Abstract
The Robetta server (http://robetta.bakerlab.org) provides automated tools for protein structure prediction and analysis. For structure prediction, sequences submitted to the server are parsed into putative domains and structural models are generated using either comparative modeling or de novo structure prediction methods. If a confident match to a protein of known structure is found using BLAST, PSI-BLAST, FFAS03 or 3D-Jury, it is used as a template for comparative modeling. If no match is found, structure predictions are made using the de novo Rosetta fragment insertion method. Experimental nuclear magnetic resonance (NMR) constraints data can also be submitted with a query sequence for RosettaNMR de novo structure determination. Other current capabilities include the prediction of the effects of mutations on protein–protein interactions using computational interface alanine scanning. The Rosetta protein design and protein–protein docking methodologies will soon be available through the server as well.
INTRODUCTION
Robetta is an Internet service that provides automated structure prediction and analysis tools that can be used to infer protein structural information from genomic data. The server uses the first fully automated structure prediction procedure that produces a model for an entire protein sequence in the presence or absence of sequence homology to protein(s) of known structure. Robetta parses input sequences into domains and builds models for domains with sequence homology to proteins of known structure using comparative modeling, and models for domains lacking such homology using the Rosetta de novo structure prediction method. Domain predictions and molecular coordinates of models spanning the full-length query are given as results. The server can also utilize nuclear magnetic resonance (NMR) constraints data provided by the user to determine protein structures using the RosettaNMR (1–3) protocol. These tools can be used in conjunction with current structural genomics initiatives to help accelerate structure determination and gain structural insight for targeted open reading frames (ORFs). Additionally, since multidomain proteins are often difficult to crystallize and many are too large for NMR structure determination, domain prediction using Robetta can aid structural genomics efforts by expanding the pool of targets from which structures can be determined. The Structural Genomics of Pathogenic Protozoa (SGPP; http://www.sgpp.org) consortium is currently using an in-house version of Robetta to identify fragments that express and crystallize from ORFs that do not express as a full chain, and to aid structure refinement. Robetta also provides the ability to identify energetically important side-chains involved in the interface of protein–protein complexes using ‘computational interface alanine scanning’ (4,5). The ultimate goal for Robetta is to provide structural information of sufficient quality to aid research, infer function and assist drug design. Comparative models are already being used to infer function and guide experimental efforts, and the research field as a whole continues to improve as shown in the Critical Assessment of Structure Prediction (CASP-5, and CAFASP-3 for ‘Fully Automated’) experiments (6,7). Robetta was among the top performers in these assessments.
METHODS USED
Robetta uses a fully automated implementation of the Rosetta software package for protein structure prediction. The Rosetta method is described in detail in references (7–9) and the use of Rosetta in CASP-5 and CAFASP-3 is described in references (6,7).
