Abstract
Thermogenic fat cells that convert chemical energy into heat are present in both mice and humans. Recent years have witnessed a great advancement in our understanding of the regulation of these adipocytes and an increased appreciation of the potential these cells have to counteract obesity. Here we summarize recent efforts to understand the formation of these fat cells and critically review genetic models and other experimental tools currently available to further investigate the development and activation of both classical brown and inducible beige fat cells. We also discuss recent discoveries about the epigenetic regulation of these adipocytes, and finally present emerging evidence revealing the metabolic impacts of thermogenic fat in humans.
Keywords: Obesity, brown fat, beige fat, adaptive thermogenesis
Fighting fat with fat
Obesity affects one in three persons globally and constitutes an increasing burden on health care systems and an urgent challenge for the biomedical research community [1, 2]. Effective prevention and treatment of obesity may drastically reduce the occurrence of comorbidities such as type 2 diabetes, cardiovascular disease, and other serious health problems, including many types of cancer. As a multifactorial disorder, obesity can be prevented through combinations of approaches that target different aspects of metabolism to decrease energy surplus. Much of energy homeostasis depends on the activity and function of adipose tissue. Two major types of adipocytes exist in mammals, white fat and thermogenic fat. The primary function of white adipose cells is to store energy and subsequently secrete hormones in response to nutritional signals. By contrast, thermogenic adipocytes defend against hypothermia and obesity through adaptive thermogenesis, mediated by regulated expression and activity of mitochondrial uncoupling protein 1 (UCP1). Thermogenic adipocytes have also been classified into so-called “classical” brown fat cells and newly identified beige adipocytes (discussed below).
It was long assumed that thermogenic fat was only present in humans at the infant stage. In 2009, however, thermogenic adipocytes were shown to exist in human adults [3–5], drawing intense interest as a potential target to increase energy expenditure and counteract obesity. These research efforts have led to significant advances of our knowledge of these cells [6–8]. It is becoming increasingly evident that thermogenic fat significantly influences whole body metabolism in humans. To leverage the full potential of the metabolic benefits of these cells, it is essential to thoroughly understand the developmental history and distinct regulatory mechanisms of different types of thermogenic fat. In this review, we discuss our current view of how these fat cells are regulated, particularly focusing on the distinguishing features of developmental formation and environmentally stimulated activation of thermogenic fat.
Thermogenic fat formation
The early developmental origins of fat remains an important question that is currently being intensively investigated [9, 10]. It has been proposed that certain adipocytes arise from endothelial [11] or hematopoietic lineages [12, 13]. Until recently, brown fat observed in the rodent interscapular depot and human infants was widely believed to share a common developmental origin with the rest of the fat cells throughout the body. These brown fat cells have high mitochondrial content, the iron in which gives them their eponymous color. A unique mitochondrial protein, UCP1, functions as a proton leak, which effectively “uncouples” ATP synthesis and oxidative phosphorylation through the electron transport chain (ETC). The inherent inefficiency of the biochemical reactions leads to the conversion of electrochemical energy into heat, so that mammals (small mammals in particular) can defend against hypothermia [14]. Despite these unique features, thermogenic fat cells share many characteristics with their white fat counterparts. They normally express most of the adipocyte-specific markers at comparable levels to white fat, such as peroxisome proliferator-activated receptor gamma (PPARG), adiponectin, and fatty acid binding protein 4 (FABP4, also referred as aP2). Both white and thermogenic fat cells contain intracellular lipid droplets, albeit unilocular morphology (one lipid droplet) is normally observed in white adipocytes and multilocular (many lipid droplets) in thermogenic fat.
