Abstract
The use of genetic engineering has vastly improved our capabilities to create animal models relevant in preclinical research. With the recent advances in gene-editing technologies, it is now possible to very rapidly create highly tunable mouse models as needs arise. Here, we provide an overview of genetic engineering methods, as well as the development of humanized neonatal Fc receptor (FcRn) models and their use for monoclonal antibody in vivo studies.
1 Introduction
Currently, the mouse is still the preferred animal model used in drug discovery and development. The advance of the mouse as a primary scientific tool really began with the ability to generate inbred strains. In contrast to outbred, inbred strains can provide repeatedly genetically consistent animals, where each mouse within a strain is an essentially genetically identical unit over place and time. Inbred strains were developed about 1909 by C.C. Little, with DBA being the first created in 1929/1930, leading to two of the still most widely used inbred strains DBA/1 and DBA/2 [1]. Since then, more than 450 inbred strains have been established, with many more substrains covering a vast genetic diversity. The use of inbred strains in experimental systems enables the experimenter to distinguish between genetic influences, versus environmental effects, providing a highly controlled and defined experimental system. Further, the resulting genetic uniformity offered within each strain simplifies their use and experimental interpretation in drug discovery, development, and toxicological studies. This is exemplified by the work of Michael Festing, who has demonstrated that using multiple inbred strains versus outbred strains provides superior toxicological data, which can be used to unravel underlying genetic factors and improve therapeutic options or approaches [2, 3]. In drug discovery, there is a long history of taking advantage of inbred strains, each with its unique phenotype and disease predispositions. Prime examples include DBA2/J, which develop glaucoma and the NOD/ShiLtJ strain, which becomes type 1 diabetic. These and many other inbred strains as models of disease have yielded valuable insights in understanding human disease [4–6].
With the recent striking advances of genetic engineering and assisted reproductive sciences (ARTs), it has become possible to routinely generate transgenic mice, with modifications ranging from transgenic animals with randomly integrated DNA to the precise tailoring of their genome. The creation of transgenic mice was first achieved in the 1970s using viral transfection; however, this approach was often hampered due to silencing of introduced transgenes by de novo DNA methylation post-insertion [7]. With the development of DNA pronuclear injection techniques in the early 1980s, the field took off, initiating the development of thousands of transgenic models expressing foreign genes, including the introduction of many human gene constructs into the mouse genome [8–11]. The next major breakthrough in this field was the development of embryonic stem (ES) cells combined with gene targeting approaches developed by Capecchi and Smithies, facilitating the precise manipulation of genes and the creation of animals transmitting these [12, 13]. Initially, these modifications were limited to DNA deletions but this was soon followed by precise DNA insertion or replacement. Further progress in this field included the development of tissue-specific expression systems and inducible gene expression systems (e.g. Cre/loxP, TET-system, CRE-ERT2 system) [14–16]. The strength of ES cell-derived transgenic animals is that this allows the pre-screening of the molecular events in cell culture and the characterization and confirmation of cell clones carrying the desired genetic changes. By this method, only ES cell clones with the desired genetic manipulation are selected to create mice. This latter process involves creating chimeric animals made by combining ES cells with host embryos, and then breeding these chimeras to test for germline transmission of the introduced ES cells with its specific genetic change.
However, recently a series of novel strategies have been developed allowing precise genetic engineering to be carried out directly in the fertilized oocyte with high efficiency, sidestepping strain and time constraints intrinsic to the ES cell route. These recent additions to the genetic engineering arsenal include zinc finger nucleases (ZFN), transcription activator-like (TAL) effectors and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9), each of which is briefly discussed below [17–30]. Collectively, this means that we now have a powerful toolbox allowing the direct manipulation of the genome of mice, providing the tailoring of their genome to specific experimental needs upon demand.
In this review, we highlight an example of a genetically modified mouse, centered on neonatal Fc receptor (FcRn) biology, and discuss how this has been achieved to date, focusing especially on its uses in pharmacokinetic studies. The FcRn is responsible for recycling of immunoglobulins G (IgG) and albumin, and provides the observed long half-life in vivo. FcRn belongs to the major histocompatibility complex (MHC) class I proteins forming a heterodimer with beta-2 microglobulin light chain [31, 32]. The Fc fragment (fragment crystallizable) of IgGs bind to FcRn at low pH (~pH 6.0), but is released upon neutral pH ~7.2 [33–36]. In vivo, extracellular proteins, including IgGs, enter cells by fluid phase endocytosis as part of the normal protein turnover. Within the cell at low pH, IgG complexes with FcRn and is recycled to the plasma membrane where the IgGs are released at neutral pH. This recycling process essentially protects IgG from catabolic degradation. The exact mechanisms involved have been extensively reviewed [36–39]. In a similar way, FcRn also recycles albumin [40]. Other roles for FcRn include transepithelial transport, antigen presentation, plus cross-presentation in macrophage and dendritic cells [39, 41–45].
In 2001, Ober et al. described significant species differences in FcRn between human and mouse, highlighting limitations when mice are used as models to study human IgG half-life [46]. Their main finding was that human FcRn does not bind mouse IgGs, while, in contrast, mouse FcRn binds human IgG as well as mouse, and in fact actually shows a higher affinity for human IgG than does human FcRn. As such, standard mice are not a good predictor of pharmacokinetics when used to analyze the serum half-life of human IgGs or when assaying their efficacy. To address this issue, genetic engineering approaches were used to create a set of humanized transgenic mice to more closely mimic the human condition [47–52]. These models are discussed in further detail in the following sections.