Domain prediction
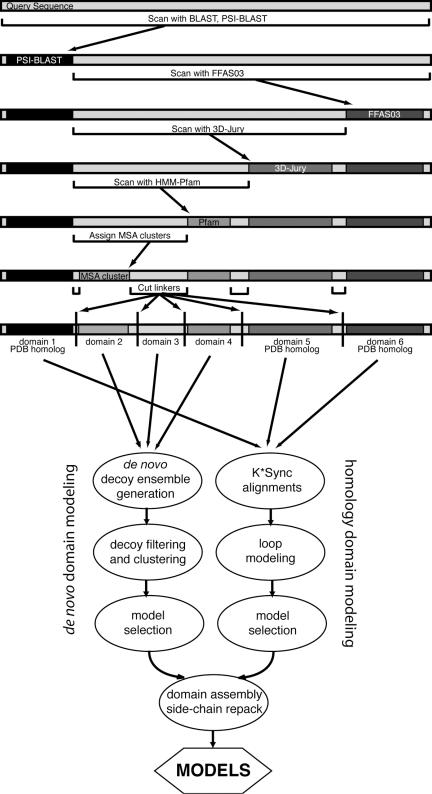
In an attempt to predict structures for full-length protein sequences, Robetta uses a domain prediction method called ‘Ginzu’ (6) as the initial step for structure prediction. Ginzu is a hierarchical screening procedure that first uses BLAST, PSI-BLAST (10), FFAS03 (11,12) and 3D-Jury (13,14) to detect regions in the query sequence that are homologous to experimentally determined structures, and then proceeds with multiple sequence alignment (MSA) based methods to predict putative domains (Figure 1). The procedure is ordered by the reliability of each method, starting with the most reliable method (BLAST), followed by the next method in terms of confidence level (PSI-BLAST), and so forth. If a match is found, the remaining unmatched portion of the sequence is used as input for the next step. Regions that are homologous to sequences with known structures are modeled using our comparative modeling protocol. Unassigned regions are either treated as domain linkers if they are less than 50 residues, or are searched against Pfam-A (15) using HMMER (16) for regions that are likely to be domains. The final step attempts to identify putative domains in the remaining uncovered sequence by using an MSA of the full-length target derived from a PSI-BLAST search against the NCBI non-redundant (NR) protein sequence database. The most populated non-overlapping clusters of sequences in the MSA are assigned as domains, and the final cut points are determined in unassigned regions at positions that have a high incidence of sequence termini, a strong loop prediction using PSIPRED (17) and a reduced occupancy of aligned residues. A weak positional preference is also used to place cuts near the middle of unassigned regions. Putative domains from Pfam-A and the MSA-based method are marked for de novo structure prediction. These domains may be parsed even further to satisfy size limitations of our de novo protocol (∼200 residue limit). A significant fraction of domains in the PDB (Protein Data Bank) are within this length (18).
Figure 1.
Ginzu domain parsing hierarchy and modeling protocol. The query sequence is scanned for matches to known structures and regions that are likely to be domains. Sections that are not covered but are large enough to be a domain (at least 50 residues) are passed on as input for the next step. The order is based on the relative accuracy of each step. Homologous structure searches are performed first, followed by a search against Pfam-A and then parsing based on MSA sequence clusters. Boundaries are assigned so that putative domains without homologous structures are within size limits accessible to the Rosetta de novo protocol. Regions that are homologous to sequences with known structures are used for comparative modeling. Matches to Pfam-A, MSA cluster domains and remaining uncovered regions of sufficient size are subjected to de novo structure prediction. Domain models are assembled into a full model in the last step.
Comparative modeling
The comparative modeling method is described in greater detail in a previously published paper (6). Rather than simply taking the alignment available from the method used to detect the structural homolog, Robetta attempts to obtain an improved alignment by employing a method, called K*Sync (D. Chivian, manuscript in preparation), that utilizes residue profile–profile comparison, secondary structure prediction and information about elements that are obligate to the fold to produce a single default alignment by dynamic programming (19). K*Sync also parametrically generates an alignment ensemble from which decoy models are selected. Unaligned regions are treated as loops, which are then modeled in the context of the fixed template using the Rosetta fragment insertion method (20). An energy function is used that includes a gap closure term to ensure continuity of the peptide backbone. Short and medium loops (<17 amino acids) are assembled first, the lowest scoring set of loops is added to the template, and long loops are then built. Multiple independent simulations are carried out, and the lowest scoring conformation is selected as the loop combination appropriate to a given alignment. Four models are selected from this ensemble using different variants of the Rosetta energy function, and returned with the default K*Sync alignment-derived model.
