional role for rs6677604 in complement activation and more importantly, aroused our interest in the complement activation mechanism in IgAN.
Undoubtedly, multiple factors contribute to complement activation in IgAN. Other than rs6677604, we found that circulating IgA, Gd-IgA1, and C3 levels were also associated with mesangial C3 deposition in IgAN. The variation in Gd-IgA1 levels paralleled that of mesangial C3 deposition, suggesting that elevated Gd-IgA1 initiated complement activation and induced subsequent mesangial C3 deposition in IgAN. Because circulating IgA consists mostly of IgA1,30 the same trend was observed in circulating IgA. It is worth noting that decreased circulating C3 levels in autoimmune disease are probably caused by C3 consumption because of systemic complement activation. Our observation of a negative correlation between circulating C3 and mesangial C3 deposition, therefore, suggested that systemic complement activation contributed to generating C3-containing immune complexes that may have been subsequently deposited in the mesangium. Although local complement activation is widely accepted in IgAN, other studies have also observed systemic complement activation, which is consistent with our findings.20,31,32 On the basis of these studies together with our understanding of a multihit mechanism of IgAN pathogenesis,7 we propose a hypothesis regarding systemic complement activation in IgAN. Gd-IgA1 molecules bound by antiglycan autoantibodies form circulating IgA1-containing immune complexes. These immune complexes induce systemic complement activation and ultimately, deposit in the glomerular mesangium, inducing glomerular injury (Figure 6). According to our finding of an association between rs6677604 genotype and mesangial C3 deposition, we speculate that variants in CFH (rs6677604) affect mesangial C3 deposition through the regulation of systemic complement activation in IgAN.
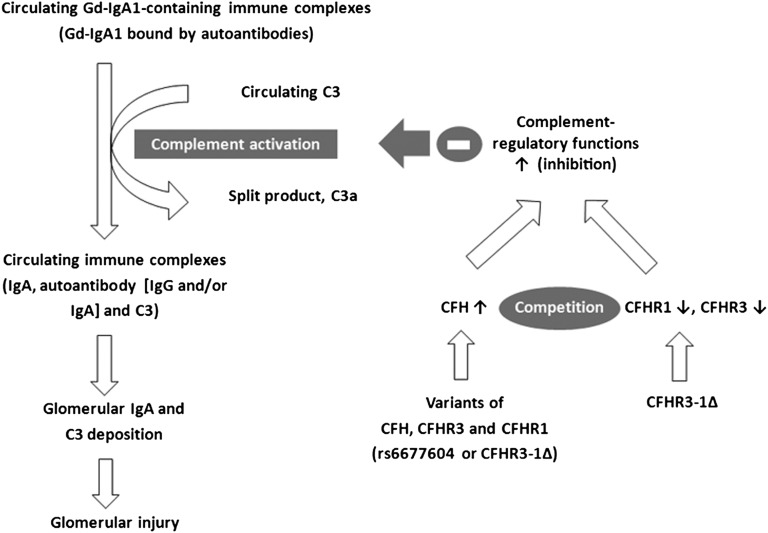
Figure 6.
A hypothesis for a potential mechanism by which variants of CFH, CFHR3, and CFHR1 genes affect complement activation in IgAN. Previous studies revealed the pathogenesis of IgAN, in which Gd-IgA1 molecules bound by antiglycan autoantibodies form circulating IgA1-containing immune complexes that ultimately deposit in the glomerular mesangium, inducing glomerular injury.45 Meanwhile, complement C3 was observed in both circulating and deposited immune complexes.46 In this study, we found that variants in CFH, CFHR3, and CFHR1 (rs6677604-A allele or CFHR3–1Δ) were associated with higher CFH levels. Because genetic deletion of CFHR3 and CFHR1 was reported to lead to absence of these proteins10 and because CFHR1 was a competitive antagonist of CFH to modulate complement activation,33 higher CFH levels together with the absence of antagonist (CFHR1 protein) resulted in the robust complement inhibition represented by higher circulating C3 and lower C3a. Furthermore, the associations between mesangial C3 deposition and circulating CFH levels as well as genetic variants suggested that variants in CFH, CFHR3, and CFHR1 influenced the formation and deposition of pathogenic immune complexes in IgAN.
