Abstract
Telomere extension has been proposed as a means to improve cell culture and tissue engineering and to treat disease. However, telomere extension by nonviral, nonintegrating methods remains inefficient. Here we report that delivery of modified mRNA encoding TERT to human fibroblasts and myoblasts increases telomerase activity transiently (24–48 h) and rapidly extends telomeres, after which telomeres resume shortening. Three successive transfections over a 4 d period extended telomeres up to 0.9 kb in a cell type-specific manner in fibroblasts and myoblasts and conferred an additional 28 ± 1.5 and 3.4 ± 0.4 population doublings (PDs), respectively. Proliferative capacity increased in a dose-dependent manner. The second and third transfections had less effect on proliferative capacity than the first, revealing a refractory period. However, the refractory period was transient as a later fourth transfection increased fibroblast proliferative capacity by an additional 15.2 ± 1.1 PDs, similar to the first transfection. Overall, these treatments led to an increase in absolute cell number of more than 1012-fold. Notably, unlike immortalized cells, all treated cell populations eventually stopped increasing in number and expressed senescence markers to the same extent as untreated cells. This rapid method of extending telomeres and increasing cell proliferative capacity without risk of insertional mutagenesis should have broad utility in disease modeling, drug screening, and regenerative medicine.—Ramunas, J., Yakubov, E., Brady, J. J., Corbel, S. Y., Holbrook, C., Brandt, M., Stein, J., Santiago, J. G., Cooke, J. P., Blau, H. M. Transient delivery of modified mRNA encoding TERT rapidly extends telomeres in human cells.
Keywords: nucleoside modified mRNA, telomerase, senescence, proliferative capacity
Telomeres comprise tandem DNA repeats that, with associated proteins collectively named shelterin, protect chromosome ends from acting as damaged DNA (1, 2). Telomeres shorten over time due in part to incomplete chromosomal end replication as well as other factors, including oxidative damage (3, 4). Telomerase is a ribonucleoprotein that extends telomeres and consists of a protein component, TERT, which complexes with an RNA component (TERC) (5–8). Telomerase is active in many cell types including stem and progenitor cells, yet over the lifetime of an individual telomeres shorten in most tissues (9–11).
When telomeres become sufficiently short, tumor suppressor p53 and DNA damage response pathways are activated, levels of the transcriptional regulator PPARγ coactivator 1-α and -β (PGC1-α and -β) are reduced leading to mitochondrial dysfunction, chromosome-chromosome fusions can occur leading to malignancy, and cells may apoptose or senesce (12–16). Genetic mutations in TERT and other genes involved in telomere length maintenance result in diseases such as aplastic anemia and dyskeratosis congenita (17, 18). Further, we recently showed that shortened telomeres underlie the progression of Duchenne muscular dystrophy (DMD) (16, 19) and that telomere extension averts endothelial cell senescence, which is associated with atherosclerosis and hypertension (20, 21).
A means to safely extend telomeres would benefit cell and tissue engineering by increasing the number of PDs and cumulative cell numbers achieved in culture (3, 22). This need is underscored by reports that short telomeres destabilize stem cell differentiation (23) and inhibit reprogramming via p53 activation (24–26). Short telomeres also limit replicative capacity essential to cell therapies using transplanted hematopoietic stem cells, cardiac progenitors, and induced pluripotent stem cell (iPSC)-derived retinal pigment epithelial cells (27–30). We found that myoblasts (progenitors) from teenage DMD patients and stem cells from the DMD mouse model were limited in their regenerative capacity as they typically underwent only a few divisions in culture before entering replicative senescence. This is in stark contrast to the extensive PDs typical of myoblasts or stem cells from normal age-matched controls (19, 31). iPSC telomere lengths are short compared with embryonic stem cells (32, 33). Furthermore, iPSCs derived from patients with diseases mediated by impaired telomere maintenance exhibit reduced self-renewal and survival (34, 35). Moreover, due to a body of literature linking telomere shortening to several genetic and age-related diseases, several investigators have proposed the use of telomere extension as a preventive or therapeutic intervention (17, 22, 36–42). Clearly, there is an unmet need for an efficacious and safe way to extend telomeres.
For cell therapy applications, avoiding the risk of cell immortalization is of paramount importance. To this end, transient, rather than constitutive, telomerase activity may be advantageous for safety, especially if the elevated telomerase activity is not only brief but extends telomeres sufficiently to overcome the need for continuous treatment. Current methods of extending telomeres include viral delivery of TERT under the control of an inducible promoter, delivery of TERT using vectors based on adenovirus and adeno-associated virus, and small molecule activators of telomerase (22, 40, 43–48). Here we provide an alternative that offers the benefits of transient telomerase activation combined with rapid telomere extension.
Modified nucleoside-containing mRNA is nonintegrating and has recently been used by others to transiently elevate levels of diverse proteins encoded by the mRNA (49–51). Here we show in two cell types that delivery of modified mRNA encoding TERT to human cells avoids immortalization, yet transiently increases telomerase activity, rapidly extends telomeres, delays expression of senescence markers, and increases proliferative capacity.
MATERIALS AND METHODS
mRNA template generation and synthesis
To generate modified mRNA encoding GFP, TERT, and catalytically inactive (CI) TERT, their respective open reading frames (ORFs) were inserted into the MCS of a starting plasmid containing the T7 promoter, the 5′-UTR of human β-globin (HBB), the MCS, the 3′-UTR of HBB, a 151 bp poly-A sequence, and a restriction site for linearization with a type IIS restriction enzyme following the poly-A sequence. TERT-3xFLAG had 3 FLAG sequences inserted at the C terminus of TERT. The resulting intermediate plasmids were sequenced, linearized, and transcribed using the buffer and RNA polymerase from the MEGAscript T7 Kit (Ambion, Austin, TX, USA), and a custom mix of canonical and noncanonical nucleotides (TriLink BioTechnologies, San Diego, CA, USA) in which the final nucleotide concentrations per 40 µl IVT reaction were 7.5 mM for each of ATP, 5-methylcytidine-5′-triphosphate (m5C), and pseudouridine-5′-triphosphate (Ψ), 1.5 mM for GTP, and 6 mM for the cap analog (ARCA) (New England Biolabs, Ipswitch, MA, USA), or a molar ratio of ATP:m5C:Ψ:GTP:ARCA of 1:1:1:0.2:0.8. The IVT products were treated with Antarctic Phosphatase (New England Biolabs). The size and integrity of the mRNA products were verified using denaturing agarose gel electrophoresis (Supplemental Fig. 4). The wild-type human TERT ORF used to generate the DNA templates for mRNA synthesis is identical to the NCBI human TERT transcript variant 1 (reference sequence NM_198253.2). The ORF was generated from the pBABE-neo-hTERT plasmid (52) (plasmid 1774, Addgene, Cambridge, MA, USA). The pBABE-neo-hTERT plasmid had a nonsilent mutation at residue 516 in the QFP motif of TERT, a motif associated with multimerization and TERT interaction with TERC RNA, and thus to avoid the possibility of artifacts due to this mutation, we made the sequence identical to the NCBI reference sequence by correcting the mutation with the change G516D. The CI TERT mutant was generated from the TERT sequence by introducing the mutation D712A.
