Abstract
We present a www server for AUGUSTUS, a novel software program for ab initio gene prediction in eukaryotic genomic sequences. Our method is based on a generalized Hidden Markov Model with a new method for modeling the intron length distribution. This method allows approximation of the true intron length distribution more accurately than do existing programs. For genomic sequence data from human and Drosophila melanogaster, the accuracy of AUGUSTUS is superior to existing gene-finding approaches. The advantage of our program becomes apparent especially for larger input sequences containing more than one gene. The server is available at http://augustus.gobics.de.
INTRODUCTION
The first step in genome annotation is to predict all gene structures in a given genomic sequence. The development of gene-finding methods is, therefore, an important field in biological sequence analysis. For eukaryotes this problem is far from trivial, since eukaryotic genes usually contain large introns, i.e. non-coding regions. Most gene-prediction programs are based on stochastic models such as Hidden Markov Models (HMMs). These models describe the statistical features of different regions and signals in genomic sequences, such as introns, coding exons, UTRs, promoters, etc. A large number of gene-finding programs have been proposed since the 1980s, e.g. GENIE (1), GENSCAN (2) and GENEID (3). GENSCAN is widely used and has been found in earlier studies (4,5) to be one of the most accurate gene-prediction programs. All these tools are routinely used for automatic genome annotation. Despite considerable efforts in the bioinformatics community, the performance of existing gene-prediction tools is still not satisfactory. A study by Guigó et al. (6) has shown that these tools are accurate if applied to rather short sequences that contain single genes together with short flanking intergenic regions. However, their performance drops dramatically if they are applied to long input sequences. Experiments with semi-artificial sequences showed that GENSCAN tends to predict many more genes than are actually present in genomic sequences.
A major problem in gene prediction is the correct modeling of the intron length distribution for a given organism. Other HMM-based gene-finding programs, such as GENSCAN (2), GENIE (1), DOUBLESCAN (7) and TWINSCAN (8), can only model a geometric intron length distribution, in which the probabilities decline exponentially with the length. This approach is computationally more efficient than explicitly modeling the actual non-geometric length distribution.
However, the assumed geometric intron length distribution is the reason why a single gene is often split into two or more predicted genes (1) and a reason why large introns are very unlikely to be correctly identified.
AUGUSTUS—A NEW APPROACH TO HMM-BASED GENE PREDICTION
AUGUSTUS is based on a generalized Hidden Markov Model (GHMM). This model defines probability distributions for the various sections of genomic sequences. Introns, exons, intergenic regions and so on correspond to states in the model, and each state is thought to create DNA sequences with certain pre-defined emission probabilities. Like other HMM-based gene finders, AUGUSTUS finds an optimal parse of a given genomic sequence, i.e. a segmentation of the sequences into states that is most likely according to the underlying statistical model. The default version of the model consists of 47 states, of which 23 states model genes on the reverse strand and are symmetric copies of corresponding states which model genes on the forward strand. We probabilistically model separately the sequence around the splice sites, the sequence of the branch point region, the bases before the translation start, the coding regions, the non-coding regions, the first coding bases of a gene, the length distribution of single exons, initial exons, internal exons, terminal exons, intergenic regions, the distribution of the number of exons per gene and the length distribution of introns.
In our intron length model, which is described in (9,10), we combine explicit length modeling with a geometric distribution. For introns shorter than a few hundred bases (human 584, Drosophila 929), we use explicit length modeling. Only for introns exceeding this length does the probability decline exponentially, but at a slower rate than if the whole distribution was geometric. In the explicitly modeled part of the distribution, intron lengths have probabilities that have been estimated from observed frequencies. This way, our program is computationally efficient but is able to model intron lengths much more realistically than standard approaches do.
Our model parameters have been estimated using training sequences with known genes. For the human version we used 1284 single-gene training sequences; for the Drosophila version we used 400 single-gene training sequences. For each species, we use one of 10 different sets of parameters according to the average GC content of the input sequence.
The performance of AUGUSTUS has been extensively evaluated on sequence data from human and Drosophila (9,10). These studies showed that, especially for long input sequences, our program is considerably more accurate than existing approaches. Table 1 shows the prediction accuracy of AUGUSTUS, GENEID and GENIE on the Drosophila Adh region, which has been carefully annotated and has been used in the Genome Annotation Assessment Project (11).
