Abstract
Hydrogen peroxide (HP) or cyanide (CN) are bacteriostatic at low-millimolar concentrations for growing Escherichia coli, whereas CN+HP mixture is strongly bactericidal. We show that this synergistic toxicity is associated with catastrophic chromosomal fragmentation. Since CN-alone does not kill at any concentration, while HP-alone kills at 20 mM, CN must potentiate HP poisoning. The CN+HP killing is blocked by iron chelators, suggesting Fenton’s reaction. Indeed, we show that CN enhances plasmid DNA relaxation due to Fenton’s reaction in vitro. However, mutants with elevated iron or HP pools are not acutely sensitive to HP-alone treatment, suggesting that, in addition, in vivo CN recruits iron from intracellular depots. We found that part of the CN-recruited iron pool is managed by ferritin and Dps: ferritin releases iron on cue from CN, while Dps sequesters it, quelling Fenton’s reaction. We propose that disrupting intracellular iron trafficking is a common strategy employed by the immune system to kill microbes.
Keywords: cyanide, hydrogen peroxide, Fenton’s reaction, iron chelators, Dps, ferritin
Introduction
Hydrogen peroxide (HP, H2O2) is an uncharged stable molecule in which oxygen is reduced halfway between the ground state molecular oxygen O2 and the fully reduced oxygen of water H2O. Due to its uncharged character and generally low reactivity with organic compounds, HP moves freely from outside to inside the cell. However, the cell tries to keep intracellular concentrations of HP, as well as superoxide O2− (a reactive oxygen species halfway between O2 and H2O2 and easily reducible to HP) as low as possible, because intracellular iron makes HP extremely toxic (Imlay, 2008).
For E. coli, HP saturates cellular defenses at low millimolar concentrations (1-10 mM), becoming bacteriostatic during short treatments (Imlay & Linn, 1986, Imlay & Linn, 1987), while bactericidal and clastogenic (degrading chromosomal DNA) upon prolonged exposures (Brudzynski et al., 2011). HP shows rapid toxicity in E. coli at concentrations higher than 20-30 mM (Imlay & Linn, 1986, Imlay & Linn, 1987). In fact, the famous 3% hydrogen peroxide first aid antiseptic (~1 M concentration) kills any kind of bacteria within a few minutes (Dittmar et al., 1930, Rutala et al., 2008).
The extreme intracellular HP toxicity is attributed to its catalytic cycle with free cytoplasmic iron that produces hydroxyl radicals (first used in the famous Fenton’s reagent (Fenton, 1894, Koppenol, 1993)) (Fig. 1A, right): Fe2+ + H2O2 → Fe3+ + OH· + OH− (Fe(III) is then returned to Fe(II) by metabolites like free flavins). The generated hydroxyl radicals react with organic compounds at diffusion rates (Buxton et al., 1988). Because of this amplified reactivity in the presence of free cytoplasmic iron, HP and superoxide are classified as “reactive oxygen species”, even though they themselves do not show reactivity with typical organic biomolecules.
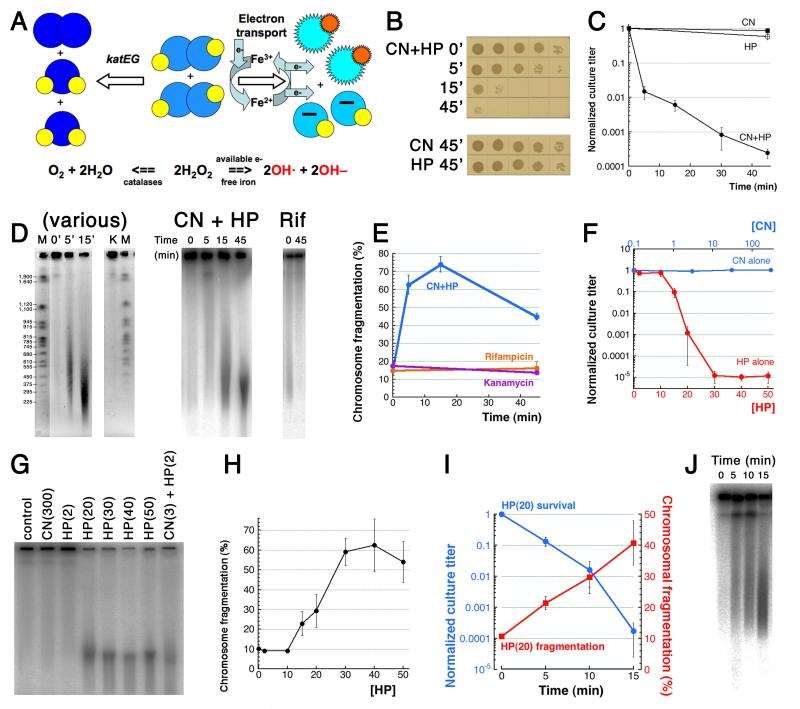
Fig. 1. CN+HP toxicity is due to CN-potentiation of HP-promoted catastrophic chromosome fragmentation.
A. A scheme of in vivo HP toxicity and detoxification. Big balls in shades of blue, oxygen atoms (the lighter the shade, the more reactive the compound), small yellow or orange balls, hydrogen atoms. B. Spotting of serial dilutions (10 μl) of treated WT cultures to demonstrate a representative kinetics. Time is in minutes. Here and everywhere: unless indicated otherwise, “CN” means 3 mM KCN, while “HP” means 2 mM hydrogen peroxide. C. Kinetics of CN+HP killing (and HP-only stasis) of exponential WT cultures. Here and for the rest of the paper, the values are means of 3 or more independent measurements ± SEM. D. Catastrophic chromosome fragmentation triggered by CN+HP treatment, as detected by pulsed-field gels. Left, ethidium bromide-stained gel (reversed image) to show the size distribution of the fragmentation smear (M, the yeast chromosome markers with their length indicated in kbp). Lanes are: 0′, 5′, 15′ time of CN+HP treatment in minutes; K, 50 μg/ml kanamycin for 45 minutes (survival 1.5×10−5). Center and right, direct radioactivity scanning of 32P-labeled chromosomal DNA. Time in minutes is duration of CN+HP treatment (center) or of 100 μg/ml rifampicin treatment (survival 2.8 × 10−3) (right). E. Quantification of CN+HP- or kanamycin- or rifampicin-induced fragmentation from several gels like in “D-right”. Here and for the rest of the paper, chromosome fragmentation is quantified within a particular lane by dividing the signal in the gel by the combined signal in the gel plus well (total signal) and multiplying the product by 100. F. The nature of CN+HP toxicity revealed by CN-alone or HP-alone dose-dependence of survival. All treatments were carried out for 45 minutes. Note that the upper X axis (CN-alone) is logarithmic, and the starting point of it is, actually, “0.1”. G. A representative pulsed-field gel demonstrating the catastrophic nature of HP-alone-induced chromosome fragmentation. All treatments were carried out for 45 minutes. Concentrations in parentheses are in mM. H. Quantification of the dose-dependence of chromosomal fragmentation by HP-alone, from several gels like in “G”. HP concentrations are in mM. I. Kinetics of killing and of chromosome fragmentation by HP(20). The latter values are from several gels like in “J”. J. A representative pulsed-field gel showing kinetics of fragmentation by HP(20).
The cell minimizes Fenton’s reaction by actively reducing intracellular concentrations of HP, superoxide and free iron. The efficiency of HP scavenging is demonstrated by the fact that cells completely deficient in it cannot grow once the endogenous HP reaches ~0.5 μM (Park et al., 2005), while the wild type (WT) cells can still grow even in the presence of 1 mM exogenous HP (Brudzynski et al., 2011). In E. coli, HP is efficiently degraded by a two-step enzyme system. At concentrations up to 20 μM in E. coli, HP is mostly degraded by the copious alklyperoxidase AhpCF (Imlay, 2008), while higher HP concentrations are scavenged by induction of two catalases, encoded by katE and katG genes in E. coli (Fig. 1A, left) (Goerlich et al., 1989). At the same time, cytoplasmic superoxide is efficiently removed by two superoxide dismutases, SodA and SodB (Imlay, 2008). Finally, the amount of free cytoplasmic iron is tightly controlled by the ferric uptake regulator Fur (Hantke, 2001). Because of low availability of soluble iron (Fe(II)) in the oxidizing environment, the cells have to secrete various iron-(Fe(III))-chelating molecules, called siderophores, and then collect their iron-loaded forms back, retrieving the iron for internal use (Wandersman & Delepelaire, 2004). Since iron is expensive to procure, and frequently limited in the environment, surplus cytoplasmic iron is not excreted into the environment, but is stored in association with the specialized proteins of the ferritin superfamily (Smith, 2004). Typical ferritins function as iron distribution centers, taking in or releasing iron according to metabolic needs of the cell, while the mini-ferritin Dps is induced in response to HP-stress and in the stationary cells, to sequester “reactive” iron from iron-HP complexes.
A peculiar phenomenon is potentiation of HP toxicity by some simple substances, which by themselves do not kill, and some of them are not even bacteriostatic. For example, aminoacids cysteine and histidine in high, but still physiological, concentrations potentiate the effect of bacteriostatic HP concentrations, killing the cells (Berglin & Carlsson, 1985). Ascorbic acid has also been long known to potentiate HP toxicity (Miller, 1969). Nitric oxide (NO) is by itself bacteriostatic in high enough concentrations (Spek et al., 2001), but addition of NO to otherwise bacteriostatic concentrations of HP makes it strongly bactericidal (Klebanoff, 1993). In fact, synergistic toxicity of NO+HP is used by our immune cells to kill invading microbes (Brunelli et al., 1995, Pacelli et al., 1995), but the mechanisms behind the microbial death are not entirely clear (Woodmansee & Imlay, 2003).
Synergistic toxicity of HP with cyanide (CN) was also noted before (Imlay et al., 1988) and explored by Woodmansee and Imlay, who proposed that CN-block of respiration leads to an increase in Fe(III)-reduction potential of the major siderophore reductase Fre, resulting in faster Fe(III)→Fe(II) cycling, thereby accelerating Fenton’s reaction (Woodmansee & Imlay, 2002). The proposed mechanism was later confirmed for a similar case of NO+HP co-toxicity (Woodmansee & Imlay, 2003). Even though CN may seem like an artificial potentiator, it is in fact produced by many plants, most fungi and even by some microbes (Knowles, 1976), in addition to being generated by our activated immune cells (But et al., 2002, Furtmüller et al., 1998, Stelmaszynska, 1985, Thomas & Fishman, 1986).