De novo structure prediction and Mammoth
Robetta uses a slightly modified version of the de novo structure prediction protocol that has been described previously (6). Modifications to the original method were made to run queries within reasonable timescales for a public server. Like the original protocol, Robetta generates three- and nine-residue fragment libraries that represent local conformations seen in the PDB, and then assembles models by fragment insertion using a scoring function that favors protein-like features. Robetta generates 10 000 decoys for the original query and 5000 decoys for up to two sequence homologs. Then 2000 query decoys and 1000 decoys of each homolog are selected based on score and on whether they pass filters that eliminate decoys having too many local contacts or unlikely strand topologies. The selected decoys are then clustered based on Cα root-mean-square deviation (RMSD) over all ungapped positions. The top 9 cluster centers are chosen as the top ranked models, and the best scoring model that passed the filters described above is chosen as the 10th model. Searches with these final models are then carried out against a representative set of PDB chains to find similar structures using Mammoth (21) to identify potential similarities to proteins of known structure. Mammoth identifies the longest structural superposition between the model and proteins in the PDB and reports a Z-score (22), which represents the likelihood of getting a similar length match between similarly sized proteins by chance. The results of these searches will be used in a confidence function that will be added in a future version.
Domain assembly and side-chain packing
If a query is parsed into multiple domains, the final step in our structure prediction procedure is to assemble the domain models into a continuous full-length structure. Robetta uses an iterative domain assembly protocol that starts with the N-terminal domain, and attempts domain association by fragment insertion in the putative linker region assigned by Ginzu using the same scoring method as in our de novo protocol. If the chain contains more than two domains, the third domain is added to the previously assembled model, and the procedure continues until the whole chain is assembled. Although we are working on improving this method, this final step must be considered as an aesthetic treatment since it was benchmarked using high-resolution crystal structures, and is likely to be inaccurate for low-resolution models. Once the chain is completely assembled, the side-chains of the final model are repacked using a Monte Carlo algorithm (23) with a backbone-dependent side-chain rotamer library (24).
RosettaNMR
A user can provide experimental NMR constraints data for RosettaNMR structure determination. The RosettaNMR method is described in already published papers (1–3). The protocol used by Robetta is slightly different from the published methods. Robetta uses the same method as RosettaNMR to generate fragment libraries that are consistent with chemical shifts, NOE constraint data and, if sufficient data exist, residual dipolar couplings. The fragment libraries are then used with the same RosettaNMR de novo fragment insertion method that utilizes the constraints data in its scoring function to generate decoys. However, at this point, the protocols diverge. Robetta continues its de novo protocol of clustering decoys, and selecting cluster centers and the lowest scoring decoy for the final models, instead of choosing just the top scoring models followed by model refinement as RosettaNMR does. Because of this, results with Robetta are likely to be slightly less accurate.
Interface alanine scanning
Robetta includes a ‘computational interface alanine scanning’ (4) method that predicts the effects of truncation mutations on the stability of protein–protein complexes as described in references (4,5). In short, the procedure identifies residues that are involved in the protein–protein interface, and uses a simple free energy function to calculate the changes in the binding free energy upon single substitutions of each side-chain to alanine. In a test set of 233 mutations in 19 protein–protein complexes, 79% of the residues identified as energetically important and 68% of neutral residues were correctly predicted.
INPUT, OUTPUT AND OPTIONS
Registration
Users must register (http://robetta.bakerlab.org/register.jsp) before submitting jobs to Robetta.
Structure prediction server
Sequences submitted to the structure prediction server must be in one-letter amino acid format. They can either be pasted into the submission form, or uploaded from a file. Users have the option to submit a sequence for either domain identification or full structure prediction. A user also has the option to specify the PDB id and chain for comparative modeling.
For RosettaNMR submissions, a user must upload experimental NMR constraints data (chemical shifts, NOE data and/or residual dipolar couplings). The required input format for each type of data is described at http://robetta.bakerlab.org/documents/data_formats.jsp.