Table 1. Accuracy results on a 2.9 million bp long sequence from the Drosophila Adh region.
| Program | Base level | Exon level | Gene level | |||
|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |
| AUGUSTUS | 98 | 93 | 85 | 65 | 68 | 38 |
| GENEID | 96 | 92 | 71 | 62 | 47 | 33 |
| GENIE | 96 | 92 | 70 | 57 | 40 | 29 |
For each of the three programs the sensitivity was measured using a set of annotations, called std1, which contains 38 genes. The specificity was measured using another set of annotations, called std3, which contains 222 genes. For testing we used release 2 of AUGUSTUS and version 1.1 of GENEID. The results for GENIE were taken from (11).
To make our tool available for the research community, we set up a www server at GOBICS (Göttingen Bioinformatics Compute Server), where AUGUSTUS is accessible through a user-friendly interface.
WEB SERVER DESCRIPTION
The AUGUSTUS web server allows a DNA sequence to be uploaded in FASTA format or as multiple sequences in multiple FASTA format or by pasting a sequence into the web form. It is also possible to paste the sequence part of the GENBANK format (which follows the ORIGIN keyword) into the web form because spaces and digits are ignored by the program.
The maximal total length of the sequences submitted to the server is 3 million bp. Currently, AUGUSTUS has two specially trained parameter sets that can be chosen on the web site: human and Drosophila. We can generate parameter sets for other species automatically from annotated GENBANK files of these species and plan to add them to the web site. For the moment, we recommend using the human version also for other vertebrates.
AUGUSTUS reports predicted genes of the input DNA sequence on the forward strand, the reverse strand or on both strands, depending on the user's choice. Usually the default version of the program is the best choice, but in some cases additional evidence about the gene structure suggests deviating from the default program behavior. For these cases the user has two ‘expert options’.
The first ‘expert option’ is a choice by radio button from one option from the following list:
predict any number of (possibly partial) genes,
only predict complete genes,
only predict complete genes—at least one,
predict exactly one gene.
The first of these options is the default setting. AUGUSTUS may predict no gene at all, one gene or more than one gene. Here, the first and the last predicted gene may be partial. ‘Partial’ means that the gene is incomplete and not all of the exons of the gene are contained in the input sequence. The last three options assume that the boundaries of the input sequence lie in the intergenic region and, thus, AUGUSTUS predicts only complete genes including both the start and stop codon. When the second option is chosen AUGUSTUS predicts zero or more complete genes. When the third option is chosen, AUGUSTUS is forced to predict at least one gene if possible. However, predicted genes may be filtered out if the coding sequence is unrealistically short. The last option forces AUGUSTUS to predict one gene and not more than one gene. If it is known that the boundaries of the input sequence are within an intergenic region, then choosing the option ‘only predict complete genes’ can significantly increase the prediction accuracy as Table 2 shows. In particular, the gene-level accuracy increases. This is because in sequences where the first exon of a gene is close to a sequence boundary often this first exon is missed with the default setting and the gene is predicted as a partial gene.
Table 2. Comparison of prediction accuracy on 178 human single-gene sequences.
| Program | Base level | Exon level | Gene level | |||
|---|---|---|---|---|---|---|
| Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | |
| AUGUSTUS, default | 93 | 90 | 80 | 81 | 48 | 47 |
| AUGUSTUS, complete | 92 | 91 | 82 | 83 | 58 | 58 |
| GENSCAN | 97 | 86 | 83 | 75 | 40 | 36 |
The first line shows the results with the default settings of AUGUSTUS. The second line shows the results with the option ‘only predict complete genes’, which are much better on the gene level. For comparison with the default version of AUGUSTUS (release 2) the results of GENSCAN (version 1.0), which may predict partial genes, are shown.