To further investigate this hypothesis, we enrolled group 2. In group 2, we found individuals with the rs6677604-A allele had increased CFH levels. Recently, Ansari et al.12 reported that rs6677604 and CFHR3–1∆ were strongly correlated with plasma CFH concentration, which is in accordance with our findings. Moreover, the CFHR3–1∆ resulted in the absence of CFHR1 protein, which was recently shown to function as a competitive antagonist of CFH.33 Therefore, higher CFH levels, together with the absence of antagonist (CFHR1 protein), resulted in the robust complement inhibition, which is represented by the higher circulating C3 and lower C3a that we observed in patients with IgAN with the rs6677604-AA genotype and CFHR3–1Δ. In accordance with our findings, Yang et al.34 reported that rs3753394 in CFH was associated with circulating C3 levels. Because rs3753394 and rs6677604 are in LD, the study by Yang et al.34 provides independent validation for our findings. Meanwhile, by showing that plasma CFH was positively correlated with circulating C3 levels and negatively correlated with mesangial C3 deposition, our results suggest that rs6677604 regulated systemic complement activation and the subsequent mesangial C3 deposition in IgAN through its influence on CFH. Together with our previous hypothesis regarding systemic complement activation in IgAN, we provide a possible functional explanation for the association between IgAN and rs6677604. We revealed that rs6677604 in CFH affected circulating complement activation through its effect on CFH function, suggesting a potential genetic mechanism for the predisposition to IgAN caused by variants in CFH, CFHR3, and CFHR1 (Figure 6).
After our investigation of IgAN susceptibility, we further explored the influence of various factors on IgAN severity. We found that mesangial C3 deposition rather than rs6677604 was associated with severe histopathologic features, which was shown by Oxford M, S, and T scores. Our results support a role for mesangial C3 deposition, which might be associated with immune complex deposition, in kidney injury in IgAN. In accordance with this finding, previous studies showed that IgA1-containing immune complexes from patients with IgAN could activate mesangial cells to secrete a number of inflammatory factors that mediated renal injury.35–38 However, rs6677604-A and rs6677604-G alleles showed no significant association with clinical manifestations or histopathologic features. This finding could be explained as follows. Variation in CFH (rs6677604) was just one of multiple contributors to the deposition of IgA1-containg immune complexes. Thus, rs6677604 alone cannot predict the severity of IgAN, which is in accordance with the multifactorial nature and complex characteristics of IgAN pathogenesis.
On the basis of the involvement of rs6677604 in IgAN susceptibility and mesangial C3 deposition in IgAN severity, it is important to evaluate the predictive value of rs6677604 as well as mesangial C3 deposition for long-term renal outcome in IgAN. Recently, Kim et al.39 showed that mesangial C3 deposition predicted renal outcome in patients with IgAN. However, owing to the slowly progressive nature of IgAN and our relatively small follow-up population, we failed to draw any convincing conclusions regarding the predictive value of rs6677604 and mesangial C3 deposition in our study.
Our study had several limitations. First, because high levels of LD were observed between rs6677604 and CFHR3–1∆, we cannot distinguish which one is the causal variant. It is likely that rs6677604, located in the intron region, is a marker linked to another functional variant. Additional in-depth functional studies on a biologic model with genetic CFHR3–1∆ are required. Second, we attempted to assess local complement activation by using a semiquantification analysis of CFH expression in kidney biopsy samples. However, the results were not successful because of poor sensitivity. Third, the sample size for the follow-up population was limited. IgAN has a slowly progressive course. Thus, our follow-up cohort had limited power for survival analysis, especially regarding rs6677604, for which the minor allele frequency was low. Additional evaluation studies in other large and long-term follow-up cohorts are required in the future.
In conclusion, our study identified a series of significant associations of variants of CFH, CFHR3, and CFHR1 with circulating CFH levels and mesangial C3 deposition in patients with IgAN. These associations suggested a regulatory effect of these variants on complement activation in IgAN and provided a possible genetic mechanism for the predisposition to IgAN caused by variants in CFH, CFHR3, and CFHR1.