We encountered the co-toxicity with HP while characterizing the surprising killing potential of old hydroxyurea solutions, that turned out to be due to a mixture of NO, CN and HP (Kuong & Kuzminov, 2009). Here we are reporting our investigation on the nature of the CN+HP co-toxicity, of its potential targets and of its chromosomal effects. Theoretically, there are two possible explanations of a co-toxicity. The most obvious one is “redundancy”; according to the redundancy explanation, both CN and HP are toxic, and both toxicities are counteracted by the same mechanism. Therefore, adding them together inactivates an essential cellular function or saturates anti-toxicity mechanisms, killing the cell. For example, both CN (Jurtshuk et al., 1975) and HP (Bickar et al., 1982) are known to bind cytochrome oxidase enzymes, so it is possible that this inactivation could be the cause of death by the CN+HP double treatment.
The second explanation of co-toxicity is “potentiation”, according to which only one of the two chemicals is toxic, but normally there are efficient mechanisms to prevent or neutralize its toxicity, while the second chemical targets these anti-toxicity mechanisms without being toxic itself. Since HP is known to be toxic in high concentrations (Imlay & Linn, 1986, Imlay & Linn, 1987), its toxicity at low concentrations could be potentiated by CN, for example, by inactivating catalases, which are heme-containing enzymes (heme iron is a well-known intracellular CN target). One of the initial objectives of this work was to distinguish between these two explanations for CN+HP co-toxicity. The second objective was to identify targets of CN and HP poisoning. The third objective was to explore chromosomal aspects of CN+HP toxicity.
Results
CN+HP treatment causes catastrophic chromosomal fragmentation
The CN(3)+HP(2) toxicity in our standard conditions (3 mM CN, 2 mM HP) is at least four orders of magnitude within one hour, in line with the previous report (Woodmansee & Imlay, 2002) (Fig. 1BC). At the same time, CN(3)-alone or HP(2)-alone treatments are bacteriostatic (Fig. 1BC). The CN(3)+HP(2) toxicity is robust, as demonstrated by step-wise lowering concentrations of one reagent, while keeping the other one constant: the extent of the killing is significantly reduced only by 10-fold reduction of either reagent (Fig. S1). In fact, the killing of the wild type strain used in these experiments can be stopped immediately by diluting the treated culture 10-fold, a useful stopping technique during a short time course.
Woodmansee and Imlay detected significant DNA damage in CN(3)+HP(2.5)-treated cells by quantitative PCR (Woodmansee & Imlay, 2002), whereas we have earlier reported detectable chromosomal fragmentation caused by unspecified low levels of CN+(NO+)HP (Kuong & Kuzminov, 2009). Therefore, we expected to observe comparatively higher levels of chromosomal fragmentation after our standard CN(3) +HP(2) treatment. Nevertheless, we were still surprised to find that CN+HP killing in our conditions is associated with extraordinary levels of chromosomal fragmentation, detected by pulsed-field gel electrophoresis (Fig. 1DE). We use the term “catastrophic fragmentation” to represents the violent, rapid and complete chromosome demise that CN+HP treatment induces. Since massive fragmentation is already observed after only one minute of treatment (see below), it is likely the cause, rather than the consequence, of death. As a negative control, we found no chromosomal fragmentation after treatment with lethal concentrations of transcription inhibitor rifampicin or translation inhibitor kanamycin (Fig. 1DE), — this also contradicts the reports of massive oxidative damage associated with various mechanisms of antibiotic-caused bacterial cell death (Kohanski et al., 2010).
After reaching a maximum of 65% (signal over background) around 15 minutes, the apparent level of CN+HP-caused fragmentation goes down at the later time point (Fig. 1E). Since the average molecular weight of the chromosomal fragments continues to decrease with time of the treatment (Fig. 1D), the observed decrease in fragmentation must be an artifact of the smallest DNA fragments migrating out of the gel. We conclude that, over the period of 45 minutes, CN+HP treatment kills E. coli cells continuously by inducing double-strand breaks in the chromosomal DNA. Moreover, since in other experimental systems, strong lethality is already observed with chromosomal fragmentation levels of 15-20% (see the Discussion), the catastrophic fragmentation induced by CN+HP treatment should leave cells no chance of survival, — an expectation supported by the 10,000-fold drop in the viable counts (Fig. 1BC).
The nature of the co-toxicity
One could distinguish between the two explanations for CN+HP co-toxicity (redundancy vs potentiation) by checking whether CN-alone or HP-alone would kill at higher concentrations. If both single treatments are toxic at high concentrations, then the redundancy explanation would be possible (potentiated toxicity is always a possibility). However, if only one of the single treatments is toxic at high concentrations, then the corresponding substance is the poison, while the other one acts as a potentiator, by enabling the poisoning mechanism or by disabling a resistance mechanism.
We found that CN-alone, even at very high concentration of 300 mM, neither kills, nor causes any chromosomal fragmentation (Fig. 1FG). In contrast, HP-alone kills at concentrations of 15 mM (HP(15)) or higher, by inducing catastrophic chromosomal fragmentation (Fig. 1FG). Remarkably, HP(10) does not kill or cause chromosomal fragmentation (Fig. 1H), whereas HP(20) already shows significant killing/fragmentation potential. In other words, transition HP(10)→HP(20) does not translate into a simple 2-fold increase in DNA damage; rather, it brings about a qualitative shift, as if a potentiator like CN has been added. Apparently, at high concentrations, HP functions as a self-potentiator, — for example, by partially converting into a substance that acts like CN. The kinetics of killing by HP(20) is similar to the one by the standard CN(3)+HP(2) co-treatment (compare Fig. 1I versus 1C) and is accompanied by a similar magnitude of catastrophic chromosomal fragmentation (Fig. 1J). Therefore, we conclude that 1) HP kills by causing catastrophic chromosomal fragmentation, while CN potentiates this killing by increasing the effective intracellular HP concentrations at least 10-fold; 2) at high concentrations, HP may self-potentiate its own poisoning; 3) the killing target of HP is the chromosomal DNA, while CN targets are unclear.
Genetic analysis of CN potentiation of the HP killing: the expected phenotypes and their interpretation
To identify the targets of CN potentiation, as well as CN-potentiation-counteracting processes, if any, we quantified HP-alone and CN+HP sensitivity of select mutants. Formally, the original phenotypes of WT cells are: “HP-alone” treatment is bacteriostatic, while CN+HP treatment is strongly bactericidal (Fig. 1C and 2A). Four distinct changes from these WT effects are possible in mutants, with specific interpretations (Fig. 2B-E)
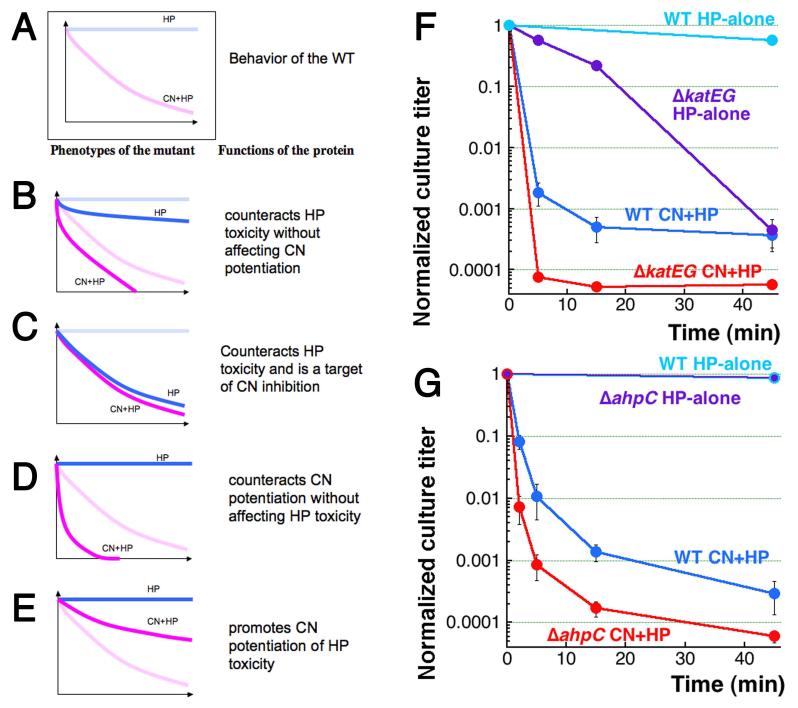
Fig. 2. Formal analysis of the expected kinetics of mutant killing by HP or CN+HP treatments, and the phenotypes of mutants in HP-neutralizing functions.
A. WT kinetics (cf. Fig. 1C). B. Increased sensitivity to both HP and CN+HP treatments. C. Increased sensitivity to HP only, approaching the unchanged sensitivity to CN+HP. D. Increased sensitivity to CN+HP only. E. Decreased sensitivity to CN+HP. F. Kinetics of killing by the two treatments of the double katEG catalase-deficient mutant, compared to WT. G. Kinetics of killing by the two treatments of the ahpC mutant in alkylperoxidase, compared to WT.
The first (perhaps the most intuitive) mutant phenotype is similarly increased sensitivity to both HP-alone and CN+HP (Fig. 2B). Interpretation: the corresponding protein counteracts HP toxicity without affecting CN potentiation of it. In other words, such a mutant identifies an independent branch of HP-toxicity that acts in parallel with the CN-potentiated branch.
Another mutant phenotype is a similar sensitivity to both HP-alone and CN+HP (that is, HP-alone sensitivity drops to the level of CN+HP sensitivity) (Fig. 2C). Interpretation: the corresponding protein counteracts HP toxicity and is a target of CN potentiation. Identification of such targets is the primary objective of our mutant search.
A third expected mutant phenotype is no HP-alone sensitivity (like in WT), but a deeper CN+HP sensitivity (Fig. 2D). Interpretation: the corresponding protein counteracts CN potentiation without affecting HP toxicity.
Finally, the fourth effect is again no sensitivity to HP-alone (like in WT), but this time a shallower sensitivity to CN+HP (Fig. 2E). Interpretation: the corresponding protein promotes CN potentiation of HP-toxicity.
HP-removing enzymes counteract CN-potentiation
Consideration of the scheme of HP toxicity/detoxification (Fig. 1A) for possible targets of CN potentiation leads to a perplexing possibility that tight CN binding to the heme iron inhibits both sides of the process, resulting in a larger intracellular HP pool on the one hand, yet a less efficient hydroxyl radical production on the other. Indeed, the CN-vulnerability of the hydroxy-radical-generating side of the scheme (Fig. 1A right) is due to the fact that CN blocks the electron transport chain, via binding to hemes of cytochrome oxidases (Jurtshuk et al., 1975), and at the same time via complexing highly reactive free iron into mildly reactive ferrocyanide. However, the “catalase” side of the scheme (Fig. 1A left) should be equally CN-vulnerable, as both E. coli catalases are also heme-containing enzymes (Chelikani et al., 2004), making them ideal targets for CN-potentiation of HP toxicity.
