Abstract
Magnetic resonance imaging (MRI) is routinely used to obtain anatomical images that have greatly advanced biomedical research and clinical health care today, but the full potential of MRI in providing functional, physiological, and molecular information is only beginning to emerge. In this work, we sought to provide a gene expression marker for MRI based on bacterial magnetosomes, tiny magnets produced by naturally occurring magnetotactic bacteria. Specifically, magA, a gene in magnetotactic bacteria known to be involved with iron transport, is expressed in a commonly used human cell line, 293FT, resulting in the production of magnetic, iron-oxide nanoparticles by these cells and leading to increased transverse relaxivity. MRI shows that these particles can be formed in vivo utilizing endogenous iron and can be used to visualize cells positive for magA. These results demonstrate that magA alone is sufficient to produce magnetic nanoparticles and that it is an appropriate candidate for an MRI reporter gene.
Keywords: SPIO, molecular imaging, contrast agent, gene expression, reporter gene
Synthetic superparamagnetic iron-oxide (SPIO) nanoparticles have been widely used for targeted molecular imaging applications (1–5). One major application is in vivo tracking of stem cells (6,7) and tumor progression (5). Labeling nonphagocytic cells in culture using modified particles, followed by transplantation or transfusion into living organisms, has made it possible to monitor cellular distribution in vivo, including cell migration and trafficking.
A limitation of using synthetic SPIO is the need to label cells in vitro with presynthesized nanoparticles prior to cell transplant. As a result, particle concentration within cells decreases over time as the cells grow and divide, and particles cannot be readily linked directly to in vivo gene expression. One way to overcome this is to utilize a genetic approach. Green fluorescent protein (GFP) is perhaps the most well-known genetic marker for optical imaging. Magnetic resonance spectroscopy (MRS) has been used to detect creatine kinase (8) and chemical shift imaging (CSI) to observe beta-galactosidase (9) activity. MRI gene expression strategies thus far include detection of beta-galactosidase activity (10,11), frequency-selective targeting of amide protons of expressed proteins (12), and expression of natural iron homeostasis proteins such as the transferrin receptor (13) and ferritin (14,15). For the transferrin receptor approach, administration of exogenous transferrin coupled to magnetic particles is required. Thus far, only ferritin exists as a purely in vivo superparamagnetic MRI marker. Although the relaxivity of ferritin is dependent on factors such as iron loading, data obtained on solutions of iron-containing materials suggest iron-oxide particles could provide higher relaxivity (16). In the present work, we report the gene-mediated cellular production of magnetic iron-oxide nanoparticles of the same composition as synthetic SPIO preparations using a gene present in magnetotactic bacteria, making it a possible MRI gene reporter.
Magnetotactic bacteria, a diverse set of Gram-negative bacteria that exhibit motility thought to be directed by the earth’s magnetic field (17), produce magnetosomes which are naturally synthesized intracellular magnetic structures (18). Each species of magnetotactic bacteria has a different, but specific, type and shape of magnetosome that can affect the MRI signal in the same way as synthetic SPIO nanoparticles (19). It is likely that natural magnetosome production requires multiple genes. Recent work has identified a 98-kb genomic island (20), among other regions, in the Magnetospirillum magneticum AMB-1 strain related to magnetosome regulation, and iron response studies have revealed 464 upregulated and 263 downregulated genes (21). Our efforts here focus on magA, a gene known to be involved in production of magnetosomes in M. magneticum (18,22,23). Nakamura et al. (22,23) found that magA encodes a protein with high sequence homology to cation-efflux proteins, KefC, a K+-translocating protein in E. coli, and NapA, a putative Na+/H+ antiporter from Enterococcus hirae. With magA expressed in E. coli, inverted membrane vesicles prepared from these cells were shown to transport Fe(II) in an energy-dependent manner, leading to accumulation of Fe(II) in the vesicle. These observations indicate MagA functions as an H+/Fe(II) antiporter. Consistent with this conjecture, a recent full genomic analysis found deletions in magA among a set of nonmagnetic mutants (24).
In this work, magA was expressed in mammalian cells and its effect was evaluated in vitro and in vivo. It was shown that: 1) its expression leads to the formation of magnetic nanoparticles that strongly affect MRI signal; and 2) its expression in vivo generates readily detectable MR contrast.