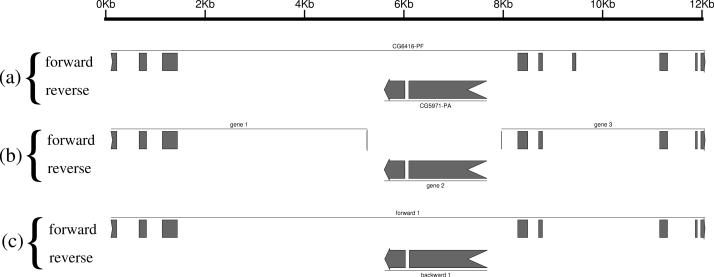
The other ‘expert option’ is a checkbox that, if checked, tells AUGUSTUS to ignore conflicts between the gene structures of the two strands. By default this option is not chosen and AUGUSTUS assumes that genes on opposite strands do not overlap (as well as genes on the same strand). This assumption is usually satisfied, and making it helps to avoid finding ‘shadow genes’, i.e. false positive genes on a certain strand, at a position where the true gene is actually on the other strand. In some cases the assumption is not satisfied, and a gene is contained in an intron of a gene on the other strand as in Figure 1a. In this case the default setting cannot produce the correct prediction. In the case of the particular pair of nested genes of Figure 1, the default version of AUGUSTUS correctly predicts the included gene but splits the including gene into two predicted genes as shown in Figure 1b. In a case with evidence about nested genes, e.g. derived by expressed sequence tag (EST) alignments, the ‘ignore conflicts’ option should be chosen. With this option the predictions are made independently on the two strands. In this example the two genes are then predicted almost correctly (Figure 1c).
Figure 1.
An example where the option ‘ignore conflicts with other strand’ helps. The lines in (a) show two nested Drosophila genes as annotated in FlyBase (12). The nine-exon gene on the forward strand includes a two-exon gene on the reverse strand within a long intron. The lines in (b) show the prediction with the default parameters. The gene on the forward strand is split into two genes by introducing two very short false positive exons so that the three predicted genes do not overlap. The lines in (c) show the prediction with the option ‘ignore conflicts with other strand’, which is identical to the annotation except for a short missed exon. This graphic has been obtained using gff2ps (13) from http://genome.imim.es/software/gfftools/GFF2PS.html.
When one of the ‘expert options’ is changed from the default setting the maximal total sequence length is 400 kb. This limit will be suspended soon. The running time for a 200 kb input sequence is approximately 30 s when the server is otherwise idle.
OUTPUT DESCRIPTION
AUGUSTUS outputs its results in both graphics and text format. The results page of the web server shows for each sequence a clickable thumbnail which links to a postscript image similar to the one in Figure 1. The pictures are generated with the program gff2ps (13) from the text output. The text output is in the ‘General Feature Format’ (GFF) proposed by Richard Durbin and David Haussler. The Sanger Institute lists at http://www.sanger.ac.uk/Software/formats/GFF a large number of tools which work with the GFF. In this format the results contain one line for each exon with data fields separated by a TAB character. These data fields include the start and end positions of the exon, a name for the sequence, a name for the gene and whether it is on the forward or reverse strand. A detailed description of the output is in the Supplementary Materials to this article.
FUTURE WORK
Currently, the AUGUSTUS web server makes its predictions ab initio, i.e. without making use of external evidence about the gene structure of the input sequence. However, a natural and flexible generalization of the GHMM of AUGUSTUS that allows the integration of uncertain extrinsic information from various sources has already been developed (10). This has been tested with extrinsic information which the program AGRIPPA (14) has constructed from the results of searching the input DNA sequence against protein and EST databases. The approach also allows such user constraints as ‘This interval of the sequence must be part of an exon’ to be set. A publication presenting the promising results of the integration of EST and protein database search results is in preparation.
During recent years, a number of comparative gene-finding tools have been proposed (15–19). These tools work by comparing genomic sequences from related organisms to each other, e.g. human and mouse. They use the phylogenetic footprinting principle, i.e. they exploit the fact that functionally important parts of sequences are usually more conserved than non-functional parts of the genome. Comparative methods try to identify evolutionarily conserved parts of the sequences and then search for signals such as splice sites near these conserved sequences.