Despite these common characteristics, however, the notion of a common developmental origin for all fat cells was disproved in 2008. After the discovery that PRD1-BF-RIZ1 homologous domain containing 16 (PRDM16) is a key regulator of the thermogenic program in brown fat [15], loss-of-function assays of PRDM16 were conducted in primary brown fat cells. These shPRDM16 brown preadipocytes differentiated into muscle-like cells instead of the expected white fat cells [16]. Cell fate mapping experiments showed that some UCP1+ fat cells from several depots, most noticeably the interscapular depot, arise from a Myf5+ lineage, which also gives rise to skeletal muscle [16]. This discovery, together with the rediscovery of thermogenic fat cells in human adults [3–5], which was reported one year later, significantly changed how we view thermogenic fat and its potential role in metabolism.
Not all UCP1+ fat cells come from the Myf5+ lineage, however [16]. Upon cold exposure, UCP1+ multilocular cells are detectable in many white adipose depots, most prominently, in subcutaneous depots like the inguinal depot in mice [14]. These “inducible” thermogenic fat cells, together with white fat cells, come from one or more Myf5− lineage(s) [16]. To investigate the developmental origin and molecular identity of these Myf5− lineage derived UCP1+ adipocytes, clonal stable cell lines were generated from the subcutaneous depot, and unbiased analysis of transcriptional profiling revealed that a subset of these cell lines are functionally more similar to classical brown fat than the rest of the lines from the subcutaneous depot. This provides direct evidence that these “inducible” thermogenic fat cells (so-called beige fat cells) may be fundamentally dissimilar from the other fat cells of the inguinal depot, even at the precursor stage (Figure 1) [17]. The exact developmental lineage of this new type of fat cell is under intensive investigation. Using a ribosome-profiling approach, it was shown that an enriched expression of a smooth muscle gene signature is present in beige fat cells but not in brown fat cells. Cell fate mapping experiments with a Myh11-driver (a smooth muscle marker) revealed that at least a subset of beige fat cells arise from a shared lineage with smooth muscle [18]. Other studies identified so-called “brite” fat (brown in white), a distinguished subpopulation of adipocytes from the visceral depot expressing UCP1 upon rosiglitazone treatment [19]. Rosiglitazone is a commonly used thiazolidinedione (TZD), a PPARγ agonist, which has been shown to induce activation of thermogenic gene expression in adipose tissue and cells [20–24].
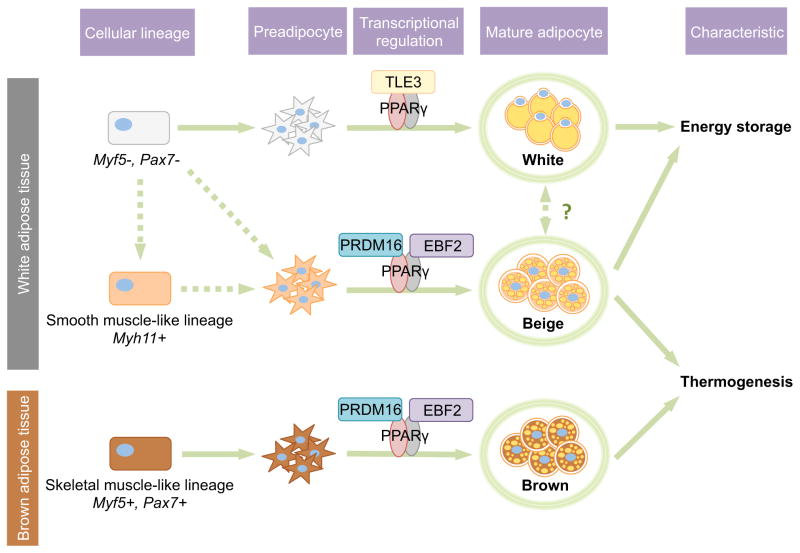
Figure 1. Development of three types of fat cells.