Cell culture and treatment
Human primary fetal lung MRC5 fibroblasts were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA) at passage 14. ATCC does not indicate the PD number; thus, our PD values cited herein refer to the number of PD after receipt of cells from ATCC. MRC5 cells were cultured in DMEM with 20% fetal bovine serum and penicillin-streptomycin. Human 30-yr-old primary skeletal muscle myoblasts (Lonza, Allendale, NJ, USA) were cultured in SkGM-2 medium (Lonza) according to the vendor’s instructions. PDs were calculated as the base 2 log of the ratio between cells harvested and cells plated at the previous passaging and were considered to be 0 if fewer cells were harvested than plated. Cells were transfected with modified TERT mRNA using Lipofectamine RNAiMax (Life Technologies, Grand Island, NY, USA) prepared in Opti-MEM Reduced Serum Media (Life Technologies) and added to the cells in a 1:5 v:v ratio with their normal medium to achieve the final concentrations indicated herein. GM847 cells were obtained from SACRI Antibody Services, University of Calgary.
Telomerase activity measurement
Twenty-four hours after the start of the transfection period, cells were harvested and lysed in CHAPS buffer. The telomeric repeat amplification protocol (TRAP) assay was performed using a modified version of the TRAPeze kit (EMD Millipore, Billerica, MA, USA), in which the primers and polymerase were added after, rather than before, the step during which the artificial telomere substrate is extended. The PCR program was 94°C 30 s/59°C 30 s/72°C 45 s for 30 cycles, and the products were run on a 15% polyacrylamide gel in 0.5× TBE stained with SYBR Gold Nucleic Acid Gel Stain (Life Technologies). The time course of telomerase activity was performed using the TRAPeze RT kit (EMD Millipore).
Western blot
Protein was harvested by washing cells once with PBS and then lysing cells in RIPA buffer (Cell Signaling Technology, Danvers, MA, USA). Protein was run on NuPAGE Novex Tris-Acetate Gels (Life Technologies), transferred to PVDF or nitrocellulose membrane for 2 h at 35 V, then hybridized to anti-α-tubulin (Sigma-Aldrich, St. Louis, MO, USA) at 1:10,000 or GAPDH (Cell Signaling Technology) at 1:2000 and anti-TERT antibody (Abcam, Cambridge, MA, USA, 32020 at 1:1000; or Rockland Immunochemicals, Gilbertsville, PA, USA, 600-401-252S at 1:500) or anti-FLAG antibody (Sigma-Aldrich) at 1:1000 and incubated overnight at 4°C or for 1 h at room temperature. Detection was performed using infrared (680 and 800 nm) antibodies (LI-COR, Lincoln, NE, USA) and the Odyssey imager (LI-COR). Total intensity of each band was quantified using ImageJ (National Institutes of Health, Bethesda, MD, USA). The intensity of each anti-TERT band was normalized by its corresponding α-tubulin band.
Flow cytometry
Cells were harvested 24 h after transfection with the indicated doses (Supplemental Fig. 1a–c) of TERT mRNA and stained with anti-TERT antibody 32020 (Abcam) at 1:500.
Telomere length measurement
Genomic DNA was extracted using phenol chloroform and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies). Telomere length analysis was performed at SpectraCell Laboratories Inc. (Houston, TX, USA) using a Clinical Laboratory Improvement Amendments-approved, high-throughput qPCR assay, essentially as described by Cawthon et al. (53, 54). The assay determines a relative telomere length by measuring the factor by which the sample differs from a reference DNA sample in its ratio of telomere repeat copy number to singe gene (36B4) copy number. This ratio (T/S ratio) is thought to be proportional to the average telomere length. All samples were run in at least duplicate with at least 1 negative control and 2 positive controls of 2 different known telomere lengths (high and low) and an average variance of up to 8% was seen. The results were reported as a telomere score equivalent to the average telomere length in kilobases.
Telomere length measurement by monochrome multiplex qPCR method
Telomere length was measured using a modified version of the monochrome multiplex qPCR (MMqPCR) protocol developed by Cawthon (54) with the following changes. Additional PCR preamplification cycles were added to make the telomere product amplify earlier, widening the gap between telomere and single-copy gene signals; a mixture of 2 Taq polymerases was experimentally determined to result in better PCR reaction efficiencies than each on its own; reducing the SYBR Green concentration from 0.75× to 0.5× resulted in earlier signal. Genomic DNA was isolated from cells using the PureGene kit (Qiagen, Germantown, MD, USA) with RNase digestion, quantified using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA), and 10–40 ng was used per 15 μl qPCR reaction performed in quadruplicate using a LightCycler 480 PCR System (Roche, Basel, Switzerland). A serial dilution of reference DNA spanning 5 points from 100 to 1.23 ng/μl was included in each assay to generate a standard curve required for sample DNA quantification. The final concentrations of reagents in each 15 μl PCR reaction were: 20 mM Tris-HCl pH 8.4, 50 mM KCl, 3 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphate, 1 mM DTT, 1 M betaine (Affymetrix, Santa Clara, CA, USA), 0.5× SYBR Green I (Life Technologies), 0.1875U Platinum Taq (Life Technologies), 0.0625× Titanium Taq (Clontech Laboratories, Mountain View, CA, USA), and 900 nM each primer (telg, telc, hbgu, and hbgd primer sequences specified by Cawthon) (54). The thermal cycling program was 2 min at 95°C; followed by 6 cycles of 15 s at 95°C, 15 s at 49°C; followed by 40 cycles of 15 s at 95°C, 10 s at 62°C, 15 s at 72°C with signal acquisition, 15 s at 84°C, and 10 s at 88°C with signal acquisition. The Roche LightCycler 480 software was used to generate standard curves and calculate the DNA concentrations of telomere and single-copy genes for each sample. T/S ratios were calculated for each sample replicate, and the results averaged to yield the sample T/S ratio, which was calibrated using blinded replicate samples of reference cells sent to SpectraCell as described above. The independently obtained relative values of T/S ratios measured using MMqPCR and by SpectraCell for the same samples were highly consistent (correlation coefficient = 0.97, P < 0.001).
Telomere length analysis by quantitative FISH
Quantitative FISH (Q-FISH) staining was performed on metaphase spreads prepared by exposing cells to colcemid for 2–6 h, swelling them in 75 mM KCl for 30 min at room temperature, gradually adding Carnoy’s fixative (3:1 methanol:acetic acid), centrifuging and resuspending in fixative dropwise 3 times, and dropping on slides, which were then stained with AlexaFluor 555 telomere PNA probe and AlexaFluor 647 Centromere probe. Cells were mounted in SlowFade Gold (Life Technologies) in DAPI. Three-dimensional image stacks of cells were acquired at 200 nm intervals on a DeltaVision photosensor-compensated microscope using a ×100 oil objective. Telomere intensities were quantified using 3-dimensional Q-FISH using custom software written as an ImageJ plug-in. For each telomere or centromere in a cell outlined by the user, the software identifies the image in the 3-dimensional stack of images in which the telomere or centromere is in best focus, and integrates the intensity of the telomere spot in that image.