There is a disagreement over whether inactivation of catalases sensitizes cells to HP-alone treatment in E. coli: Pueyo and colleagues report that the double katEG mutant is extremely sensitive to 20′ treatments with HP (Prieto-Alamo et al., 1993), while Imlay and colleagues claim their double catalase mutant is not generally sensitive to HP (~10-fold deeper viability loss compared to WT after 15′ treatment) (Imlay & Linn, 1987). In our hands, the ΔkatEG mutant is killed by CN+HP faster and deeper than WT cells (Fig. 2F), but it is the unusual kinetics of HP-alone sensitivity, which fails to plateau within 45 minutes of treatment, that makes the overall pattern of the double catalase deletion mutant complex. Indeed, at 15 minutes it corresponds best to Fig. 2B, but then matches Fig. 2C pattern at 45 minutes. If we are to ignore the earliest time point, catalases counteract HP toxicity (as they should), without affecting CN potentiation at first, but then they gradually become targets of CN inhibition. Still, the dramatic difference between the first 15 minutes of CN+HP versus the HP-alone sensitivity curves excludes catalases from being the potential targets of immediate CN inhibition.
While catalases are responsible for defending cells against millimolar concentrations of exogenous HP, E. coli employs alkylperoxidase AhpCF to remove micromolar concentrations of endogenous HP (Imlay, 2008). Alkylperoxidase does not have any metal that cyanide could complex and is too weak to counter 2 mM HP of our treatment anyway, so the ahpC mutant was expected to be killed similar to the WT with the CN+HP treatment. Surprisingly, we observed increased CN+HP sensitivity in the ahpC mutant with no increase in HP-alone sensitivity (Fig. 2G). Thus, unexpectedly, if the first 5 minutes of the two treatments are considered, both ahpC and katEG results are generally consistent with the “Fig. 2D” scenario, suggesting that both enzymes somehow counteract CN-potentiation of HP-toxicity, with either no or delayed effect on HP-alone toxicity. Since the existence of a specific intracellular pool of HP that is preferentially used for CN potentiation of Fenton’s reagent is unlikely (uncharged and small HP molecules diffuse freely), the unexpected phenotype of the katEG and ahpC mutants suggest tight trafficking of iron inside the cell and, perhaps, even physical proximity of HP-removing enzymes to the iron hubs.
The electron transport connection
Interestingly, Woodmansee and Imlay have already reported a major target of CN potentiation of HP toxicity, working in the “electron transport” area in the Fig. 1A scheme (expanded in Fig. 3A) (Woodmansee & Imlay, 2002). Nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) is the main reducing power of E. coli cells grown aerobically in rich medium, its electrons being funneled into the aerobic respiratory chain by two NADH dehydrogenase (NDH) enzymes (one coded by the nuo gene cluster, the other by the ndh gene) through quinones to the three terminal cytochrome oxidases, bo, bd-I and bd-II (Fig. 3A) (Borisov et al., 2011). Woodmansee and Imlay confirmed an earlier report (Imlay et al., 1988) that the ndh mutants are sensitive to HP-alone treatment, and this sensitivity is not further increased by CN (Woodmansee & Imlay, 2002). At the same time, the nuo mutants behaved like WT in these assays. The equal and high sensitivity of ndh mutants to both HP-alone and CN+HP (“Fig. 2C” pattern) was a clear genetic indication that the electron transport chain is a target of cyanide potentiation of the HP killing (Woodmansee & Imlay, 2002).
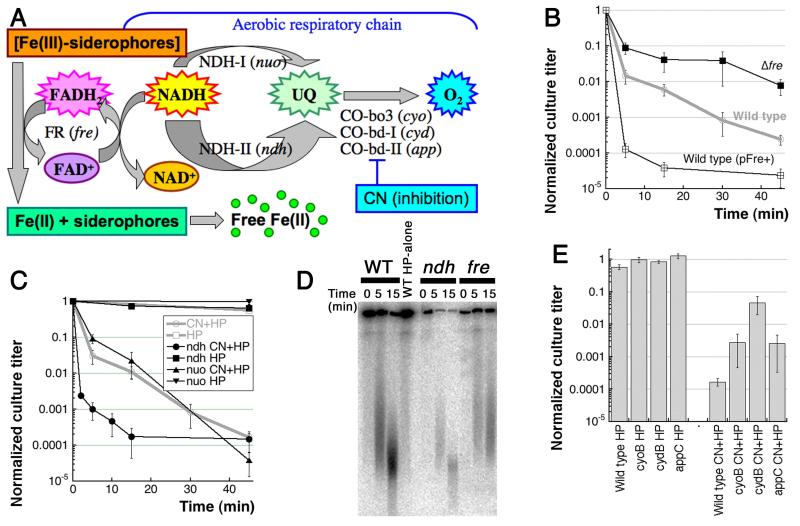
Fig. 3. The effect of mutations in NADH-users.
A. The expanded view of the “electron transport” part of the Fig. 1A scheme. B. Kinetics of CN+HP killing: the fre mutant shows partial resistance, while Fre overproduction elevates CN+HP sensitivity of WT cells. The pFre+ plasmid is pES1. C. Kinetics of killing by the two treatments of ndh and nuo mutants. The nuo mutant behaves like WT, whereas ndh is hyper-sensitive to CN+HP, but shows no sensitivity to HP-alone. D. Comparison of WT chromosome fragmentation kinetic pattern to the ones in the ndh and fre mutants. E. The two treatment survival of the cytochrome oxidase mutants. For clarity, sensitivity is shown at a single time point of 60 minutes (HP-alone) or 45 minutes (CN+HP).
Woodmansee and Imlay also reported that the fre defect in the flavin mononucleotide (FMN) reductase has an opposite effect, by mostly preventing CN-potentiation of the HP killing (“Fig. 2E” pattern) (Woodmansee & Imlay, 2002). The FMN reductase Fre reduces free flavins using excess of NAD(P)H (Fieschi et al., 1995). Interestingly, it is the major ferrisiderophore reductase activity in E. coli, reducing ferric citrate or other iron-siderophores so that they release ferrous iron (Coves & Fontecave, 1993). Woodmansee and Imlay proposed that Ndh action prevents buildup of excess of NAD(P)H that would otherwise be used by Fre to reduce ferric iron (Fe3+ ⇒ Fe2+), thereby promoting Fenton’s reaction (Fig. 3A) (Woodmansee & Imlay, 2002).
We have confirmed that the fre mutant has a reduced sensitivity to CN+HP treatment, whereas Fre overproduction dramatically increases this sensitivity (Fig. 3B). Thus, Fre is indeed partially responsible for fueling Fenton’s reagent in E. coli cells, probably via reduction of insoluble Fe(III) to soluble Fe(II). We have also confirmed the WT behavior of the nuo mutant (Fig. 3C). However, using ndh alleles both from Imlay’s collection and from the E. coli Genetic Stock Center, we failed to observe the expected sensitivity of ndh mutants to HP-alone (Fig. 3C). Even more surprisingly, while being resistant to HP-alone, the ndh mutants in our hands turned out to be hyper-sensitive to CN+HP (Fig. 3C). This opposite effect of ndh inactivation, compared to the effect of fre inactivation, leads to faster chromosomal fragmentation in the ndh mutants and slower fragmentation in the fre mutants (Fig. 3D). Remarkably, the fact that Ndh, like HP-targeting enzymes above, counteracts CN-potentiation without affecting HP-toxicity (“Fig. 2D” pattern) means that Ndh counteracts CN-potentiation directly, rather than through the elevated NADH concentration boost to the overall Fenton’s reaction — otherwise the ndh mutants would have increased sensitivity to both HP-alone and CN+HP treatments (Fig. 2B). The general idea of tight intracellular iron control and its possible disruption by CN is consistent with this overall interpretation.
The possibility of NADH-independent nature of the ndh mutant effect was further supported by the phenotypes of the cyo, cyd and app mutants, deficient in the three cytochrome oxidases that catalyze the final step in electron transport from NADH to O2 in aerobic respiration (Borisov et al., 2011). According to the scheme (Fig. 3A), inactivation of these enzymes should lead to accumulation of NADH, elevating sensitivity to both HP-alone and CN+HP. Looking at it from a different perspective: since all cytochrome oxidases have heme as a critical cofactor that binds CN tightly, they should be targets of CN potentiation, meaning that all three mutants should be equally sensitive to HP-alone or CN+HP (“Fig. 2C” pattern). However, contrary to this strong prediction, all three cytochrome oxidase mutants showed the phenotype of the fre mutant: no HP-alone sensitivity and at the same time a shallower CN+HP sensitivity (Fig. 3E), as if CN utilizes the cytochrome oxidase molecules themselves to promote HP toxicity.
The disagreement among these results within the confines of the NADH-centered scheme (Fig. 3A) suggests that the overall aerobic respiratory chain has no direct role in CN-potentiation of HP toxicity, even though individual activities of (or around) this chain do contribute to the phenomenon. Specifically, if cyanide potentiates formation of Fenton’s reagent in vivo at low HP concentrations, NADH dehydrogenase II (Ndh) counteracts this potentiation, while flavin reductase and cytochrome oxidases promote this potentiation.
The role of DNA and iron in CN+HP toxicity
Our simplistic in vitro modeling of CN-potentiation of HP toxicity, by measuring the effect of cyanide on the standard in vitro Fenton’s reaction (Imlay et al., 1988), expectedly revealed a complete block, the stoichiometry of inhibition suggesting formation of Fe(CN)6 as the inert species (Fig. 4A). The iron-cyanide complexes Fe(CN)6 are known to be significantly less reactive than free iron, and this inhibition of Fenton’s reaction in vitro emphasizes the paradoxical phenomenon of the CN+HP toxicity in vivo (Fig. 1), indicating complicated nature of the underlying mechanisms. At face value, the complete shutdown of Fenton’s reaction by CN in vitro predicts that complexing of iron by CN in vivo should similarly prevent production of hydroxyl radicals and should save the cells from HP toxicity. To make sure that CN+HP killing is due to Fenton’s reaction in vivo, we have confirmed the previous report (Woodmansee & Imlay, 2002) that the presence of in vivo iron chelators, either deferoxamine or dipyridyl, completely blocks CN+HP killing (Fig. 4B). The two iron chelators also block chromosomal fragmentation (Fig. 4C). The dramatic effect of iron chelators implies that, in contrast to the formation on inert iron-cyanide complexes in vitro, the intracellular iron is somehow recruited by CN to promote Fenton’s reaction.