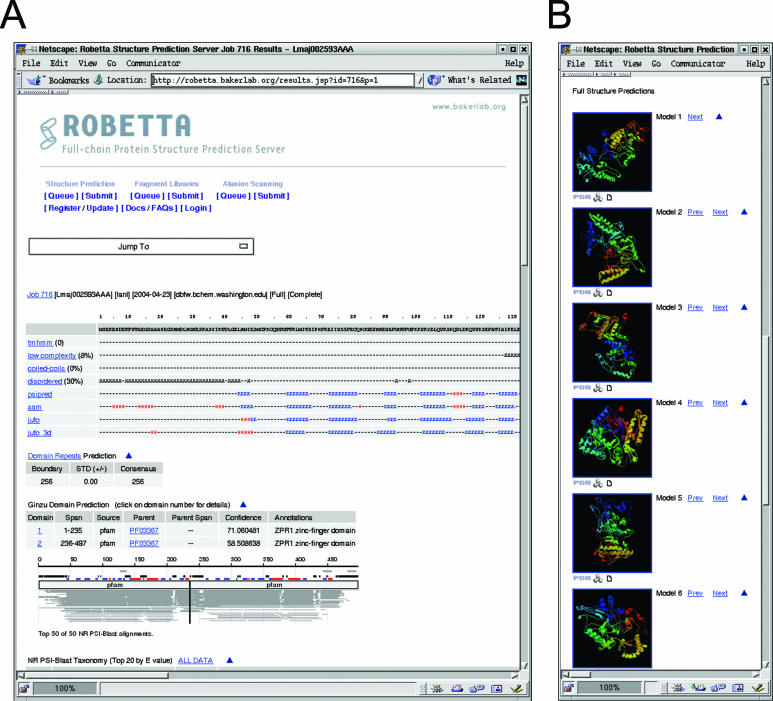
Results for a specific job are provided through the web interface (Figure 2) by clicking on the job id listed in the queue table (http://robetta.bakerlab.org/queue.jsp). For full structure predictions, coordinates are also emailed to the user. For added insight, the following results are displayed along with the predicted models (Figure 2A):
Figure 2.
An example of structure prediction results. Screen shot of the top of the web page (A) and the first 6 of 10 structure predictions located at the bottom of the web page (B).
the prediction of transmembrane helices using TMHMM (25,26);
low-complexity regions assigned by the program SEG (27);
coiled-coils prediction using COILS (28);
the prediction of disordered regions using DISOPRED (29);
secondary structure predictions using PSIPRED (17), SAM-T99 (30,31), Jufo and Jufo3D (32);
the results listed above, domain predictions and the NR PSI-BLAST multiple sequence alignment used for the last step in the domain prediction protocol condensed into an image to help corroborate the domain prediction results;
domain repeats prediction using REPRO (33,34); predicted boundaries are given if repeats are detected;
the top NR PSI-BLAST results and annotations for the top 20 species determined by lowest E-values.
The models for the full query are displayed as images at the bottom of the page (Figure 2B). The coordinates for these models can be downloaded from the web site by clicking on the icons represented below each model image.
Specific results are also provided for each domain by clicking on the domain number listed in the Ginzu domain prediction results table. For comparative models, the K*Sync alignment used for modeling is displayed. For de novo models, the Mammoth (21) structure-model comparison results are displayed for the top 10 matches with Z-scores >4.5. The actual Mammoth structure-model alignment can be downloaded by clicking on the Z-score and viewed for further inspection using a molecular viewer such as RasMol. Users can download domain models by clicking on the icons below each domain model image.
Interface alanine scanning
A detailed description and directions on how to use the web interface for computational interface alanine scanning have already been published (4). As input, a user must supply the PDB coordinates of the protein–protein complex and define which chains belong to the partners of the binding interface to be scanned. The user also has the option to supply a list of interface residues to consider. If a list is not provided, all side-chains involved in the interface are used. Results are emailed as a list of calculated binding free energy changes (ΔΔGbind) for each interface side-chain considered.
Fragment libraries
Robetta includes a fragment library server to accommodate research groups who are running the Rosetta software package locally but do not have the ability to generate fragment libraries. For Rosetta fragment libraries, a user must submit a query sequence in single-letter amino acid format. NMR constraints data can also be uploaded to generate fragment libraries for use with RosettaNMR. A URL to download results is emailed to the user when the job completes. Since the fragment libraries are large (on the order of megabytes), they will be removed from the server a week after the job completes.