MATERIALS AND METHODS
Molecular Biology
The magA gene (provided by L.E. Bertani, California Institute of Technology) was cloned into a doxycycline-inducible lentiviral vector to control the expression of magA. In this way, MRI changes could be correlated with gene expression via doxycycline administration. magA was constructed downstream of a tetracycline response element (TRE) under the control of a mini cytomegalovirus (miniCMV) promoter activable by rtTA2s-M2 (obtained from Dr. W. Hillen). Replication defective lentivirus was generated by cotransfection of this vector, pΔ8.9 (composed of structural genes for virion assembly; obtained from Dr. C. Lois, MIT) and pVSV-G (Invitrogen, Carlsbad, CA, USA) into 293FT packaging cells (Invitrogen). Cell culture medium consisted of Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum (heat-inactivated at 60°C for 60 min) and supplemented with 50 U penicillin/50 μg streptomycin/ml and 2 mM glutamine. Culture media was collected at 48 h posttransfection, and the resulting virus was used to infect 293FT cells. After several passages to ensure stable integration, single cells were picked and expanded to create clonal cell lines. Polymerase chain reaction (PCR) was performed to ascertain the presence of the magA gene to identify positive lines. A total of three clonal lines were evaluated for imaging and found to produce MRI signal changes. For the following experiments, one of these clonal cell lines, named 2B5, was used.
MRI of Cells in Culture
Four plates were prepared for each cell line: 1) no doxycycline, no iron supplement; 2) no doxycycline, but with iron supplement; 3) with doxycycline, without iron; and 4) with doxycycline, with iron. The appropriate cell samples were incubated with 2 μg/ml doxycycline and/or 200 μM Fe (from ferric citrate) for 4 d. For dosing experiments, 2B5 cells were initially cultured under normal conditions. Doxycycline was added to media (0.5, 1.0, and 2.0 μg/ml) for 24 h, after which media was replaced with fresh media supplemented with 200 μM Fe for 1–4 d. For iron dosing, cells were cultured with 0.5 μg/ml doxycycline for 24 h, after which, media was replaced with fresh media with varying iron concentration for 4 d.
In all cases, cells were trypsinized and collected. A total of 107 cells in ~100-μl pellets were imaged using a Siemens 3T Trio MR scanner (Siemens Medical Solutions, Malvo, PA, USA). T2 and R2 (R2 ≡ 1/T2) were calculated by fitting decay curves produced from a Carr-Purcell-Meiboom-Gill (CPMG) sequence. For dosing experiments, three measurements were made, and error bars are ± 1 SD. Imaging parameters: TE = 20–400 ms in increments of 20 ms, TR = 1500 ms, FOV = 128 × 128 mm, in-plane resolution = 0.5 mm, and a slice thickness = 1 mm. Image processing and analysis were performed using MATLAB and Excel.
Iron Uptake Determination
Iron uptake was determined using an o-phenanthroline procedure modified from a previously reported method (25). 2B5 cells were grown under conditions of 0, 0.5, and 2.0 μg/ml doxycycline and 200 μM Fe for 4 d. Original 293FT cells were cultured in the presence of 200 μM Fe for 4 d to serve as baseline. A total of 2 × 106 cells were counted, pelleted, resuspended in water, and vortexed to rupture cell membranes. 0.15% v/v mercaptoacetic acid was added to each sample and incubated overnight prior to analysis. o-Phenanthroline was added, reaching a final concentration of 0.0075% w/v. Samples and standards both contained 0.2% hydroxylamine hydrochloride to maintain iron in the 2+ state and sodium citrate (150 μl/ml of iron solution) to maintain acidic pH for Fe-phenanthroline complex formation. Absorbance was read at 510 nm. Iron uptake due to magA was determined by subtracting total iron in the 293FT sample from the 2B5 samples. Samples were measured in triplicate and error bars are ±SD.
Real-Time (RT)-PCR
2B5 cells were grown under conditions of 0, 0.5, and 2.0 μg/ml doxycycline and 200 μM Fe for 4 d. First strand complementary deoxyribonucleic acid (cDNA) was created from DNase I-treated ribonucleic acid (RNA) using Superscript III (Invitrogen). Subsequent PCR used Taq DNA polymerase (Invitrogen) over 35 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Primers were as follows: forward, 5′-catcccgaactgacctatgc-3′; and reverse, 5′-acgaacagcagcatcagc-3′, resulting in a 200–base pair amplicon.
Electron Microscopy
2B5 cells were cultured with 2 μg/ml doxycycline and 200 μM Fe for 4 d. The cells were then fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer followed by 1% OsO4 in the same buffer. Cells were then dehydrated and embedded in Eponate resin. Ultrathin sections (60–70 nm) were observed on a Hitachi H-7500 transmission electron microscope (TEM) without counterstaining.
