Abstract
Life has semiotic nature; and as life forms differ in their complexity, functionality, and adaptability, we assume that forms of semiosis also vary accordingly. Here we propose a criterion to distinguish between the primitive kind of semiosis, which we call “protosemiosis” (following Prodi) from the advanced kind of semiosis, or “eusemiosis”. In protosemiosis, agents associate signs directly with actions without considering objects, whereas in eusemiosis, agents associate signs with objects and only then possibly with actions. Protosemiosis started from the origin of life, and eusemiosis started when evolving agents acquired the ability to track and classify objects. Eusemiosis is qualitatively different from protosemiosis because it can not be reduced to a small number of specific signaling pathways. Proto-signs can be classified into proto-icons that signal via single specific interaction, proto-indexes that combine several functions, and proto-symbols that are processed by a universal subagent equipped with a set of heritable adapters. Prefix “proto” is used here to characterize signs at the protosemiotic level. Although objects are not recognized by protosemiotic agents, they can be reliably reconstructed by human observers. In summary, protosemiosis is a primitive kind of semiosis that supports “know-how” without “know-what”. Without studying protosemiosis, the biosemiotics theory would be incomplete.
1. Introduction
The discipline of biosemiotics is an ambitious attempt to describe sign relationships across the whole evolutionary tree of life (Hoffmeyer 1996). The major assumption of biosemiotics is that life has semiotic nature (Anderson et al. 1984; Sharov 1992). Because life forms differ in their complexity, functionality, and adaptability, it is natural to expect that forms of semiosis and the complexity of signs also vary accordingly. The scope of variability of life forms has multiple dimensions. First, there is a phylogenetic tree of life which includes prokaryotes (bacteria and archae), unicellular eukaryotes and multicellular eukaryotes (plants, fungi, and animals). Second, there is a structural hierarchy of life that includes organisms, organs, tissues, cells, cellular organelles, and functional molecular complexes. Each of these structural levels has specific signaling pathways (e.g., molecular signaling, growth factors, hormones, neural signals). More structural levels are found above the organism level and they include families or colonies of social animals as well as inter-species communities at each structural level. Communication of living systems at various structural levels should be analyzed using semiotic terms in order to represent their functional aspects and association with goals (Hoffmeyer 2008).
Because of the enormous variability of sign processes in living systems, one of the important tasks of biosemiotics is to develop criteria that help to classify sign processes in various living systems (Kull 2009). The most common way of classifying semiotic processes has been based on how they occur at various structural levels or in systematic groups. For example, zoosemiotics referred to animal semiosis (Sebeok 1963) and phytosemiotics to plant semiosis (Krampen 1981). In parallel, Thomas Sebeok coined the terms endosemiotics and exosemiotics (Sebeok 1976). Endosemiotics studies internal communications within organisms, whereas exosemiotics refers to the communication of organisms with their environments, including the other organisms. In addition, vertical semiosis, which designates the transfer of hereditary messages between successive generations, was distinguished from horizontal semiosis, which refers to both endo- and exo-semiotic interactions of individual organisms (Hoffmeyer 1996).
Terrence Deacon (1997) and more recently Kalevi Kull (2009) have suggested to relate the types of semiosis with Charles Peirce’s classification of signs. In particular, Kull distinguished vegetative semiosis, which is presumably based on iconic signs, animal semiosis, which is based on indices, and cultural semiosis that is respectively based on symbols. However, the proposed terminology and criteria are problematic. Terms “vegetative” and “animal” remind of phylogenetic branches of plants and animals, to which they are not directly connected. Our approach in this paper makes the general application of Peirce’s term “icon” to vegetative semiosis questionable. In particular, the distinction between icon, index, and symbol is based on the type of the relation between a sign and its object, and the result of such association creates an interpretant-sign of the object in the interpreting mind. But molecular signals (that are in the domain of Kull’s vegetative semiosis) appear to control actions of specific cell components directly without any internal reference to either an object or mental interpretant. Most cellular components seem to have no capacity to handle and classify objects.
In this paper we discuss two major modes of semiosis: the primitive “mindless” semiosis, which we call “protosemiosis” and the more genuine advanced kind of semiosis, or “eusemiosis” which requires at least a minimal mind capable of tracking and classifying objects. Eusemiosis becomes possible in the sphere of Kull’s animal and cultural semiosis and protosemiosis corresponds roughly to the vegetative semiosis. We think that at least in protosemiotics, the Peircean logical concept of sign should be substituted by a more general one, by a concept where signs are not associated with objects.
2. Signs in Biosemiotics and Their Relation to Theories of Peirce, Uexküll, Sebeok, and Barbieri
Sebeok based his zoosemiotics on the Peircean tradition of triadic signs and emphasized its advantages in comparison to Saussurian-based semiological structuralism (Sebeok 1976). Because Saussure’s main intention was a study of language and he understood language primarily as the medium of social communication, the signs in his theory are viewed as arbitrary dyadic relations between linguistic signifiers and mental signifieds, and such signs form a static synchronic system (Saussure et al. 1986). Peirce, instead, took signs principally as a medium of cognition or increasing knowledge that gives them a more dynamic and processual character (Vehkavaara 2007). He defined signs as follows: “a sign is something which stands to somebody for something in some respect or capacity” (Peirce 1931–1935: CP 2.2281). Peircean signs are not restricted to linguistic signs and include also types of signs whose relation to their referents is not purely conventional (e.g., icons and indices). Because of their dynamic character, signs get a triadic structure, representamen – object – interpretant. The basic idea is that when a (first) thing or event is cognized as a representamen of some sign, the cognizer recognizes it as referring to another (second), thing or event, which is the object of that sign. This act of recognition is manifested by the production of a (third) thing or event in the mind of the recognizer, the interpretant of the sign (Peirce 1998: EP 2:493–494). This means that there is a difference between direct sensing and sign-mediated recognition of an object. In order to recognize a representamen as a sign of some object, the representamen must first be perceived as an object (with some characters that connect it to the object of that sign). Also the object of the sign must be familiar beforehand (Peirce 1931–1935: CP 8.178–179), although the sign may provide some additional information about it.
One of the founders of biosemiotics, Jakob von Uexküll considered meanings and signs as products of learning living organisms (Uexküll 1982). According to Uexküll, animals act in their world according to their species-specific Umwelt, the internal model of the world constructed by the animal on the basis of its previous experience and/or heredity. Umwelt is constituted of perceptible objects that are recognized and associated with functional meanings such as “food”, “nest”, or “enemy”. Although Uexküll’s approach neglects evolutionary considerations, it introduces an important methodological perspective. His idea was to describe animal subjectivity from the animal point of view but in terms available in the Umwelt of a human observer. When studying Umwelten of animals, however, humans should not populate them with objects of the human Umwelt unless these objects are equally cognizable by animals. Instead, it is important to focus only on those aspects of the world that are perceived by organisms and recognized in the context of living functions, including potential novel functions within adjacent possible. Because various organisms differ in their functions and interaction with the world, it makes sense to follow the methodology of Uexküll and study species-specific Umwelten.
A scientist, thus, gets a double access to the objects perceived and manipulated by animals: a direct one through scientist’s Umwelt (a meta-agent perspective) and the indirect one through the study of the cognitive and behavioral capacities of the animal (an object-agent perspective) (Vehkavaara 2002: 299–300). This double access opens up a possibility for a scientist to compare the representations of the same object (fixed within the meta-agent perspective) in the Umwelten of different species of animals. Species of animals differ in the depth of representation of the same object. For example, a wolf hunting a deer follows the trail of smell and starts searching actively for a visual pattern specific to a deer. When the deer is found, the wolf keeps track of its movements and selects the shortest path for its attack; its movements are dynamically adjusted based on the localization of the deer as well as on its “body language”. In contrast, ticks that prey on the same deer have small eyes not capable of capturing the visual image of the deer. Instead, they are guided by the odor of butyric acid emitted by deer (Uexküll 1957). In response to butyric acid, ticks drop down from vegetation. We cannot say that the object (deer) is entirely absent in this sign relationship, because after dropping down the tick expects to land on a warm and furry surface of an animal and is prepared for a certain sequence of actions. This object is, however, substantially reduced in its content compared to the wolf’s perception of the same animal, and includes specific elements for a parasite (e.g., response to butyric acid, attraction to warm surface, and biting behavior on the skin).