STRUCTURE PREDICTION PERFORMANCE AND LIMITATIONS
The performance of Robetta has been evaluated in the CASP-5 and CAFASP-3 experiments, and is continually benchmarked using LIVEBENCH (http://bioinfo.pl/LiveBench) (35). Robetta's performance in CASP-5 and CAFASP-3 has been discussed in previous publications (6,7). In general, Robetta compared quite favorably with the other servers for the homology modeling and fold recognition targets, and did a reasonable job with its de novo protocol for the remote fold recognition and new fold targets. A thorough evaluation of the comparative modeling protocol used by Robetta for LIVEBENCH will be discussed in another manuscript (D. Chivian, manuscript in preparation). Robetta's performance on de novo predicted domains in LIVEBENCH 7 and 8 is summarized in Table 1. Since de novo structure predictions can still not be done with high accuracy, we use two relatively generous definitions of correct predictions in the table. First, a domain is considered to be correctly modeled if at least 1 of the 10 models has a Mammoth alignment of 50 or more residues with an RMSD of 4 Å or less to the native structure (definition I, upper part of Table 1), or second, a Mammoth Z-score of 6 or greater to the native structure (definition II, lower part of Table 1). Under both definitions, Robetta produces correct predictions for more than half of the domains, and does particularly well with domains that consist of either all-alpha or alpha-beta secondary structure. Domains with all-beta secondary structure containing anti-parallel β–sheets and within the length range of 151–200 residues were also modeled well. Domains with <100 residues were usually incorrect due to errors in domain predictions.
Table 1. Robetta's performance on de novo predicted domains in LIVEBENCH 7 and 8.
| Length: | <100 residues | 100–150 residues | 151–200 residues | >200 residues | ||||
|---|---|---|---|---|---|---|---|---|
| ss | Correct | Total | Correct | Total | Correct | Total | Correct | Total |
| Correct if at least 50 residues have an RMSD of 4 Å or less to the native structure | ||||||||
| α | 2 (22%) | 9 | 14 (56%) | 25 | 9 (90%) | 10 | 2 (67%) | 3 |
| β | 0 | 2 | 3 (38%) | 8 | 4 (67%) | 6 | 0 | 2 |
| αβ | 2 (18%) | 11 | 33 (83%) | 40 | 16 (64%) | 25 | 4 (80%) | 5 |
| Correct if the Mammoth MaxSub Z-score is 6 or greater to the native structure | ||||||||
| α | 3 (33%) | 9 | 13 (52%) | 25 | 7 (70%) | 10 | 1 (33%) | 3 |
| β | 1 (50%) | 2 | 3 (38%) | 8 | 4 (67%) | 6 | 0 | 2 |
| αβ | 2 (18%) | 11 | 28 (70%) | 40 | 10 (40%) | 25 | 2 (40%) | 5 |
ss, native secondary structure.
There are many limitations to consider when using Robetta, as with all structure prediction methods. The de novo protocol is optimized for small single domain proteins (<120 residues). Within this limit, models are frequently around 3–7 Å RMSD to more than half of the native structure. Above this limit, models are still likely to have at least 50 residues within 4 Å RMSD, as shown in Table 1. For comparative modeling, the quality of the model is greatly dependent on the correct selection of the best possible parent template and alignment. Because of these factors, results are highly dependent on the accuracy of the domain assignments. A general rule to follow is that BLAST, PSI-BLAST, FFAS03 and 3D-Jury parent detections should be considered the most reliable, in that order. Domains predicted from Pfam-A and the MSA should be treated with caution, particularly for longer domains and also those that were assigned solely by the MSA.