2 Mouse as Model
The mouse is a well established model for research, drug discovery, and development, with a continually increasing number of genetically engineered models being made available. With international efforts such as the Knockout Mouse Project (KOMP) as part of the International Knockout Mouse Consortium (IKMC), model development has received major funding, supporting the creation of ES cells with targeted knockout mutations for all genes, and the development of mice from such [53–56]. By August 2013, over 18,000 ES cells with targeted events have been made available, from which more than 2,400 mouse strains have been created. This ultimately will lead to an abundance of genetically engineered mice covering all known genes. However, when choosing mouse models for use in drug development, a number of properties need to be considered, including: (i) homology of gene and protein between mouse and human, (ii) similarity of pathway, (iii) translation of the disease or aspects of it from mouse to human, and (iv) experimental design. Further, when considering using models from any organism, it must be remembered that a model is just a model, at best a partial reflection of reality, and that the results and how far they are applicable depend on the validity and individual properties of each model. However, with the advent of new genetic engineering technologies, we now have available a highly versatile arsenal of tools enabling the generation of new models and/or the further enhancement of current models, allowing sequential improvement and building upon existing models [30, 57].
2.1 Genetic Engineering of Mice
The current toolbox to genetically engineer mice includes standard random transgenics generated by injection of DNA into fertilized oocytes, and ES-cell derived transgenics, through to sophisticated direct gene editing systems using nucleases.
2.1.1 Transgenic Mice
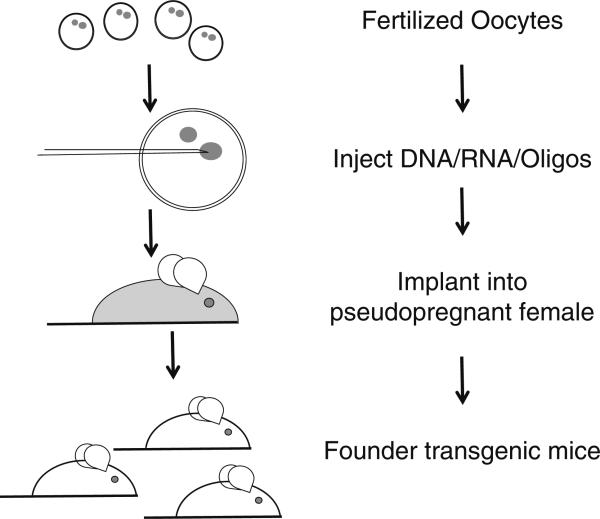
Transgenic mice were first generated in 1982, and the basic technology has changed little since [11]. For this approach, DNA is microinjected into the pronucleus or cytoplasm of fertilized oocytes. After bringing to term, the resulting offspring are screened for the presence and then expression of the injected DNA gene constructs (Fig. 1). Founder animals are then crossed to establish either hemizygous or homozygous mouse lines.
Fig. 1.
Process for creating transgenic mice, by injection of DNA, RNA, or oligonucleotides into the pronuclei or cytoplasm of fertilized oocytes. This process can be used for random transgenics, ZFN, TALEN, or CRISPR. CRISPR Clustered Regularly Interspaced Short Palindromic Repeats, TALEN transcription activator-like effector nucleases, ZFN zinc finger nucleases
A key advantage of this method is that it is applicable to DNA covering a vast range of sizes; e.g. from oligonucleotides to bacterial artificial chromosomes (BACs). Further, it can be practiced on practically any available mouse strain where oocytes can be isolated. With the availability of tissue-specific and inducible promoters (e.g. Cre/loxP, tetracycline/doxycycline-inducible systems), expression of genes can also be directed and specific aspects of a gene function studied [58]. Commonly, many transgenic mice are made using complementary DNA (cDNA) from the gene of interest, linking it either to a ubiquitous or tissue-specific promoter. This approach was used, for example, when generating the Tg276 humanized FcRn mouse (see below). For gene expression to more closely resemble normal physiology it is often necessary to use large genomic fragments, e.g. BACs or cosmids, which include their often poorly defined but functional regulatory elements. It was this approach that was used to generate the Tg32 humanized FcRn mice. A drawback of this serendipitous random transgenesis approach is that gene expression levels are not predictable due to position integration effects. Further, as is the nature of the approach, there is also no control of how many copies integrate (in tandem or throughout the genome). Lastly, random transgene integration can lead to disruption of endogenous genes, resulting in phenotypes independent on transgene expression. However, due to the simplicity of this approach and associated speed in generating a potential new model, pronuclear DNA microinjection is still a widely used method to generate new animal models.
2.1.2 Knockout Mice via ES cells
The development of ES cells in the 1980s, with their capability to contribute to the germline of the mouse embryo, more precise genetic manipulation became available [59]. The major breakthrough in 1987 was the advent of homologous recombination in ES cells, whereby it now became possible to precisely delete selected sequences and create knockout mice [12, 13]. The ES field then rapidly developed, with the defined introduction of DNA into chosen locations, enabling replacement of mouse sequences with, for example, human, creating so-called knock-ins.
The overall procedure became well established and moderately reliable. First, a DNA construct or vector containing the desired modification and selection or resistance marker was assembled. This is electroporated into ES cells, which are selected for DNA uptake using a resistance gene, for example, neomycin with the selection drug G418 (geneticin). Surviving ES clones are screened by polymerase chain reaction (PCR) and/or Southern blot to detect the desired homologous recombination event. Correctly targeted ES clones are then expanded for blastocyst injection or morula aggregation. The host embryos carrying ES cells are then implanted into pseudopregnant animals, which are brought to term to generate chimeric mice, which are bred for germline transmission of the ES cells. Often, ES cell germline transmission is observed by the use of coat color, taking advantage of the difference in coat color of ES cells versus host embryo. The ES-derived offspring are then genotyped to determine which are carrying the selected genetic modification and are then bred to homozygosity (Fig. 2).
Fig. 2.