Reverse transcription qPCR
Twenty-four hours after start of treatment, cells were washed 3 times with PBS before harvesting in Buffer RLT (Qiagen). RNA was converted to cDNA using High Capacity RNA-to-cDNA Master Mix (Life Technologies). Primers were designed using Primer3 (55) and were as follows. Endogenous TERT mRNA was amplified using a forward primer GCCCTCAGACTTCAAGACCA in the ORF of TERT (NM_198253.2) and reverse primer GCTGCTGGTGTCTGCTCTC in the 3′-UTR of endogenous TERT mRNA, resulting in a 74 bp product. Exogenous TERT mRNA was amplified using a forward primer GTCACCTACGTGCCACTCCT in the ORF of TERT mRNA and a reverse primer AGCAAGAAAGCGAGCCAAT in the 3′-UTR of HBB present in our exogenous TERT and CI TERT mRNA, but not in endogenous TERT mRNA, resulting in a 162 bp product. Relative levels were calculated using the Pfaffl method. Reference genes were RPL37A (using primers specified by Greber et al.) (56) and GAPDH (forward primer CAATGACCCCTTCATTGACC and reverse primer TTGATTTTGGAGGGATCTCG, producing a 159 bp product), neither of which exhibited a significant change in Ct value in control or treated cells.
Senescence-associated β-galactosidase staining and cell size scoring
β-Galactosidase staining was performed using the Senescence β-galactosidase staining kit (Cell Signaling Technology). At least 50 cells per population were scored in duplicate. Cell diameter was scored manually after trypsinization on a hemocytometer grid (57).
Statistics
Student’s t-tests and Pearson correlation coefficient calculations were performed using Microsoft Excel (Microsoft, Redmond, WA, USA). Error bars represent the mean ± sem.
RESULTS
Increase in TERT protein levels following modified TERT mRNA transfection
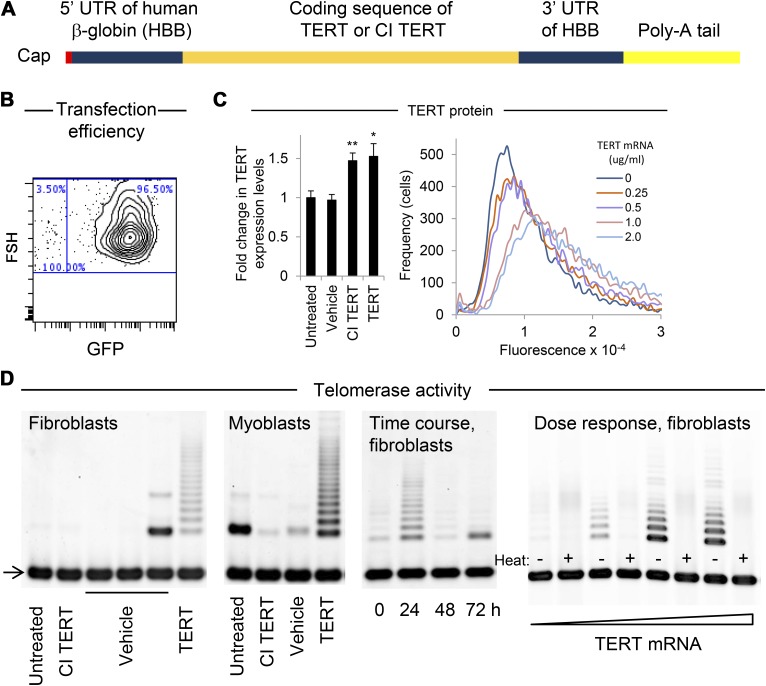
To test the hypothesis that modified TERT mRNA could substantially increase telomere lengths and cell proliferative capacity, we synthesized and delivered modified mRNA comprising pseudouridine and 5-methylcytidine. To increase mRNA stability, the full-length human TERT ORF was flanked by the 5′- and 3′-UTRs of HBB, a 5′ cap, and a 151 nt 3′ poly-A tail (Fig. 1A) (58). The mRNA was transfected via a cationic lipid into primary human fibroblasts and myoblasts, cells known to have limited proliferative capacity (31, 59, 60). Transfection efficiency was determined using flow cytometric single-cell quantitation of fluorescence following delivery of GFP mRNA, which showed that most cells (>90%) were transfected even at relatively low concentrations of modified mRNA (0.1 µg/ml) (Fig. 1B and Supplemental Fig. 1a–c). Treatment of cells with concentrations of exogenous TERT mRNA or mRNA encoding a CI form of TERT equal to each other (1 µg/ml) resulted in internalization of similar amounts of mRNA (Supplemental Fig. 1d), as measured by RT-qPCR 24 h after the first treatment. CI TERT has a substitution mutation at one of the triad of metal-coordinating aspartates at the catalytic site of the reverse transcriptase domain of TERT. As a result, CI TERT cannot add nucleotides to telomeres, yet remains structurally intact and able to bind template DNA and exhibits stability comparable to wild-type TERT in reticulocyte lysates (61). Neither TERT nor CI TERT mRNA treatment affected levels of endogenous TERT mRNA relative to untreated cells as measured by RT-qPCR (Supplemental Fig. 1e). Transfection with a 1 µg/ml concentration of either TERT or CI TERT mRNA resulted in equivalent increases (P < 0.05 and < 0.01, respectively) in the amount of protein detected by anti-TERT antibody (Abcam clone Y182) in fibroblasts (Fig. 1C, left panel) (62, 63). Although we confirmed the ability of the Y182 TERT antibody to recognize TERT using cell types that are known to express low and high levels of TERT, respectively (64, 65) (Supplemental Fig. 2), the Y182 TERT antibody was recently found to cross-react with another endogenous protein with a molecular weight similar to TERT (66). Therefore to confirm that the amounts of TERT protein resulting from both TERT and CI TERT mRNA delivery were similar, we treated fibroblasts with modified mRNA encoding FLAG-tagged TERT and CI TERT (TERT-3xFLAG), and again found similar increases in the amount of protein of the size of TERT in cells receiving either construct, as detected by Western blot using anti-FLAG antibody (Supplemental Fig. 2d). These results are consistent with the similar transfection efficiencies of TERT and CI TERT mRNA as measured by RT-qPCR (Supplemental Fig. 1d). Treatment with increasing amounts of TERT mRNA resulted in a dose-dependent increase in protein recognized by the Y182 anti-TERT antibody as measured in single-cell assays by flow cytometry (Fig. 1C, right panel).
Figure 1.