THROUGHPUT
Robetta is computationally demanding and requires significantly sized computer clusters for structure predictions to be made on a reasonable timescale. Therefore, to try to meet demand, Robetta was designed to run on computer clusters that may be distributed regionally as mirrors. It takes Robetta around 4–6 h to run a 150 residue query on a single cluster of about 80 CPUs. With two mirrors of similarly sized clusters, Robetta is currently able to process around 10 normally sized jobs per day. Because of this computational demand, users are only allowed to submit one sequence to the job queue at a time. Once a job is submitted for structure prediction, the time it takes to complete the job depends on the length of the query, the number of jobs already queued and the number of available CPUs. This time is estimated in days for every job and is listed in the queue table to give users an idea of when jobs will finish. Domain predictions in the absence of 3D-Jury and fragment libraries are far less demanding, and take around 15–30 min on a single processor. Computational alanine scanning is a quick procedure that takes just a few minutes to run on a single CPU.
FUTURE WORK
We plan to broaden the scope of the Robetta server by adding protein design (36) and protein–protein docking (37) capabilities which have been developed in our laboratory as part of the Rosetta software package. We are also planning to add confidence values for our de novo structure prediction results derived from a function similar to one used in a previous study (38). Comparative modeling options will be added that will allow users to provide their own starting templates. We hope to improve Robetta's throughput by expanding the distributed network of cluster mirrors.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Charlie Strauss and Richard Bonneau for providing the cluster computing resources that make up the Robetta mirrors. We thank Carol Rohl and Tanja Kortemme for allowing their RosettaNMR and Interface Alanine Scanning methods, respectively, to be provided among the Robetta services. We also thank all the software contributors who have provided various components used by Robetta, and Keith Laidig and Formix for designing, implementing and administering the Robetta hardware architecture. We additionally appreciate the useful feedback we have received from Adam Godzik and Leszek Rychlewski, who have also generously allowed Robetta to utilize information from their servers, as well as Sean Eddy for the use of HMMER, Richard George and Jaap Heringa for the use of REPRO, David Jones for the use of PSIPRED and DISOPRED, Kevin Karplus for the use of the SAM software, Anders Krogh for the use of TMHMM, Andrei Lupas for the use of COILS, Jens Meiler for the use of Jufo, Angel Ortiz for the use of Mammoth, and John Wootton and Scott Federhen for the use of SEG. This work was supported by NIH grant No. P50 GM64655 and the Howard Hughes Medical Institute.
REFERENCES
- 1.Bowers P.M., Strauss,C.E. and Baker,D. (2000) De novo protein structure determination using sparse NMR data. J. Biomol. NMR, 18, 311–318. [DOI] [PubMed] [Google Scholar]
- 2.Rohl C.A. and Baker,D. (2002) De novo determination of protein backbone structure from residual dipolar couplings using Rosetta. J. Am. Chem. Soc., 124, 2723–2729. [DOI] [PubMed] [Google Scholar]
- 3.Meiler J. and Baker,D. (2003) Rapid protein fold determination using unassigned NMR data. Proc. Natl Acad. Sci. USA, 100, 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kortemme T., Kim,D.E. and Baker,D. (2004) Computational alanine scanning of protein–protein interfaces. Sci. STKE, 2004, pl2. [DOI] [PubMed] [Google Scholar]
- 5.Kortemme T. and Baker,D. (2002) A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl Acad. Sci. USA, 99, 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chivian D., Kim,D.E., Malmstrom,L., Bradley,P., Robertson,T., Murphy,P., Strauss,C.E., Bonneau,R., Rohl,C.A. and Baker,D. (2003) Automated prediction of CASP-5 structures using the Robetta server. Proteins, 53 (Suppl. 6), 524–533. [DOI] [PubMed] [Google Scholar]
- 7.Bradley P., Chivian,D., Meiler,J., Misura,K.M., Rohl,C.A., Schief,W.R., Wedemeyer,W.J., Schueler-Furman,O., Murphy,P., Schonbrun,J. et al. (2003) Rosetta predictions in CASP5: successes, failures, and prospects for complete automation. Proteins, 53 (Suppl. 6), 457–468. [DOI] [PubMed] [Google Scholar]