Cytotoxicity Assays
A total of 103 cells/well were plated in a 96-well plate and incubated for 5 d under various conditions of doxycycline and iron supplement as indicated. For the glucose-6-phosphate dehydrogenase (G6PD) assay, which measures release of cytosolic G6PD (increased release indicates cytotoxicity), three wells per condition were measured, along with a control in which cells were lysed to release all G6PD into the assay medium (for quantification). Vendor protocol was followed (Molecular Probes, Eugene, OR, USA). For the tetrazolium salt ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide [MTT]) assay, four wells per condition were measured according to vendor protocol (ATCC, Manassas, VA, USA). For the apoptosis assay, three wells per condition were measured according to vendor protocol (Promega, Madison, WI, USA) with the modification of using Hoescht (5 min at room temperature), rather than propidium iodide to stain all cells (both apoptotic and nonapoptotic). Superoxide anion was assessed according to vendor protocol (Sigma, St. Louis, MO, USA). Analysis was performed with Microsoft Excel, and error bars are ±SD. Note that due to cell division, it is not possible to examine the same cell for periods longer than 5 d.
In Vivo Animal Experiments
For in vivo experiments, a clonal cell line 2B5R was established by infection of 2B5 with a lentivirus containing DsRed, a fluorescence label, under constitutive control of the CMV promoter, so cells could be also tracked via fluorescence imaging. Single cells were plated to produce the clonal cell line, 2B5R. For cell transplantation, burr holes were made through the dorsal skull using coordinates chosen for injection into the striatum (+0.4 mm anterior and ±1.75 mm medial/lateral relative to bregma, and −2.5 mm ventral from the dorsal surface of the brain). Uninduced 2B5R cells were transplanted into the left mouse striatum (right side of image). The contralateral side was transplanted with a 293FT cell line inducible for GFP, but not magA. In each case, transplantation was made using a stereotaxic platform fitted with an automated injector and Hamilton syringe and using 105 cells suspended in a total volume of 4 μl in phosphate buffered saline (PBS). Cells were allowed to grow for 5 d, during which doxycycline was administered in drinking water (5 mg/ml) and in the food (200 ppm). On day 5, mice were imaged and sacrificed for histology. A total of three mice were studied with this protocol. Control animals (N = 3) were performed following the same protocol, but without addition of doxycycline.
Animal MRI was performed under 1.5% isoflurane on a 9.4T Bruker system fitted with a heated mouse cradle. For T2*-weighting, fast low-angle shot (FLASH) was used with TE = 6.0 ms, TR = 50 ms, FOV = 256 × 256 mm, in-plane resolution = 88 μm, and slice thickness = 0.450 mm, and for T2-weighting, a spin-echo sequence was applied with TE = 10.21 ms, TR = 1000 ms, FOV = 256 × 256 mm, in-plane resolution = 86 μm, and slice-thickness = 0.654 mm. Animals were sacrificed postimaging and brains collected for histology. Brains were fixed in 4% paraformaldehyde for 1 d, followed by 2 d in 30% sucrose. Sections of 40 μm were cut on a Leica CM3050S Cryostat (Nussloch, Germany). Sections were washed with PBS, stained with Hoechst for five min, and washed twice again with PBS, prior to mounting. Imaging was performed on an Olympus BX51 microscope fitted with a Metamorph Imaging System. Brightfield images were also obtained without any additional staining.
RESULTS AND DISCUSSION
Images and relaxivities of 2B5 cells cultured with different conditions are shown in Fig. 1a. 2B5 is a monoclonal cell line derived from 293FT that expresses the magA gene under the control of the doxycycline-inducible promoter. The results demonstrate no change in R2 without induction of gene expression and a significant increase in R2 upon induction and iron supplementation. This cell line has been passaged continuously over 8 months with stable integration of magA as verified by PCR. The cells have stable growth rates when compared with 293FT and have exhibited consistent relaxation rates at all passages.
FIG. 1.