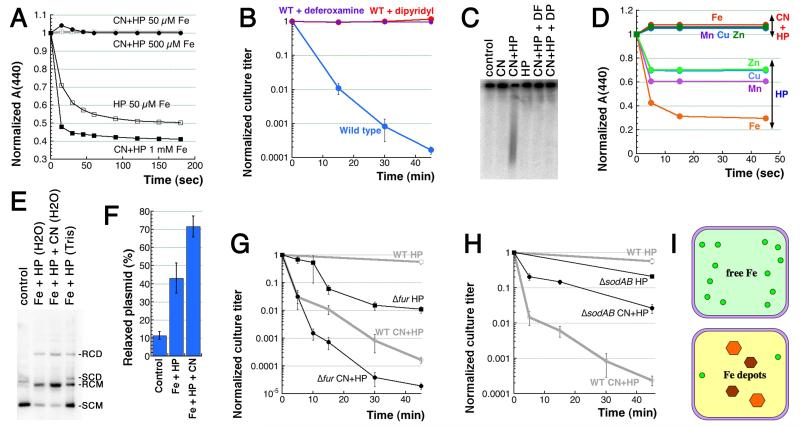
Fig. 4. The complex role of iron in CN+HP toxicity.
A. CN inhibits the standard Fenton reaction in vitro via formation of Fe(CN)6. The reaction, which is started by addition of FeSO4 to the indicated concentration to solution containing 2 mM HP, is followed spectrophotometrically (absorbance at 440 nm), by disappearance of a colored substance, p-nitrosodimethylaniline (p-NDA). When added, CN is at 3 mM. B. Pre-treatment of the cultures with iron chelators, such as 20 mM deferoxamine or 2 mM dipyridyl, for five minutes before the treatment blocks CN+HP killing. C. Pre-treatment with the iron chelators like in “B” similarly blocks CN+HP-induced chromosome fragmentation. D. CN inhibits Fenton-like in vitro reactions with other transition metals. The reactions contained 50 μM EDTA and 250 μM of the indicated metal and were ran like in “A”. The values are means of two independent repetitions. E. CN stimulates in vitro Fenton’s reaction with free iron in water or a Tris buffer and plasmid relaxation as a readout. SCD, supercoiled dimer; SCM, supercoiled monomer; RCD, relaxed circular dimer; RCM, relaxed circular monomer. Concentrations were: 250 μM FeSO4, 2 mM HP, 3 mM CN, 40 mM Tris-HCl pH 8.0. F. Quantification of several gels like in “E”. The values are (RCM/RCM+SCM)×100. G. Kinetics of killing by the two treatments of the fur mutant. H. Kinetics of killing by the two treatments of the sodAB double mutant. I. Our original idea about free cytoplasmic iron (top) and its current evolution (bottom). Small green circles, DNA-accessible Fe(II); orange and brown hexagons, Fe(III)-depots.
This surprising discrepancy could be explained if, instead of iron, another common transition metal in the cell is responsible for CN-potentiation of Fenton’s reaction. The result with iron chelators would still be consistent with this idea, as both deferoxamine and dipyridyl are known to chelate other transition metals (Cini & Orioli, 1981, Keberle, 1964). To test the involvement of other transition metals in CN-potentiation of in vivo Fenton’s reagent, we assembled standard Fenton’s reactions in vitro with 2 mM HP and 250 μM of the following transition metals: chromium, manganese, (iron as a positive control), cobalt, nickel, copper and zinc. We found that some of the metals did promote weaker Fenton-like reactions, but these reactions were all inhibited by addition of 3 mM CN, just like the iron-promoted Fenton’s reaction (Fig. 4D), making contribution of other transition metals to CN-potentiation of HP toxicity unlikely.
Since iron was the best transition metal to promote Fenton’s reaction in vitro and apparently in vivo, we tested the possibility that CN-stimulation of Fenton’s reaction in vivo was specific to our readout, which was (chromosomal) DNA. Plasmid relaxation is a convenient and sensitive DNA-based readout for Fenton’s reaction in vitro (Kang, 2010, Park & Imlay, 2003, Surguladze et al., 2004). Remarkably, in contrast to CN-inhibition of the classic in vitro Fenton’s reaction (Fig. 4A), we have observed a robust CN-stimulation of Fe+HP-promoted plasmid relaxation in vitro (Fig. 4EF). Since DNA readily binds both Fe(II) and Fe(III) iron (Ouameur et al., 2005), DNA-iron complexes can act as a self-targeting Fenton’s reagent (Luo et al., 1994) (“self-targeting” in the sense that DNA, by binding iron readily, serves not only as a platform for Fenton’s reagent, but also as a proximal target for the generated hydroxyl radicals). Remarkably, when cyanide joins this DNA-iron complex, it further potentiates Fenton’s reaction, instead of inhibiting it.
Mutants with increased “free” cytoplasmic iron
With this better understanding of iron’s role in the DNA-self-targeted Fenton’s reaction in vivo, we tested the effect of increased free intracellular iron in certain mutants on the CN-potentiation of HP toxicity. The Fur protein is a ferric ion uptake regulator that functions as either a repressor or activator (or sometimes both) for a number of genes (Hantke, 2001). In aerobically-growing fur mutant cells, the amount of free cytoplasmic iron is elevated ~7-fold (Keyer & Imlay, 1996). We found that the fur mutant shows delayed, yet significant sensitivity to HP and a matching additional sensitivity to CN+HP (Fig. 4G). In other words, potentiation of HP-alone toxicity by fur inactivation is independent of CN potentiation of the same toxicity. Thus, the Fur regulator is not a target of CN potentiation, but counteracts HP toxicity directly (“Fig. 2B” pattern). Apparently, the extra free cytoplasmic iron in fur mutants neither contributes to CN potentiation of HP killing, nor is “neutralized” by CN, like it is in vitro (Fig. 4A). In other words, there appear to be at least two subpopulations of iron in the cell: the one that can be recruited by CN for potentiation of HP toxicity, and the other that is neither “recruitable”, nor “neutralizable” by CN, yet fuels its own Fenton’s reagent.
Besides fur, a similar ~8-fold increase in pool of free intracellular iron in E. coli is detected in the double sodAB mutant lacking cytoplasmic superoxide dismutases (Keyer & Imlay, 1996, McCormick et al., 1998). Surprisingly, in contrast to the fur mutant behavior (Fig. 4G), the sodAB mutant showed only a slight sensitivity to HP-alone treatment and at the same time a greatly-reduced sensitivity to CN+HP (Fig. 4H), as if the “increased free iron” in this mutant is not only poorly-recruited by CN to fuel Fenton’s reagent, but also is not accessible to HP-alone for the same purpose. At a minimum, these unexpected effects in mutants with increased free cytoplasmic iron argue against the existence of a significant pool of DNA-accessible cytoplasmic iron outside the control of the iron-trafficking system (Fig. 4I, top). Instead, the bulk of cytoplasmic iron must be distributed among specific depots (Fig. 4I, bottom), with CN apparently recruiting it from a subset of these depots.
The role of iron-depository Dps
Therefore, we tested the idea that CN recruits iron for Fenton’s reagent from two major intracellular depots of the bound iron. One of such iron depository is Dps, a major nucleoid-organizing protein of stationary cells. Its dodecamers form ball-like structures that bind DNA non-specifically and organize it in a paracrystalline form for preservation during non-growth state (Frenkiel-Krispin et al., 2004), as well as offering long-term storage of iron by converting it into oxidized form within the balls (Chiancone & Ceci, 2010) (Fig. 5A). Although accumulated in massive numbers only in stationary cells, Dps is also present in 5-10-times fewer copies in exponentially-growing cells (Ali Azam et al., 1999), while its synthesis is induced in growing cells by oxidative damage or starvation (Altuvia et al., 1994). We found that exponential cultures of dps mutants are not different from WT in their lack of sensitivity to HP-alone (suggesting no general increase in free iron), but they are killed deeper by CN+HP (Fig. 5B), showing that Dps counters CN-potentiation in exponential cells (“Fig. 2D” pattern), perhaps by sequestering the CN-recruited iron.
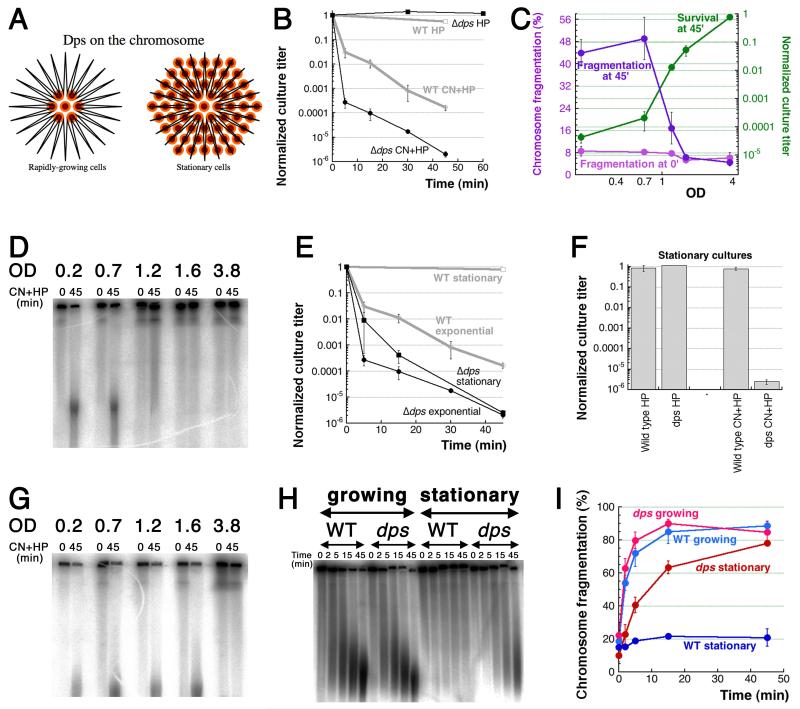
Fig. 5. Resistance of stationary cells to CN+HP is due to Dps.