Concise Methods
Ethics Statement
This study was approved by the medical ethics committee of Peking University, and informed written consent was obtained from every participant.
Study Population
To investigate the clinical meaning of genetic variants identified in the GWAS (rs6677604 and CFHR3–1∆), we first recruited 1178 subjects with IgAN and available GWAS data (group 1). This group was almost the same as the Beijing Discovery Cohort reported in our previous IgAN GWAS,10 except for 16 patients with IgAN who were diagnosed by histochemical identification of IgA deposition rather than immunofluorescence (IF). In group 1, all of the patients had cross-sectional clinical data at the time of renal biopsy, but only 196 patients were regularly followed up in our hospital. Next, to further explore the underlying mechanism and the predictive value of rs6677604 for IgAN prognosis, we enrolled all of the northern Chinese patients with IgAN with available plasma samples and long-term follow-up data in our hospital to form group 2. Thus, 196 patients with IgAN with regular follow-up data in group 1 were also recruited into group 2 to make a total of 365 patients (group 2). The diagnosis of IgAN was verified by immunostaining showing granular deposition of IgA in the glomerular mesangium as well as ultrastructural examination showing the deposition of electron-dense materials in mesangium. Patients with Henoch–Schonlein purpura, SLE, and chronic hepatic diseases were excluded by detailed clinical and laboratory examinations. Additionally, plasma samples from 117 healthy controls from the GWAS population were used for circulating CFH detection.
For patients with IgAN in group 1, information regarding the rs6677604 genotype was extracted from our previous GWAS data.
Because measurements of circulating Ig and complement concentration were routine tests for patients undergoing renal biopsy in our hospital, we were able to collect circulating IgA and C3 levels from medical records for the majority of patients (including 1029 patients for IgA and 1008 patients for C3). Moreover, Gd-IgA1 levels were detected in 480 patients with IgAN in group 1. In our hospital, renal biopsy samples were processed in a standardized manner. C3 deposition in the mesangial area was detected by FITC-labeled polyclonal rabbit anti-human C3c complement antibody (Dako Cytomation, Glostrup, Denmark) and semiquantified using IF staining. IF findings were graded on a scale of 0–4 as follows: 0 (lack of deposits), 1+ (trace), 2+ (weak), 3+ (moderate), and 4+ (strong) (Supplemental Figure 1). Three successive pathologists were responsible for pathologic diagnosis of renal biopsy in our hospital, each one working for several years. All were trained by the same senior special pathologist to maintain consistency. For this study, the glomerular IF findings recorded in the biopsy reports were retrospectively reviewed.
For patients in group 2, information regarding the rs6677604 genotype was either extracted from GWAS data or detected by TaqMan allele discrimination assays. Genotyping of CNVs of CFHR3–1∆ was performed in a subgroup of group 2 including 83 patients with IgAN and 88 healthy controls.
Clinical manifestations at the time of renal biopsy, including creatinine level, 24-hour urine protein excretion, and BP, were collected from medical records. The eGFR of patients with IgAN was calculated by the modified GFR-estimating equation for the Chinese.40 Circulating CFH was measured in all 365 patients with IgAN in group 2, and circulating C3a levels were also measured in 68 of these patients.
As in group 1, information on mesangial C3 deposition was obtained from biopsy reports. Additionally, with the exception of 2 patients with IgAN with less than eight glomeruli in biopsy samples, 363 patients were graded according to the Oxford classification for evaluating pathologic lesions.41
All patients in group 2 were regularly followed up for at least 1 year, with a mean follow-up time of 57.25±31.24 months. During follow-up, patients were prospectively treated with uniform therapy, including optimal BP control (target<130/80 mmHg), renin-angiotensin-aldosterone system inhibition, and intensive treatment with steroids or other immunosuppressive agents for patients with persistent proteinuria. Clinical and pathologic data are summarized in Table 1.