Process for creating mice via the ES cell route, by injecting ES cells into blastocysts. ES embryonic stem
This approach, although functional, has a number of shortcomings, including possible low ES cell targeting frequency, requiring the screening of many ES cell clones. Further and highly time consuming processes involved in germline transmission of genetically modified ES cells is not a guarantee of success. ES cells are often maintained in culture for many passages, leading to a loss of germline competency. Although this can be overcome by selecting several correctly targeted ES cell clones or using a number of different ES cell lines, this is resource and time consuming. In comparison with the development of DNA microinjected transgenics, the ES cell route to heterozygous mice is longer due to this need to generate chimeras, and then the chance that these fail germline transmission. This can add easily 6 months to a project and, worse, a level of uncertainty and complete failure. However, perhaps more crucially, the variety and number of ES backgrounds available, although improving, is still a major limiting factor that can only be mitigated by time-consuming backcrossing to the desired strain background necessary (Table 1).
Table 1.
Genetic engineering—approximate timelines. Note, shorter timeline assumes access to an efficient core facility
| Random transgenesis | ES cell-mediated transgenesis | ZFN | TALEN | CRISPR | |
|---|---|---|---|---|---|
| Time frame to heterozygous animal or founder animal | 6–9 months | 12–18 months | 6–9 months | 6–9 months | 5–9 months |
| Construct | Easy | Easy | Difficult | Easy | Very easy |
| Cell type | Fertilized oocytes | ES cells | Fertilized oocytes | Fertilized oocytes | Fertilized oocytes |
| Controlled copy number integration | No | Yes | Yes | Yes | Yes |
| Random insertion | Yes | Yes | n/a | n/a | n/a |
| Targeted deletion | No | Yes | Yes | Yes | Yes |
| Targeted integration | No | Yes | Yes | Yes | Yes |
| Strain background | All | Very limited | All | All | All |
| Cost | $ | $$$$ | $$$ | $$ | $$ |
CRISPR Clustered Regularly Interspaced Short Palindromic Repeats, ES embryonic stem, TALEN transcription activator-like effector nucleases, ZFN zinc finger nucleases
2.1.3 Zinc Finger Nuclease Technology
ZFN proteins are composed of two sequence-specific DNA binding domains that are linked to a DNA cleavage domain, FokI, which is functionally an obligate dimer, requiring both DNA binding domains to be active. These are introduced by microinjection into the fertilized oocytes as messenger RNA (mRNA), encoding the ZFN directed towards a selected sequence, where the mRNA is translated to DNA binding proteins, which find their target sequence and, upon dimerization of the FokI monomers, cause a double-stranded break at the preselected sequence site [30, 60]. The cell, or in this case the fertilized oocyte, responds to the break by initiating an error prone DNA repair process known as non-homologous end joining (NHEJ) repair, leading occasionally to the addition of one to two bases at the site of the break, or more often to the deletion of one to many hundreds of bases. When using mouse oocytes, this event can be found in 20–75 % of offspring [22] and appears relatively refractive to genetic background. Additionally, if homologous DNA is present during this process, e.g. by co-microinjection of a DNA oligonucleotide or a targeting vector, it can, albeit at a lower frequency incorporate into the target region and provide transgenic animals containing the targeted insertion. Reported frequencies for targeted integration events have been from 1.7 to 4.5 % for mouse oocytes [28, 61]. The process of generating mice follows the same scheme as shown in Fig. 1. However, ZFNs have some disadvantages, including difficulty to design and assemble, and therefore generally need to be purchased from a proven provider, such as Sigma, with their attached limited terms of use. As such, the timeline to produce them is a function of an external supplier; however, once acquired, the microinjection into oocytes is fast, with founder animals being ready to screen a few weeks post-birth. However, at the time of writing, ZFNs appear to have limitations in the DNA regions where they can successfully target compared with more recent developments such as TAL and CRISPR/Cas9.
2.1.4 TAL/TALEN Technology
TAL effector nucleases (TALEN) essentially operate in a similar way to ZFN. However, TALE are characterized by a central repeat domain of 34 residues, which recognizes a DNA target sequence and can be easily assembled to target specific DNA loci.
By combining a pair of TALE each with a FokI nuclease monomer, specific DNA sequences can be targeted. TALEN are designed to bind 14–18 bp of the target sequence with an intervening spacer sequence of 16–19 bp, followed by a further 14–18 bp, the target sequence. A major benefit over ZFN includes their ease of construct using web-available computer software and from readily available units in a matter of 3–7 days (plus a further week to quality control and make mRNA for injection). Further, they are available from a variety of providers. However, their key advantage is that almost any DNA region can be targeted [30, 62–64]. Upon microinjection of mRNA encoding TALEN into mouse fertilized oocytes NHEJ events can range from 4.8 % [29] to in excess of 80 % (Wiles MV, unpublished data). Such high rates of double-stranded breaks greatly facilitate homologous recombination when donor DNA is present, leading to the replacement of specific DNA sequences at or near the target site (Wiles MV, unpublished data) [29]. In summary, TALEN can be relatively easily produced and can target practically any site in the genome. The production of TALENs is faster than of ZFNs, and the timelines from injection into zygotes are the same. To date, the concern regarding off-target events has not generated a major obstacle.
2.1.5 CRISPR/Cas Technology
At the time of writing, the first reports of genetic engineering directly in mouse oocytes using CRISPR have been published. Their mode of operation is strikingly different from ZFN and TALEN, using an RNA with 18–20 bases defining the targeting or ‘guide’ (sgRNA) to direct a nonspecific nuclease (Cas9) to the desired target DNA. These have been used with great success directly in mouse fertilized oocytes with a high frequency of NHEJ events (i.e. gene disruption) [20]. Further, these double-stranded breaks are amenable to homologous recombination if donor DNA is present [21, 65, 66]. Like the TALEN, they are also simple to construct, and in fact may be simpler, being amiable to direct synthesis approaches (i.e. from inception to sgRNA for injection could be 3 days or less). Such simple technology is also considerably cheaper and faster than either TALEN or ZFN. However, their range of targeting is slightly less than TALEN due to the need for a ‘proto-spacer adjacent motif’ (PAM) with a 3 nucleotide conserved sequence. There is also a suggestion that, due to the more limited length of the targeting guide RNA, there are potentially increased off-target effects, i.e. double-stranded breaks at other sites [67]. At this time, it appears that the generation of CRISPR is even faster, simpler, and cheaper than ofTALEN, and therefore may become the preferred method. However, with the limited published data, it is too early to say which of these technologies will become the front-runner, or if they will be quickly superseded by other approaches.