Increased TERT protein and telomerase activity following modified TERT mRNA delivery. A) Schematic of modified mRNA comprising the coding sequence of the full-length functional form of TERT or a CI form of TERT, flanked by UTRs of HBB and a 151 nt poly-A tail, synthesized using modified nucleotides pseudouridine and 5-methylcytidine. B) Transfection efficiency of myoblasts (n = 2,000) treated with 0.8 µg/ml modified mRNA encoding GFP measured by flow cytometry 24 h post-transfection exceeded 95% (additional plots in Supplemental Fig. 1a). C, left) Levels of protein recognized by anti-TERT antibody (clone Y182) were measured by Western blot (panel C, left; Supplemental Fig. 2). Quantification of levels of protein recognized by anti-TERT antibody 24 h after transfection with 1 µg/ml of either TERT or CI TERT mRNA (n = 3). An endogenous protein similar in size to TERT has also been detected in WI 38 and BJ human fibroblasts using a different TERT antibody (64). C, right) Quantification of protein recognized by anti-TERT antibody in response to various doses of mRNA was measured at the single-cell level by flow cytometry (n = 10,000). *P < 0.05, **P < 0.01 compared with untreated cells. Error bars represent sem. D) Detection of telomerase activity in fibroblasts and myoblasts transfected with modified TERT mRNA, as measured using the TRAP. Arrow indicates internal controls for PCR efficiency. Left to right: Delivery of 1 μg/ml TERT but not CI TERT mRNA increased telomerase activity in fibroblasts and myoblasts, and in fibroblasts telomerase activity returned to baseline levels by 48 h after treatment. Fibroblasts electroporated with the following concentrations of TERT mRNA (0, 10, 40, or 80 μg/ml) exhibited telomerase activity in a dose-dependent manner.
Telomerase activity is transiently increased
To test whether modified TERT mRNA delivery resulted in the generation of functional TERT protein, telomerase activity was quantified using a gel-based TRAP assay. Telomerase activity was detected in fibroblasts and myoblasts treated with 0.25–2.0 μg/ml modified mRNA encoding TERT and was not detected in untreated cells or cells treated with either vehicle only or modified mRNA encoding CI TERT, even at the highest dose of 2.0 µg/ml (Fig. 1D). Telomerase activity levels were dose dependent, and a time course revealed that telomerase activity peaked at 24 h and returned to baseline levels within 48 h after a single transfection. The half-life of human telomerase depends on cell type and conditions (67), and TERT is subject to multiple modes of posttranslational regulation, including by targeted degradation and interaction with factors that affect its catalytic activity (68).
Cell type-dependent increases in proliferative capacity
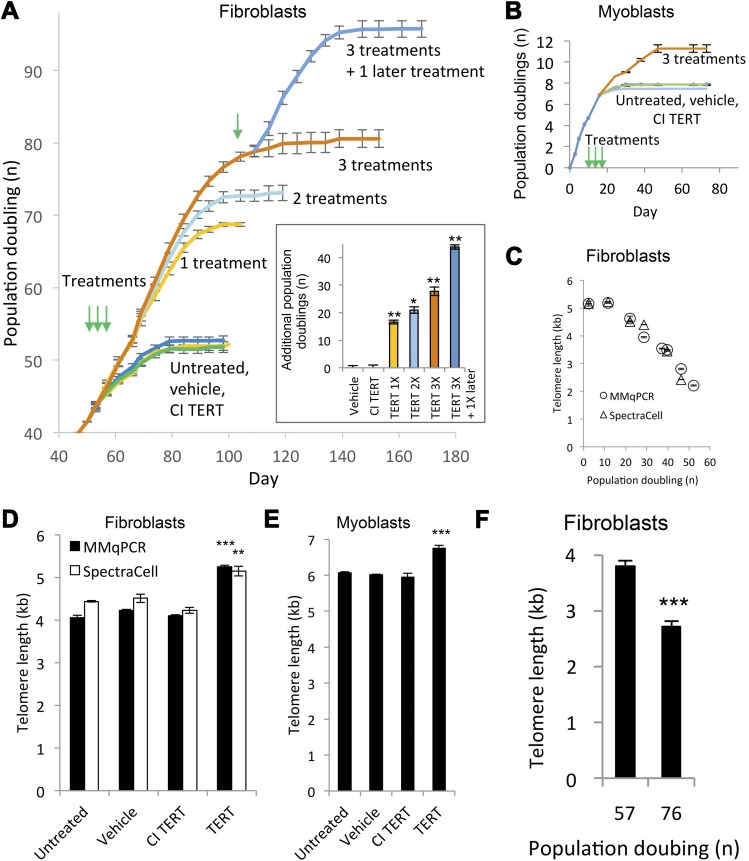
To test the effect of modified TERT mRNA delivery and consequent telomere extension on cell proliferative capacity, we transfected human fibroblasts either once, twice, or 3 times in succession. Treatments were delivered at 48 h intervals. Untreated, vehicle only-treated, and CI TERT mRNA-treated fibroblasts exhibited an equivalent plateau in cell number after approximately 50–60 PDs, whereas cells treated 3 times with TERT mRNA continued to proliferate for a finite additional 28 ± 1.5 PDs with an overall increase in cell number of 2.7 × 108 beyond untreated cells (Fig. 2A). The effect was dose-dependent with each additional treatment conferring additional PD. The incremental increase in proliferative capacity was greater with the first treatment than with the second or third treatments that followed in close succession. However, when an interval of 8 wk followed the initial transfection, an additional transfection at PD 78 increased proliferative capacity by an additional 15.2 ± 1.1 PDs, similar to the 16 ± 0.6 PDs resulting from the initial transfection. These experiments reveal that a refractory period follows the initial transfection during which additional transfections are less effective than a transfection delivered at a later time. Human myoblasts treated 3 times in succession every 48 h gained 3.4 ± 0.4 PDs, equivalent to a 10-fold increase in cell number compared with untreated or vehicle-treated controls (Fig. 2B). Such differences in PD between myoblasts and fibroblasts are not unexpected, as prior studies found similar limited effects of TERT overexpression to a few PDs and showed that this limitation was due to a p16-mediated growth arrest in human myoblasts, in contrast to fibroblasts (22, 69). In both fibroblasts and myoblasts, vehicle only or CI TERT mRNA had no effect on proliferative capacity compared with untreated controls. These data show that delivery of modified TERT mRNA is an effective method for increasing PD in culture. The absence of a proliferative effect in the CI TERT mRNA-treated fibroblasts and myoblasts indicates that the proliferative effect in the modified TERT mRNA-treated cells is due to the catalytic activity of telomerase and rules out nontelomerase activity effects of TERT. Importantly, all of the treated cells studied exhibited a significant increase in cell numbers, but eventually reached a plateau in their growth curves, suggesting absence of immortalization.
Figure 2.
Increased proliferative capacity and telomere length following modified TERT mRNA delivery. A) Growth curves of fibroblasts transfected with 1 µg/ml TERT mRNA, CI TERT mRNA, or vehicle only, once, twice, or 3 times in succession at 48 h intervals starting at PD 42, with green arrows indicating treatment times. Growth curves were repeated twice with each population cultured in triplicate. Full growth curve of untreated cells is shown in Supplemental Fig. 3. (Inset) Proliferative capacity increased in a dose-dependent manner. The additional treatment at PD 78 conferred an additional 15.2 ± 1.1 PDs, an increase similar to that of the initial transfection (16 ± 0.6 PDs). *P < 0.05, **P < 0.01 compared with cells treated only with vehicle. B) Proliferation capacity of myoblasts, treated as in (A) (green arrows). Growth curves were repeated twice, with each population cultured in triplicate. All data are presented as means ± sem. C) Mean telomere lengths in untreated fibroblasts decreased over time in culture as measured by MMqPCR and by SpectraCell (correlation coefficient 0.97, P < 0.001). Experiment was repeated twice with 4 technical replicates each. D) Mean telomere lengths in fibroblasts treated 3 times in succession at 48 h intervals starting at PD 25. Experiment was repeated twice with 4 technical replicates each. **P < 0.01, ***P < 0.001 compared with cells treated only with vehicle. E) Mean telomere lengths in myoblasts treated as in (A). Experiment was repeated twice with 4 technical replicates each. ***P < 0.001 compared with cells treated with vehicle only. F) Telomeres shorten after treatment with TERT mRNA. Telomeres in fibroblasts treated 3 times with TERT mRNA at 48 h intervals starting at PD 42 exhibited telomere shortening as measured by MMqPCR (n = 3 technical replicates per biologic sample). The cell population stopped expanding at PD 80. ***P < 0.001 compared with cells at PD 57.