A. Dodecameric Dps protein forms spheres that bind DNA non-specifically and accumulate up to 500 iron atoms per sphere. Orange/brown small circles, Dps dodecamers; black line in the shape of a star, chromosomal (duplex) DNA. B. Kinetics of killing by the two treatments of Δdps mutant. WT curves from Fig. 1C are shown in grey for comparison. C. OD-dependence of CN+HP killing and fragmentation of wild type cells. Aliquots of the same culture were challenged with standard CN+HP treatment at various ODs of the culture, and their normalized survival or fragmentation were plotted as a function of OD. In our growth conditions, the maximal density (fully stationary cultures) of WT cells does not exceed 4.0. D. A representative pulsed-field gel of OD-dependence of fragmentation in WT cells after CN+HP treatment. E. Kinetics of CN+HP killing of exponential vs stationary cultures of wild type cells and Δdps mutant. F. Stationary cultures killing by HP-alone or CN+HP 45 minute treatment. G. OD-dependence of fragmentation in Δdps mutants: a representative gel, similar to the one in (D). Cultures were treated without prior dilution. H. A representative pulsed-field gel of CN+HP treatment kinetics to show that Δdps stationary cultures undergo robust chromosomal fragmentation if diluted 10× into a fresh medium right before the treatment. I. Kinetics of chromosome fragmentation in wild type and Δdps cells, either in growing or stationary/diluted cultures. This is quantification of several independent runs like in “H”.
Since the major role of Dps is during the stationary phase, we next tested the sensitivity to the two treatments of wild type cultures sampled at different densities from rapid growth all the way to complete saturation. We found that, as cells enter stationary phase, their sensitivity to CN+HP treatment completely disappears (Fig. 5C, the green curve), in parallel with disappearance of chromosomal fragmentation (Fig. 5C, the purple curve and Fig. 5D), suggesting that CN+HP-induced chromosomal fragmentation requires active metabolism. Similarly, starving the exponentially growing cultures for two hours in M9 salts protects them against the killing (Fig. S2). In contrast, stationary dps cultures, while still resistant to HP, remain fully sensitive to CN+HP (Fig. 5EF), while CN+HP still induces catastrophic chromosomal fragmentation in dps mutants in early stationary phase (Fig. 5G), further stressing the link between chromosomal fragmentation and the loss of viability. At first, we detected no CN+HP-induced fragmentation in the fully stationary dps mutant cells (Fig. 5G and Fig. S3 and S4), even though they were killed by CN+HP just like exponential dps mutant cells (Fig. 5E), suggesting a major discrepancy between the two readouts. However, diluting fully-stationary cultures 10-fold into a fresh medium just before the treatment makes Δdps mutants again susceptible to CN+HP-induced fragmentation (wild type stationary cells continue to be resistant) (Fig. 5HI), suggesting an activation step. Since the energy production is blocked by CN in these cells, we are not sure about the nature of this activation, but the chromosomal fragmentation is blocked by iron chelator deferoxamine (Fig. S4), suggesting participation of free iron. Diluting stationary cultures 10-fold into a “conditioned” medium failed to revive fragmentation in the Δdps mutant (Fig. S5), ruling out cell density as the fragmentation-inhibiting factor. We conclude that the resistance of stationary WT cells to CN+HP is due to Dps organizing the chromosome and/or sequestering CN-mobilizable iron, but even in exponentially-growing cells, Dps already offers considerable protection against CN+HP treatment.
The role of iron-depot ferritin
In fact, Dps is a member of the ferritin family of proteins, present in all organisms from bacteria to mammals. All ferritins have a highly-conserved structure of thick-walled spheres accumulating insoluble ferric iron. Besides Dps, there are two more ferritins in E. coli: the ferritin proper (homologous to all ferritins from other kingdoms) and bacterioferritin (a heme-containing ferritin found only in bacteria) (Smith, 2004). In contrast to the smaller 12-mer Dps spheres, which hold up to 500 iron atoms, the larger 24-mer ferritin spheres can hold up to 4,500 iron atoms (Smith, 2004). Another important difference from Dps, which functions as a long-term iron depository, is that ferritins act as dynamic iron distribution centers, both accumulating and releasing iron (Fig. 6A), prompted by interactions with iron-containing proteins or by some simple chemical cues (Reif, 1992). Although CN was never reported to release iron from ferritin, we decided to use ferritin mutants to test if CN could be one of those “iron-release” cues.
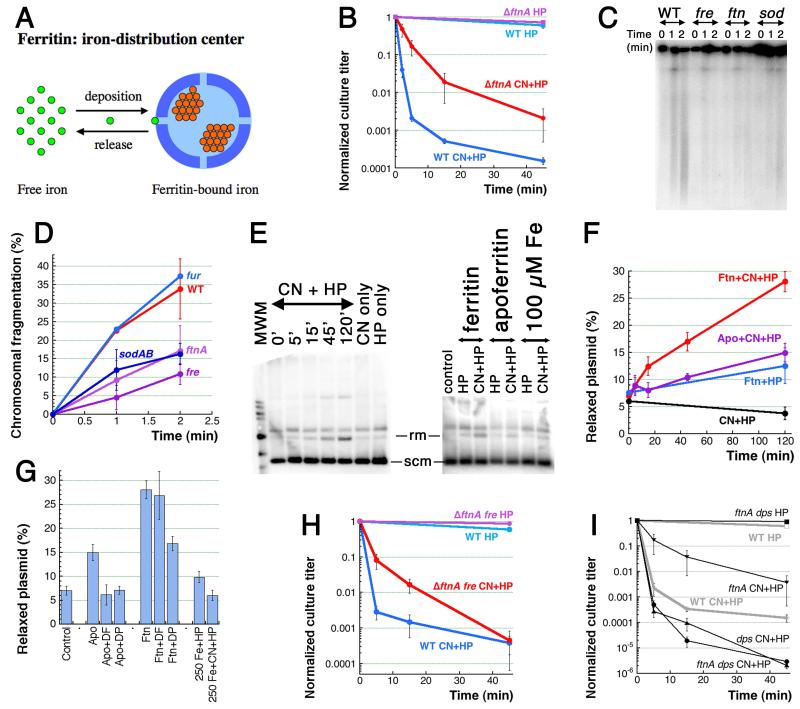
Fig. 6. Ferritin is one source of CN-recruited iron.
A. Ferritin spheres function as iron-distribution centers. Small bright-green circles, Fe(II) iron; small orange circle, Fe(III) iron. B. Kinetics of killing by the two treatments of the ftnA mutant. C. Early chromosomal fragmentation is slower in the ftnA, fre and sodAB mutants — a representative pulsed-field gel of 1′ and 2′ CN+HP treatments. D. Quantification of the early fragmentation from three gels like in “C”. In this case, fragmentation values at 0 time point are subtracted as a background. E. in vitro Fenton reaction with ferritin as a source of iron and plasmid relaxation as a readout. The left panel, kinetics of ferritin + CN + HP, with CN-only and HP-only controls (both at 120 minutes). All reaction have 5 μg of ferritin. The right panel, the apoferritin (also 5 μg, 120 minutes) and the pure iron (the indicated amount, 2 minutes) controls. Rm, relaxed monomer; scm, supercoiled monomer. F. Qauntitative plasmid relaxation kinetics from several gels like in “E”. G. The influence of iron chelators on the plasmid relaxation by ferritin and apoferritin (120 minutes). DF, deferoxamine; DP, dipyridyl. Plasmid relaxation by pure iron in the reaction conditions (treatment length is 2 minutes) is also shown. H. Kinetics of killing by the two treatments of the ftnA fre double mutant. I. Kinetics of killing by the two treatments of the ftnA dps double mutant.
The bfr mutant lacking bacterioferritin shows essentially the wild type behavior in HP or CN+HP treatments (Fig. S5A), but the ftnA mutant lacking regular ferritin shows the “Fig. 2E” pattern of reduced sensitivity to CN+HP (Fig. 6B), suggesting that ferritin does participate in CN-potentiation of HP toxicity, perhaps by releasing its iron upon CN cue. The ftnA bfr double mutant shows an intermediate phenotype of some CN+HP resistance (Fig. S5B). Although no difference is seen in the level of CN+HP-induced fragmentation at 5 or 15 minutes of treatment (see above), the level of chromosomal fragmentation at 1 or 2 minutes of the treatment in all the mutants showing resistance to CN+HP (ftnA, fre, sodAB) is only half of the level in wild type cells (Fig. 6CD), corroborating our earlier conclusion that the early fragmentation is the cause of death.
In order to use the plasmid relaxation assay to monitor ferritin-driven Fenton’s reaction in vitro, we introduced EDTA in the reaction buffer to block the background reaction with free iron (Fig. 6EFG). We found that iron-loaded (horse spleen) ferritin does not promote Fenton’s reaction with HP-alone, but does promote it with CN+HP (Fig. 6EF). Apoferritin (the same protein shell, but supposedly lacking iron) is much less active in this nicking reaction (Fig. 6EF). Moreover, addition of iron chelators completely eliminate this residual activity of apoferritin (Fig. 6G), indicating that this activity is due to traces of iron still present in apoferritin preparations. Not only Fenton’s reagent is not formed by free iron under these reaction conditions, but also it is not stimulated by CN (Fig. 6EG). Thus, the in vitro evidence with horse spleen ferritin is consistent with the genetic evidence that CN recruits iron from ferritin to fuel Fenton’s reagent.
The partial resistance of the ftnA mutants to CN+HP is not further increased by additional fre defect (Fig. 6H), suggesting that ferritin and flavin reductase work together in CN-mobilization of reduced iron. On the other hand, the double ftnA dps mutant is as sensitive to CN+HP as the single dps mutant (Fig. 6I), suggesting that, in the absence of the iron depository Dps, CN-recruited iron from sources other than ferritin could still kill. We conclude that CN potentiation of HP toxicity is in part due to iron recruitment from ferritins by CN, perhaps even with direct delivery of the recruited iron to DNA. The CN-recruited iron then either fuels the DNA-self-targeting Fenton’s reagent or is removed from DNA by Dps, the long-term iron depository of E. coli.
Discussion
Our investigation into the spectacular co-toxicity of cyanide and hydrogen peroxide, which by themselves are bacteriostatic at the corresponding concentrations, yielded the following original findings: 1) rapid exponential cell death is accompanied by catastrophic chromosomal fragmentation; 2) CN potentiates HP killing, but does not kill by itself in any concentration, while HP kills in 10-fold higher concentrations, apparently via self-potentiation; 3) in vitro CN blocks non-DNA Fenton’s reaction, but promotes DNA-self-targeting Fenton’s reaction, while iron chelators block CN+HP-induced chromosomal fragmentation in vivo, suggesting importance of iron-DNA complexes in CN+HP toxicity; 4) the weak/no sensitivity to HP-alone of mutants that cannot remove HP (katEG, ahpC) or that elevate “free” cytoplasmic iron (fur, sodAB) suggests a strict system of iron distribution inside the cell, while frequent hyper-sensitivity of these mutants to CN+HP suggests that CN targets the iron-distribution system; 5) the iron depository Dps partially protects against CN+HP killing during growth and offers absolute protection during stationary phase, indicating that Dps neutralizes the CN-recruited iron; 6) at the same time, the iron depot ferritin may be one source of CN-recruited iron that works together with flavin reductase to fuel Fenton’s reagent in vivo.