Assay for Gd-IgA1
Gd-IgA1 was detected by lectin ELISA as previously reported.42,43 Briefly, F(ab')2 fragments of goat anti-human IgA (Jackson ImmunoResearch Laboratories, West Grove, PA) were coated onto high-binding MaxiSorp 96-well plates (Nalge-Nunc, Rochester, NY) at 4°C overnight. After blocking with 1% BSA (Sigma-Aldrich, St. Louis, MO), 2-fold dilutions (from 1:2000 to 1:16,000) of serum samples and standards were added and incubated overnight at room temperature. A polymeric Gd-IgA1 protein isolated from a patient with multiple myeloma was used as a standard. Incubation with 1 milliunit per well neuraminidase (Roche Diagnostic, Indianapolis, IN) for 3 hours at 37°C was then performed to remove terminal sialic acid. Terminal N-acetylgalactosamine was detected by biotin-labeled HAA (Sigma-Aldrich) followed by horseradish peroxidase–ExtrAvidin (Sigma-Aldrich). The reaction was developed using the peroxidase chromogenic substrate o-phenylenediamine–H2O2 (Sigma-Aldrich) and stopped with 1 mol/L sulfuric acid before the absorbance was measured at 490 nm. The amount of Gd-IgA1 in each sample was calculated using the DeltaSoft II program (BioMetallics, Princeton, NJ) by interpolating the ODs on calibration curves constructed using standard Gd-IgA1 myeloma protein. The results were expressed in units per milliliter, in which 1 unit Gd-IgA1 was defined as 1 mg standard Gd-IgA1 myeloma protein.
SNP Genotyping
For 169 patients with IgAN in group 2 who were not included in group 1, TaqMan allele discrimination assays (Applied Biosystems, Foster City, CA) were used to determine the genotype of rs6677604 according to the manufacturer’s instructions. The variants were detected using an ABI Prism 7500 Sequence Detection System (Applied Biosystems).
MLPA Genotyping
CNVs of the CFH–CFHRs region were detected using the SALSA MLPA KIT P236-A2 ARMD mix-1 according to the manufacturer’s instructions (MRC-Holland, Amsterdam, The Netherlands). Fragment analyses were performed on an ABI Prism Model 3130xl Genetic Analyzer (Applied Biosystems). For each sample, 200 ng genomic DNA at a concentration of 40 ng/ml was used for MLPA genotyping.
Circulating CFH Detection
On the morning of renal biopsy, plasma (EDTA anticoagulated) was collected from patients with IgAN, divided into aliquots, and stored at −80°C pending the measurement of circulating CFH. Circulating CFH was assessed by ELISA as previously described.44 Briefly, goat anti-human CFH antibody (Calbiochem, Darmstadt, Hessen, Germany) was diluted to 10 µg/ml with 0.05 mol/L bicarbonate buffer (pH 9.6) and coated onto one half of the wells of a microtiter plate (Nunc Immunoplate, Roskilde, Denmark), whereas the wells in the other one half were coated with bicarbonate buffer alone to exclude nonspecific binding. After each step, the plates were washed with 0.01 M PBS containing 0.1% Tween 20. The plasma samples were diluted in PBS containing 0.1% Tween 20 to 1:4000 and incubated in the plates for 1 hour at 37°C. Binding of CFH was examined using mouse anti-human CFH antibodies (US Biologic, Swampscott, MA), which detected only CFH and not FHL-1– or CFH-related proteins (Supplemental Figure 2). After addition of alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma-Aldrich), the reaction was developed using 1 mg/ml p-nitrophenyl phosphate (Sigma -Aldrich) dissolved in substrate buffer (0.1% diethanolamine, 1.1 mM MgCl2, and milliQ H2O), and the absorbance value at 405 nm was measured. Serial dilutions of highly purified human CFH (Calbiochem) were used to establish the standard curve, which was then used to measure circulating CFH.
Circulating C3a Detection
Circulating C3a levels were detected using commercial ELISA kits according to the manufacturer’s specifications (Quidel, San Diego, CA).