Lengthening of telomeres
Telomere lengths in untreated fibroblasts declined over time (3 mo) as expected (70) (Fig. 2C) and was quantified using two different methods. We used the MMqPCR method to assess length and validated our measurements independently with a qPCR method performed by SpectraCell Laboratories, Inc. (correlation coefficient 0.97, P < 0.001). Delivery of TERT mRNA 3 times in succession at 48 h intervals to fibroblasts or myoblasts starting at PD 25 and 6, respectively, extended telomeres by 0.9 ± 0.1 kb (22 ± 3%), and 0.7 ± 0.1 kb (12 ± 2%), respectively (Fig. 2D, E). The average rate of telomere lengthening between the start of treatment and cell harvesting was 0.13 ± 0.02 kb/PD. Treatment with vehicle only or CI TERT mRNA had no significant effect on telomere length relative to untreated cells. Telomeres resumed shortening after the end of treatment and continued to shorten up to replicative senescence (Fig. 2F).
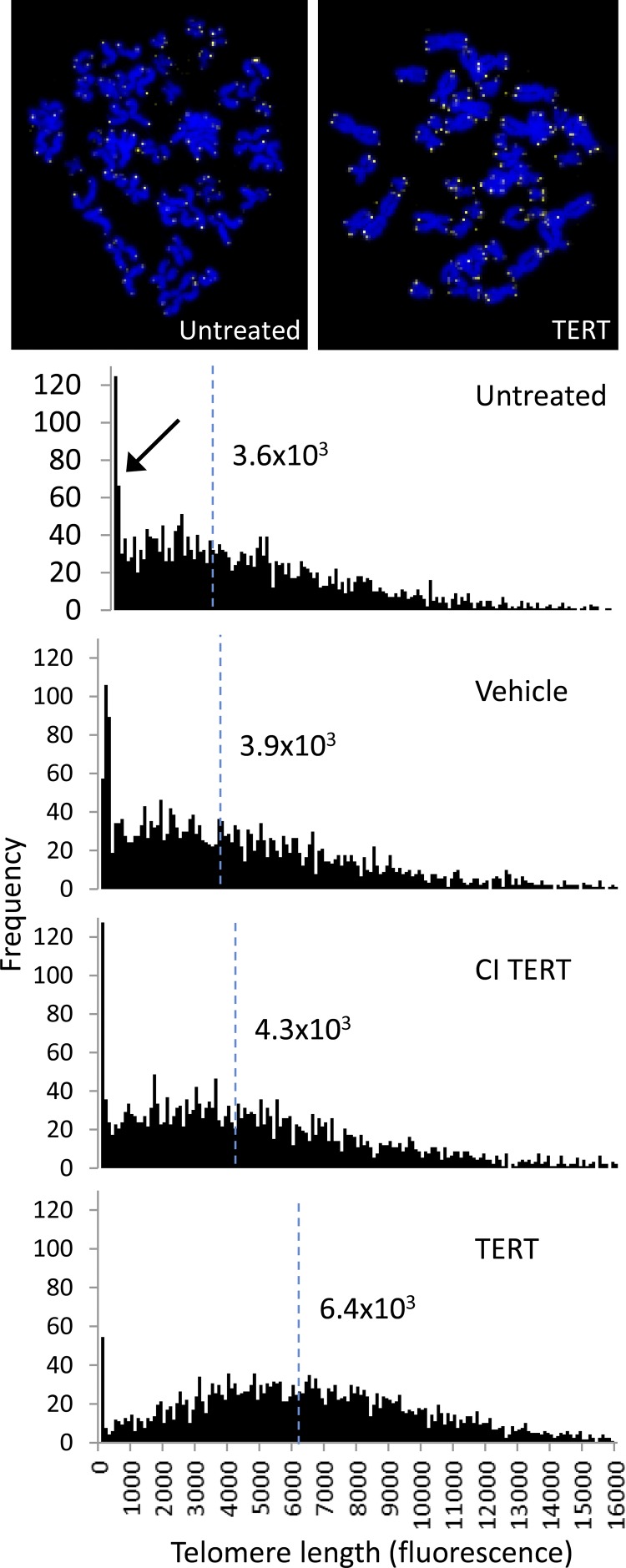
Shift in the frequency distribution of telomere lengths
Q-FISH analysis showed a shift in the frequency distribution of telomere lengths following treatment with TERT mRNA but not CI TERT or vehicle, compared with untreated controls (Fig. 3). The distribution was bimodal, with a left peak at short lengths (arrow), and a broader right peak encompassing longer telomere lengths.
Figure 3.
Telomere length analysis of fibroblasts treated with TERT mRNA. Representative images of metaphase spreads in which yellow represents the signal from the telomere probe and blue is DAPI. Telomere lengths for each population are shown as frequency distributions of telomere signal intensities. Median telomere signal intensities are indicated as vertical dashed blue lines. The arrow indicates the portion of the bimodal distribution corresponding to shorter telomeres.
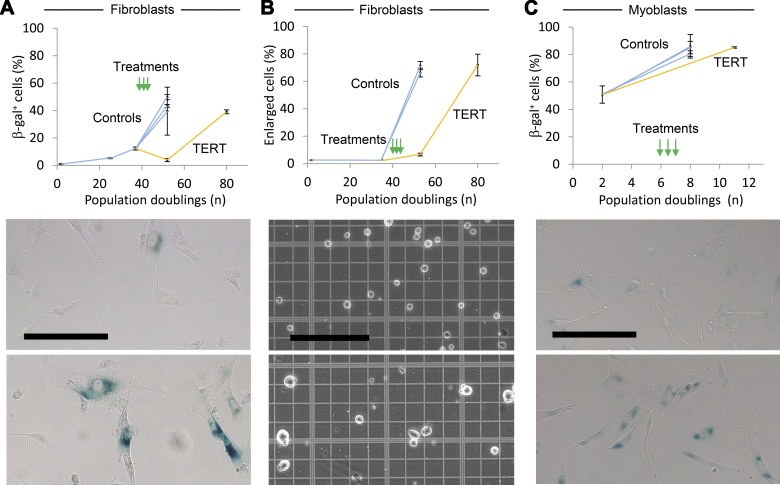
Transient reduction in markers of senescence
As the fibroblast populations stopped growing, they exhibited markers of senescence including senescence-associated β-galactosidase (β-gal) staining and enlarged size (Fig. 4A–C) (57, 71–73). These changes were transiently reduced in fibroblasts treated with TERT mRNA relative to untreated cells and cells receiving CI TERT mRNA or vehicle only. In accordance with findings by others, not all cells in the populations that had entered a growth plateau expressed β-gal at detectable levels (73, 74). However, TERT mRNA-transfected fibroblasts and myoblasts expressed β-gal to the same degree as the control cells of each type after the two populations reached a growth plateau. These data suggest that cells treated with TERT mRNA eventually and predictably cease division and express markers of senescence, and are therefore unlikely to be transformed.