Organisms with a low level of functional organization (e.g., with small number of sensors and/or weak information processing) tend to have reduced representations of objects around them. Thus, it seems reasonable to expect that there is some threshold of functionality below which objects are no longer represented by organisms (Bickhard 1998: 196–198, Sharov 2013; Vehkavaara 2003: 579–582). Instead, such organisms (e.g., bacteria) respond to external and internal molecular signals with specific functional actions. Bacteria do not anticipate any object standing for ligand molecules that interact with receptors; they simply utilize signals for better regulation of their living functions. We define objects here as distinct components of the environment which can be addressed selectively and repeatedly by agents for sensing and action purposes. Because protosemiotic agents do not seem to be able to interact with the environment in this way (i.e., they select actions but not objects), we think that the notion of object is not applicable. Thus, responses of bacteria to molecular signals do not fit well to the Peirce’s triadic sign2, although other kinds of triadic relations may exist in molecular interactions. For example, Hoffmeyer discussed triadic relations between DNA, fertilized egg, and ontogenetic trajectory or between DNA, lineage, and ecological niche (Hoffmeyer 1996: 19–24), whereas Alexander discussed the triadic relation between sign, response, and objective of self-affirmation and/or self-preservation (Alexander 2013). A better strategy would be not to apply Peirce’s concepts to simple molecular signaling because natural substitutes for objects and interpretants do not seem to exist at this level of organization.
To describe molecular signaling, Barbieri proposed a new signification model, which he called “organic code” or “code model of semiosis” (Barbieri 2003, 2009: 21). He defined code as a mapping rule between a set of signs and a set of meanings. For example, the genetic code maps triplets of nucleotides of mRNA to aminoacids or stop signal. Specific agents called “codemakers” (i.e., ribosomes together with the set of tRNA and aminoacyl tRNA synthetase) convert the encoded message (mRNA) in to its meaning (polypeptide) (Barbieri 2009: 21). In addition, another kind of agents, “copymakers” support replication of DNA and transcription (i.e., synthesis of mRNA on the DNA template). The advantage of the code model of semiosis is that it is well tailored to molecular processes and does not require objects and interpretants. To describe various kinds of sign processes in living organisms, Barbieri developed the concept of “three types of semiosis” (Barbieri 2009), which includes manufacturing semiosis (e.g., protein synthesis), signaling semiosis (molecular signaling), and interpretative semiosis that is based on constructing the internal representations of the world. The former two types of semiosis are based on coding as opposed to interpretation. The major weakness of the concept of code-based semiosis is that it does not capture the meaning of molecular signs in terms of cellular functions. For example, the functionality of the protein products is not discussed in Barbieri’s model. Also, there are problems with terminology: the term “meaning” is unexpectedly applied to a material molecule – a synthesized polypeptide, whereas the term “interpretation” is used narrowly as referring merely to conscious representations.
To characterize primitive forms of signaling here we use the term “protosemiosis”, which was earlier suggested by Giorgio Prodi in relation to molecular processes (Prodi 1988). Similar to Barbieri, he pursued a materialistic understanding of meaning: “Meaning in nature is thus the relations of correspondence between material states which appear as triggers for change” (Prodi 1988: p. 195). But he clearly attributed semiosis to living systems: “This semiotics or (proto-semiotics) is the basic feature of the whole biological organization (protein synthesis, metabolism, hormone activity, transmission of nervous impulse, and so on).” (Prodi 2010 p. 329). However, Prodi did not formulate criteria that separate protosemiosis from the advanced sign processes in the minds of animals and humans.
Here we propose a new definition of protosemiosis as a kind of sign processing, where agents3 (i.e., active systems guided by natural self-interest) initiate or modify their functional activities in response to incoming signs directly, rather than by associating signs with objects. In contrast to the concept of code-based semiosis, which is narrowly focused on a mappings between signs and meanings, our analysis of protosemiosis attempts to uncover signaling networks that support these mappings and evaluate their functional roles within a cell or entire organism. A sign is considered here in the context of its functional role within the agency, which is not the same as its physical nature. Of course, the role of signaling molecules should be consistent with their physical properties. But the agent provides specific and unusual local contexts for the action of signaling molecules; and therefore, the functional roles of signs are not reduced to their physical properties.
We further exploit the prefix “proto-“ and apply it to various semiotic terms to indicate their involvement in protosemiosis (e.g., proto-sign, proto-language). Protosemiosis is opposed here to “eusemiosis” where signs are associated with objects and interpretants. Eusemiosis roughly matches to the “interpretative semiosis”, as defined by Barbieri, as well as to the animal and social semiosis, as described by Kull. Eusemiosis is possible only in agents capable of tracking and classifying objects, which can be viewed as the core functions of the “minimal mind” (Sharov 2013; 348, 354). Human mind has many additional components such as learning, reason, logic, motivation, emotion, and attention (Premack and Woodruff 1978), which are mostly beyond the scope of this paper. Learning is an optional feature of the “minimal mind” because the capacity of organisms to track and classify objects can emerge and improve solely via genetic selection. But genetic selection is inefficient in adjusting the functions of the minimal mind, and thus, the emergence of rewritable memory (e.g., epigenetic memory) and adaptive learning4 marks an important qualitative step in the progressive evolution of mind (Sharov 2013: 352–354).
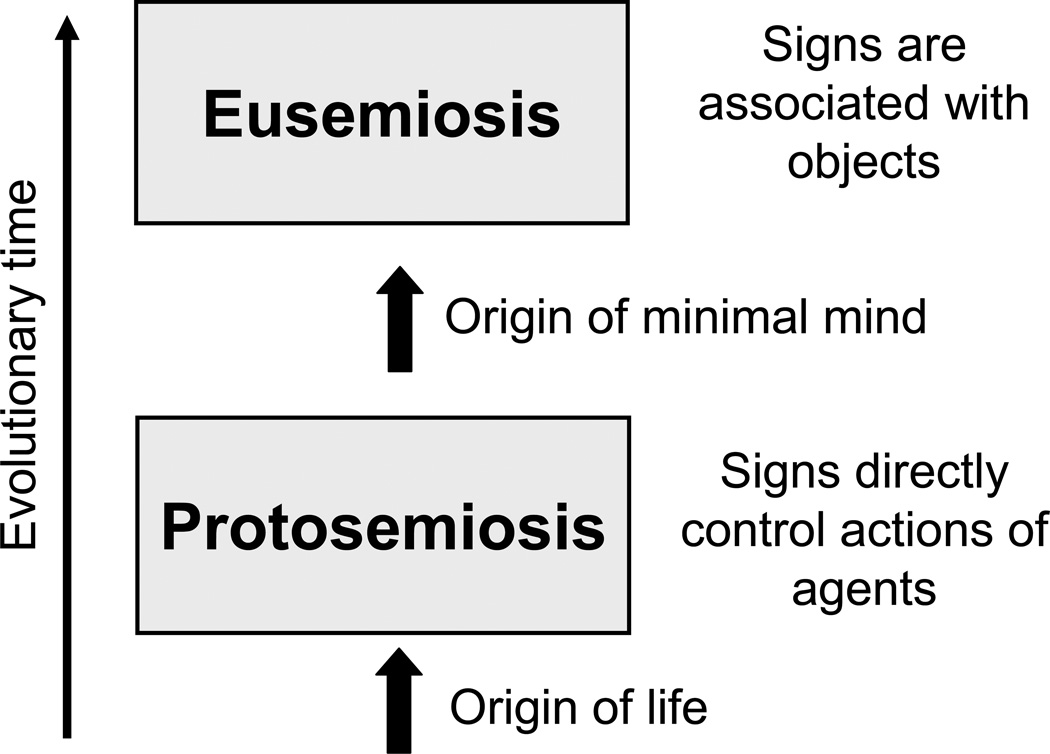
Thus, we consider two major transitions in the evolution of semiosis: the emergence of protosemiosis coincided with the origin of life, whereas eusemiosis started from the origin of minimal mind (Fig. 1). The evolutionary timing of the emergence of eusemiosis is not clear, but there are some indications that this transition may have occurred already in eukaryotic single-cell organisms (Sharov 2013: 352). However, only animals with nervous system achieved higher levels of eusemiosis that operated with complex associative and dynamic models of objects. But eusemiosis did not replace protosemiosis; instead both types of semiosis coexist. Protosemiosis operates mostly at the molecular level in cells, whereas eusemiosis supports higher-level cellular functions and cell interactions in various organs, especially in the neural system.
Fig. 1.
Major transitions in the evolution of semiosis.