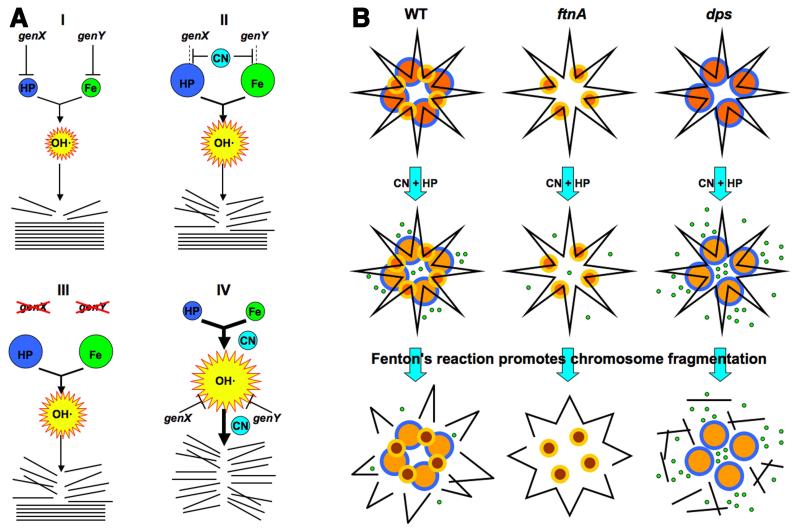
Our overall results are surprising not only in what we have found, but also in what we failed to find. One of the original objectives of this study was to find targets of CN-inhibition that would explain CN-potentiation of HP toxicity. Such targets would be identified by mutants that would show a characteristic pattern of similar toxicities of HP-alone treatment versus CN+HP treatment (Fig. 2C). Remarkably, we failed to find a single example of Fig. 2C pattern, instead encountering several examples of Fig. 2D pattern (Figs. 2F (5 minute time point), 2G, 3C, 5B). This means that CN-potentiation does not increase the intracellular concentrations of the two components of Fenton’s reagent, HP and iron (Fig. 7A, II), but stimulates Fenton’s reaction directly (Fig. 7A, IV). This stimulation could be either at the level of hydroxyl radical formation, or at the level of bringing these radicals to DNA, or both. In fact, CN potentiation could accomplish both by promoting formation of iron-DNA complexes.
Fig. 7. The interpretation scheme and the chromosome fragmentation scheme.
A. Interpretation of genetic results. The intracellular HP, iron and OH· pools are shown by spheres whose size reflect the relative size of the pools. Stack of straight lines at the bottom, chromosomal DNA; broken lines, broken DNA. (I) Our initial HP-only treatment scenario, in which both HP and iron pools are actively limited by dedicated cellular functions, genX and genY. (II) Our initial expectation of a typical CN-potentiation route (inactivation of the HP- or iron-limiting functions). (III) The expected phenotypes of the corresponding mutants (in functions that are CN-targets) in HP-only treatment. (IV) An alternative scenario suggested by the most frequently observed phenotypes of mutants (no HP-only sensitivity, increased CN+HP sensitivity), according to which CN stimulates Fenton’s reaction near DNA. B. The proposed role of ferritin-like proteins in CN potentiation of HP poisoning via catastrophic chromosome fragmentation, based on phenotypes of the corresponding mutants. Top row: yellow/orange smaller circles, Dps dodecamers; blue/orange bigger circles, ferritin 24-mers; black line in the shape of a star, chromosomal (duplex) DNA. Middle row: small green circles, released ferrous iron (note a lighter color of the ferritin circles). In the absence of ferritin, some ferrous iron is still coming from other (unknown) sources. Bottom row: broken black lines, chromosome fragments. The released iron is taken into Dps spheres (note their darker color). For clarity, DNA fragments are shown dissociated from ferritin and Dps.
With this clarification in mind and on the basis of our new findings, we propose that CN potentiates HP toxicity by fueling Fentons’ reagent in at least two ways: 1) by recruiting iron from depots like ferritin directly to DNA; 2) by further activating iron-DNA complexes for DNA-self-targeting Fenton’s reaction. At least two activities, NADH dehydrogenase II and the iron depository Dps, counteract this CN potentiation, the former probably indirectly, via decreasing the pool of reduced flavins, the latter apparently by sequestering CN-recruited iron. The proposed hypothetical interplay of ferritin and Dps in iron sequestration and CN-recruitment vis-à-vis chromosome fragmentation is schematically presented in Fig. 7B. We will discuss our specific findings below.
Iron distribution inside the cell and its subversion by CN
One general conclusion from our results is that distribution of iron is tightly controlled inside the cell. The pool of free cytoplasmic iron in the cell that mixes with freely-diffusing HP and forms hydroxyl radicals throughout the cytoplasm must be limited by iron sequestration in depots (Fig. 4I, bottom). At the least the pool of free cytoplasmic iron is limited in the vicinity of DNA. This conclusion mostly follows from our frequent finding that a particular mutant, expected to be sensitive to HP-alone treatment, either due to higher intracellular HP concentrations or increased free cytoplasmic iron, failed to show increased sensitivity in at least the first 10 minutes of HP treatment (Fig. 2FG, 3C, 4GH and 5B). Since most of these mutants show increased sensitivity to CN+HP, the interpretation is that CN disrupts the overall distribution of iron or the restrictions on iron movement around DNA, allowing Fenton’s reaction to happen in the vicinity of DNA.
One specific mechanism we propose is direct iron recruitment to DNA from ferritin depots that are themselves in the physical proximity to DNA. Direct iron-DNA interactions are important for DNA damage during in vivo Fenton’s reaction (Luo et al., 1994), but is there any evidence that ferritins are close to DNA inside the cell? It is not a widely-appreciated fact that ferritin readily binds DNA, both in vivo (Thompson et al., 2002) and in vitro (Surguladze et al., 2004). Moreover, in vitro in the presence of ferritin, a supercoiled plasmid DNA is gradually nicked, this nicking being dependent on the internal iron, because it is blocked by iron chelators (Surguladze et al., 2004). Interestingly, reduced flavin mononucleotides facilitate iron release from ferritin in vitro (Funk et al., 1985, Melman et al., 2013), consistent with our (indirect) conclusion about the role of FMN reductase in the ferritin-mediated fueling of Fenton’s reagent in vivo.
Catastrophic chromosomal fragmentation
Chromosomal fragmentation does not have to be dramatic to kill. In fact, a single unrepaired double-strand break kills organisms of various levels of complexity, from phages and bacteria to lower and higher eukaryotes (Bonura & Smith, 1975, Dahm-Daphi et al., 2000, Freifelder, 1965, Iliakis, 1991, Kouzminova & Kuzminov, 2012, Resnick, 1976). Interestingly, one break per chromosome in growing bacterial cultures, where chromosome replicates continuously, will not be even detectable by our physical analysis. Indeed, the chromosome fragmentation detectable in growing bacterial cells (cultures) by pulsed-field gels as a smear below the compression zone reflects far more than a single break, because the detected sub-chromosomal fragments must have no branches to be able to enter the gel. In other words, in order for random double-strand breaks to generate detectable sub-chromosomal fragments in a theta-replicating chromosome, more than one break needs to happen in the same chromosomal arc. Therefore, quantifying chromosome fragmentation from the smear within pulsed-field gels seriously underappreciates the true density of double-strand breaks in the chromosome. In support of this argument, we reported before that even a 5% level of spontaneous fragmentation corresponds to severe growth inhibition in strains that experience it (Kouzminova et al., 2004), with 10% spontaneous fragmentation usually meaning complete block of growth (Budke & Kuzminov, 2010, Kouzminova et al., 2004, Rotman et al., 2014). In fact, the maximal levels of spontaneous fragmentation of ~15% in the ligA recBC mutants (Kouzminova & Kuzminov, 2012) and ~20% in the dut recBC mutants (Kouzminova & Kuzminov, 2006, Kouzminova et al., 2004) are associated with deep lethality.
Even higher levels of fragmentation can be achieved in E. coli cells after DNA damaging treatments — up to 25% fragmentation during thymineless death (Kuong & Kuzminov, 2010), up to 30% fragmentation after UV irradiation (Khan & Kuzminov, 2012), up to 40% fragmentation after phleomycin treatment (Khan & Kuzminov, 2013). In all cases, cells were excision-repair-proficient, and there were essentially no survivors. Finally, the maximum level of induced fragmentation approaching 50% was observed in repair-defective polA mutants after UV (Khan & Kuzminov, 2013). But never before have we encountered such a swift and utter chromosome demise as in the case of CN+HP treatment, which we therefore termed “catastrophic chromosomal fragmentation”. The actual fragmentation over the background peaks around 70% in this case, but only because some chromosomal DNA is chopped to pieces small enough to migrate out of the gel, while ~10% of the total orthophosphate label in the well is a non-DNA species (probably fragments of the cell wall) (Kuong & Kuzminov, 2009). Such a local catastrophic chromosome fragmentation within an eukaryotic nucleus could be the initial event behind chromothripsis, which is a complete randomization of a particular chromosome or one of its arms, observed with some frequency in certain cancers (Jones & Jallepalli, 2012).
There are currently more questions than answers about the possible mechanisms behind this catastrophic fragmentation. Is it dependent on DNA replication, transcription or translation? Does it happen randomly over the chromosome or is concentrated around the origin, for example? Is it associated with Bayer’s patches (chromosome’s attachments to the envelope) or with the proteinaceous core of the nucleoid (its central part)? Does the nucleoid structure collapse or does it mostly survive during fragmentation? Are double-strand breaks direct or are they preceded by primary (one-strand) DNA lesions? Are these primary DNA lesions repaired by excision repair? Are the double-strand breaks repaired by recombinational repair? Finally, why do fragmentation levels in various mutants poorly correlate with differential survival of these mutants? We would be addressing these questions in our future studies.