2.2 Humanized FcRn Mice
The humanized FcRn mice were established on the C57BL/6J background in a sequential manner, starting with the creation of a mouse strain carrying a deletion in the mouse FcRn gene, followed by introduction of the human FcRn gene. Currently, there are two major mouse lines used: so-called Tg276 and Tg32 (The Jackson Laboratory stock number 004919 and 014565). With further backcrossing and sequential alterations, the number of lines increase, and it is important to properly reference such in publications to ensure that results will be reproducible.
2.2.1 FcRn-null Mice
To generate the null FcRn strain, a targeting vector was constructed to delete 1588 nucleotides of the mouse FcRn gene, including the promoter sequence 5’ at the transcriptional start site, exon 1 and most of exon 2. The ES cell line ESV/J-1182, derived from the mouse strain 129X1/SvJ, was used. After characterization of ES cell clones by Southern blot analysis, a correct clone was chosen for injection into C57BL6/J (B6) host blastocysts to generate chimeras. Male chimeras were crossed to B6 females to obtain germline, establishing mice heterozygous for the FcRn knockout. As these mice had a mixed background consisting of 129X1/SvJ and C57BL6/J, the FcRn knockout carrier mice were backcrossed 11 times to B6 mice, resulting in over 99.8 % identity to C57BL6/J. These were then crossed to generate a homozygous FcRn null mouse strain. The FcRn knockout is viable and shows no overt phenotype with the exception of low IgG and albumin serum levels due to the expected lack of FcRn recycling [48, 51].
2.2.2 Humanized Tg276 FcRn Mice
To create the humanized FcRn (hFcRn) mouse strain Tg276, human cDNA was cloned and put under the control of a human cytomegalovirus immediate early promoter/enhancer plus a chicken beta-actin/rabbit beta-globin hybrid promoter (CAG). This construct was pronuclear microinjected into B6 fertilized oocytes and the resulting offspring screened for expression of the transgene. One of the founder animals expressing hFcRn, Tg276, was inter-crossed with FcRn-null mice, generating a strain lacking mouse FcRn but expressing hFcRn [48, 50]. In this strain, as predicted from the use of the strong ubiquitous CAG promoter driving hFcRn expression, hFcRn is almost ubiquitously expressed (Roopenian DC, unpublished data). Clinical chemistry analysis of this mouse revealed that albumin levels have been restored to normal; however, endogenous IgG levels are low and similar to the FcRn-null mice [51]. This mouse strain is called B6.Cg-Fcgrt<tm1Dcr>Tg(CAG-FCGRT)276Dcr/DcrJ with The Jackson Laboratory stock number 004919. This strain is commonly referred to as Tg276. This strain can be either homozygous or hemizygous for the hFcRn transgene, and shows an expression dose effect for hFcRn expression. To date, in most studies for comparing Fc-engineered monoclonal antibodies (mAbs), the hemizygous version has been used.
2.2.3 Humanized Tg32 FcRn Mice
The creation of the Tg32 hFcRn mouse used a human 33 kb cosmid clone containing the complete human FCGRT gene, including approximately 10 kb of 5’ flanking sequences and 10 kb of 3’ flanking sequences, pronuclear microinjected into fertilized B6 mouse oocytes. The offspring were analyzed for the presence and expression of the transgene. Several lines were established and then backcrossed to the FcRn-null mice. Several mouse lines expressing hFcRn while lacking mouse FcRn were developed. One of the lines further described here is Tg32 hFcRn [47, 48, 50]. In this strain, albumin levels are slightly above the B6 wild-type level, probably due to the higher hFcRn expression level, and endogenous IgG levels are low and similar to the FcRn-null mice [51]. As transgene expression is driven by the human FcRn promoter, a more restrictive expression pattern is predicted and has been observed resembling the human expression profile (Roopenian DC, unpublished data). The official name for this mouse strain is B6.Cg-Fcgrt<tm1Dcr>Tg(FCGRT)32Dcr/DcrJ (The Jackson Laboratory stock number 014565). This strain can be either homozygous or hemizygous for the hFcRn transgene, and again shows a dose effect of the transgene regarding hFcRn expression. In most studies, the hemizygous version has been used.
2.2.4 Immunodeficient hFcRn Models
Both hFcRn mouse models Tg276 and Tg 32 have been backcrossed to immune-deficient mice to allow their use for xenograft studies. For the severe combined immunodeficiency (SCID) mice, the strain B6.CB17-Prkdc<scid>SzJ mice (The Jackson Laboratory stock number 001913) was used. The new immunodeficient strains have the official names B6.Cg-Fcgrt<tm1Dcr>Prkdc<scid>Tg(CAG-FCGRT)276Dcr/DcrJ (The Jackson Laboratory stock number 021146) and B6.Cg-Fcgrt<tm1Dcr>Prkdc <scid>Tg(FCGRT)32Dcr/DcrJ (The Jackson Laboratory stock number 018441).
In addition, Tg276 was backcrossed to Rag1-null mice (The Jackson Laboratory stock number 002216), and the new resulting strain is called B6.Cg-Rag1<tm>Mom<Fcgrt>tm1Dcr[Tg(CAG-FCGRT)276Dcr/DcrJ (The Jackson Laboratory stock number 16919) [52].