3. Normative Criterion of Semiosis Delineates Life from Non-Living Systems
The essential quality common to all living systems is that their structure and function cannot be described satisfactorily without some normative semiotic concepts. If we consider physical systems in general, their dynamics fits mostly into two categories: passive fluctuations near stable equilibrium (or closed trajectory) and active maintenance of a metastable state far away from the equilibrium state. The latter category of dynamics exists in all living organisms as well as in some non-living systems, such as flames, tornadoes, and hurricanes. If these systems fail to perform self-maintenance activities, they fall towards the thermodynamical equilibrium and lose their structural and dynamic properties (Bickhard 2001). The difference between life and non-life depends on whether their metastable state can emerge spontaneously in natural environments. Non-living metastable systems emerge naturally, whereas living systems require some input from other living systems in the form of construction and/or activation. Thus, living systems persist in recursive evolutionary lineages, where each system is a product of parental system(s) of the same kind. The extinction of a lineage is historically irreversible, whereas non-living metastable systems keep re-emerging in certain natural conditions. This fundamental historic asymmetry is necessary for the existence of normativity. In particular, evolving living systems have natural self-interest in the sense that any heritable (or reproducible) trait that contributes to the maintenance of the living state and its reproduction is functional and beneficial for the system5 (Bickhard 2001; Vehkavaara 2003: 564).
Because the living state of organisms (1) does not emerge by chance in natural environments, and (2) is constructed/activated by other living system(s) to maintain the evolutionary lineage, it can be viewed as artificial conditions or state. In comparison, Barbieri wrote that “life is artifact making” (Barbieri 2008 p. 579). Because artificial conditions that support life have to be preserved to avoid rapid dissipation, they are separated from the environment, which accounts for the second fundamental internal/external asymmetry supported by various boundary structures (e.g., membranes, walls) or physical attachment (Hoffmeyer 2008, p.34). Although the living state is normally supported by self-maintenance, it may also persist passively to survive short periods of harsh environment or absence of resources (e.g., in a frozen or desiccated state). In addition, evolving living systems acquired a capacity to change their mode of self-maintenance in response to the change of environment or internal state of the system, which has been called recursive self-maintenance (Bickhard 1993). All living organisms have sensor subsystems for reading environmental (and usually also internal) inputs, and effectors for executing or modifying actions in relation to sensorial signals and internal context of the whole system (Bickhard 2001). Obviously, interactions with the environment, have to be heritable in order to persist through generations (Sharov 2010).
Because the living state is artificial, its existence cannot be fully explained by the laws of physics which are designed to describe the natural dynamics (i.e., changes that are not deliberately manipulated by living beings). From the perspective of physics, the living state acts as boundary conditions or constraints on the physical laws (Pattee 2008). But physics ignores the active role of agents in creating and maintaining the living state. The only way to contemplate the existence of living systems is to use evolutionary reasoning and assume that the living state first emerged in a very simple form and then gradually became more complex and “more artificial” in the course of evolution. Because organism bodies are perishable, functions can be preserved and accumulated in a sequence of generations only if they are encoded as copyable signs (Hoffmeyer and Emmeche 1991; Sharov 2010). Thus, evolving living systems require internally encoded representations of their functions, and these representations have semiotic nature.
The ability of living systems to transfer their functions to the following generation depends on their own success in persisting in the living state and producing progeny organisms. Successful strategies for survival and reproduction tend to propagate in the sequence of generations, whereas unsuccessful strategies gradually vanish unless they become useful again in some novel conditions. This, however, does not mean that the presence of any part of a living organism is reducible to the goal of survival (Gould and Lewontin 1979). Since the time when a part had emerged in evolution, it may had lost its original functionality but remained in the context of the whole system because the functions of other parts became dependent on its presence.
Natural self-interest can be viewed as the most primitive kind of normativity, which is a universal criterion of life applicable to both protosemiotic and eusemiotic agents. This idea is consistent with the “pragmatic turn” in studying life processes (Witzany 2010: chapter 1). Semiosis can be said to be goal-directed at least in a negative sense that it should fulfill the goal of avoiding failure. More advanced forms of normativity emerged later in evolution and included the ability of organisms to select and pursue positive goals, as represented by internally classifiable states of the brain or immune system. But protosemiotic agents (e.g., bacteria) do not seem to have representations of their goals as objects. Each molecular process contributes to the survival of bacteria only on condition that all other processes are fully functional. Thus, the natural self-interest is a property of the whole network, which can be analyzed by humans, but apparently has no internal representation within bacteria themselves.
Our use of a normative semiotic criterion to delineate life from non-life may contradict the attempts of some interpreters of Peirce to apply semiotics to non-living systems (e.g. Christiansen 2002; Deely 1990; Taborsky 2003). In these views, either signs can exist even without life (physiosemiosis or pansemiosis), or all existing nature is considered as alive to certain extent (hylozoism) (Brier and Joslyn 2013). In biosemiotics, instead, it is common to emphasize a qualitative difference between life and non-life and to focus on such apparently semiotic processes as inheritance, memory, and learning that can be attributed only to living organisms and their artifacts (e.g. computers). The major difference is that in physiosemiosis the normativity of the process (i.e. possibility to misinterpretation or failure) seems to be lost (Vehkavaara 2006: 304–305).
4. Criteria for Distinguishing Protosemiosis from Eusemiosis
In this section we discuss criteria of protosemiosis, which explain why protosemiotic signs (shortly, proto-signs) are not associated with objects. We (humans) consider objects as anything visible or tangible and relatively stable in its form, which can be studied, manipulated, or constructed either physically or mentally. However, this approach is too broad because we identify only those objects, which we have learned to detect, construct, or detach from the environment. It is better to say that humans are equipped with certain tools for detecting and handling objects, and the results of application of these tools are reproducible. It is possible to re-examine the same object using these tools, and find additional properties or errors in earlier judgments concerning the object. Repeated interaction with the same object is based on the capacity of object tracking, which requires specific tools, senses, and information-processing units. Besides individual objects, we consider object types and use specific names for these types (e.g., apples, berries, leaves). We perceive many more properties of individual objects than what is needed to associate them with specific types. Thus, classification of objects includes the abstraction from details and is based on the categorization6 procedure, which has been learned from previous experience and implemented as an epistemic tool in the mind.
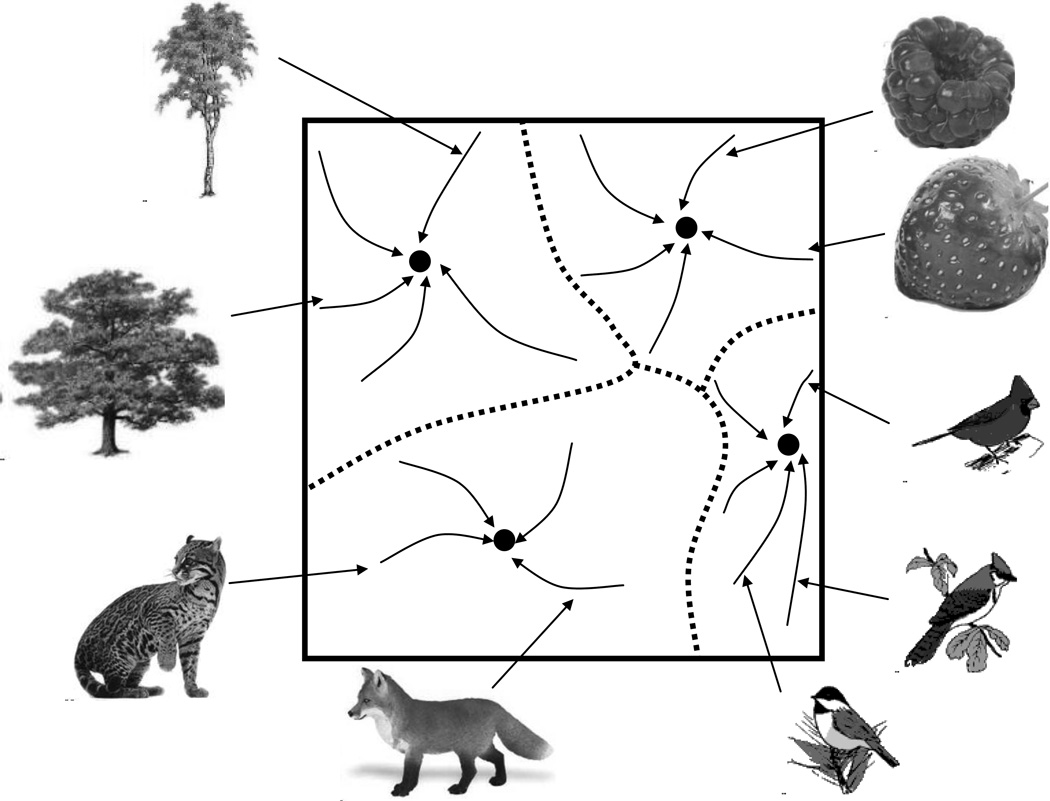
The categorization toolbox can be visualized as a set of fixed point attractors in the phase space of mind, where attractors represent object categories (Fig. 2). Each trajectory in this space corresponds to a process of recognition that leads from the initial sensory input to some attractor: tree, berry, bird, or predator (Fig. 2). Ontologically, each category corresponds to a type of objects whose perception by humans converges to the same attractor. This kind of recognition algorithm, explains the categorization capacity of the brain and can be implemented in computational neural networks (Amit 1989).