Potentiated toxicity
Potentiated toxicity is a useful practical concept, because it allows an organism to reduce dangers associated with generation and handling of immediately toxic concentrations of a poisonous substance, like HP. The transition from bacteriostatic (10 mM) to strongly bactericidal (20 mM) concentrations of HP in our experiments is quite abrupt (Fig. 1FG), immediately justifying development of ways to potentiate this toxicity as a strategy to keep the working concentrations of the poison, HP, in the static range for the organism that produces it. As mentioned in the introduction, one well-studied example of how HP toxicity is potentiated in real life is offered by immune cells, that use nitric oxide to potentiate killing of pathogenic bacteria with HP (Brunelli et al., 1995, Pacelli et al., 1995). The proposed mechanism of this potentiated toxicity is similar to the one proposed before for CN+HP toxicity: block of respiration increases NADH pools, accelerating Fe(III) → Fe(II) reduction by Fre (Woodmansee & Imlay, 2003). In the light of this work, it would be interesting to test if NO potentiates HP-induced catastrophic chromosomal fragmentation by the same kind of iron recruitment from intracellular depots, especially since NO is known to cause iron release from ferritins (Reif, 1992).
Another potentiator of HP toxicity is the amino acid cysteine, the only redox-active amino acid. Cysteine is always present in the cell, but its normal levels are not enough to cause problems in combination with low millimolar HP treatment. However, when cells are forced to take in too much cysteine from the environment, they become transiently sensitive to low mM HP concentrations (Park & Imlay, 2003). Cysteine directly stimulates Fenton’s reaction in vitro (Park & Imlay, 2003), but the in vivo effectors of cysteine potentiation may also include hydrogen sulfide (Berglin & Carlsson, 1985). Interestingly, sulfide is known to release iron from mammalian ferritins (Cassanelli & Moulis, 2001). Besides cysteine, another amino acid that potentiates HP toxicity is histidine (Berglin & Carlsson, 1985). Finally, the fact that 10 mM HP is bacteriostatic, while 20 mM is already strongly bactericidal, suggests that HP self-potentiates at higher concentrations. However, our preliminary experiments show that 20 mM HP still kills wild-type cells in the presence of iron-chelator deferoxamine, suggesting a distinct, iron-unrelated mechanism in this case.
Conclusion
Concentrations of HP in excess of 20 mM efficiently kill by causing catastrophic chromosomal fragmentation, yet it is metabolically hard and physiologically dangerous to produce high concentrations of HP as a bio-weapon. Potentiation with an unrelated substance, like CN described in this work, allows organisms to kill their enemies using physiologically-achievable concentrations of HP, which may be indeed quite modest inside the cells (Imlay, 2009, Slauch, 2011, Winterbourn et al., 2006). Cells of our immune system use NO as such a potentiator, but bacteria scavenge NO with several dedicated enzymes (Stern & Zhu, 2014), decreasing efficiency of this potentiation route. At the same time, CN is readily generated from serum thiocyanate by myeloperoxidase of our immune cells (But et al., 2002, Furtmüller et al., 1998, Stelmaszynska, 1985, Thomas & Fishman, 1986), and bacteria typically have no capacity to degrade CN. Therefore, as we have argued before, our immune cells may also use CN as another or co-potentiator of HP toxicity (Kuong & Kuzminov, 2009). This makes CN+HP, NO+HP and similar mixtures the binary bio-weapons. We propose that the iron depot ferritin plays a dual role in CN-potentiation: first, ferritin releases small amount of iron on CN cue; second, since ferritin is a DNA-associated protein, the released iron is instantly complexed by DNA, which allows CN to potentiate DNA-self-targeting Fenton’s reaction. The overall validity and the details of this complicated potentiation scheme will have to be addressed in future studies.
Experimental procedures
Strains and Plasmids
Escherichia coli strains used are all K-12 BW25117 derivatives (Baba et al., 2006). Alleles were moved between strains by P1 transduction (Miller, 1972). The mutants were all deletions from the Keio collection (Baba et al., 2006), purchased from the E. coli Genetic Stock Center, and were verified by PCR (and, whenever possible, phenotypically). For double mutants construction, the resident kanamycin-resistance cassette was first removed by transforming the strain with pCP20 plasmid (Datsenko & Wanner, 2000). The plasmid pMTL20 (Chambers et al., 1988) was used for all in vitro plasmid relaxation assays.
Enzymes and Reagents
Ferritin and apoferritin from equine spleen, catalase from bovine liver, hydrogen peroxide, deferoxamine mesylate, 2,2′-Bipyridyl and N,N-Dimethyl-4-nitrosoaniline (p-nitrosodimethylaniline) were purchased from Sigma. Potassium Cyanide was purchased from Fisher-Scientific. Yeast chromosome pulsed-field gel electrophoresis markers were from New England Biolabs.
Growth conditions and Viability assay
To generate killing kinetics, fresh overnight cultures were diluted 500-fold into LB medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 250 μl of 4 M NaOH per liter (Miller, 1972)) and were shaken at 37°C for about two and a half hours or until they reached exponential phase (OD600 approximately 0.3). At this point, the cultures were made 3 mM for CN and/or 2 mM for HP (or the indicated treatment) and the shaking at 37°C was continued. Viability of cultures was measured at the indicated time points by spotting 10 μL of serial dilutions in 1% NaCl on LB plates (LB medium supplemented with 15 g of agar per liter). The plates were developed overnight at 28 °C, the next morning colonies in each spot were counted under the stereomicroscope. All titers have been normalized to the titer at time 0 (before addition of the treatment).
Measuring chromosomal fragmentation by pulsed-field gel electrophoresis
This generally follows our previous protocols (Khan & Kuzminov, 2013, Kouzminova et al., 2004). All strains were grown in LB medium; overnight cultures were diluted 500-fold and grown with 1-10 μCi of 32P-orthophosphoric acid per ml of culture for 2.5 h at 37 °C (OD600 approximately 0.3) before addition of 3 mM CN + 2 mM HP (or the indicated treatment). The reactions were stopped by addition of 315 μg of catalase, and aliquots of the culture were taken at the indicated times to make plugs. Cells of the aliquot were spun down, resuspended in 60 μl of TE buffer and put at 37°C. 2.5 μl of proteinase K (5 mg/ml) was added, immediately followed by 63 μl of molten 1.2% agarose in the lysis buffer (1% sarcosine, 50 mM Tris-HCl, and 25 mM EDTA) held at 70°C. The mixture was pipetted a couple of times before being poured into a plug mold and let solidify for 2 minutes at room temperature. The plugs were then pushed out of the molds and incubated overnight at 60°C in 1 ml of the lysis buffer. Half plugs were loaded into a 1.0% agarose gel in 0.5× Tris-borate-EDTA buffer and run at 6.0 V/cm with the initial and the final switch times of 60 and 120 s, respectively, at 12°C in a Bio-Rad CHEF-DR II PFGE system for 20-22 hours. The gel was vacuum-dried at 80°C and then exposed to a PhosphorImager screen overnight. The resulting signals were measured with a PhosphorImager (Fuji Film FLA-3000).
In vitro non-DNA Fenton’s reaction
The reaction containing 40 μM N,N-Dimethyl-4-nitrosoaniline, 2 mM HP and/or 3 mM CN in 2 ml water (Imlay et al., 1988) was supplemented with various concentrations of up to 1 mM of stable salts of the indicated transition metals. The reaction was followed spectrophotometrically by measuring absorbance at 440 nm.
Plasmid relaxation assay
About 100-200 ng of plasmid (1-2 μl) was incubated with 5 μM of either ferritin or apoferritin with 3 mM CN and 2 mM HP in 1× TAE buffer (40 mM Tris-HCl, 20 mM acetic acid, 1 mM EDTA, pH 8) at 37°C. At specified times, aliquots were removed and the reaction was stopped by adding 45 μg of catalase. Since apoferritin was in 50% glycerol (an inhibitor of Fenton’s reagent), both apoferritn and ferritin preparations were first changed into 1% NaCl solution by using 100K Amicon ultra-0.5 ml centrifugal filter and spin column units.
When using iron instead of enzymes, the plasmid was incubated for only 2 minutes at room temperature before addition of catalase. These samples were then run on a 1.1% Agarose gel at 3 V/cm before being transferred to a nylon membrane and hybridized with pMTL20-specific radioactive probe to calculate the percentage of the relaxed plasmid form in the total plasmid.
Southern hybridization
The agarose gels were washed with 0.25 M HCl, followed by 0.5 M NaOH and, finally, with 1 M Tris HCl pH 8.0. Each wash was 40 minute long. The treated gels were then placed on Amersham Hybond N+ (GE Healthcare) nylon membrane, covered with Saran wrap, and DNA was transferred by vacuum for 1-2 hours. After that the DNA was UV-crosslinked to the membranes and probed with 32P-labelled pMTL20-specific probe. Hybridization was carried out overnight at 63°C in a 0.5M Sodium Phosphate (pH 7.4) and 5% SDS hybridization buffer. In the morning, the membranes were washed thrice with 1% hybridization buffer and rinsed with water just before covering them with Saran wrap and exposing to a PhosphorImager screen. The resulting signals were measured with a PhosphorImager (Fuji Film FLA-3000).
Supplementary Material
Acknowledgements
We are grateful to Jim Imlay (Microbiology, UIUC), Elena Kouzminova (this lab) and Bénédicte Michel (CNRS) for many helpful suggestions on the project and the manuscript. We would like to thank Murat Saparbaev (Institut Gustave Roussi), Kawai Jessica Kuong (University of Washington) and all the members of this laboratory for enthusiastic discussion of our results. This work was supported by grant # GM 073115 from the National Institutes of Health.
Abbreviations
- CN
cyanide
- HP
hydrogen peroxide
Footnotes
The authors have no conflict of interest to declare.