MRI of magA cell line. a: Transverse relaxation rates (R2) of cell cultures. Cell line 2B5 shows no change in MR relaxivity without induction and subsequent increase in R2 upon induction. Error bars are ± SEM. Inset shows T2-weighted images taken from a CPMG sequence. These images show cell line 2B5 is capable of producing a visible contrast by MRI. At the shortest echo time (TE), all cells samples are bright, but as TE increases, cells expressing magA exhibit attenuated signal in contrast to normal cells. The arrow indicates the sample with the greatest relaxivity, 2B5 induced with doxycycline and supplemented with iron. b: Iron uptake in pg Fe/cell for 2B5 cells grown in various concentrations of doxycycline. Error bars are ± SD. These differences are significant at P < 0.05. Inset shows results of reverse-transcription PCR (RT-PCR) detecting messenger RNA (mRNA) transcript of magA in 2B5 cells incubated with no doxycycline, and 0.5 μg/ml and 2.0 μg/ml doxycycline. c: Cell culture experiments indicating that a maximum R2 is achieved with a combination of 1.0 μg/ml doxycycline and incubation in iron for 2 d. Error bars are ± SD. d: Iron-dosing results showing strong dependence on iron concentration below 200 μM Fe and less dependence above 200 μM for in vitro cell culture. Error bars are ± SD.
Figure 1b indicates iron uptake of 0.59 ± 0.14 pg Fe/cell for cells incubated with 2 μg/ml doxycycline and 200 μM Fe for 4 d. This value fell to 0.19 ± 0.03 pg Fe/cell for cells incubated with 0.5 μg/ml doxycycline and 0.04 ± 0.01 pg Fe/cell for cells incubated without doxycycline. There is a correlation between iron uptake and magA expression detected with RT-PCR shown in the inset of Fig. 1b.
Figure 1c shows R2 data from dosing experiments. At 0.5 μg/ml doxycycline, the relaxivity reached a plateau of 12 s−1 after 1-d incubation with iron. With 1.0 and 2.0 μg/ml doxycycline, relaxivities increased to ~20 s−1, reaching a plateau after 2 d incubation with iron. According to these data, a maximal effect is reached with a combination of doxycycline at 1 μg/ml and 2-d incubation with iron. This, taken together with iron uptake measurements (Fig. 1b), translates to a cellular relaxivity of R2 = 15.6 ± 1.8 (mM Fe)−1 s−1 at 3T. The value of 0.59 ± 0.14 pg Fe/cell found here is slightly lower than, but comparable to that for transferrin-mediated monocrystalline iron oxide nanocompounds (MION) uptake (range = 0.61–1.54 pg Fe/cell) reported previously (3). Figure 1d shows results of iron dosing response with 0.5 μg/ml doxycycline. R2 increased with iron concentration below 100 μM but leveled off beyond 200 μM. These experiments indicate that both gene expression level and iron availability are important factors in inducing R2 increase.
Electron micrographs of the cell line after 4 d of induction and incubation with 200 μM Fe are shown in Fig. 2. Because iron-oxide is not as electron-dense as gold and silver particles often used in electron microscopy, the iron-oxide particles exhibited a moderate contrast in the micrographs. The particles are approximately 3–5 nm in diameter, spherical in shape, and homogeneous in size. Although magnetotactic bacteria tend to form magnetosomes in chains, here the particles do not appear in such a configuration. This can be expected, as the anchoring of magnetosome vesicles to filaments within the cell, thereby creating chain-like formations, is likely to be mediated by other magnetosome-related genes (26,27). Although formation of chains and conversion of the signal provided by magnetic force into directed cell motility are functions needed for magnetotactic bacteria, they are not needed for MRI and other applications.
FIG. 2.
TEM images of cell line 2B5. Nanoparticles can be found within membrane enclosed vesicles. These vesicles, resembling endosomes, can be found individually (a) or in groups (b), with varying numbers of particles within, as can be seen when comparing vesicles in (b) and (d). b: Arrows show compartmentalization resembling the multivesicular bodies of the cellular degradation pathway (29). c: Particles can also be seen outside the vesicles, although they are less visible individually. e: Uninduced cell line 2B5, lacking these magnetic particles.
As shown in Fig. 2, although particles can be found throughout the cell, they are most easily identified within membrane-enclosed structures. This clustering, leading to effectively larger magnetic particles, may in fact enhance the transverse relaxation effect on the MRI signal (28). In Fig. 2, these structures are often found near one another and often near the cell membrane with a varying number of particles within. As marked in Fig. 2b, many of these structures appear to be compartmentalized, resembling endosomes known as multivesicular bodies (29). These compartments are known to indicate the sorting of material to be degraded (30), suggesting that particles here may be directed toward a cellular degradation pathway. It should be pointed out that not all endosomes are destined to migrate into a degradation pathway, and the exact fate of particles produced here remains unknown. After MRI, cells incubated for an additional 10 d without doxycycline or iron showed a return to control values for relaxivity (data not shown). Measurement of cellular iron content after cessation of gene induction showed a return to control values at day 6. It is possible that, in the absence of new particle formation, already-formed particles have simply been diluted by continued cell division. It is also possible that low pH, such as that present in maturing endosomes, breaks down the nanoparticles, causing release of iron from the cells. In fact, lowering pH to dissolve synthetic iron-oxide particles is often the first step in determining iron concentration (25). This possibility raises concern of potential adverse affects on cellular iron homeostasis and cell viability.