Fig. 2.
Portion of the phase space of mind with four attractors (black dots).
Now let us compare human perception with sensing in primitive organisms, such as bacteria. Bacterial cells use protein receptors inserted into cytoplasmic membrane as tools to differentiate between environment states. Receptors have high specificity in binding certain molecules (e.g., glucose), and after binding they activate the intracellular signaling molecules, which then trigger specific actions, such as rotation of flagella (i.e., external protein appendages) to propel the bacterial cell forward. This process is qualitatively different from human perception for the following reasons. Human sensorial input (i.e., a starting point of a trajectory in Fig. 2) is represented by thousands or even millions of different kinds of signals, each handled by a separate signaling pathway. For example, visual images perceived by eye are composed of ~100 million “pixels”. These signals are then processed to extract objects each comprised of a large number of ”pixels”. Finally, the image of each object is recognized as shown in Fig. 2 and becomes associated with a certain object category. In contrast, bacterial receptors generate only one kind of internal signal after sensing glucose. Although glucose molecule has many components (e.g., atoms and electrons), bacteria has no separate signaling pathways to evaluate each of these components. There is no object extraction from the signaling field and no categorization. Of course, bacteria have some tools for signal processing, such as threshold-based filtering or simple logical gates to combine a few signals (Bruni 2008). But these tools have no association with external molecules as objects because bacteria cannot track molecules and cannot verify the presence or qualities of those molecules that were bound to receptors.
Considering that bacteria cannot perceive molecules as objects, we can ask if they can perceive larger objects such as sources of nutrients. In fact, bacteria can swim against a gradient of glucose (chemotaxis), which seems to support the idea that they can track the sources of nutrients. Bacteria Escherichia coli has large arrays of transmembrane chemoreceptors that respond to glucose binding by methylation of multiple glutamic acid residues at the inner part of the receptors (Porter et al. 2011). Methylation of receptors is reversible and it is maintained in a dynamic equilibrium so that the number of methylated glutamates corresponds to the change of concentration of the glucose over time, rather than to a single glucose molecule or the current actual concentration. The level of receptor methylation then affects the activity of specific kinase proteins, which move freely in the cytoplasm. As kinase molecules reach the base of flagella, they can modify the direction of rotation. When the kinase is in its active state, it causes the flagella filaments to rotate clock-wise (CW). As a result, flagella remain separated from each other, and the cell tumbles.
However, if the kinase in not active, then flagella rotate counter-clock-wise (CCW), which causes them to form a bundle and propel the bacterial cell forward. Human observers explain the benefits of this mechanism as follows: increasing concentration of glucose (as detected by chemoreceptors) indicates that the bacterium is moving towards a source of glucose. In this case, extension of movement in the same direction (via CCW rotation of flagella) will rapidly bring the bacterium towards the source of glucose. However, if there is no increase in the concentration of glucose along the trajectory of a moving bacterium, then there is no reason for moving forward. In this case, a bacterium tumble randomly (via CW rotation of flagella) until it finds a direction where the concentration increases.
But if bacteria could talk and reason they would explain their behavior in a different way. First, they do not “know” that glucose exists outside. Their only observable is the level of methylation of transmembrane receptors. The chemoreceptor system can be said to function as a differentiator that distinguishes between the different types of its possible interactions with the current local environment. The end state of the interaction of a differentiator functions as an internal sign, which further modifies the function of effectors, i.e. the rotation of flagella (Bickhard 1998). Also bacteria “know” (in the sense of possessing a heritable habit) that association of internal signal with rotation of flagella somehow facilitates their metabolism. There is no evidence that bacteria are aware of their spatial locations or directions of movement. Receptor output can be seen as a measurement of the environment, but the only way a bacterium can partition its environment is by the types of its own receptors. This kind of sensory system is not sufficient for categorization of objects, which requires a dynamic convergence of a large number of signaling pathways to stable attractors (Fig. 2). Instead a bacterium responds to the signal directly by modifying its actions. We can say that bacteria categorize the types of whole environment, not the objects in it. Thus, analysis of chemotaxis does not support the idea that bacteria perceive sources of nutrients as objects.
Next, let us examine if bacteria can perceive their own states (e.g., complex conditions, needs, or stress) as objects. If bacteria could integrate large quantities of heterogeneous signals into discrete attractors, then we should indeed admit the existence of a categorization toolbox. However, available biochemical information indicates that bacterial signaling pathways are poorly integrated. The majority of feedback loops are restricted to a single signaling pathway; even genes related to a pathway are cotranscribed and cotranslated from the same operon. But molecular processes in eukaryotic cells may include elements of eusemiosis because they can (1) categorize complex patterns of signaling in different pathways, including spatially- and temporally-structured signals (Bruni 2008), and (2) track macro-objects via cytoskeleton connections or encapsulation in tagged membrane-bound vesicles.
Finally, even if individual bacteria cannot perceive and handle objects, it may be argued that populations of bacteria respond to environmental challenges with certain reproducible genetic changes that can be viewed as signs representing those challenging environments. In particular, bacterial populations swiftly develop resistance to antibiotics by modifying their multifunctional enzymes capable of degrading toxic molecules. Due to random mutations, these enzymes may increase their capacity to bind and break antibiotic molecules. These mutations are then favored by genetic selection and lead to the antibiotic resistance. However, mutations in genes encoding detoxicating enzymes do not represent the antibiotic molecule as an object. Instead, they work as mere differentiators that somehow help the cells to counteract the toxic effect of antibiotics. But a human observer can connect the dots and identify causal relations between mutations, structural and functional properties of the enzyme, and antibiotic resistance.
Our conclusion that sensing in bacteria does not refer to objects contradicts to the views of Hoffmeyer and Emmeche (1991) and Bruni (2008) who believe that categorization as well as Peircean triadic relationship between sign vehicle, object, and interpretant are universal for semiotic processes in all living organisms. However, this disagreement possibly results from differences in terminology. For example, Bruni views stereochemical key-and-lock binding of a ligand to receptor as an example of categorization, where a digital output is produced from analogical input (i.e., spatial pattern). We do not consider this process as categorization because bacteria do not perceive the spatial pattern of a ligand. Instead they use a receptor, which is a tool refined in evolution by genetic selection, to handle the ligand (in a holistic way) and produce a signal. We reserve the notion of categorization for only those processes, where the analog input is already internalized in the form of a large number of heterogeneous signals. For example, visual perception integrates signals that come from millions of photoreceptors which “digitize” the visual field.