Statistical Analyses
Statistics were performed using SPSS software (version 13.0). For continuous variables, an unpaired t test or ANOVA between groups was used if the data were a normal distribution; otherwise, a Mann–Whitney U test or Kruskal–Wallis H test was performed. Data with a normal distribution were expressed as means±SDs, whereas other data were expressed as median (first and third quartiles). Categorical variables were analyzed by the chi-squared test. For survival analysis, a composite end point of ESRD or all-cause mortality was used. ESRD was defined as a GFR<15 ml/min per m2 or the need for RRT. The Kaplan–Meier method and the log-rank test were used to assess and compare cumulative incidences of developing end points among patients within different groups. A P value<0.05 was considered statistically significant.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by Major State Basic Research Development Program of China 973 Program Grant 2012CB517700, Natural Science Foundation for Innovation Research Group of China Grant 81321064, National Science Foundation for Youths of China Grant 81000297, Beijing Natural Science Foundation Grant 7131016, and Capital of Clinical Characteristics and Applied Research Fund Grant Z141107002514037.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014010096/-/DCSupplemental.
References
- 1.Zhou FD, Zhao MH, Zou WZ, Liu G, Wang H: The changing spectrum of primary glomerular diseases within 15 years: A survey of 3331 patients in a single Chinese centre. Nephrol Dial Transplant 24: 870–876, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Levy M, Berger J: Worldwide perspective of IgA nephropathy. Am J Kidney Dis 12: 340–347, 1988 [DOI] [PubMed] [Google Scholar]
- 3.D’Amico G: The commonest glomerulonephritis in the world: IgA nephropathy. Q J Med 64: 709–727, 1987 [PubMed] [Google Scholar]
- 4.Kiryluk K, Julian BA, Wyatt RJ, Scolari F, Zhang H, Novak J, Gharavi AG: Genetic studies of IgA nephropathy: Past, present, and future. Pediatr Nephrol 25: 2257–2268, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu SI, Ramirez SB, Winn MP, Bonventre JV, Owen WF: Evidence for genetic factors in the development and progression of IgA nephropathy. Kidney Int 57: 1818–1835, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Beerman I, Novak J, Wyatt RJ, Julian BA, Gharavi AG: The genetics of IgA nephropathy. Nat Clin Pract Nephrol 3: 325–338, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, Wyatt RJ, Scolari F, Mestecky J, Gharavi AG, Julian BA: The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22: 1795–1803, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA, Brooks LD, Collins FS: A HapMap harvest of insights into the genetics of common disease. J Clin Invest 118: 1590–1605, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, Sun LD, Sim KS, Li Y, Foo JN, Wang W, Li ZJ, Yin XY, Tang XQ, Fan L, Chen J, Li RS, Wan JX, Liu ZS, Lou TQ, Zhu L, Huang XJ, Zhang XJ, Liu ZH, Liu JJ: A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44: 178–182, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, Kumar A, Peden JF, Maxwell PH, Morris DL, Padmanabhan S, Vyse TJ, Zawadzka A, Rees AJ, Lathrop M, Ratcliffe PJ: HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 21: 1791–1797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansari M, McKeigue PM, Skerka C, Hayward C, Rudan I, Vitart V, Polasek O, Armbrecht AM, Yates JR, Vatavuk Z, Bencic G, Kolcic I, Oostra BA, Van Duijn CM, Campbell S, Stanton CM, Huffman J, Shu X, Khan JC, Shahid H, Harding SP, Bishop PN, Deary IJ, Moore AT, Dhillon B, Rudan P, Zipfel PF, Sim RB, Hastie ND, Campbell H, Wright AF: Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum Mol Genet 22: 4857–4869, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Wu H, Khosravi M, Cui H, Qian X, Kelly JA, Kaufman KM, Langefeld CD, Williams AH, Comeau ME, Ziegler JT, Marion MC, Adler A, Glenn SB, Alarcón-Riquelme ME, Pons-Estel BA, Harley JB, Bae SC, Bang SY, Cho SK, Jacob CO, Vyse TJ, Niewold TB, Gaffney PM, Moser KL, Kimberly RP, Edberg JC, Brown EE, Alarcon GS, Petri MA, Ramsey-Goldman R, Vilá LM, Reveille JD, James JA, Gilkeson GS, Kamen