References
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglin EH, Carlsson J. Potentiation by sulfide of hydrogen peroxide-induced killing of Escherichia coli. Infect. Immun. 1985;49:538–543. doi: 10.1128/iai.49.3.538-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickar D, Bonaventura J, Bonaventura C. Cytochrome c oxidase binding of hydrogen peroxide. Biochemistry. 1982;21:2661–2666. doi: 10.1021/bi00540a013. [DOI] [PubMed] [Google Scholar]
- Bonura T, Smith KC. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J. Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Murali R, Verkhovskaya ML, Bloch DA, Han H, Gennis RB, Verkhovsky MI. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. U S A. 2011;108:17320–17324. doi: 10.1073/pnas.1108217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudzynski K, Abubaker K, St-Martin L, Castle A. Re-examining the role of hydrogen peroxide in bacteriostatic and bactericidal activities of honey. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00213. article 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli L, Crow JP, Beckman JS. The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch. Biochem. Biophys. 1995;316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- Budke B, Kuzminov A. Production of clastogenic DNA precursors by the nucleotide metabolism in Escherichia coli. Mol. Microbiol. 2010;75:230–245. doi: 10.1111/j.1365-2958.2009.06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- But PG, Muraviev RA, Fomina VA, Rogovin VV. Antimicrobial activity of mieloperoxidase from neutrophilic peroxisome. Biology Bulletin. 2002;29:212–215. [Google Scholar]
- Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical Review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (⊕OH/⊕O−) in aqueous solution. J. Phys. Chem. Ref. Data. 1988;17:513–886. [Google Scholar]
- Cassanelli S, Moulis J. Sulfide is an efficient iron releasing agent for mammalian ferritins. Biochim. Biophys. Acta. 2001;1547:174–182. doi: 10.1016/s0167-4838(01)00182-0. [DOI] [PubMed] [Google Scholar]
- Chambers SP, Prior SE, Barstow DA, Minton NP. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Cini R, Orioli P. Ternary complexes between adenosine 5′ -triphosphoric acid, 2,2′-bipyridyl and the divalent metal ions manganese (II), cobalt (II), copper (II), and zinc (II). Preparation and physiochemical properties. J. Inorg. Biochem. 1981;14:95–105. doi: 10.1016/s0162-0134(00)80030-6. [DOI] [PubMed] [Google Scholar]
- Coves J, Fontecave M. Reduction and mobilization of iron by a NAD(P)H:flavin oxidoreductase from Escherichia coli. Eur. J. Biochem. 1993;211:635–641. doi: 10.1111/j.1432-1033.1993.tb17591.x. [DOI] [PubMed] [Google Scholar]
- Dahm-Daphi J, Sass C, Alberti W. Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int. J. Radiat. Biol. 2000;76:67–75. doi: 10.1080/095530000139023. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar HR, Baldwin IL, Miller SB. The Influence of Certain Inorganic Salts on the Germicidal Activity of Hydrogen Peroxide. J. Bacteriol. 1930;19:203–211. doi: 10.1128/jb.19.3.203-211.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton HJH. The oxidation of tartaric acid in presence of iron. J. Chem. Soc. Proc. 1894;10:157–158. [Google Scholar]
- Fieschi F, Nivière V, Frier C, Décout JL, Fontecave M. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 1995;270:30392–30400. doi: 10.1074/jbc.270.51.30392. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Mechanism of inactivation of coliphage T7 by x-rays. Proc. Natl. Acad. Sci. USA. 1965;54:128–134. doi: 10.1073/pnas.54.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Ben-Avraham I, Englander J, Shimoni E, Wolf SG, Minsky A. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 2004;51:395–405. doi: 10.1046/j.1365-2958.2003.03855.x. [DOI] [PubMed] [Google Scholar]
- Funk F, Lenders JP, Crichton RR, Schneider W. Reductive mobilisation of ferritin iron. Eur. J. Biochem. 1985;152:167–172. doi: 10.1111/j.1432-1033.1985.tb09177.x. [DOI] [PubMed] [Google Scholar]
- Furtmüller PG, Burner U, Obinger C. Reaction of myeloperoxidase compound I with chloride, bromide, iodide, and thiocyanate. Biochemistry. 1998;37:17923–17930. doi: 10.1021/bi9818772. [DOI] [PubMed] [Google Scholar]
- Goerlich O, Quillardet P, Hofnung M. Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J. Bacteriol. 1989;171:6141–6147. doi: 10.1128/jb.171.11.6141-6147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Iliakis G. The role of DNA double-strand breaks in ionising radiation-induced killing of eukaryotic cells. BioEssays. 1991;13:641–648. doi: 10.1002/bies.950131204. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Chapter 5.4.4. Oxidative Stress. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Slauch JM, Squires CL, editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C.: 2009. doi:10.1128/ecosalplus.1125.1124.1124. [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 1986;166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev. Cell. 2012;23:908–917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtshuk PJ, Mueller TJ, Acord WC. Bacterial terminal oxidases. CRC Crit. Rev. Microbiol. 1975;3:399–468. doi: 10.3109/10408417509108757. [DOI] [PubMed] [Google Scholar]
- Kang JH. Protective effects of carnosine and homocarnosine on ferritin and hydrogen peroxide-mediated DNA damage. BMB Rep. 2010;43:683–687. doi: 10.5483/BMBRep.2010.43.10.683. [DOI] [PubMed] [Google Scholar]
- Keberle H. The biochemistry of desferrioxamine and its relation to iron metabolism. Ann. N.Y. Acad. Sci. 1964;119:758–768. doi: 10.1111/j.1749-6632.1965.tb54077.x. [DOI] [PubMed] [Google Scholar]
- Keyer K, Imlay J. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SR, Kuzminov A. Replication forks stalled at ultraviolet lesions are rescued via RecA and RuvABC protein-catalyzed disintegration in Escherichia coli. J. Biol. Chem. 2012;287:6250–6265. doi: 10.1074/jbc.M111.322990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SR, Kuzminov A. Trapping and breaking of in vivo nicked DNA during pulsed field gel electrophoresis. Anal. Biochem. 2013;443:269–281. doi: 10.1016/j.ab.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff SJ. Reactive nitrogen intermediates and antimicrobial activity: role of nitrite. Free Radic. Biol. Med. 1993;14:351–360. doi: 10.1016/0891-5849(93)90084-8. [DOI] [PubMed] [Google Scholar]
- Knowles CJ. Microorganisms and cyanide. Bacteriol. Rev. 1976;40:652–680. doi: 10.1128/br.40.3.652-680.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH. The centennial of the Fenton reaction. Free Rad. Biol. Med. 1993;15:645–651. doi: 10.1016/0891-5849(93)90168-t. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA, Kuzminov A. Fragmentation of replicating chromosomes triggered by uracil in DNA. J. Mol. Biol. 2006;355:20–33. doi: 10.1016/j.jmb.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA, Kuzminov A. Chromosome demise in the wake of ligase-deficient replication. Mol. Micorbiol. 2012;84:1079–1096. doi: 10.1111/j.1365-2958.2012.08076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova EA, Rotman E, Macomber L, Zhang J, Kuzminov A. RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure. Proc. Natl. Acad. Sci. USA. 2004;101:16262–16267. doi: 10.1073/pnas.0405943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ, Kuzminov A. Cyanide, peroxide and nitric oxide formation in solutions of hydroxyurea causes cellular toxicity and may contribute to its therapeutic potency. J. Mol. Biol. 2009;390:845–862. doi: 10.1016/j.jmb.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ, Kuzminov A. Stalled replication fork repair and misrepair during thymineless death in Escherichia coli. Genes Cells. 2010;15:619–634. doi: 10.1111/j.1365-2443.2010.01405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Han Z, Chin SM, Linn S. Three chemically distinct types of oxidants formed by iron-mediated Fenton reactions in the presence of DNA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12438–12442. doi: 10.1073/pnas.91.26.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick ML, Buettner GR, Britigan BE. Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J. Bacteriol. 1998;180:622–625. doi: 10.1128/jb.180.3.622-625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman G, Bou-Abdallah F, Vane E, Maura P, Arosio P, Melman A. Iron release from ferritin by flavin nucleotides. Biochim. Biophys. Acta. 2013;1830:4669–4674. doi: 10.1016/j.bbagen.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. p. 466. [Google Scholar]
- Miller TE. Killing and lysis of gram-negative bacteria through the synergistic effect of hydrogen peroxide, ascorbic acid, and lysozyme. J. Bacteriol. 1969;98:949–955. doi: 10.1128/jb.98.3.949-955.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouameur AA, Arakawa H, Ahmad R, Naoui M, Tajmir-Riahi HA. A Comparative study of Fe(II) and Fe(III) interactions with DNA duplex: major and minor grooves bindings. DNA Cell Biol. 2005;24:394–401. doi: 10.1089/dna.2005.24.394. [DOI] [PubMed] [Google Scholar]
- Pacelli R, Wink DA, Cook JA, Krishna MC, DeGraff W, Friedman N, Tsokos M, Samuni A, Mitchell JB. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J. Exp. Med. 1995;182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Alamo MJ, Abril N, Pueyo C. Mutagenesis in Escherichia coli K-12 mutants defective in superoxide dismutase or catalase. Carcinogenesis. 1993;14:237–244. doi: 10.1093/carcin/14.2.237. [DOI] [PubMed] [Google Scholar]
- Reif DW. Ferritin as a source of iron for oxidative damage. Free Radic. Biol. Med. 1992;12:417–427. doi: 10.1016/0891-5849(92)90091-t. [DOI] [PubMed] [Google Scholar]
- Resnick MA. The repair of double-strand breaks in DNA: a model involving recombination. J. Theor. Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rotman E, Khan SR, Kouzminova E, Kuzminov A. Replication fork inhibition in seqA mutants of Escherichia coli triggers replication fork breakage. Mol. Microbiol. 2014;93:50–64. doi: 10.1111/mmi.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutala WA, Weber DJ, H. I. C. P. A. C. (HICPAC) Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008. Centers for Disease Control and Prevention; 2008. [Google Scholar]
- Slauch JM. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol. 2011;80:580–583. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 2004;30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- Spek EJ, Wright TL, Stitt MS, Taghizadeh NR, Tannenbaum SR, Marinus MG, Engelward BP. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 2001;183:131–138. doi: 10.1128/JB.183.1.131-138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmaszynska T. Formation of HCN by human phagocytosing neutrophils--1. Chlorination of Staphylococcus epidermidis as a source of HCN. Int. J. Biochem. 1985;17:373–379. doi: 10.1016/0020-711x(85)90213-7. [DOI] [PubMed] [Google Scholar]
- Stern AM, Zhu J. An introduction to nitric oxide sensing and response in bacteria. Adv. Appl. Microbiol. 2014;87:187–220. doi: 10.1016/B978-0-12-800261-2.00005-0. [DOI] [PubMed] [Google Scholar]
- Surguladze N, Thompson KM, Beard JL, Connor JR, Fried MG. Interactions and reactions of ferritin with DNA. J. Biol. Chem. 2004;279:14694–14702. doi: 10.1074/jbc.M313348200. [DOI] [PubMed] [Google Scholar]
- Thomas EL, Fishman M. Oxidation of chloride and thiocyanate by isolated leukocytes. J. Biol. Chem. 1986;261:9694–9702. [PubMed] [Google Scholar]
- Thompson KJ, Fried MG, Ye Z, Boyer P, Connor JR. Regulation, mechanisms and proposed function of ferritin translocation to cell nuclei. J. Cell Sci. 2002;115:2165–2177. doi: 10.1242/jcs.115.10.2165. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu. REv. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J. Biol. Chem. 2006;281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J. Biol. Chem. 2002;277:34055–34066. doi: 10.1074/jbc.M203977200. [DOI] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA. A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol. Microbiol. 2003;49:11–22. doi: 10.1046/j.1365-2958.2003.03530.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.