White, beige, and brown adipocytes are derived from different lineages and subpopulations of precursors. Brown adipocytes come from Myf5+/Pax7+ lineages, whereas white and beige adipocytes arise from Myf5−/Pax7− lineages. Recently, Myh11 expressing progenitors have been newly identified as a population giving rise to at least some of the “inducible” thermogenic beige adipocytes. PPARγ coordinates the adipogenesis of three types of fat cells through interactions with different transcriptional coregulators. TLE3 functions as a coactivator of PPARγ during white adipogenesis [70]. Recruitment of PRDM16 and EBF2 to PPARγ leadscells to differentiate into thermogenic beige and brown adipocytes.
The discovery of a common lineage of skeletal muscle and brown fat has drawn much attention and has been confirmed by many groups in various cell fate mapping models with different skeletal muscle markers. Experiments with an inducible Pax7 tracing model revealed that the cell fate diverging decision to become either brown fat or skeletal muscle happens between embryonic day 9.5–e11.5 during gestation [25]. Detailed mapping analysis of multiple fat depots using Myf5 and several other skeletal-muscle specific genes (MyoD and Pax3) confirmed the earlier discovery that UCP1+ cells in the interscapular fat depot are from the Myf5+ lineage and UCP1+ cells in the perigonadal (visceral) and posterior-subcutaneous (inguinal) depots are from a Myf5− lineage [26]. Additional analysis in the same study revealed that certain depots (e.g., the cervical depot) contain UCP1+ adipocytes of both Myf5+ and Myf5− lineages [26]. It has also been shown that many unilocular adipocytes within dorsal-anterior depots arise from a Myf5+/Pax3+ lineage [26]. Furthermore, the abundance of adipocytes of Myf5+ or Pax3+ lineages at different adipose depots vary between mice of different gender, age, and metabolic status [26], suggesting adipose precursors with different developmental origins may differentially contribute to the dynamic adipose tissue remodeling process in vivo.
As we learn more about how thermogenic adipocytes are regulated, clearer definitions based on molecular insights and functional significance will be possible for different types of thermogenic fat cells. For example, the terms of “beige adipocyte” and “brite adipocyte” are currently used interchangeably in most contexts. However, the beige adipocyte was first defined studying thermogenic adipocytes isolated from subcutaneous depot [17], whereas brite adipocytes were isolated from visceral depot [19]. Future studies will elucidate whether beige and brite fat cells are molecularly equivalent or if there are many different subtypes of inducible thermogenic fat cells of separate lineages and distinct functions.
To date, most studies on thermogenic fat cells have been carried out with either the interscapular or inguinal fat depot, mainly due to the substantial size of these tissues and the availability of protocols for culture and differentiation of the adipose precursors in these two depots. For the thermogenic fat cells resident in other adipose depots, particularly the ones from depots containing cells of both Myf5+ and Myf5− lineages, it is not yet known whether they are more similar to classical brown fat cells in the interscapular depot, beige fat identified in the inguinal depot, or are a completely different, new type of adipocyte. In this review, for the purpose of simplicity and clarity, we mainly discuss thermogenic adipocytes of Myf5+ lineage resident in interscapular depot (referred to as classical brown fat) and Myf5− lineage thermogenic adipocytes in subcutaneous depot (beige adipocytes).
In depth examination of the development and regulation of thermogenic fat cells further revealed a complicated role for PRDM16. Studies with Myf5-CRE mediated cell-type specific PRDM16 knockout mice revealed that it is not essential for the embryonic development of brown fat, likely due to compensation by PRDM3 [27]. As the animals age, PRDM16 plays a more prominent role in maintaining the identity of brown fat in adult and obese mice [27]. The regulatory role of PRDM16 in beige fat surfaced after studying mouse models of fat-specific ectopic expression or knockout of PRDM16 [28, 29]. Adipose-specific PRDM16 transgenic mice are protected against diet-induced-obesity and preserve glucose homeostasis much better upon challenge with a high-fat-diet compared to control mice. Mechanistic investigations revealed these beneficial effects are likely caused by significant activation of the thermogenic program in the subcutaneous depot [28]. Conversely, inguinal fat tissue from fat-specific knockout of PRDM16 mice present a “whitening” phenotype, with greatly repressed thermogenic gene expression [29]. It is tempting to speculate that diverse functions of PRDM16 in regulating brown fat cell fate and beige fat function are achieved through recruitment of cell-type-specific transcription factor(s) or cofactor(s) to the PRDM16-containing transcriptional complex.