Figure 4.
Transient reduction of senescence-associated markers following modified TERT mRNA delivery. A) Quantification of β-gal-expressing fibroblasts after modified TERT mRNA transfection 3 times in succession at 48 h intervals (green arrows). The control cells, comprising untreated cells, cells treated only with vehicle, and CI TERT mRNA-treated populations, stopped expanding at PD 53, and the TERT mRNA-treated population stopped expanding at PD 80. Each experiment was conducted twice with > 50 cells per sample scored manually. Representative images show β-gal-stained TERT mRNA-treated fibroblasts at PD 53 (top) and PD 80 (bottom). Scale bar, 200 μm. B) Quantification of enlarged cells associated with replicative senescence in fibroblasts transfected 3 times with modified TERT mRNA. Population plateaus are as in A. Controls are vehicle only-treated and CI TERT mRNA-treated. Each experiment was conducted twice, with >50 cells per sample scored manually. Representative images show untreated fibroblasts at PD 2 (top) and PD 53 (bottom). All data are presented as means ± sem. Scale bar, 200 μm. C) Quantification of β-gal expression in myoblasts treated as in A. Controls are as in A. The control and TERT mRNA-treated populations stopped expanding at PD 8 and PD 11, respectively. Each experiment was conducted twice, with > 50 cells per sample scored manually. Representative images show myoblasts at PD 2 (top) and TERT mRNA-treated myoblasts at PD 11 (bottom). Scale bar, 200 μm.
DISCUSSION
Here, we report that transient delivery of TERT mRNA comprising modified nucleotides transiently increases telomerase activity, telomere length, and proliferative capacity without immortalizing cells. The average rate of telomere extension in fibroblasts observed here of 0.13 ± 0.02 kb per PD is comparable to rates reported using viral methods, from 0.1 to > 0.15 kb per PD (22, 75). Modified TERT mRNA extended telomeres in fibroblasts in a few days by 0.9 ± 0.1 kb, and fibroblast telomere lengths have been reported to shorten over a human lifetime by approximately 1–2 kb on average (76). Thus, modified TERT mRNA is efficacious, yet transient and nonintegrating, overcoming major limitations of constitutively expressed viral TERT mRNA delivery.
Human cells of greatest interest are often limited in number, including stem cells for use in experimentation or regenerative medicine. This problem is currently being addressed by various methods including somatic nuclear transfer, viral methods for gene delivery, and the use of culture conditions that lessen the rate of telomere shortening (26, 27, 30). The modified TERT mRNA treatment described here provides an advantageous complement or alternative to these methods that is brief, extends telomeres rapidly, and does not risk insertional mutagenesis. The brevity of TERT mRNA treatment is particularly attractive in that it can avert the loss of stem cell phenotype that can occur over time in culture (77) and shorten the post-reprogramming stage of iPSC generation during which telomeres extend (78). The overall increase in proliferative capacity from 52.7 ± 0.6 PD to 95.7 ± 1.1 PD for fibroblasts corresponded to an increase in absolute cell number of more than 1012-fold. Notably, even this substantial increase in total cell number may not represent the ultimate limit on extending proliferative capacity of modified TERT mRNA-treated fibroblasts, which remains to be determined. Such a method of extending telomeres has the potential to increase the utility of diverse cell types for modeling diseases, screening for ameliorative drugs, and use in cell therapies.
A spectrum of effects on proliferative capacity was observed for the cell types tested, in agreement with previous studies demonstrating different effects of TERT overexpression on myoblast and fibroblast proliferative capacity (22, 69). Moreover, the amount of telomere extension did not correlate with proliferative capacity. Thus, cell context determines the efficacy of TERT expression on proliferative capacity, and an understanding of the factors mediating this effect is of interest in overcoming this limitation. Factors that have been implicated in limiting myoblast proliferative capacity upon viral TERT overexpression include p16-mediated growth arrest, cell type and strain, and culture conditions (69). More generally, the effect may be mediated by nontelomeric DNA damage, age, and mitochondrial integrity (13, 16, 42). The absence of an increase in telomere length or cell proliferative capacity in CI TERT mRNA-transfected cells is consistent with the treatment acting through the catalytic site of TERT by which nucleotides are added directly to telomeres. TERT mRNA-treated cell populations increased in number exponentially for a period of time and then eventually ceased expanding and exhibited markers of senescence to a similar degree as untreated populations, consistent with the absence of immortalization.
Although the therapeutic potential of modified TERT mRNA delivery remains to be determined, the transient nonintegrating nature of modified mRNA and finite increase in proliferative capacity observed here are likely to render it safer than currently used viral or DNA vectors. Furthermore, the method extends telomeres rapidly so that the treatment can be brief, after which the protective telomere shortening mechanism remains intact. This method could be used ex vivo to treat cell types that mediate certain conditions and diseases, such as hematopoietic stem cells or progenitors in cases of immunosenescence or bone marrow failure. Although delivery of modified mRNA to certain tissues in vivo has been achieved, it remains a challenge for most tissues (50). In summary, the method described here for transient elevation of telomerase activity and rapid extension of telomeres, which leads to delayed senescence and increased cell proliferative capacity without immortalizing human cells, constitutes an advance that will enable biologic research and medicine.
Supplementary Material
Acknowledgments
The authors thank Elizabeth Blackburn (University of California, San Francisco) for helpful discussions. They appreciate the technical assistance of Luis Batista (in the laboratory of Steve Artandi, Stanford University) and Linda Ward (SpectraCell, Inc.). They also thank Robert Weinberg for depositing the pBABE-neo-hTERT plasmid at Addgene. This work was supported by Stanford Bio-X Grant IIP5-31 to J.G.S. and H.M.B., U.S. National Institutes of Health (NIH) Director’s Transformative Research Award R01AR063963 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases to H.M.B., NIH National Heart, Lung, and Blood Institute (NHLBI) Grant U01HL100397 to H.M.B. and J.P.C, NIH National Institute on Aging Grant AG044815-01 to H.M.B.., Federal Ministry of Education and Research Grant BMBF 01EO1003 to M.B., University of Maryland, Baltimore Subaward SR00002307 (on NIH NHLBI Grant U01HL099997) to H.M.B. and J.G.S., and funding from the Baxter Foundation to H.M.B. J.R., E.Y., J.P.C., and H.M.B. are inventors on patents for use of modified mRNA for telomere extension.