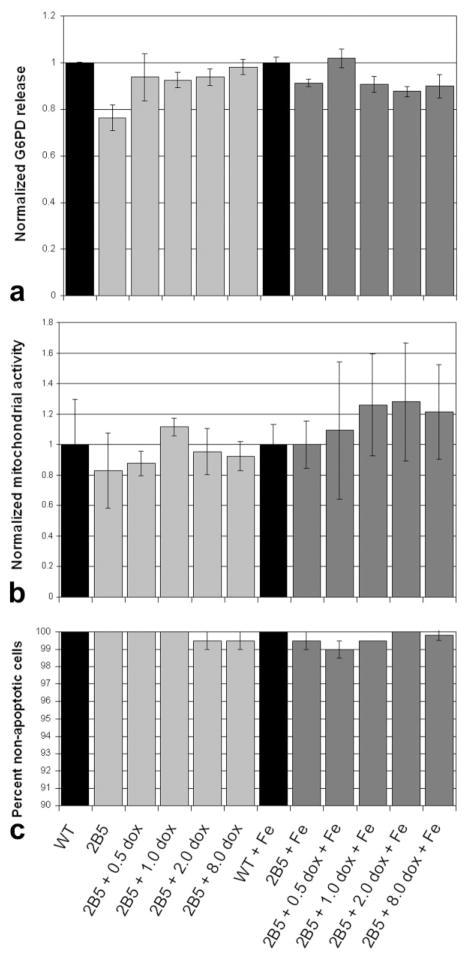
To address such concerns, we performed a series of proliferation and cytotoxicity assays. No difference in release of cytosolic G6PD was found among the different conditions (Fig. 3a); as increased cytotoxicity would mean increased G6PD release, this result indicates absence of cytotoxicity with magA expression and iron uptake. Mitochondrial activity showed no statistically significant changes in cell proliferation due to magA expression and iron uptake (Fig. 3b), and a direct assay for the presence of the reactive oxygen species, superoxide anion, was negative for cells incubated with and without iron and magA expression. In addition, magA expression and iron uptake did not increase apoptosis (Fig. 3c).
FIG. 3.
Results of cytotoxicity assays on 2B5 cell line. a: Results of G6PD assay showing no increased cytotoxicity as measured by G6PD release into medium. b: Results of MTT assay showing no significant decrease in cell proliferation. c: Results of apoptosis assay showing no increase in cellular apoptosis. Error bars are ± SD.
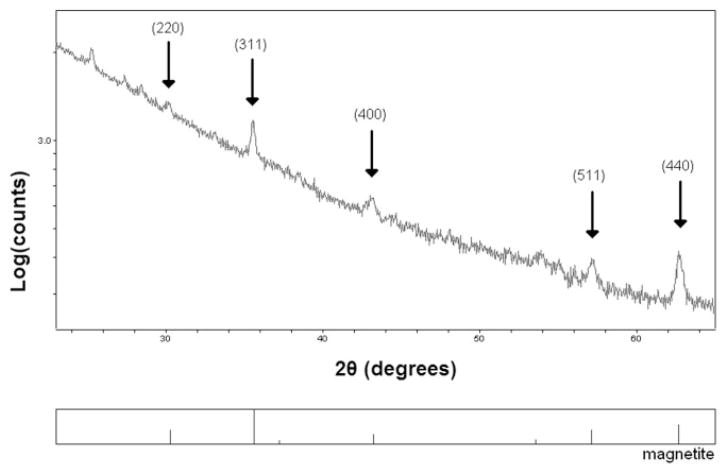
To further characterize the particles seen with microscopy, they were isolated from cells via magnetic separation and analyzed with X-ray powder diffraction (XRD). The results (Fig. 4) show that the particles consist primarily of magnetite (Fe3O4), consistent with the iron-oxide magnetosomes produced by magnetotactic bacteria.
FIG. 4.
XRD analysis of isolated nanoparticles. Arrows indicate important magnetite peaks. Numbers in parentheses are Miller indices of the reflections for major peaks of magnetite. The data show the particles consist primarily of magnetite (Fe3O4), which is the form of iron-oxide found in the magnetosomes of naturally occurring magnetotactic bacteria. Note that raw data, without baseline correction, is shown and the vertical scale is logarithmic making peak heights not readily comparable.