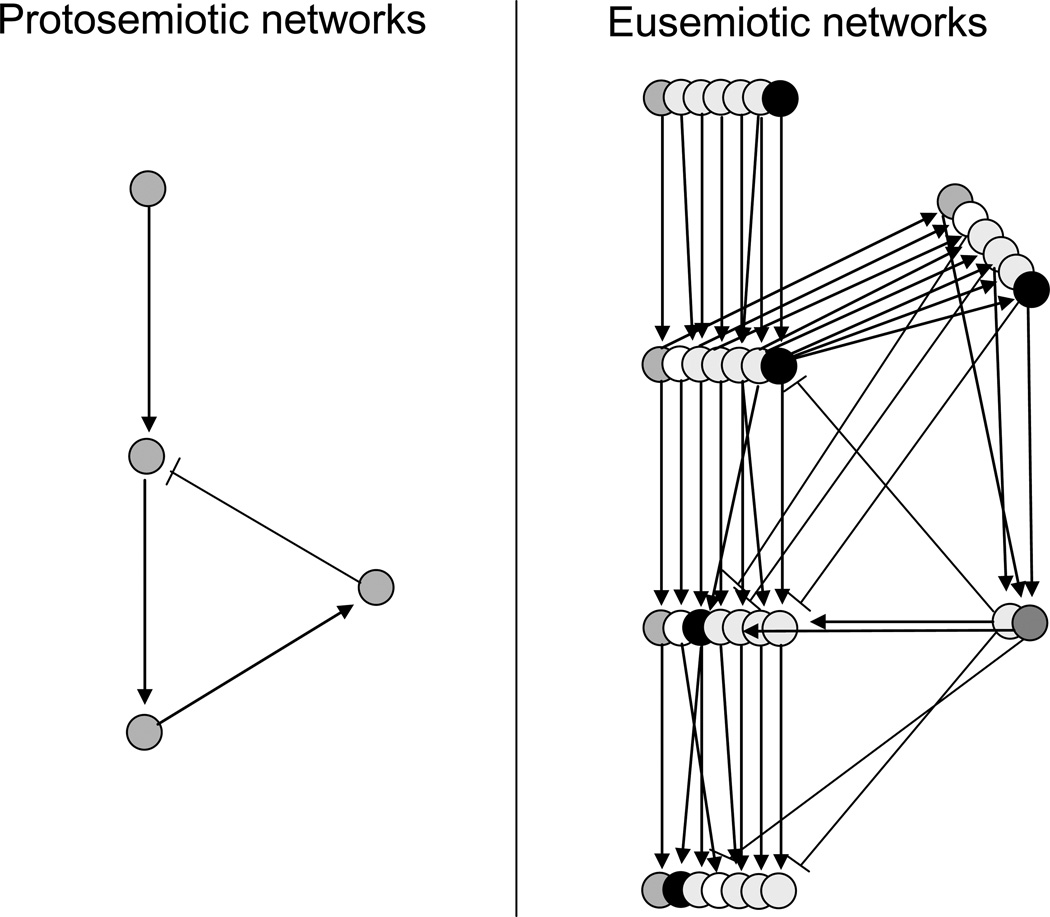
The qualitative difference between protosemiosis and eusemiosis follows from the fact that eusemiosis cannot be reduced to a few molecular signaling pathways. In contrast to relatively simple and non-redundant protosemiotic signaling networks, eusemiosic networks are substantially more complex and highly redundant (Fig. 3). As a result, signals can travel via entirely different pathways in each particular case, but the system outcomes remain the same. Thus, the topology of attractors (Fig. 2) appears more important for understanding eusemiosis than information on specific signaling pathways. Note that attractors may also exist in simple protosemiotic signaling networks, but only in eusemiotic networks, attractors are not reducible to molecular signaling.
Fig. 3.
Molecular signaling networks in protosemiosis and eusemiosis.
Another suggested criterion of a triadic relationship between sign, object, and interpretant is the possibility of mistakes in sign processing (Hoffmeyer and Emmeche 1991). Indeed, glucose receptors in bacteria can be “fooled” by responding to similarly-shaped chemicals that cannot be metabolized. For example, bacterial chemoreceptors stimulated by saccharin may lead the cell away from areas with abundant glucose. Bacteria also have internal gauges that detect glucose shortage and activate alternative metabolic pathways (see section 6 for details), and this signaling pathway can be interpreted as a detection of “error” (Vehkavaara 2003: 577–579). But bacterial response to glucose shortage does not rewire the chemotaxis, which keeps malfunctioning in the presence of saccharin. Thus, the mistake is recognized by a human observer rather than by a bacterium itself. There is no evidence that bacteria can detect such events as mistakes and modify signaling networks accordingly. Instead, bacteria utilize a compensatory mechanism that helped ancestral organisms to avoid starvation.
Mistake (or error) is a logical term, which seems relevant only if an agent has capacities to assign truth values (Boolean or fuzzy) to propositions or hypotheses. In the case of bacteria it is more relevant to talk about success and failure in surviving and reproduction as consequences of associating certain signs with certain actions. Human observers interpret molecular proof-reading (e.g., during synthesis of DNA, RNA, or protein) or refolding of damaged proteins, as error-correction. However, there is no indication that molecular subagents in bacteria can perceive one molecule as a damaged version of another molecule. Most likely, each kind of molecules is detected separately, and in this case, sub-cellular agents have no representation of “error” as a category or truth value.
The qualitative difference between protosemiosis and eusemiosis should not be confused with another qualitative difference between life and non-life (see previous section). For example, it was assumed that normativity (i.e., formal and final causation) required triadic sign relationships that include objects (Bruni 2008; Hoffmeyer 1996). According to this view, there are no signs without objects and no protosemiosis. This argument, however, confuses two different capacities of agents: the capacity to associate signs with objects and normativity. Natural self-interest, which is the most primitive kind of normativity, does not require that agents under study are aware of the objects. Nevertheless, the responses of bacteria are not determined by physical and chemical properties of signaling molecules. Instead, these responses are shaped by agent’s semiotic architecture, which has been refined by genetic selection to achieve increased survival and reproduction of bacteria. Thus, protosemiosis includes normativity, in contrast to physics and chemistry, which study things as they are without intervention of agents.
Protosemiosis is based on automatic sign processing and, in this respect, it resembles computation, which is often excluded from semiosis because computers do not process the meaning of signs (except perhaps artificial intelligence projects). For example, Barbieri wrote: “A computer contains codes but is not a semiotic system” (Barbieri 2008 p. 594). Hoffmeyer considered computers non-semiotic because “these have not (at least yet) been constructed to depend on the creative activity of an analogly coded version interacting with real world processes in such a way as to test the fitness of the digital specifications necessary for its own construction” (Hoffmeyer 1998 p. 34). However, automatic sign processing is not limited to computers. For example a ribosome in a living cell is a nano-scale programmed robot which makes polypeptides based on the mRNA sequence.
There are several reasons why automated processing of signs should be viewed as a special kind of semiosis. First, simplicity and speed are the main advantages of automatic responses; thus, they are often used by organisms as a default way of sign processing. Second, automatic processing of signs does not imply determinism, because automatic responses of living organisms are products of evolution and adjusted to the environment and/or internal context (Kawade 2009). Although automatic processing of signs does not include learning, it emerged historically via either adaptive evolution or prior learning of some agents. For example, the structure and function of the peptidyl transferase center in ribosomes apparently evolved before the availability of proteins within RNA-world organisms and was later reused for protein synthesis (Fox 2010). Similarly, computers and robots emerged via human learning and technological evolution. Based on these considerations we have to accept a broader concept of semiotic agency, where agents differ in the degree of their autonomy and automation of sign processing (Sharov 2010). Agents can manufacture or recruit slave-agents which are then programmed to perform specific functions in an automated way. For example, cells manufacture and utilize non-coding RNA subagents for editing of the RNA and DNA sequences, which may have long-term evolutionary consequences (Witzany 2010: chapter 7).
In summary, categorization and object tracking are two main criteria that distinguish eusemiosis from protosemiosis. Although protosemiosis is automated, it is not reduced to chemical reactions because the responses of agents to proto-signs are products of adaptive evolution driven by natural self-interest. These responses are agent-specific and often context-dependent.
5. Types of Proto-Signs
Proto-signs can be classified into three categories according to their immediate interactions with partner agents (Sharov 2010). Though the classification is inspired by Peirce’s division of iconic, indexical, and symbolic signs, the similarity is somewhat illusory, because here we consider the type of mechanism according to which the functional effect of sign is produced, whereas Peirce considered the relation of signs to their objects. This classification of proto-signs is an example of a meta-agent approach to protosemiosis, because proto-semiotic agents have no capacity for any kind of classification. Categories of proto-signs are discussed here in the order of increasing complexity, where the more simple signs appear to be components of more complex signs.