Glossary
- ATCC
American Type Culture Collection
- β-gal
senescence-associated β-galactosidase
- CI
catalytically inactive
- DMD
Duchenne muscular dystrophy
- HBB
human β-globin
- iPSC
induced pluripotent stem cells
- m5C
5-methylcytidine-5′-triphosphate
- MMqPCR
monochrome multiplex qPCR method of telomere length measurement
- ORF
open reading frame
- PD
population doubling
- Q-FISH
quantitative FISH
- TERC
RNA component that complexes with telomerase reverse transcriptase
- TERT
telomerase reverse transcriptase
- TRAP
telomeric repeat amplification protocol
- Ψ
pseudouridine-5′-triphosphate
- T/S ratio
ratio of telomere repeat copy number to singe gene (36B4) copy number
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Szostak J. W., Blackburn E. H. (1982) Cloning yeast telomeres on linear plasmid vectors. Cell 29, 245–255 [DOI] [PubMed] [Google Scholar]
- 2.De Lange T. (2009) How telomeres solve the end-protection problem. Science 326, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shay J. W., Wright W. E. (2001) Telomeres and telomerase: implications for cancer and aging. Radiat. Res. 155, 188–193 [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp P. M. (2005) Major cutbacks at chromosome ends. Trends Biochem. Sci. 30, 388–395 [DOI] [PubMed] [Google Scholar]
- 5.Greider C. W., Blackburn E. H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 6.Greider C. W., Blackburn E. H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337 [DOI] [PubMed] [Google Scholar]
- 7.Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 8.Artandi S. E., DePinho R. A. (2010) Telomeres and telomerase in cancer. Carcinogenesis 31, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takubo K., Aida J., Izumiyama-Shimomura N., Ishikawa N., Sawabe M., Kurabayashi R., Shiraishi H., Arai T., Nakamura K. (2010) Changes of telomere length with aging. Geriatr. Gerontol. Int. 10(Suppl 1), S197–S206 [DOI] [PubMed] [Google Scholar]
- 10.Sahin E., Depinho R. A. (2010) Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464, 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Signer R. A. J., Morrison S. J. (2013) Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 12, 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn E. H. (2005) Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579, 859–862 [DOI] [PubMed] [Google Scholar]
- 13.Sahin E., Colla S., Liesa M., Moslehi J., Müller F. L., Guo M., Cooper M., Kotton D., Fabian A. J., Walkey C., Maser R. S., Tonon G., Foerster F., Xiong R., Wang Y. A., Shukla S. A., Jaskelioff M., Martin E. S., Heffernan T. P., Protopopov A., Ivanova E., Mahoney J. E., Kost-Alimova M., Perry S. R., Bronson R., Liao R., Mulligan R., Shirihai O. S., Chin L., DePinho R. A. (2011) Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackburn E. H. (2011) Walking the walk from genes through telomere maintenance to cancer risk. Cancer Prev. Res. (Phila.) 4, 473–475 [DOI] [PubMed] [Google Scholar]
- 15.Calado R. T., Cooper J. N., Padilla-Nash H. M., Sloand E. M., Wu C. O., Scheinberg P., Ried T., Young N. S. (2012) Short telomeres result in chromosomal instability in hematopoietic cells and precede malignant evolution in human aplastic anemia. Leukemia 26, 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourkioti F., Kustan J., Kraft P., Day J. W., Zhao M.-M., Kost-Alimova M., Protopopov A., DePinho R. A., Bernstein D., Meeker A. K., Blau H. M. (2013) Role of telomere dysfunction in cardiac failure in Duchenne muscular dystrophy. Nat. Cell Biol. 15, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calado R., Young N. (2012) Telomeres in disease. F1000 Med. Rep. 4, 8 Available at http://www.ncbi.nlm.nih.gov/pmc/issues/204091/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armanios M., Blackburn E. H. (2012) The telomere syndromes. Nat. Rev. Genet. 13, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacco A., Mourkioti F., Tran R., Choi J., Llewellyn M., Kraft P., Shkreli M., Delp S., Pomerantz J. H., Artandi S. E., Blau H. M. (2010) Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushita H., Chang E., Glassford A. J., Cooke J. P., Chiu C. P., Tsao P. S. (2001) eNOS activity is reduced in senescent human endothelial cells: preservation by hTERT immortalization. Circ. Res. 89, 793–798 [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Rivero G., Ruiz-Torres M. P., Rivas-Elena J. V., Jerkic M., Díez-Marques M. L., Lopez-Novoa J. M., Blasco M. A., Rodríguez-Puyol D. (2006) Mice deficient in telomerase activity develop hypertension because of an excess of endothelin production. Circulation 114, 309–317 [DOI] [PubMed] [Google Scholar]
- 22.Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., Wright W. E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 [DOI] [PubMed] [Google Scholar]
- 23.Pucci F., Gardano L., Harrington L. (2013) Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 12, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marión R. M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M. A. (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460, 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le R., Kou Z., Jiang Y., Li M., Huang B., Liu W., Li H., Kou X., He W., Rudolph K. L., Ju Z., Gao S. (2014) Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell 14, 27–39 [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann S., Martens U. M. (2008) Telomeres, senescence, and hematopoietic stem cells. Cell Tissue Res. 331, 79–90 [DOI] [PubMed] [Google Scholar]
- 28.Suhr S. T., Chang E. A., Rodriguez R. M., Wang K., Ross P. J., Beyhan Z., Murthy S., Cibelli J. B. (2009) Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE 4, e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokkinaki M., Sahibzada N., Golestaneh N. (2011) Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells 29, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohsin S., Khan M., Nguyen J., Alkatib M., Siddiqi S., Hariharan N., Wallach K., Monsanto M., Gude N., Dembitsky W., Sussman M. A. (2013) Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circ. Res. 113, 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster C., Blau H. M. (1990) Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat. Cell Mol. Genet. 16, 557–565 [DOI] [PubMed] [Google Scholar]
- 32.Vaziri H., Chapman K. B., Guigova A., Teichroeb J., Lacher M. D., Sternberg H., Singec I., Briggs L., Wheeler J., Sampathkumar J., Gonzalez R., Larocca D., Murai J., Snyder E., Andrews W. H., Funk W. D., West M. D. (2010) Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen. Med. 5, 345–363 [DOI] [PubMed] [Google Scholar]
- 33.Allsopp R. (2012) Telomere length and iPSC re-programming: survival of the longest. Cell Res. 22, 614–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista L. F. Z., Pech M. F., Zhong F. L., Nguyen H. N., Xie K. T., Zaug A. J., Crary S. M., Choi J., Sebastiano V., Cherry A., Giri N., Wernig M., Alter B. P., Cech T. R., Savage S. A., Reijo Pera R. A., Artandi S. E. (2011) Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 474, 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade L. N. de S., Nathanson J. L., Yeo G. W., Menck C. F. M., Muotri A. R. (2012) Evidence for premature aging due to oxidative stress in iPSCs from Cockayne syndrome. Hum. Mol. Genet. 21, 3825–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harley C. B. (2002) Telomerase is not an oncogene. Oncogene 21, 494–502 [DOI] [PubMed] [Google Scholar]
- 37.Harley C. B. (2005) Telomerase therapeutics for degenerative diseases. Curr. Mol. Med. 5, 205–211 [DOI] [PubMed] [Google Scholar]
- 38.Blasco M. A. (2005) Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 [DOI] [PubMed] [Google Scholar]