That a single bacterial gene can lead to formation of magnetic nanoparticles in a mammalian cell is an unexpected finding, raising questions to be answered. For instance, the exact process of particle formation remains unknown, although data in the literature may shed some light in this regard. It is known that MagA protein is present in both the cytoplasmic and magnetosome membranes of M. magneticum strain AMB-1 (23), and recent work showing that magnetosome vesicles are initially formed as invaginations of the cell membrane appears to confirm this (27). The uptake of iron into the cell, however, is likely to be dependent on the cell type hosting the magA gene. Tissue culture studies of human cells (31) show direct transport of ferrous iron, Fe(II), which is soluble at physiological pH but rapidly oxidized to ferric iron in an aerobic environment. For the magnetotactic bacterium, M. magneticum AMB-1, it was found that ferrous sulfate and ferric gallate as iron sources enhanced magnetosome yield as compared with ferric quinate, an iron chelate often used, and it has been postulated that size regulation of magnetite could be controlled by coprecipitation of Fe(II) and Fe(III) via alkalization, much as can be found in in vitro methods (32). The difference in iron environment of 293FT cells and magnetotactic bacteria is therefore one possible reason for the smaller size of particles created here. In magnetotactic bacteria, magnetosomes are tens of nanometers in size whereas particles here are only a few nanometers in diameter.
Figure 5 shows results of in vivo cell-transplantation experiments. In Fig. 5a, a T2*-weighted image at 5 d post-transplantation in one representative animal is shown. On the right side of the figure (white arrow), magA positive cells induced significant signal loss while the GFP inducible 293FT cells, not expressing magA, did not exhibit significant contrast on the contralateral side. The fluorescence histology in Fig. 5b shows that GFP was in fact induced in control cells, confirming delivery of doxycycline to the cells. The red fluorescence on the right side of Fig. 5b confirms the presence of magA-positive cells in conjunction with the location of these cells as seen by MRI.
FIG. 5.

MRI of magA cells induced in vivo. a: T2*-weighted image of mouse brain with transplanted magA cells (right) and GFP control cells (left) after 5 d of induction. These cells were neither induced nor incubated with iron supplement prior to transplantation. The magA cells (white arrow) exhibit significantly lower MRI signal, reflecting an increase in R2, suggesting that magA cells are able to use endogenous iron sources. The control cells on the left do not show such an effect. b: MRI of same mouse brain showing magA cells can still be readily seen by T2-weighted imaging, which is less sensitive than T2* with regard to magnetic nanoparticles. The fluorescence histology (not exactly registered to the MR image) confirms the presence of control (green) and magA-positive (red) cells. Magnified green and red channels are also shown. c: Brightfield histology section of the brain in (b), indicating the site of transplanted cells. d: T2*-weighted image of control animal following the same protocol as for (a) but without the addition of doxycycline. The lack of MRI signal change suggests that background expression from the doxycycline promoter is not seen in vivo.
T2*-weighted imaging is often used for cell tracking because it is more sensitive (19), but it can exaggerate the spatial extent of cells. In Fig. 5b, a T2-weighted image of the same slice as shown in Fig. 5a is presented. The 2B5R cells produce sufficient particles in vivo to be detectable by T2-weighted imaging, which is presumably a more accurate representation of the spatial extent of the cells.
The needle tracks, likely containing edema due to the trauma of the injection, can be seen in all cases but exhibit much less contrast than the cells containing particles. Although bleeding due to the injection could potentially mimic iron accumulation, this is unlikely the cause of contrast for the right side of Fig. 5a and b, as similar contrast is not found in the contralateral transplants, and additional histology (Fig. 5c) shows a lack of bleeding at the site of cell transplantation.
There is a great interest in the development of gene reporters for MRI, and the use of ferritin as a reporter gene has been previously described (14,15). Even though a definitive comparison between the ferritin approach and magA approach cannot be made due to differences in experimental conditions, some comparisons can be made with the present approach. With regard to structure, ferritin is a ubiquitous iron storage protein, capable of storing up to 4500 Fe(III) atoms in a ferrihydrite core with varying degrees of crystallinity (33). In contrast, the magA expressed here produces particles of magnetite, with similar particles estimated to contain 2064 iron atoms (34). With magA expression, cell pellets consistently showed an increase in R2 of approximately three- to four-fold, comparing to an increase in R2 of approximately 2.5-fold for ferritin expression (14).