DL, Freedman BI, Anaya JM, Merrill JT, Criswell LA, Scofield RH, Stevens AM, Guthridge JM, Chang DM, Song YW, Park JA, Lee EY, Boackle SA, Grossman JM, Hahn BH, Goodship TH, Cantor RM, Yu CY, Shen N, Tsao BP, BIOLUPUS Network. GENLES Network : Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet 7: e1002079, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boon CJ, van de Kar NC, Klevering BJ, Keunen JE, Cremers FP, Klaver CC, Hoyng CB, Daha MR, den Hollander AI: The spectrum of phenotypes caused by variants in the CFH gene. Mol Immunol 46: 1573–1594, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Zipfel PF: Complement and immune defense: From innate immunity to human diseases. Immunol Lett 126: 1–7, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Skerka C, Zipfel PF: Complement factor H related proteins in immune diseases. Vaccine 26[Suppl 8]: I9–I14, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Hälbich S, Mihlan M, Schlötzer-Schrehardt U, Zipfel PF, Skerka C: Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114: 2439–2447, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Köhl J, Zipfel PF, Weber BH, Skerka C: An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet 19: 4694–4704, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Haas M: Histology and immunohistology of IgA nephropathy. J Nephrol 18: 676–680, 2005 [PubMed] [Google Scholar]
- 20.Onda K, Ohi H, Tamano M, Ohsawa I, Wakabayashi M, Horikoshi S, Fujita T, Tomino Y: Hypercomplementemia in adult patients with IgA nephropathy. J Clin Lab Anal 21: 77–84, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JJ, Jiang L, Liu G, Wang SX, Zou WZ, Zhang H, Zhao MH: Levels of urinary complement factor H in patients with IgA nephropathy are closely associated with disease activity. Scand J Immunol 69: 457–464, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HP, Charbel Issa P, Cano M, Brandstätter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ: Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 478: 76–81, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Barricarte R, Pianetti G, Gautard R, Misselwitz J, Strain L, Fremeaux-Bacchi V, Skerka C, Zipfel PF, Goodship T, Noris M, Remuzzi G, de Cordoba SR, European Working Party on the Genetics of HUS : The complement factor H R1210C mutation is associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 19: 639–646, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gale DP, Pickering MC: Regulating complement in the kidney: Insights from CFHR5 nephropathy. Dis Model Mech 4: 721–726, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid-Kubista KE, Tosakulwong N, Wu Y, Ryu E, Hecker LA, Baratz KH, Brown WL, Edwards AO: Contribution of copy number variation in the regulation of complement activation locus to development of age-related macular degeneration. Invest Ophthalmol Vis Sci 50: 5070–5079, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Ricklin D, Hajishengallis G, Yang K, Lambris JD: Complement: A key system for immune surveillance and homeostasis. Nat Immunol 11: 785–797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Daha MR, Kallenberg CG: The complement system in systemic autoimmune disease. J Autoimmun 34: J276–J286, 2010 [DOI] [PubMed] [Google Scholar]
- 28.Mestecky J, Raska M, Julian BA, Gharavi AG, Renfrow MB, Moldoveanu Z, Novak L, Matousovic K, Novak J: IgA nephropathy: Molecular mechanisms of the disease. Annu Rev Pathol 8: 217–240, 2013 [DOI] [PubMed] [Google Scholar]
- 29.Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F, Snyder HJ, Choi M, Hou P, Scolari F, Izzi C, Gigante M, Gesualdo L, Savoldi S, Amoroso A, Cusi D, Zamboli P, Julian BA, Novak J, Wyatt RJ, Mucha K, Perola M, Kristiansson K, Viktorin A, Magnusson PK, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boland A, Metzger M, Thibaudin L, Wanner C, Jager KJ, Goto S, Maixnerova D, Karnib HH, Nagy J, Panzer U, Xie J, Chen N, Tesar V, Narita I, Berthoux F, Floege J, Stengel B, Zhang H, Lifton RP, Gharavi AG: Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet 8: e1002765, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morell A, Skvaril F, Noseda G, Barandun S: Metabolic properties of human IgA subclasses. Clin Exp Immunol 13: 521–528, 1973 [PMC free article] [PubMed] [Google Scholar]