2.3 Pharmacokinetic Studies Using hFcRn Mice
2.3.1 Monoclonal Antibodies
The main use of these mouse models is for pharmacokinetic studies, especially for Fc-engineered antibodies, where half-life extension is one of the desired features in engineered mAbs. A study published by Petkova et al. [49] elegantly demonstrated the importance of model selection and where an improper model, for example in this case wild-type B6 mice, can lead to the wrong conclusion, and therefore incorrect prioritization when ranking preclinical candidates. We compared the half-life of wild-type trastuzumab and three Fc-engineered variants in B6 mice, FcRn-null, Tg276 (homozygous and hemizygous) and Tg32 (hemizygous) mice. In this case, the best discrimination between the variants was observed in the Tg276 model, while the wild-type B6 mice did not differentiate between the mAbs tested. In another example, Zalevsky et al. used the Tg276 hemizygous hFcRn model for ranking of Fc-engineered antibodies derived from bevacizumab and cetuximab. They observed good discrimination between the variants, allowing them to rank these according to in vivo serum half-life and further showed a high correlation to non-human primates [52]. More recently Tam et al. [68] published an extensive study comparing seven mAbs in the Tg32 hemizygous hFcRn model, and correlated this to non-human primate and human clinical data. These studies clearly establish the robustness of these mouse models for PK analysis and critically, their translation to non-human primates and humans.
2.3.2 Fc-Fusion Proteins
With the understanding of FcRn recycling, the idea of prolonging half-life by fusing short-lived proteins or small molecules to the Fc domain of antibodies is apparent. It is here, in the development of these new and powerful therapeutics that these humanized FcRn mice will be of use in the investigation of the pharmacokinetics and efficacy of Fc-fusion molecules. One published example uses the recombinant factor VIII-Fc fusion protein, and shows a twofold increase in serum half-life for the factor VIII Fc-fusion protein versus factor VIII protein [69]. With further development of Fc-containing compounds, these mice strains will increasingly act as powerful predictors for serum pharmacokinetics in humans.
2.3.3 Albumin
In humans, albumin has a serum half-life of ~3 weeks, making it an attractive conjugate candidate to extend the half-life of short-lived drug compounds, such as small molecules, peptides, and proteins. This approach can use intact albumin protein or albumin fragments capable of binding to FcRn or albumin mimetics [70–77]. To understand this, it needs to be remembered that binding affinities and kinetics of human FcRn versus mouse FcRn are different, and that endogenous mouse albumin can interfere with in vivo studies when using the currently available humanized FcRn models [78]. For example, Andersen et al. observed, in hFcRn transgenic mice, that mouse serum albumin interacts strongly with hFcRn. They estimated that this interaction can be up to 100-fold stronger when compared with mouse serum albumin binding to mouse FcRn [78]. This could lead to human serum albumin compounds being outcompeted by the presence of endogenous mouse albumin. To improve this situation further, development of the current hFcRn models are underway, deleting the murine albumin and adding the human albumin gene to provide a more appropriate model for the class of albumin-based therapeutics. This will use the new genetic engineering techniques, allowing the sequential modification of the current models, i.e. we can build on the current models, keeping the historic data obtained to date, allowing a rapid translation and comparison with the new results. Overall, this will allow a swifter building of stronger preclinical models, advancing drug development.
3 Efficacy Models
While the hFcRn mice are mostly used for pharmacokinetic studies, it should not be overlooked that these models are easily adaptable for efficacy testing. This is possible now that mAbs can be assayed in a better hFcRn context, reflecting the physiological half-life of the mAb tested instead of an (artificially) extended half-life in mice expressing mouse FcRn.
3.1 Cancer Xenograft Models
Tumor xenograft studies using cell lines or primary tumor material in immunodeficient mice are a well established model to test efficacy of anti-tumor agents. This has to be conducted in the right FcRn context for Fc-based or albumin-based compounds, the Tg276 and Tg32 mice have been backcrossed to the immunodeficient SCID background (Prkdc<scid>/SzJ) mice. The derived immunodeficient strains B6.Cg-Fcgrt<tm1Dcr>Prkdc<scid>Tg (CAG-FCGRT)276Dcr/DcrJ (Stock number 021146) and B6.Cg-Fcgrt<tm1Dcr>Prkdc<scid>Tg(FCGRT)32Dcr/DcrJ (Stock number 018441). For Tg276, a Rag1-null variant is also available, with the strain name B6.Cg-Rag1<tm1-Mom>Fcgrt<tm1Dcr>Tg(CAG-FCGRT)276Dcr/DcrJ and stock number 016919.
Xenograft studies have been performed with 016919 mice hemizygous for the Tg276 hFcRn transgene, testing a set of anti-vascular endothelial growth factor (VEGF) mAbs and anti-epidermal growth factor receptor (EGFR) mAbs. Zalevsky et al. [52] compared two well studied antibodies, bevacizumab and cetuximab, to Fc-engineered versions developed using the Xtend technology, which increases the affinity to FcRn. Here, xenografts with the human ovarian carcinoma SKOV-3 cell line were established for subsequent treatment with the wild-type and Xtend variant of bevacizumab, and human epidermoid carcinoma A431 cells for treatment with the wild-type and Xtend variant of cetuximab [52]. When comparing the Xtend variants with wild-type mAbs, the Xtend-engineered mAbs showed superior efficacy along with their superior half-life. This demonstrates the utility of this model approach.