Although not necessary for the use of magA as a reporter, we chose to use a doxycycline-inducible promoter to aid in the study of the effects of magA expression. These promoters, however, are known to allow for some background expression. Figure 1 shows that incubation in the presence of iron without addition of doxycycline does not cause an increase in relaxivity, suggesting negligible background magA expression. Figure 5d confirms this for the in vivo case, showing that without doxycycline induction, cells did not exhibit the same contrast. With a controllable system, uninduced cells could be allowed to grow, migrate, divide, and differentiate over a period of time in the absence of nanoparticles, minimizing their interference with normal cellular processes, before induction of magA expression, to allow particles to form, just prior to imaging.
The ability to produce magnetic nanoparticles in a novel host cell type opens up the possibility of other biotechnological applications. For instance, many molecular imaging applications rely on targeting contrast agents to specific receptors or cell surface ligands and disease-specific markers. Synthetic particles are often nonuniform, nonhomogeneous in composition, and difficult to disperse evenly in solution, and production of magnetosomes for biotechnological applications has been limited by the sensitivity of magnetotactic bacteria to oxygen, making these strains difficult to isolate and maintain in culture (18). Particles formed by expression of magA are highly uniform, and could potentially replace these sources of iron-oxide particles. Additionally, particles produced by immune-matched host cells will have less potential for triggering immune responses because they will not display foreign molecules to the subject of the imaging procedure. This is an advantage over simply isolating and using magnetosomes from bacterial cells, as the coating derived from bacteria would contain molecules foreign to other animals and humans in which the contrast agent may be used. It may also be possible to modify the particle for targeting applications with genetic manipulation, rather than chemical modification, by expressing magA in cell lines that also express desired ligands/receptors on their membranes so that isolated particles would contain these molecules on their surface, thus avoiding the need to chemically attach such molecules to the contrast agent.
CONCLUSIONS
In summary, the present work opens up a new avenue for in vivo cellular and molecular imaging, including applications such as cell tracking, gene expression reporting by coexpressing magA with any gene of interest, and in vivo sensing of iron concentration. Since MRI is noninvasive and particles can be regenerated, magA-based cell tracking and gene reporting allow new, longer-term molecular imaging experiments not possible with traditional forms of contrast agents.
Acknowledgments
Grant sponsor: National Heart, Lung, and Blood Institute (NHLBI); Grant number: UO1HL89711; Grant sponsor: National Institute of Biomedical Imaging and Bioengineering (NIBIB), Grant number: F31EB005928; Grant sponsors: National Institutes of Health; Georgia Research Alliance; Alzheimer Research Consortium.
We thank L.E. Bertani for providing the magA gene, W. Hillen for providing the rtTA2s-M2 fragment, Hong Yi for assistance with electron microscopy, Michael Haluska for assistance with X-ray diffraction analysis, Shijun Zhu and Shang-Hsun Yang for assistance with RT-PCR, Jin-Jing Yang for assistance with the production of high-titer lentivirus, and GuoFu Fang and the Emory Center for Neurodegenerative Disease and Center for Behavioral Neurosciences Viral Vector Core for assistance in constructing the inducible lentiviral vector. All animal procedures were approved by the Emory University Animal Care and Biosafety Committees. This work was supported by grants from the National Institutes of Health (to O.Z., X.H, and A.W.S.C.), Georgia Research Alliance (to X.H.), and Alzheimer Research Consortium (to A.W.S.C.).