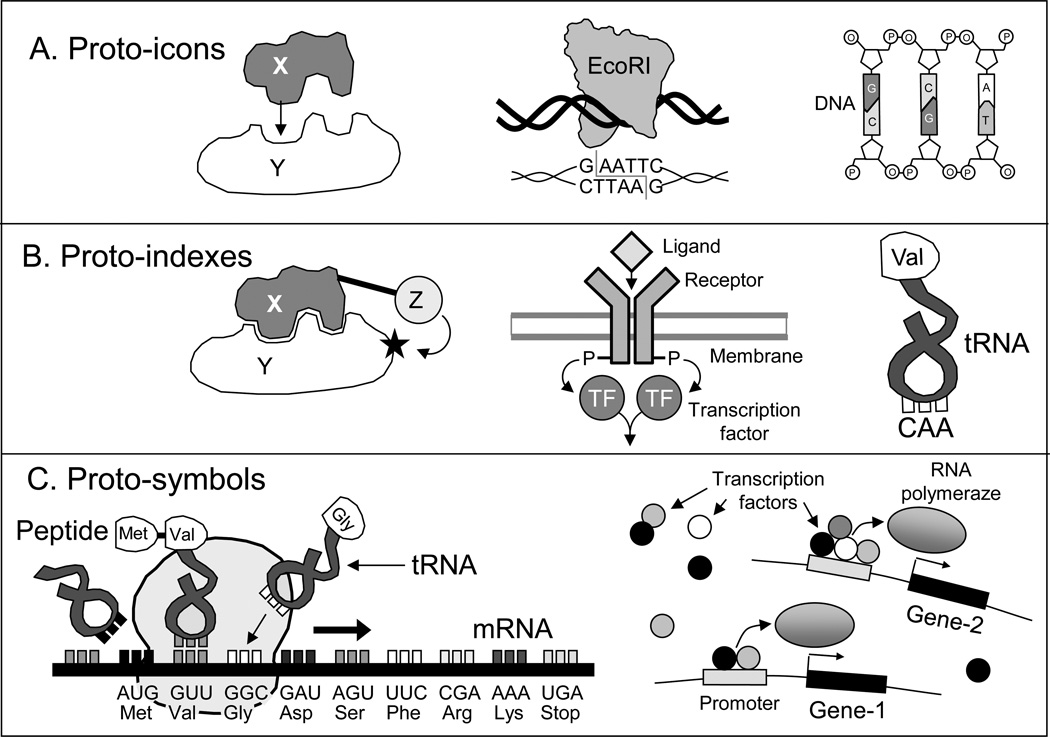
The most simple molecular signs, which we call proto-icons, interact with recipient molecules via one specific action, such as binding of a signal molecule X to another molecule Y via key-and-lock matching of their surfaces (Fig. 4A). Such bindings have to be functional for the cell, and thus, they are followed by some downstream actions or changes that are expected to be beneficial in a given situation. Downstream effects may include the production, activation, or inactivation of another signal molecule with different properties or translocation to another cell compartment.
Fig. 4.
Three types of proto-signs in living cells
Signal molecules that function as proto-icons either come from the environment (e.g., attractants or repellents) or become fabricated within a cell. Examples of the latter kind of signs include chemical labels, such as phosphate, methyl, or ubiquitin, which are attached to various proteins by molecular agents and later detected by other kind of agents or receptors. Leading peptide is a label that serves as an address for protein transportation (e.g., to the nucleus or endoplasmic reticulum). Leading peptide is not attached via protein modification, but instead synthesized together with the protein (Lodish et al. 2000). Some proto-icons are catalytically active, and thus, can modify the partners to which they bind. However, their capacity to bind and catalyze are inseparable and combined in a single action (because otherwise it would be a proto-index rather than proto-icon). For example, a type II restriction enzyme EcoRI in bacteria binds DNA and cuts it at GAATTC palindromic sites (Pingoud and Jeltsch 2001) (Fig. 4A). The DNA molecule is an example of a composite proto-icon which is build as a sequence of elementary blocks-nucleotides (Fig. 4A). Although it includes multiple components, the binding action between DNA strands is still single and thus satisfies the criterion of a proto-icon.
Proto-index carries two or more independent functional subunits, which are involved in different (and often sequential) actions (Fig. 4B). For example, type I restriction enzymes in bacteria include three subunits: HsdR is a nuclease which cuts DNA, HsdS binds a specific DNA motif, and HsdM (methyltransferase) can methylate the DNA molecule (Murray 2000). Membrane receptors are proto-indexes that combine an external sensing domain for recognition of signal molecules (ligands) with internal domain for activating secondary messengers (e.g., cAMP or phosphorylated transcription factor) (Fig. 4B). Proto-indexes may also function as adaptors to interconnect unrelated substrates. For example, tRNA has the anticodon loop that binds to mRNA and the acceptor stem that is used by acyl-transferase to attach an aminoacid. A transcription factor is also a proto-index because it combines a DNA-binding domain with activation domain. The DNA-binding domain is needed for sequence-specific binding to the promoters of target genes, whereas the activation domain interacts with RNA-polymerase either directly or indirectly and activates the transcription.
Proto-indexes often combine a catalytic domain with a regulatory domain, which modifies the level of catalytic activity. For example, the transcription activity of RNA-polymerase II is repressed if its CTD domain is not sufficiently phosphorylated. Upon double phosphorylation of Ser2,5 repeats within the CTD by a specific kinase, the RNA-polymerase II becomes active and successfully transcribes genes (Phatnani and Greenleaf 2006).
Proto-indexes can interconnect different processes within cells, and such connections comprise the regulatory network that grounds the meaning of molecular signals. Recently, Hoffmeyer introduced the term “semiotic scaffolding” to describe the network of semiotic controls that are tuned to the needs of the system and support meaningful interpretation of signs (Hoffmeyer 2007). At the molecular level, semiotic scaffolding is represented by signaling networks welded via proto-indexes. For example, a P300 protein is an important component of enhancers that control the expression of nearby genes. It can bind to various transcription factors, including steroid receptors, CREB, MYB, p53, and interferon receptors (Vo and Goodman 2001). In addition, P300 acetylates histones and opens the chromatin, which makes DNA accessible to various transcription factors. Obviously, P300 is a hub in the cellular signaling network that integrates multiple incoming signals, although many aspects of its function are still unknown.
Finally, proto-symbols are members of a family of similarly-structured molecular signs that are processed uniformly by the same kind of subagents and the same set of heritable adapters (proto-indexes). For example, mRNA sequences are processed by ribosomes and a set of aminoacyl-tRNA adapters in order to make proteins (polypeptides) (Fig. 4C). As a ribosome moves along the mRNA (each step by 3 nucleotides), a triplet of nucleotides is bound by a tRNA molecule that carries a specific aminoacid, and the ribosome links the incoming aminoacid into a growing polypeptide chain. But it would be wrong to consider aminoacid as an object for a sign represented by a triplet of nucleotides (in the sense of a triadic sign relation). A ribosome does not recognize aminoacids as objects. Instead, a triplet of nucleotides induces a combined action of a ribosome together with corresponding tRNA, and this action results in the addition of an aminoacid to the polypeptide. The relationships between proto-symbols and its products are not fixed but can be modified by cellular subagents. For example, before mRNA molecules leave the nucleus they are processed by spliceosomes, which cut out segments (introns) that are usually marked by nucleotides GU at the start and AG and the end. This process (i.e., splicing) is not deterministic and can be modified on demand by regulatory factors. Thus, many genes have multiple alternative splicing patterns, and can produce different proteins from the same gene sequence (Lodish et al. 2000).
Another example of proto-symbols is a family of transcription factors that control the activity of various genes. Their action is mediated by a common agent, RNA polymerase, and by a set of adaptors represented by promoters of genes that carry DNA motifs, to which transcription factors bind with high affinity. In contrast to protein synthesis, where the set of adaptors is nearly universal in the whole tree of life, promoter sequences are conserved only within evolutionary-related species of organisms. Thus, transcription factors may not work properly if introduced into cells of unrelated species.
The relationship between proto-symbols and their products is contingent because it is based on a heritable accident in evolution, which resulted in the emergence of a given set of adapters. This “conventional” property makes proto-symbols similar to symbols used in human communication. However, in contrast to cognitive symbols, the processing of proto-symbols is based on a “heritable convention” rather than socially-mediated convention. There is even a deeper similariry between the genetic code and human language: both have a hierarchical structure (i.e., nucleodite, codon, cistron, promoter, enhancer, chromatin loops), they have syntax and “generative grammar” (García 2005), and both make a network of mutually-defined meanings. Thus, the genetic code can be viewed as a kind of language system (Pattee 2008). Because genetic communication belongs to protosemiosis it is logical to call it “proto-language”7 (Sharov 2010), following our convention to add prefix “proto” to protosemiotic notions.