3.2 Arthritis Model
To create this model, the hFcRn Tg32 model was inter-crossed with the Fc-gamma-RIIB-null mice to create mice lacking mouse FcRn and Fc-gamma-RIIB expression, while expressing hFcRn. Mice are injected over 7 days with plasma isolated from a patient with active rheumatoid arthritis [49, 79]. Arthritis can be scored using well established factors, including ankle and joint swelling, histopathology, and inflammatory biomarkers. Petkova et al. treated such mice with high levels of Hu4D5 (trastuzumab) wild-type and two Fc-engineered variants of Hu4D5, three or five times over 8 days or 14 days, respectively, as it has been known for high doses of IgG (e.g. 20 mg per 20 g body weight) to saturate the FcRn protection pathway, resulting in relief of the pathogenic activity of IgG auto-antibodies [80]. In this experiment, one Fc-engineered variant, the triple amino acid exchange T307A/E380A/N434A antibody, showed the most effective reduction in ankle swelling, with superior performance when compared with the wild-type mAb, correlating with the increased half-life of this antibody, and improved efficacy of FcRn blocking can be achieved [49].
4 Conclusion
The examples of the humanized FcRn mouse models described here show how genetic engineering has and will continue to open the field, creating superior models for preclinical analysis. Further, using these novel approaches, it is now possible to further fine-tune well established models directly in the zygote without losing the genetic and historical context. Previously, genetic engineering approaches required extensive backcrossing and intercrossing, often introducing genomic regions from other strain backgrounds, which are then difficult to correct, especially when such are linked to genetic changes made originally in ES cells. It is well known in the field that, even after extensive backcrossing, for example, 129 genomic sections from the ES cell genome are persistent in the congenic strain and as such may influence the phenotype of such models.
The herein described hFcRn mouse models are currently the only rodent models available to perform predictive pharmacokinetic studies for Fc-based compounds, and especially human/ized mAbs. These models alleviate the screening of Fc-engineered proteins regarding their optimal half-life, and can serve as predictors of pharmacokinetics to guide human clinical studies. Further refinement will also make them valuable for albumin-based substances. With these new tools, we can expect an explosion of genetically modified mice in the near future. With careful characterization, these will yield more appropriate preclinical models, supporting more rapid and relevant drug discovery and development.
Acknowledgments and Disclosures
Derry C. Roopenian is an inventor on patent applications containing some of the subject matter described in this report; the patent applications are assigned to The Jackson Laboratory.
Footnotes
Publisher's Disclaimer: Your article is protected by copyright and all rights are held exclusively by Springer International Publishing Switzerland. This eoffprint is for personal use only and shall not be self-archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: “The final publication is available at link.springer.com”.
References
- 1.Murray WS. The Breeding Behavior of the Dilute Brown Stock of Mice (Little dba). Am J Cancer. 1934;20(3):573–93. doi:10.1158/ajc.1934.573. [Google Scholar]
- 2.Festing MF. Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicol Pathol. 2010;38(5):681–90. doi: 10.1177/0192623310373776. doi:10.1177/0192623310373776. [DOI] [PubMed] [Google Scholar]
- 3.Festing MF. Improving toxicity screening and drug development by using genetically defined strains. Methods Mol Biol. 2010;602:1–21. doi: 10.1007/978-1-60761-058-8_1. doi:10.1007/978-1-60761-058-8_1. [DOI] [PubMed] [Google Scholar]
- 4.Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24(1):23–5. doi: 10.1038/71641. doi:10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 5.Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30(1):81–5. doi: 10.1038/ng794. doi:10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 6.Serreze DV, Chapman HD, Varnum DS, Gerling I, Leiter EH, Shultz LD. Initiation of autoimmune diabetes in NOD/Lt mice is MHC class I-dependent. J Immunol. 1997;158(8):3978–86. [PubMed] [Google Scholar]
- 7.Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci USA. 1976;73(4):1260–4. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214(4526):1244–6. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 9.Costantini F, Lacy E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature. 1981;294(5836):92–4. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 10.Wagner EF, Stewart TA, Mintz B. The human beta-globin gene and a functional viral thymidine kinase gene in developing mice. Proc Natl Acad Sci USA. 1981;78(8):5016–20. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinster RL, Chen HY, Warren R, Sarthy A, Palmiter RD. Regulation of metallothionein–thymidine kinase fusion plasmids injected into mouse eggs. Nature. 1982;296(5852):39–42. doi: 10.1038/296039a0. [DOI] [PubMed] [Google Scholar]
- 12.Doetschman T, Maeda N, Smithies O. Targeted mutation of the HPRT gene in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1988;85(22):8583–7. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44(3):419–28. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 14.Nishijima H, Yasunari T, Nakayama T, Adachi N, Shibahara K. Improved applications of the tetracycline-regulated gene depletion system. Biosci Trends. 2009;3(5):161–7. [PubMed] [Google Scholar]