- 31.Zwirner J, Burg M, Schulze M, Brunkhorst R, Götze O, Koch KM, Floege J: Activated complement C3: A potentially novel predictor of progressive IgA nephropathy. Kidney Int 51: 1257–1264, 1997 [DOI] [PubMed] [Google Scholar]
- 32.van Es LA, van den Wall Bake AW, Valentijn RM, Daha MR: Composition of IgA-containing circulating immune complexes in IgA nephropathy. Am J Kidney Dis 12: 397–401, 1988 [DOI] [PubMed] [Google Scholar]
- 33.Goicoechea de Jorge E, Caesar JJ, Malik TH, Patel M, Colledge M, Johnson S, Hakobyan S, Morgan BP, Harris CL, Pickering MC, Lea SM: Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci U S A 110: 4685–4690, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Sun J, Gao Y, Tan A, Zhang H, Hu Y, Feng J, Qin X, Tao S, Chen Z, Kim ST, Peng T, Liao M, Lin X, Zhang Z, Tang M, Li L, Mo L, Liang Z, Shi D, Huang Z, Huang X, Liu M, Liu Q, Zhang S, Trent JM, Zheng SL, Xu J, Mo Z: Genome-wide association study for serum complement C3 and C4 levels in healthy Chinese subjects. PLoS Genet 8: e1002916, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu L, Zhang Q, Shi S, Liu L, Lv J, Zhang H: Synergistic effect of mesangial cell-induced CXCL1 and TGF-β1 in promoting podocyte loss in IgA nephropathy. PLoS ONE 8: e73425, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai KN, Leung JC, Chan LY, Saleem MA, Mathieson PW, Tam KY, Xiao J, Lai FM, Tang SC: Podocyte injury induced by mesangial-derived cytokines in IgA nephropathy. Nephrol Dial Transplant 24: 62–72, 2009 [DOI] [PubMed] [Google Scholar]
- 37.Lai KN, Leung JC, Chan LY, Saleem MA, Mathieson PW, Lai FM, Tang SC: Activation of podocytes by mesangial-derived TNF-alpha: Glomerulo-podocytic communication in IgA nephropathy. Am J Physiol Renal Physiol 294: F945–F955, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Chan LY, Leung JC, Tsang AW, Tang SC, Lai KN: Activation of tubular epithelial cells by mesangial-derived TNF-alpha: Glomerulotubular communication in IgA nephropathy. Kidney Int 67: 602–612, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Kim SJ, Koo HM, Lim BJ, Oh HJ, Yoo DE, Shin DH, Lee MJ, Doh FM, Park JT, Yoo TH, Kang SW, Choi KH, Jeong HJ, Han SH: Decreased circulating C3 levels and mesangial C3 deposition predict renal outcome in patients with IgA nephropathy. PLoS ONE 7: e40495, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma YC, Zuo L, Chen JH, Luo Q, Yu XQ, Li Y, Xu JS, Huang SM, Wang LN, Huang W, Wang M, Xu GB, Wang HY: Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol 17: 2937–2944, 2006 [DOI] [PubMed] [Google Scholar]
- 41.Roberts IS, Cook HT, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, Cattran DC, Coppo R, D’Agati V, D’Amico G, Emancipator S, Emma F, Feehally J, Ferrario F, Fervenza FC, Florquin S, Fogo A, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Li LS, Li PK, Liu ZH, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H, Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The Oxford classification of IgA nephropathy: Pathology definitions, correlations, and reproducibility. Kidney Int 76: 546–556, 2009 [DOI] [PubMed] [Google Scholar]
- 42.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J: Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int 71: 1148–1154, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Zhao N, Hou P, Lv J, Moldoveanu Z, Li Y, Kiryluk K, Gharavi AG, Novak J, Zhang H: The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int 82: 790–796, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang FM, Yu F, Tan Y, Song D, Zhao MH: Serum complement factor H is associated with clinical and pathological activities of patients with lupus nephritis. Rheumatology (Oxford) 51: 2269–2277, 2012 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czerkinsky C, Koopman WJ, Jackson S, Collins JE, Crago SS, Schrohenloher RE, Julian BA, Galla JH, Mestecky J: Circulating immune complexes and immunoglobulin A rheumatoid factor in patients with mesangial immunoglobulin A nephropathies. J Clin Invest 77: 1931–1938, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.