Abstract
The SledgeHMMER web server is intended for genome-scale searching of the Pfam database without having to install this database and the HMMER software locally. The server implements a parallelized version of hmmpfam, the program used for searching the Pfam HMM database. Pfam search results have been calculated for the entire Swiss-Prot and TrEmbl database sequences (∼1.2 million) on 256 processors of IA64-based teragrid machines. The Pfam database can be searched in local, glocal or merged mode, using either gathering or E-value thresholds. Query sequences are first matched against the pre-calculated entries to retrieve results, and those without matches are processed through a new search process. Results are emailed in a space-delimited tabular format upon completion of the search. While most other Pfam-searching web servers set a limit of one sequence per query, this server processes batch sequences with no limit on the number of input sequences. The web server and downloadable data are accessible from http://SledgeHmmer.sdsc.edu.
INTRODUCTION
Searching for conserved domains has become an integral part of several bioinformatics data analysis pipelines. The Pfam protein families database [(1); http://pfam.wustl.edu] contains the single largest public collection of conserved functional domains and hence is integrated into many bioinformatics resources. The current version (release 12) of the Pfam-A database contains 7316 HMM domains, making it an indispensable resource for protein functional annotation (2). However, the size of the Pfam database is quite large (∼600 MB) and genome-scale searching of protein sequences against this database is a highly compute-intensive task. Hence, with the exception of the Sanger Center web server (http://www.sanger.ac.uk/Software/Pfam), all other Pfam-searching servers permit submission of only one protein sequence at a time (http://pfam.wustl.edu, http://pfam.jouy.inra.fr, http://pfam.cgb.ki.se). Currently, the Sanger Center web server limits batch submission to 1000 sequences at a time. Other than this, for batch searching, users are required to install the Pfam database and the HMMER software [(3); http://hmmer.wustl.edu) locally. Given the size and dynamic nature of this database (14 updates in the last two years), it is not convenient to maintain the latest versions of these tools locally. Moreover, the hmmpfam program used for searching the Pfam database is very slow and memory intensive (on Solaris machines, the processing speed is about 10 sequences per 1 h of CPU time), making this process a bottleneck in several data analysis pipelines.
Web servers that enable batch searching of the Pfam database are extremely beneficial to the user community, especially to those without local access to high-end computational power, memory and disk space. To this end we have developed a parallelized version, as well as an optimized single-processor version, of hmmpfam from release 2.3.2 of the HMMER software, as described in the next section. Using these programs, we present the SledgeHMMER web server (http://SledgeHmmer.sdsc.edu), which is capable of batch searching a large number of protein sequences concurrently.
DESIGN AND IMPLEMENTATION
The current web service is mainly intended for genome-scale searching of the Pfam-A database. Searches are possible for different Pfam-searching modes such as local, glocal and merged, using either gathering or E-value thresholds. To expedite the response time, we developed a database containing pre-calculated Pfam search results. Query sequences are matched against the pre-calculated entries using hexadecimal hashing methods from the MD5 Perl module, and those without matches are separated out. Results for matching sequences are retrieved from the pre-calculated database while, for other sequences, a new search process is initiated using our improved hmmpfam program. Results are emailed in a space-delimited tabular format upon completion of the search.
Parallelizing the hmmpfam algorithm
Computing batch searches against the Pfam database is a two-dimensional (2D) problem, i.e. searching N query sequences against M Pfam HMMs. Thus, this problem can be parallelized across any dimension or across both dimensions. A minor disadvantage of parallelizing across the M dimension is that a global gather needs to be performed at the end of the computation to identify the best matched HMM models. Along the N dimension the computation is completely independent, however; due to the variation in the query sequence lengths, the traditional message passing interface (MPI) technique of splitting the file into equal numbers of sequences may result in load imbalance. Hence, the best way to parallelize code along the N dimension is to stripmine the query sequences as the processors finish working on their previous ones. This can be achieved using an MPI technique in which one processor reads the query sequence file and sends the sequences to all others.
Here, we have parallelized the HMMER code along the N dimension (query sequences) using a Unix-based file-locking technique. This implementation requires all the processors, nodes and computers working on a single problem to have access to a file system that honors Unix file locking. So far, we have tested the implementation on AIX, Linux, NFS and GPFS file systems and found our program to work correctly. The advantages of parallelizing with file locking are that (i) new processes can join or leave the process pool in the middle of computing; (ii) almost perfect load balancing can be achieved; (iii) processors with different clock speeds running different operating systems can be part of the same pool; and (iv) systems do not need to have MPI or parallel virtual machine (PVM) installed. Since the parallelized subroutines are written in a separate file as function calls, very little change is needed to the original HMMER code. These parallel routines are generic and, hence, can be used to parallelize other programs with similar computational requirements, such as the BLAST suite of programs. A brief description of the parallel function calls is provided below.
All the processes joining the work pool will call
iteratorInit(int strt, char* lockFile);
The first process that calls this subroutine will create the lockFile and write the starting index. This is the index from which sequence stripmining happens. The rest of the processes open the file stream to the lockFile. Then, inside the main work loop, every process calls
int iteratorNext();
resulting in every process getting a unique next available job (index). Each process computes the homology search for the sequence index matching the unique index obtained. These steps are iterated until no more sequences are left in the query sequence file. Outside the work loop at the end, all the processes call
iteratorStop();
which will close the lockFile. The user has to ensure a unique lockFile for each new query sequence file. If for some reason the work pool is stopped in the middle of a query sequence file, it can continue from the same point by using the same lockFile. In this version of the hmmpfam program, all processes write search results to a single output file. To minimize synchronization overheads, each process writes the output into a temporary buffer and writes once per sequence into the output file. For the sake of convenience, additional synchronization routines are provided for locking and unlocking.