Contemporary organisms use all three kinds of proto-signs for their molecular communication, however primordial living systems had no proto-symbols according to proposed scenarios of life origin. For example, the RNA-world scenario assumes that life started from self-replicating RNA molecules and did not include protein synthesis (Schuster 1993). After spontaneous self-folding, RNA acquired catalytic properties and were able to support metabolism and replication of other RNA molecules. Another scenario of the origin of life starts with even more simple coenzyme-like molecules, which inhabited oil microspheres in water and encoded surface properties of these microspheres (Sharov 2009: 1843). Such systems could have evolved by accumulating additional heritable and functional coenzyme-like molecules, which eventually became organized into RNA-like polymers with template-based replication. Interestingly, both scenarios require proto-indexes to interconnect otherwise separate processes. For example, coenzyme-like molecules had to combine a catalytic part capable of oxidizing hydrocarbons (alkenes) with a binding part to anchor the molecule to the surface of an oil microsphere. These molecules may also need additional components to facilitate their autocatalytic self-assembly. Similarly, in the RNA-world scenario, RNA molecules had to support several kinds of interactions. They served as a template for replication, guided a unique way of self-folding, and finally played the role of catalysts. Proto-icons alone cannot make a functional living system because they have no capacity to organize a semiotic scaffold.
6. How to Study Protosemiosis
Recognition of protosemiosis as a primitive kind of sign relationship prompts the question on how to study it, and what benefits can we expect from the use of semiotic terminology? The latter question is especially important because most biologists and biochemists feel that semiotics only renames things that are already known. The immediate sign relationships (i.e., signal-response effects) in protosemiosis are rather simple and, in most cases, already well studied in molecular biology. Thus, we think it is more interesting to focus on the “extended” sign relationships that include dynamic downstream signaling, contribution to system goals, and evolutionary aspects of cell signaling.
To explore these options we propose a heuristics, which can be called “protosemiotic projection”: human rational solution for a specific task/function of a cell may help to understand the real solution of this task as it is implemented in living cells. This approach provides a projection of a bacterial protosemiosis into a human knowledge-base while keeping a clear distinction between the bacterial and human “points of view”. Protosemiotic projection does not mean that any ad-hoc hypothesis immediately explains the mechanism and evolution of a given protosemiotic system. Instead, we think that human understanding of the phenomenon is sequentially adjusted as each hypothesis is formulated and tested.
As it has been discussed, bacteria cannot associate incoming signals with objects or goals (as objects), however human observers can reconstruct “virtual” objects and goals associated with bacterial signs. For example, cAMP is used by bacteria as a switch to alternative metabolic pathways in the case of glucose depletion. Bacteria, however, have no abstract notion of glucose and its depletion. Instead, the signal comes from the protein that normally brings glucose into the cell. If this protein appears idle (i.e., in the absence of glucose), then it activates the synthesis of cAMP (Bettenbrock et al. 2007). In turn, cAMP binds to a specific intra-cellular cAMP receptor protein (CRP), which activates the lac operon responsible for lactose processing. Because this mechanism helped generations of bacteria to survive in nutrient-poor environments, cAMP is not a sign of glucose depletion (from the bacterial perspective) but a sign that differentiates between “well-being” and “dying” states Vehkavaara 2003; 576–579). The human observer, however, can interpret the action of cAMP sign in terms of a triadic sign relation, where the depletion of glucose is an object of the cAMP sign, and the goal of the metabolic switch (interpretant) is to prevent the interruption of energy supply. Such interpretation of proto-semiotic sign can be viewed as a hypothesis that requires experimental validation. For example, we can check if cAMP signaling indeed correlates well with the deficiency of glucose. It may happen that some other conditions also induce cAMP signaling, and therefore our assumption that cAMP always stands for glucose depletion may appear incorrect. Another possible experiment is to disrupt cAMP signaling with antagonists (Van Haastert et al. 1984) or by the knockdown of CPR receptor. Then we can test if the disruption of cAMP signaling would cause a lower rate of population increase in glucose-depleted environments. Finally, we can set experiments with unusual combinations of resources and check if the cAMP signaling pathway is able to keep cells alive. The results of these experiments should be evaluated and used to refine the hypothesis of the cAMP action. Considering that human intuition plays an important role in such iterative method of inquiry, we cannot guarantee that it leads to a reproducible object and goal of molecular action. However, it may generate reasonable explanations with testable consequences and promote further analysis of cell biology. This example shows that biosemiotics should include both object-agent perspective and meta-agent perspective, as defined in (Vehkavaara 2002: 299–303), in studying sign processes in living organisms.
The contribution of sign relations to the “natural self-interest” (i.e., survival and reproduction) or derivative goals (e.g., capturing resources) has been called “value” (Sharov 1992: 354–356). We believe that studying pragmatic values of proto-signs would help to explain the function and evolution of molecular processes. Theoretically, the value of a sign relationship can be quantified by disrupting the signaling pathways (e.g., by antagonist molecules) and monitoring the success rate of cells in reaching specific goals. However, in practice, such measurements may yield ambiguous results because the interruption of signaling can be immediately compensated by other processes. In addition, normal levels of signaling may be required to support structural integrity of a cell, therefore its full disruption would be non-physiological. Although the quantification of values is difficult, it may be still possible to evaluate its ranges or at least distinguish between positive and negative values.
The advantage of semiotic terminology in studying molecular interactions can be seen in the possibility to focus on large-scale goals of evolving agents. Thus, the semiotic inquiry starts from goals/tasks rather than from molecular interactions (as it is common in biology). We can ask a question: how does it feel to be a bacterium? What kind of inputs does it get from the environment? What are the strategies and mechanisms for entering dormancy state in stress conditions? How does it counteract to invading bacteriophages? We have only partial answers to these questions. A comprehensive analysis requires integration of a vast literature on individual molecular signals. Moreover, new experiments have to be designed and executed.
One of the unsolved questions is whether the notion of protosemiosis can be expanded beyond molecular signaling and applied to such processes as automatic neural processing of signs in animals and humans. In practice, we do not exercise freedom in the interpretation of ordinary signs (e.g., common words or traffic signs) that we meet in everyday life. Only when a sign is unfamiliar, or appears in unusual context, then we hesitate and scan for possible alternative interpretations. However, the differentiation between protosemiotic and eusemiotic processes in complex multi-level systems is not trivial. For example, the jaw opening reflex, which is responsible for mouth opening after biting a solid particle or a tongue, is fully automated and can be compared to protosemiosis. But the underlying neural processes possibly include intracellular categorization tools and object tracking (e.g., tracking of synapses), and therefore they may belong to eusemiosis.
7. Conclusions
This paper outlines the features of the most primitive type of sign processes in nature, which we call “protosemiosis”, following Prodi. In protosemiosis, agents respond to signs by changes in their actions without prior associating them with objects. Thus, the traditional definition of sign as a representation of an object may not be universal. The reason why we consider protosemiotic signs (i.e., proto-signs) as signs, is because the response of agents is specific and contributes to their “natural self-interest” (i.e., inceases the rates of survival and self-reproduction). Protosemiosis differs from the advanced type of semiosis (i.e., “eusemiosis”) because it is not based on categorization and object tracking capacities in agents. These capacities are essential properties of the minimal mind and are necessary for associating signs with object categories. We believe that the notion of protosemiosis will help to introduce semiotic concepts into the developing fields of molecular and synthetic biology. Thus, without studying protosemiosis and the recognition of its special characters (i.e., no association with objects and no categorization), the biosemiotics theory would be incomplete.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging, and by the School of Social Sciences and Humanities, University of Tampere. The content of this paper is not endorsed or suggested by the funding agencies. We thank anonymous reviewers for valuable input about this paper.
Footnotes
For citations of Peirce we use abbreviations: CP = Collected papers, EP = Essential Peirce, and W = Writings.
Note that Peirce’s notion of object is not related to agents who interact with them. He used only the meta-agent perspective assuming that objects are the same for every agent.
Detailed analysis of the term “agent” can be found in (Sharov 2010).
Known epigenetic mechanisms can support adaptive learning within individual cells as follows from a hypothetical model (Sharov 2013).
The notion of fitness in neo-Darwinism (i.e., the relative rate of self-reproduction) provides a quantitative measure of natural self-interest but it does not capture its functional aspects. For example, mules cannot reproduce and their fitness is zero, but yet they are alive and capable of goal-directed actions at both molecular and behavioral levels. In fact, mules have inherited their goals and capacity for interpreting signs from their self-reproducing parents. But self-reproduction is necessary for the continuation of life lineages and for adaptive evolution.
By categorization we mean recognition and differentiation of objects by agents based on a large set of perceived properties, which cannot be reduced to a small number of logical gates (e.g., “AND”, “OR”) and contributes to the functional goal-directed activity of the agent (Sharov 2013).
It should not to be confused with the linguistic notion of proto-language (which is better called “ancestral language”).