- 39.Blackburn E. H., Tlsty T. D., Lippman S. M. (2010) Unprecedented opportunities and promise for cancer prevention research. Cancer Prev. Res. (Phila.) 3, 394–402 [DOI] [PubMed] [Google Scholar]
- 40.Jaskelioff M., Muller F. L., Paik J.-H., Thomas E., Jiang S., Adams A. C., Sahin E., Kost-Alimova M., Protopopov A., Cadiñanos J., Horner J. W., Maratos-Flier E., Depinho R. A. (2011) Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469, 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernardes de Jesus B., Blasco M. A. (2011) Aging by telomere loss can be reversed. Cell Stem Cell 8, 3–4 [DOI] [PubMed] [Google Scholar]
- 42.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernardes de Jesus B., Vera E., Schneeberger K., Tejera A. M., Ayuso E., Bosch F., Blasco M. A. (2012) Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol. Med. 4, 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernardes de Jesus B., Schneeberger K., Vera E., Tejera A., Harley C. B., Blasco M. A. (2011) The telomerase activator TA-65 elongates short telomeres and increases health span of adult/old mice without increasing cancer incidence. Aging Cell 10, 604–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harley C. B., Liu W., Blasco M., Vera E., Andrews W. H., Briggs L. A., Raffaele J. M. (2011) A natural product telomerase activator as part of a health maintenance program. Rejuvenation Res. 14, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eitan E., Tichon A., Gazit A., Gitler D., Slavin S., Priel E. (2012) Novel telomerase-increasing compound in mouse brain delays the onset of amyotrophic lateral sclerosis. EMBO Mol. Med. 4, 313–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mogford J. E., Liu W. R., Reid R., Chiu C.-P., Said H., Chen S.-J., Harley C. B., Mustoe T. A. (2006) Adenoviral human telomerase reverse transcriptase dramatically improves ischemic wound healing without detrimental immune response in an aged rabbit model. Hum. Gene Ther. 17, 651–660 [DOI] [PubMed] [Google Scholar]
- 48.Steinert S., Shay J. W., Wright W. E. (2000) Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun. 273, 1095–1098 [DOI] [PubMed] [Google Scholar]
- 49.Karikó K., Muramatsu H., Keller J. M., Weissman D. (2012) Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 20, 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kormann M. S. D., Hasenpusch G., Aneja M. K., Nica G., Flemmer A. W., Herber-Jonat S., Huppmann M., Mays L. E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. (2011) Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., Su H.-H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. (2013) Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 21, 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Counter C. M., Hahn W. C., Wei W., Caddle S. D., Beijersbergen R. L., Lansdorp P. M., Sedivy J. M., Weinberg R. A. (1998) Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 95, 14723–14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cawthon R. M. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cawthon R. M. (2009) Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 37, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., Rozen S. G. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greber B., Coulon P., Zhang M., Moritz S., Frank S., Müller-Molina A. J., Araúzo-Bravo M. J., Han D. W., Pape H. C., Schöler H. R. (2011) FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 30, 4874–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristofalo V. J., Kritchevsky D. (1969) Cell size and nucleic acid content in the diploid human cell line WI-38 during aging. Med. Exp. Int. J. Exp. Med. 19, 313–320 [DOI] [PubMed] [Google Scholar]
- 58.Tavernier G., Andries O., Demeester J., Sanders N. N., De Smedt S. C., Rejman J. (2011) mRNA as gene therapeutic: how to control protein expression. J. Control. Release 150, 238–247 [DOI] [PubMed] [Google Scholar]
- 59.Hayflick L., Moorhead P. S. (1961) The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 [DOI] [PubMed] [Google Scholar]
- 60.Yakubov E., Rechavi G., Rozenblatt S., Givol D. (2010) Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 394, 189–193 [DOI] [PubMed] [Google Scholar]
- 61.Wyatt, H. D. M. (2009) Structure-Function Analysis of the Human Telomerase Reverse Transcriptase. Doctoral dissertation, University of Calgary, Calgary, Alberta, Canada
- 62.Wick M., Zubov D., Hagen G. (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232, 97–106 [DOI] [PubMed] [Google Scholar]
- 63.Ahmed S., Passos J. F., Birket M. J., Beckmann T., Brings S., Peters H., Birch-Machin M. A., von Zglinicki T., Saretzki G. (2008) Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 121, 1046–1053 [DOI] [PubMed] [Google Scholar]
- 64.Wu Y.-L., Dudognon C., Nguyen E., Hillion J., Pendino F., Tarkanyi I., Aradi J., Lanotte M., Tong J. H., Chen G. Q., Ségal-Bendirdjian E. (2006) Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J. Cell Sci. 119, 2797–2806 [DOI] [PubMed] [Google Scholar]
- 65.Haendeler J., Dröse S., Büchner N., Jakob S., Altschmied J., Goy C., Spyridopoulos I., Zeiher A. M., Brandt U., Dimmeler S. (2009) Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler. Thromb. Vasc. Biol. 29, 929–935 [DOI] [PubMed] [Google Scholar]
- 66.Xi L., Cech T. R. (2014) Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 42, 8565–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holt S. E., Aisner D. L., Shay J. W., Wright W. E. (1997) Lack of cell cycle regulation of telomerase activity in human cells. Proc. Natl. Acad. Sci. USA 94, 10687–10692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cifuentes-Rojas C., Shippen D.E. (2012) Telomerase regulation. Mutat. Res. 730, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu C.-H., Mouly V., Cooper R. N., Mamchaoui K., Bigot A., Shay J. W., Di Santo J. P., Butler-Browne G. S., Wright W. E. (2007) Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 6, 515–523 [DOI] [PubMed] [Google Scholar]
- 70.Sitte N., Saretzki G., von Zglinicki T. (1998) Accelerated telomere shortening in fibroblasts after extended periods of confluency. Free Radic. Biol. Med. 24, 885–893 [DOI] [PubMed] [Google Scholar]
- 71.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., Peacocket M., Campisi J. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cristofalo V. J., Lorenzini A., Allen R. G., Torres C., Tresini M. (2004) Replicative senescence: a critical review. Mech. Ageing Dev. 125, 827–848 [DOI] [PubMed] [Google Scholar]
- 73.Lawless C., Wang C., Jurk D., Merz A., Zglinicki Tv., Passos J. F. (2010) Quantitative assessment of markers for cell senescence. Exp. Gerontol. 45, 772–778 [DOI] [PubMed] [Google Scholar]
- 74.Binet R., Ythier D., Robles A. I., Collado M., Larrieu D., Fonti C., Brambilla E., Brambilla C., Serrano M., Harris C. C., Pedeux R. (2009) WNT16B is a new marker of cellular senescence that regulates p53 activity and the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 69, 9183–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaziri H., Benchimol S. (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 8, 279–282 [DOI] [PubMed] [Google Scholar]
- 76.Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., Greider C. W., Harley C. B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 89, 10114–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gilbert P. M., Havenstrite K. L., Magnusson K. E. G., Sacco A., Leonardi N. A., Kraft P., Nguyen N. K., Thrun S., Lutolf M. P., Blau H. M. (2010) Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang F., Yin Y., Ye X., Liu K., Zhu H., Wang L., Chiourea M., Okuka M., Ji G., Dan J., Zuo B., Li M., Zhang Q., Liu N., Chen L., Pan X., Gagos S., Keefe D. L., Liu L. (2012) Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 22, 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.