Some authors have combined intrinsic and comparative gene-finding approaches (7,8,20,21). We also plan to utilize the homology information produced by the alignment program DIALIGN (22) for the above-mentioned generalization of AUGUSTUS. DIALIGN has been used in the past for genome sequence analysis; it has been shown that local sequence similarities returned by DIALIGN are highly correlated to protein-coding exons (23). A new version of the program has been implemented that is considerably faster than the original version and can therefore be applied to larger sequence data (24).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Reese M.G., Kulp,D., Tammana,H. and Haussler,D. (2000) Gene finding in Drosophila melanogaster. Genome Res., 10, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge C.B. (1997) Identification of genes in human genomic DNA. Ph.D. Thesis, ‘Stanford University’, Stanford, CA, USA.
- 3.Parra G., Blanco,E. and Guigó,R. (2000) GeneID in Drosophila. Genome Res., 10, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogic S., Mackworth,A.K. and Ouellette,F.B.F. (2001) Evaluation of gene-finding programs on mammalian sequences. Genome Res., 11, 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claverie J.-M. (1997) Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet., 6, 1735–1744. [DOI] [PubMed] [Google Scholar]
- 6.Guigó R., Agarwal,P., Abril,J., Burset,M. and Fickett,J.W. (2000) An assessment of gene prediction accuracy in large DNA sequences. Genome Res., 10, 1631–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer I.M. and Durbin,R. (2002) Comparative ab initio prediction of gene structures using pair HMMs. Bioinformatics, 18, 1309–1318. [DOI] [PubMed] [Google Scholar]
- 8.Korf I., Flicek,P., Duan,D. and Brent,M.R. (2001) Integrating genomic homology into gene structure prediction. Bioinformatics, 1(Suppl. 1), S1–S9. [DOI] [PubMed] [Google Scholar]
- 9.Stanke M. and Waack,S. (2003) Gene prediction with a hidden Markov model and new intron submodel. Bioinformatics, 19(Suppl. 2), ii215–ii225. [DOI] [PubMed] [Google Scholar]
- 10.Stanke M. (2004) Gene prediction with a hidden markov model. Ph.D. Thesis, ‘University of Göttingen’, Germany.
- 11.Reese M.G., Hartzell,G., Harris,N.L., Ohler,U., Abril,J.F. and Lewis,S.E. (2000) Genome annotation assessment in Drosophila melanogaster. Genome Res., 10, 391–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The FlyBase Consortium (2003) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 31, 172–175, http://flybase.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abril J.F. and Guigó,R. (2000) gff2ps: visualizing genomic annotations. Bioinformatics, 16, 743–744. [DOI] [PubMed] [Google Scholar]
- 14.Schöffmann O. (2003) Gewinnung extrinsischer Informationen zur Genvorhersage und Einbindung in ein Hidden Markov Modell. Diploma thesis, ‘University of Göttingen’, Germany.
- 15.Bafna V. and Huson,D.H. (2000) The conserved exon method for gene finding. Bioinformatics, 16, 190–202. [PubMed] [Google Scholar]
- 16.Batzoglou S., Pachter,L., Mesirov,J.P., Berger,B. and Lander,E.S. (2000) Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res., 10, 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinner O. and Morgenstern,B. (2002) AGenDA: gene prediction by comparative sequence analysis. In Silico Biol., 2, 195–205. [PubMed] [Google Scholar]
- 18.Blayo P., Rouzé,P. and Sagot,M.-F. (2003) Orphan gene finding—an exon assembly approach. Theor. Comput. Sci., 290, 1407–1431. [Google Scholar]
- 19.Wiehe T., Gebauer-Jung,S., Mitchell-Olds,T. and Guigó,R. (2001) SGP-1: Prediction and validation of homologous genes based on sequence alignments. Genome Res., 11, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cawley S., Pachter,L. and Alexandersson,M. (2003), SLAM web server for comparative gene finding and alignment. Nucleic Acids Res., 31, 3507–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra G., Agarwal,P., Abril,J.F., Wiehe,T., Fickett,J.W. and Guigó,R. (2003) Comparative gene prediction in human and mouse. Genome Res., 13, 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 23.Morgenstern B., Rinner,O., Abdeddaïm,S., Haase,D., Mayer,K., Dress,A. and Mewes,H.-W. (2002) Exon discovery by genomic sequence alignment. Bioinformatics, 18, 777–787. [DOI] [PubMed] [Google Scholar]
- 24.Brudno M., Chapman,M., Göttgens,B., Batzoglou,S. and Morgenstern,B. (2003) BMC Bioinformatics, 4, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.