- 8.Simons K.T., Ruczinski,I., Kooperberg,C., Fox,B.A., Bystroff,C. and Baker,D. (1999) Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins, 34, 82–95. [DOI] [PubMed] [Google Scholar]
- 9.Simons K.T., Kooperberg,C., Huang,E. and Baker,D. (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol., 268, 209–225. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaroszewski L., Rychlewski,L. and Godzik,A. (2000) Improving the quality of twilight-zone alignments. Protein Sci., 9, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rychlewski L., Jaroszewski,L., Li,W. and Godzik,A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginalski K. and Rychlewski,L. (2003) Detection of reliable and unexpected protein fold predictions using 3D-Jury. Nucleic Acids Res., 31, 3291–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginalski K., Elofsson,A., Fischer,D. and Rychlewski,L. (2003) 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics, 19, 1015–1018. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 17.Jones D.T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 18.Wheelan S.J., Marchler-Bauer,A. and Bryant,S.H. (2000) Domain size distributions can predict domain boundaries. Bioinformatics, 16, 613–618. [DOI] [PubMed] [Google Scholar]
- 19.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 20.Rohl C.A., Strauss,C.E., Chivian,D. and Baker,D. (2004) Modeling structurally variable regions in homologous proteins with Rosetta. Proteins, 55, 656–677. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz A.R., Strauss,C.E. and Olmea,O. (2002) MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci., 11, 2606–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siew N., Elofsson,A., Rychlewski,L. and Fischer,D. (2000) MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics, 16, 776–785. [DOI] [PubMed] [Google Scholar]
- 23.Kuhlman B. and Baker,D. (2000) Native protein sequences are close to optimal for their structures. Proc. Natl Acad. Sci. USA, 97, 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunbrack R.L. Jr and Cohen,F.E. (1997) Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci., 6, 1661–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 26.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 27.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 29.Jones D.T. and Ward,J.J. (2003) Prediction of disordered regions in proteins from position specific score matrices. Proteins, 53 (Suppl. 6), 573–578. [DOI] [PubMed] [Google Scholar]
- 30.Karplus K., Karchin,R., Barrett,C., Tu,S., Cline,M., Diekhans,M., Grate,L., Casper,J. and Hughey,R. (2001) What is the value added by human intervention in protein structure prediction? Proteins, 45 (Suppl. 5), 86–91. [DOI] [PubMed] [Google Scholar]
- 31.Karplus K., Barrett,C. and Hughey,R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 32.Meiler J. and Baker,D. (2003) Coupled prediction of protein secondary and tertiary structure. Proc. Natl Acad. Sci. USA, 100, 12105–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heringa J. (1994) The evolution and recognition of protein sequence repeats. Comput. Chem., 18, 233–243. [DOI] [PubMed] [Google Scholar]
- 34.Heringa J. and Argos,P. (1993) A method to recognize distant repeats in protein sequences. Proteins, 17, 391–411. [DOI] [PubMed] [Google Scholar]
- 35.Rychlewski L., Fischer,D. and Elofsson,A. (2003) LiveBench-6: large-scale automated evaluation of protein structure prediction servers. Proteins, 53 (Suppl. 6), 542–547. [DOI] [PubMed] [Google Scholar]
- 36.Kuhlman B., Dantas,G., Ireton,G.C., Varani,G., Stoddard,B.L. and Baker,D. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science, 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- 37.Gray J.J., Moughon,S., Wang,C., Schueler-Furman,O., Kuhlman,B., Rohl,C.A. and Baker,D. (2003) Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol., 331, 281–299. [DOI] [PubMed] [Google Scholar]
- 38.Bonneau R., Strauss,C.E., Rohl,C.A., Chivian,D., Bradley,P., Malmstrom,L., Robertson,T. and Baker,D. (2002) De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol., 322, 65–78. [DOI] [PubMed] [Google Scholar]