References
- 1.Weissleder R. Target-specific superparamagnetic MR contrast agents. Magn Reson Med. 1991;22:209–212. doi: 10.1002/mrm.1910220209. [DOI] [PubMed] [Google Scholar]
- 2.Remsen LG, McCormick CI, Roman-Goldstein S, Nilaver G, Weissleder R, Bogdanov A, Hellstrom KE, Hellstrom I, Kroll RA, Neuwelt EA. MR of carcinoma-specific monoclonal antibody conjugated to monocrystal-line iron oxide nanoparticles: the potential for noninvasive diagnosis. AJNR Am J Neuroradiol. 1996;17:411–418. [PMC free article] [PubMed] [Google Scholar]
- 3.Moore A, Basilion JP, Chiocca EA, Weissleder R. Measuring transferrin receptor gene expression by NMR imaging. Biochim Biophys Acta. 1998;1402:239–249. doi: 10.1016/s0167-4889(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Bulte JW, Zhang S, van Gelderen P, Herynek V, Jordan EK, Duncan ID, Frank JA. Neurotransplantation of magnetically labeled oligodendrocyte progenitors: magnetic resonance tracking of cell migration and myelination. Proc Natl Acad Sci USA. 1999;96:15256–15261. doi: 10.1073/pnas.96.26.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artemov D, Mori N, Okollie B, Bhujwalla ZM. MR molecular imaging of the Her-2/neu receptor in breast cancer cells using targeted iron oxide nanoparticles. Magn Reson Med. 2003;49:403–408. doi: 10.1002/mrm.10406. [DOI] [PubMed] [Google Scholar]
- 6.Bulte JW, Douglas T, Witwer B, Zhang SC, Strable E, Lewis BK, Zywicke H, Miller B, van Gelderen P, Moskowitz BM, Duncan ID, Frank JA. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nat Biotechnol. 2001;19:1141–1147. doi: 10.1038/nbt1201-1141. [DOI] [PubMed] [Google Scholar]
- 7.Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, Weissleder R. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 8.Auricchio A, Zhou R, Wilson JM, Glickson JD. In vivo detection of gene expression in liver by 31P nuclear magnetic resonance spectroscopy employing creatine kinase as a marker gene. Proc Natl Acad Sci USA. 2001;98:5205–5210. doi: 10.1073/pnas.081508598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodibagkar VD, Yu J, Liu L, Hetherington HP, Mason RP. Imaging beta-galactosidase activity using (19)F chemical shift imaging of LacZ gene-reporter molecule 2-fluoro-4-nitrophenol-beta-d-galactopyranoside. Magn Reson Imaging. 2006;24:959–962. doi: 10.1016/j.mri.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Louie A. Design and characterization of magnetic resonance imaging gene reporters. Methods Mol Med. 2006;124:401–417. doi: 10.1385/1-59745-010-3:401. [DOI] [PubMed] [Google Scholar]
- 11.Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 12.Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr, Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25:217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 13.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6:351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 14.Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11:450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 15.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossuin Y, Muller RN, Gillis P. Relaxation induced by ferritin: a better understanding for an improved MRI iron quantification. NMR Biomed. 2004;17:427–432. doi: 10.1002/nbm.903. [DOI] [PubMed] [Google Scholar]
- 17.Blakemore R. Magnetotactic bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 18.Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 19.Bulte JWM, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda Y, Okamura Y, Takeyama H, Matsunaga T. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 2006;580:801–812. doi: 10.1016/j.febslet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Okamura Y, Calugay RJ, Takeyama H, Matsunaga T. Global gene expression analysis of iron-inducible genes in Magnetospirillum magneticum AMB-1. J Bacteriol. 2006;188:2275–2279. doi: 10.1128/JB.188.6.2275-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura C, Burgess JG, Sode K, Matsunaga T. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J Biol Chem. 1995;270:28392–28396. doi: 10.1074/jbc.270.47.28392. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura C, Kikuchi T, Burgess JG, Matsunaga T. Iron-regulated expression and membrane localization of the magA protein in Magnetospirillum sp. strain AMB-1. J Biochem. 1995;118:23–27. doi: 10.1093/oxfordjournals.jbchem.a124884. [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 2005;12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- 25.Nitin N, LaConte LE, Zurkiya O, Hu X, Bao G. Functionalization and peptide-based delivery of magnetic nanoparticles as an intracellular MRI contrast agent. J Biol Inorg Chem. 2004;9:706–712. doi: 10.1007/s00775-004-0560-1. [DOI] [PubMed] [Google Scholar]
- 26.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 27.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 28.Perez JM, Josephson L, O’Loughlin T, Hogemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 29.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 30.Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 31.Conrad ME, Umbreit JN, Moore EG, Hainsworth LN, Porubcin M, Simovich MJ, Nakada MT, Dolan K, Garrick MD. Separate pathways for cellular uptake of ferric and ferrous iron. Am J Physiol Gastrointest Liver Physiol. 2000;279:G767–G774. doi: 10.1152/ajpgi.2000.279.4.G767. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga T, Okamura Y, Tanaka T. Biotechnological application of nanoscale engineered bacterial magnetic particles. J Mater Chem. 2004;14:2099–2105. [Google Scholar]
- 33.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 34.Shen T, Weissleder R, Papisov M, Alexei Bogdanov J, Brady TJ. Monocrystalline iron oxide nanocompounds (MION): physicochemical properties. Magn Reson Med. 1993;29:599–604. doi: 10.1002/mrm.1910290504. [DOI] [PubMed] [Google Scholar]