This parallel algorithm has been tested on several platforms and file systems including Intel IA32, IA64 Linux platforms, power3 and power4-based AIX platforms and Sparc4-based Solaris platforms. The code was run on 800 processors of power4-based datastar machines and 256 processors of IA64-based teragrid machines located at the San Diego Supercomputer Center (SDSC) to test scalability issues.
Single-processor optimization of hmmpfam
Initially, we utilized version 2.2g of HMMER. Performance analysis revealed that the main computational kernel was accessing the 2D arrays with a wrong stride. Correcting this resulted in doubling of the performance; however, the HMMER developer has rectified this issue in release 2.3.2. In addition, HMMER reads the database file from disk for every sequence it searches. This is extremely inefficient for batch searches. We have corrected this by reading the whole database into memory once and conducting searches from memory.
Performance testing
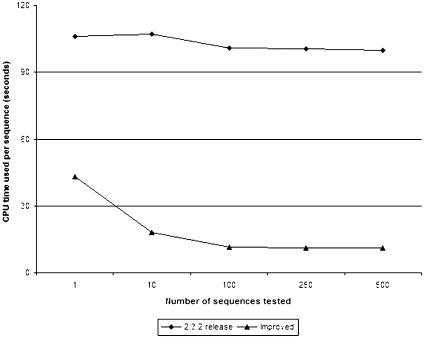
We compared our improved IA64 single-processor hmmpfam with the equivalent program from the original HMMER 2.3.2 release on an Itanium II IA64 processor (Figure 1). As expected, the per sequence CPU time for our program is reduced up to 10-fold in batch searches, while in the case of the original 2.3.2 release, it is linear to the number of sequences. Performance difference in single-sequence processing could be attributed to compilation differences. Performance differences resulting in the slope difference of the two plots may be due to the elimination of the need to reload the Pfam database from disk for every sequence in our case. Processing data from disk can be an order of magnitude slower than processing in memory.
Figure 1.
Comparison of CPU times between the original (release 2.3.2) and improved hmmpfam binaries.
Scaling tests were conducted on a 128-node Itanium II IA64 cluster. We used a fixed set of 500 sequences as the test set and ran on 1–32 processors. As expected, the time required to process a given set of sequences reduces linearly with the number of processors (data not shown). This also indicates that the parallelization overheads using our method are minimal. However, as the ratio of the number of sequences to the number of processors reduces to one, load imbalances due to the processing time differences between different sequences start to dominate, reducing parallel efficiency.
Pre-calculated results
We have generated pre-calculated Pfam search results for the entire Swiss-Prot and TrEmbl protein sequences (∼1.2 million), using our parallelized ‘hmmpfam’ on 256 processors of IA64-based teragrid machines located at the SDSC. Pfam searches have been performed separately, using either gathering thresholds or E-value cutoffs against glocal (Pfam_ls) or local (Pfam_fs) alignment models. Glocal and local search results were parsed and merged by removing all local search hits that are completely overlapped by glocal search hits.
Input and output formats
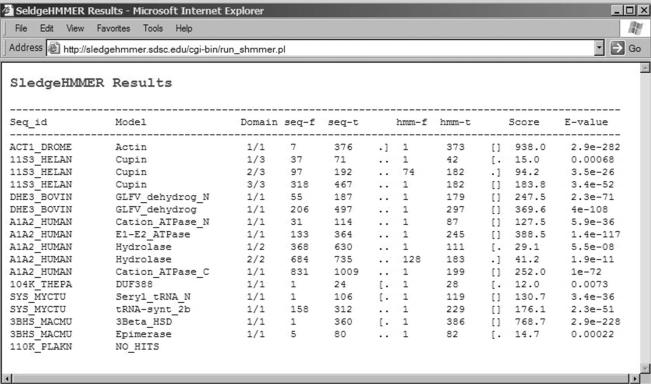
The program takes only FASTA-formatted sequences as input and searches can be done in any search mode, i.e. local, glocal or merged. The maximum E-value allowed for searches is 10. SledgeHMMER results are emailed to the user in a space-delimited, one hit per line tabular format (Figure 2). The response time for receiving results depends on the number of query sequences and the fraction of these existing in the pre-calculated entries.
Figure 2.
Screen shot showing SledgeHMMER results and output format.
DISCUSSION
The SledgeHMMER server provides a unique service to the scientific community wishing to batch search the Pfam database on the web. To the best of our knowledge, this is the only such server that has no limit on the number of input sequences. The current pre-calculated database covers ∼70% of the available non-redundant protein sequence space, and we plan to add additional new sequences from other resources such as the NR database from GenBank and SEQRES from the PDB (Protein Data Bank) in the future. Our pre-calculated database is updated for every new release of the Pfam database or SPTr databases (Swiss-Prot + TrEMBL) in order to provide access to the most current data. Single-processor optimized hmmpfam binaries and the source code can be downloaded from the current web server. In the future, we intend to make such binaries available for many additional operating systems and propagate these modifications to other HMMER programs as well.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Pfam consortium and the HMMER developers for making available such valuable tools to the scientific community.
REFERENCES
- 1.Bateman A., Coin,L., Durbin,R., Finn, R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L., Studholme,D.J., Yeats,C. and Eddy,S.R. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guda C., Fahy,E. and Subramaniam,S. (2004) MITOPRED: a genome-scale method for prediction of nuclear-encoded mitochondrial proteins. Bioinformatics, 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 3.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]