References
- Alexander V. Creativity: self-referential mistaking, not negating. Biosemiotics. 2013;6(2):253–272. [Google Scholar]
- Amit DJ. Modeling Brain Function: The World of Attractor Neural Networks. Cambridge, New York: Cambridge University Press; 1989. [Google Scholar]
- Anderson M, Deely J, Krampen M, Ransdell J, Sebeok TA, Uexküll T. von. A semiotic perspective on the sciences: Steps toward a new paradigm. Semiotica. 1984;52(1/2):7–47. [Google Scholar]
- Barbieri M. The Organic Codes: An Introduction to Semantic Biology. Cambridge, New York: Cambridge University Press; 2003. [Google Scholar]
- Barbieri M. Biosemiotics: a new understanding of life. Naturwissenschaften. 2008;95(7):577–599. doi: 10.1007/s00114-008-0368-x. [DOI] [PubMed] [Google Scholar]
- Barbieri M. Three types of semiosis. Biosemiotics. 2009;2(1):19–30. [Google Scholar]
- Bettenbrock K, Sauter T, Jahreis K, Kremling A, Lengeler JW, Gilles ED. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J Bacteriol. 2007;189(19):6891–6900. doi: 10.1128/JB.00819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhard MH. Representational content in humans and machines. Journal of Experimental and Theoretical Artificial Intelligence. 1993;5:285–333. [Google Scholar]
- Bickhard MH. Levels of representationality. Journal of Experimental and Theoretical Artificial Intelligence. 1998;10(2):179–215. [Google Scholar]
- Bickhard MH. Function, anticipation, representation. In: Dubois D, editor. Computing Anticipatory Systems. CASYS 2000 - Fourth International Conference. Melville, NY: American Institute of Physics; 2001. pp. 459–469. [Google Scholar]
- Brier S, Joslyn C. What does it take to produce interpretation? Informational, Peircean and code-semiotic views on biosemiotics. Biosemiotics. 2013;6(1):143–159. [Google Scholar]
- Bruni LE. Cellular semiotics and signal transduction. In: Barbieri M, editor. Introduction to Biosemiotics. The New Biological Synthesis. Dordrecht: Springer; 2008. pp. 365–407. [Google Scholar]
- Christiansen PV. Habit formation as symmetry breaking in the early universe. Sign Systems Studies. 2002;30(1):347–360. [Google Scholar]
- Deacon TW. The Symbolic Species: The Co-Evolution of Language and the Brain. 1st ed. New York: W.W. Norton; 1997. [Google Scholar]
- Deely JN. Basics of Semiotics (Series: Advances in Semiotics) Bloomington, IN: Indiana University Press; 1990. [Google Scholar]
- Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2(9):a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García ÁL. The Grammar of Genes: How the Genetic Code Resembles the Linguistic Code. Bern, Germany: Peter Lang; 2005. [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 1979;205(1161):581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer J. Signs of Meaning in the Universe. Bloomington, IN: Indiana University Press; 1996. [Google Scholar]
- Hoffmeyer J. Surfaces inside surfaces. Cybernetics and Human Knowing. 1998;5(1):33–42. [Google Scholar]
- Hoffmeyer J. Semiotic scaffolding of living systems. In: Barbieri M, editor. Introduction to Biosemiotics. The New Biological Synthesis. Dordrecht: Springer; 2007. pp. 149–166. [Google Scholar]
- Hoffmeyer J. Biosemiotics: An Examination into the Signs of Life and the Life of Signs. Scranton, PA: University of Scranton Press; 2008. [Google Scholar]
- Hoffmeyer J, Emmeche C. Code-duality and the semiotics of nature. In: Anderson M, Merrell F, editors. On Semiotic Modeling. Berlin: Mouton de Gruyter; 1991. pp. 117–166. [Google Scholar]
- Kawade Y. On the nature of the subjectivity of living things. Biosemiotics. 2009;2(2):205–220. [Google Scholar]
- Krampen M. Phytosemiotics. Semiotica. 1981;36(3/4):187–209. [Google Scholar]
- Kull K. Vegetative, animal, and cultural semiosis: the semiotic threshold zones. Cognitive Semiotics. 2009;4:8–27. [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Darnell J. Molecular cell biology. 4th ed. New York: W. H. Freeman and Co; 2000. [Google Scholar]
- Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64(2):412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattee HH. Physical and functional conditions for symbols, codes, and languages. Biosemiotics. 2008;1(2):147–168. [Google Scholar]
- Peirce CS. Collected Papers of Charles Sanders Peirce. 1–8. Cambridge: Harvard University Press; 1931–1935. [Google Scholar]
- Peirce CS. Writings of Charles S. Peirce: A Chronological Edition. Vol. 3. Bloomington, IN: Indiana University Press; 1986. [Google Scholar]
- Peirce CS. The Essential Peirce: Selected Philosophical Writings. Vol. 2. Bloomington, IN: Indiana University Press; 1998. [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20(21):2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29(18):3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9(3):153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Premack DG, Woodruff G. Does the chimpanzee have a theory of mind? The Behavioral and Brain Sciences. 1978;1:515–526. [Google Scholar]
- Prodi G. Material bases of signification. Semiotica. 1988;69(3/4):191–241. [Google Scholar]
- Prodi G. Signs and codes in immunology. In: Favareau D, editor. Essential Readings in Biosemiotics. Dordrecht: Springer; 2010. pp. 328–335. [1988] [Google Scholar]
- Saussure Fd, Bally C, Sechehaye A, Riedlinger A. Course in General Linguistics. LaSalle, Ill: Open Court; 1986. [1916] [Google Scholar]
- Schuster P. RNA based evolutionary optimization. Orig Life Evol Biosph. 1993;23(5–6):373–391. doi: 10.1007/BF01582087. [DOI] [PubMed] [Google Scholar]
- Sebeok TA. Communication in animals and men. Language. 1963;39:448–466. [Google Scholar]
- Sebeok TA. Contributions to the Doctrine of Signs. Vol. 5. Bloomington, IN: Indiana University Press; 1976. Studies in semiotics. [Google Scholar]
- Sharov AA. Biosemiotics: functional-evolutionary approach to the problem of the sense of information. In: Sebeok TA, Umiker-Sebeok J, editors. Biosemiotics. The Semiotic Web 1991. New York: Mouton de Gruyter; 1992. pp. 345–373. [Google Scholar]
- Sharov AA. Coenzyme autocatalytic network on the surface of oil microspheres as a model for the origin of life. International Journal of Molecular Sciences. 2009;10(4):1838–1852. doi: 10.3390/ijms10041838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Functional information: Towards synthesis of biosemiotics and cybernetics. Entropy. 2010;12(5):1050–1070. doi: 10.3390/e12051050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Minimal mind. In: Swan L, editor. Origins of Mind. Dordrecht: Springer; 2013. pp. 343–360. [Google Scholar]
- Taborsky E. The six semiosic predicates. S.E.E.D. (Semiotics, Evolution, Energy, and Development) 2003;3(2):5–23. [Google Scholar]
- Uexküll J. von. A stroll through the worlds of animals and men: A picture book of invisible worlds. In: Schiller CH, editor. Instinctive Behaviour: The Development of a Modern Concept. New York: International Universities Press, Inc; 1957. pp. 5–80. [Google Scholar]
- Uexküll J. von. The theory of meaning. Semiotica. 1982;42(1):25–82. [1940] [Google Scholar]
- Van Haastert PJ, Van Driel R, Jastorff B, Baraniak J, Stec WJ, De Wit RJ. Competitive cAMP antagonists for cAMP-receptor proteins. J Biol Chem. 1984;259(16):10020–10024. [PubMed] [Google Scholar]
- Vehkavaara T. Why and how to naturalize semiotic concepts for biosemiotics. Sign Systems Studies. 2002;30(1):293–313. [Google Scholar]
- Vehkavaara T. Natural self-interest, interactive representation, and the emergence of objects and Umwelt. Sign Systems Studies. 2003;31(2):547–587. [Google Scholar]
- Vehkavaara T. From the logic of science to the logic of the living. The relevance of Charles Peirce to biosemiotics. In: Barbieri M, editor. Introduction to Biosemiotics : the New Biological Synthesis. Dordrecht: Springer; 2007. pp. 260–262. [Google Scholar]
- Vehkavaara T. Limitations on applying Peircean semeiotic. Biosemiotics as applied objective ethics and esthetics rather than semeiotic. Journal of Biosemiotics. 2006;1(2):269–308. [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276(17):13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Witzany G. Biocommunication and Natural Genome Editing. Dordrecht: Springer; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]