- 15.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237(3):752–7. doi: 10.1006/bbrc.1997.7124. doi:10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 17.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. doi:10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrangou R. RNA-mediated programmable DNA cleavage. Nat Biotech. 2012;30(9):836–8. doi: 10.1038/nbt.2357. doi:10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- 19.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23(5):720–3. doi: 10.1038/cr.2013.46. doi:10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186(2):451–9. doi: 10.1534/genetics.110.117002. doi:10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29(1):64–7. doi: 10.1038/nbt.1731. doi:10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, et al. TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol. 2013;31(6):530–2. doi: 10.1038/nbt.2595. doi:10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies B, Davies G, Preece C, Puliyadi R, Szumska D, Bhattacharya S. Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS One. 2013;8(3):e60216. doi: 10.1371/journal.pone.0060216. doi:10.1371/journal.pone.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu Z, Liu M, Chen Z, Shao Y, Pan H, Wei G, et al. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 2013;41(11):e120. doi: 10.1093/nar/gkt258. doi:10.1093/nar/gkt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31(1):23–4. doi: 10.1038/nbt.2477. doi:10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 28.Meyer M, Ortiz O, Hrabe de Angelis M, Wurst W, Kuhn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc Natl Acad Sci USA. 2012;109(24):9354–9. doi: 10.1073/pnas.1121203109. doi:10.1073/pnas.1121203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wefers B, Meyer M, Ortiz O, Hrabe de Angelis M, Hansen J, Wurst W, et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA. 2013;110(10):3782–7. doi: 10.1073/pnas.1218721110. doi:10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013 doi: 10.1016/j.tibtech.2013.04.004. doi:10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337(6203):184–7. doi: 10.1038/337184a0. doi:10. 1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 32.Ahouse JJ, Hagerman CL, Mittal P, Gilbert DJ, Copeland NG, Jenkins NA, et al. Mouse MHC class I-like Fc receptor encoded outside the MHC. J Immunol. 1993;151(11):6076–88. [PubMed] [Google Scholar]
- 33.Raghavan M, Gastinel LN, Bjorkman PJ. The class I major histocompatibility complex related Fc receptor shows pH-dependent stability differences correlating with immunoglobulin binding and release. Biochemistry. 1993;32(33):8654–60. doi: 10.1021/bi00084a037. [DOI] [PubMed] [Google Scholar]
- 34.Burmeister WP, Gastinel LN, Simister NE, Blum ML, Bjorkman PJ. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature. 1994;372(6504):336–43. doi: 10.1038/372336a0. doi:10. 1038/372336a0. [DOI] [PubMed] [Google Scholar]
- 35.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372(6504):379–83. doi: 10.1038/372379a0. doi:10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 36.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7(9):715–25. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 37.Ghetie V, Ward ES. FcRn: the MHC class I-related receptor that is more than an IgG transporter. Immunol Today. 1997;18(12):592–8. doi: 10.1016/s0167-5699(97)01172-9. [DOI] [PubMed] [Google Scholar]
- 38.Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I- related receptor FcRn. Annu Rev Immunol. 2000;18:739–66. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 39.Tzaban S, Massol RH, Yen E, Hamman W, Frank SR, Lapierre LA, et al. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J Cell Biol. 2009;185(4):673–84. doi: 10.1083/jcb.200809122. doi:10.1083/jcb.200809122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson CL, Chaudhury C, Kim J, Bronson CL, Wani MA, Mohanty S. Perspective-FcRn transports albumin: relevance to immunology and medicine. Trends Immunol. 2006;27(7):343–8. doi: 10.1016/j.it.2006.05.004. doi:10.1016/j.it.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Kuo TT, de Muinck EJ, Claypool SM, Yoshida M, Nagaishi T, Aveson VG, et al. N-glycan moieties in neonatal Fc receptor determine steady-state membrane distribution and directional transport of IgG. J Biol Chem. 2009;284(13):8292–300. doi: 10.1074/jbc.M805877200. doi:10.1074/jbc.M805877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker K, Qiao SW, Kuo TT, Aveson VG, Platzer B, Andersen JT, et al. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b + dendritic cells. Proc Natl Acad Sci USA. 2011;108(24):9927–32. doi: 10.1073/pnas.1019037108. doi:10.1073/pnas.1019037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi K, Qiao SW, Yoshida M, Baker K, Lencer WI, Blumberg RS. An FcRn-dependent role for anti-flagellin immunoglobulin G in pathogenesis of colitis in mice. Gastroenterology. 2009;137(5):1746–56. e1. doi: 10.1053/j.gastro.2009.07.059. doi:10.1053/j.gastro.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Lu L, Yang Z, Palaniyandi S, Zeng R, Gao LY, et al. The neonatal FcR-mediated presentation of immune-complexed antigen is associated with endosomal and phagosomal pH and antigen stability in macrophages and dendritic cells. J Immunol. 2011;186(8):4674–86. doi: 10.4049/jimmunol.1003584. doi:10.4049/jimmunol.1003584. [DOI] [PubMed] [Google Scholar]
- 45.Vegh A, Farkas A, Kovesdi D, Papp K, Cervenak J, Schneider Z, et al. FcRn overexpression in transgenic mice results in augmented APC activity and robust immune response with increased diversity of induced antibodies. PLoS One. 2012;7(4):e36286. doi: 10.1371/journal.pone.0036286. doi:10.1371/journal.pone.0036286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ober RJ, Radu CG, Ghetie V, Ward ES. Differences in promiscuity for antibody-FcRn interactions across species: Implications for therapeutic antibodies. Int Immunol. 2001;13(12):1551–9. doi: 10.1093/intimm/13.12.1551. [DOI] [PubMed] [Google Scholar]
- 47.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, et al. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197(3):315–22. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, et al. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J Immunol. 2003;170(7):3528–33. doi: 10.4049/jimmunol.170.7.3528. [DOI] [PubMed] [Google Scholar]
- 49.Petkova SB, Akilesh S, Sproule TJ, Christianson GJ, Al Khabbaz H, Brown AC, et al. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18(12):1759–69. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 50.Roopenian DC, Christianson GJ, Sproule TJ. Human FcRn transgenic mice for pharmacokinetic evaluation of therapeutic antibodies. Methods Mol Biol. 2010;602:93–104. doi: 10.1007/978-1-60761-058-8_6. [DOI] [PubMed] [Google Scholar]
- 51.Stein C, Kling L, Proetzel G, Roopenian DC, de Angelis MH, Wolf E, et al. Clinical chemistry of human FcRn transgenic mice. Mamm Genome. 2012;23(3–4):259–69. doi: 10.1007/s00335-011-9379-6. doi:10.1007/s00335-011-9379-6. [DOI] [PubMed] [Google Scholar]
- 52.Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IW, Sproule TJ, et al. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2010;28(2):157–9. doi: 10.1038/nbt.1601. doi:10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd KC. A knockout mouse resource for the biomedical research community. Ann N Y Acad Sci. 2011;1245:24–6. doi: 10.1111/j.1749-6632.2011.06311.x. doi:10.1111/j.1749-6632.2011.06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayadi A, Birling MC, Bottomley J, Bussell J, Fuchs H, Fray M, et al. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome. 2012;23(9–10):600–10. doi: 10.1007/s00335-012-9418-y. doi:10.1007/s00335-012-9418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, et al. The knockout mouse project. Nat Genet. 2004;36(9):921–4. doi: 10.1038/ng0904-921. doi:10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, et al. The European dimension for the mouse genome muta-genesis program. Nat Genet. 2004;36(9):925–7. doi: 10.1038/ng0904-925. doi:10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurray F, Moir L, Cox RD. From mice to humans. Curr Diab Rep. 2012;12(6):651–8. doi: 10.1007/s11892-012-0323-2. doi:10.1007/s11892-012-0323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2(10):743–55. doi: 10.1038/35093537. doi:10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 59.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 60.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105(15):5809–14. doi: 10.1073/pnas.0800940105. doi:10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107(34):15022–6. doi: 10.1073/pnas.1009424107. doi:10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reyon D, Khayter C, Regan MR, Joung JK, Sander JD. Ausubel FM, et al., editors. Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Current protocols in molecular biology. 2012. Chapter 12. [DOI] [PMC free article] [PubMed]
- 63.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. doi:10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. doi: 10.1038/nbt.1755. doi:10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 65.Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt772. doi:10.1093/nar/gkt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. doi:10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 2013 doi: 10.1038/nbt.2623. advance online publication. doi:10.1038/nbt.2623. http://www.nature.com/nbt/journal/vaop/ncurrent/abs/nbt.2623.html. supplementary-information. [DOI] [PMC free article] [PubMed]
- 68.Tam SH, McCarthy SG, Brosnan K, Goldberg KM, Scallon BJ. Correlations between pharmacokinetics of IgG antibodies in primates vs. FcRn-transgenic mice reveal a rodent model with predictive capabilities. mABs. 2013;5(3):1–9. doi: 10.4161/mabs.23836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dumont JA, Liu T, Low SC, Zhang X, Kamphaus G, Sakorafas P, et al. Prolonged activity of a recombinant factor VIII-Fc fusion protein in hemophilia A mice and dogs. Blood. 2012;119(13):3024–30. doi: 10.1182/blood-2011-08-367813. doi:10.1182/blood-2011-08-367813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wunder A, Muller-Ladner U, Stelzer EH, Funk J, Neumann E, Stehle G, et al. Albumin-based drug delivery as novel therapeutic approach for rheumatoid arthritis. J Immunol. 2003;170(9):4793–801. doi: 10.4049/jimmunol.170.9.4793. [DOI] [PubMed] [Google Scholar]
- 71.Fiehn C. Methotrexate transport mechanisms: the basis for targeted drug delivery and ss-folate-receptor-specific treatment. Clin Exp Rheumatol. 2010;28(5 Suppl 61):S40–5. [PubMed] [Google Scholar]
- 72.Muller MR, Saunders K, Grace C, Jin M, Piche-Nicholas N, Steven J, et al. Improving the pharmacokinetic properties of biologics by fusion to an anti-HSA shark VNAR domain. mAbs. 2012;4(6):673–85. doi: 10.4161/mabs.22242. doi:10.4161/mabs.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stork R, Campigna E, Robert B, Müller D, Kontermann RE. Biodistribution of a bispecific single-chain diabody and its half-life extended derivatives. J Biol Chem. 2009;284(38):25612–9. doi: 10.1074/jbc.M109.027078. doi:10.1074/jbc.M109.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andersen JT, Pehrson R, Tolmachev V, Daba MB, Abrahmsen L, Ekblad C. Extending half-life by indirect targeting of the neonatal Fc receptor (FcRn) using a minimal albumin binding domain. J Biol Chem. 2011;286(7):5234–41. doi: 10.1074/jbc.M110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller D, Karle A, Meissburger B, Hofig I, Stork R, Kontermann RE. Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin. J Biol Chem. 2007;282(17):12650–60. doi: 10.1074/jbc.M700820200. doi:10.1074/jbc.M700820200. [DOI] [PubMed] [Google Scholar]
- 76.Dennis MS, Zhang M, Meng YG, Kadkhodayan M, Kirchhofer D, Combs D, et al. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J Biol Chem. 2002;277(38):35035–43. doi: 10.1074/jbc.M205854200. doi:10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen A, Reyes AE, 2nd, Zhang M, McDonald P, Wong WL, Damico LA, et al. The pharmacokinetics of an albumin-binding Fab (AB.Fab) can be modulated as a function of affinity for albumin. Protein Eng Des Sel. 2006;19(7):291–7. doi: 10.1093/protein/gzl011. doi:10.1093/protein/gzl011. [DOI] [PubMed] [Google Scholar]
- 78.Andersen JT, Daba MB, Berntzen G, Michaelsen TE, Sandlie I. Cross-species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. J Biol Chem. 2010;285(7):4826–36. doi: 10.1074/jbc.M109.081828. doi:10.1074/jbc.M109.081828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petkova SB, Konstantinov KN, Sproule TJ, Lyons BL, Awwami MA, Roopenian DC. Human antibodies induce arthritis in mice deficient in the low-affinity inhibitory IgG receptor Fc gamma RIIB. J Exp Med. 2006;203(2):275–80. doi: 10.1084/jem.20051951. doi:10.1084/jem.20051951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akilesh S, Petkova S, Sproule TJ, Shaffer DJ, Christianson GJ, Roopenian D. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J Clin Investig. 2004;113(9):1328–33. doi: 10.1172/JCI18838. doi:10.1172/jci200418838. [DOI] [PMC free article] [PubMed] [Google Scholar]