Functional marker-based approaches – activation versus formation
Many mouse models have been developed based on reporter-labeling or CRE-mediated deletion driven by promoters of functional genes in adipocytes. These genetic models provide powerful tools to investigate the regulation of fat cells in vivo, both at basal states and upon environmental stimulations (Table I) [30–32]. How progenitors commit to adipose cell fate and how preadipocytes develop into mature fat cells in vivo have been investigated with transgenic mouse models in which reporter (GFP or β-gal) expression is regulated by the promoters of Zfp423 (a preadipocyte marker) or Pparg [33–35].
Table 1.
Approaches for the study of three types of fat
| Approaches | Brown fat | Beige fat | White fat | Ref. | |
|---|---|---|---|---|---|
| Mouse Strains | Functional marker-labeling reporter mice |
Adipoq-Cre Adipoq-Cre BAC Adipoq-CreER(T)2 Adipoq-rTA Pparγ-rTA Ucp1-GFP Ucp1-Cre Ucp1-CreER(T)2 |
Adipoq-Cre Adipoq-Cre BAC Adipoq-CreER(T)2 Adipoq-rTA Pparγ-rTA Ucp1-GFP Ucp1-Cre Ucp1-CreER(T)2 |
Adipoq-Cre Adipoq-Cre BAC Adipoq-CreER(T)2 Adipoq-rTA Pparγ-rTA |
30–32,35–37,40 |
| Developmental marker-tracing reporter mice |
Myf5-Cre Zfp423-GFP Pax7-Cre Pax7-CreER(T)2 Ebf2-GFP Ebf2-GFP BAC Pdgfrα-Cre |
Zfp423-GFP Myh11-Cre Ebf2-GFP Ebf2-GFP BAC Pdgfrα-Cre |
Zfp423-GFP Pdgfrα-Cre |
16,18,25,26,34,47 | |
| Cellular markers | Cell surface markers | PDGFRα SCA1 CD34 PAT2 P2RX5 |
PDGFRα SCA1 CD34 CD137 TMEM26 PAT2 P2RX5 |
PDGFRα SCA1 CD34 ASC-1 |
9,17,41–44 |
| Functional markers |
Ucp1 Cidea Dio2 Fgf21 Cpt1 Cox8b Cox7a1 Prdm16 Pparα Ppargc1α |
Ucp1 Cidea Dio2 Fgf21 Cpt1 Cox8b Cox7a1 Prdm16 Pparα Ppargc1α |
Retn Lep Tcf21 Igfbp3 |
- | |
Elegant work with a model called the adipoChaser mouse showed that thermogenic fat cells induced by cold exposure within the subcutaneous depot mainly arise from precursor cells [36, 37]. This model is a doxycycline-inducible, mature adipocyte-specific tracing system, allowing researchers to pulse-label all mature fat cells at a selected time point with β-gal. A significant advantage of the adipoChaser model is that doxycycline can be removed from the system within 24 hours, in contrast to tamoxifen-mediated inducible deletion, which has been shown to cause prolonged effects, therefore rendering it unfit for in vivo pulse-chase experiments [38, 39].
Another commonly used approach to study cell proliferation is BrdU labeling. However when applied to studying adipose precursors in vivo, BrdU labeling identifies mitotically active cells but cannot be used to distinguish existing mature adipocytes from newly differentiated fat cells converted from existing precursors. It is essential to consider all the technical limitations and caveats of each genetic model when studying adipocyte development and fat tissue remodeling in vivo.
The development of thermogenic fat in vivo has also been studied with the GFP-RFP dual labeling, Ucp1-tracing mice [40]. In this model, GFP labels all cells presently expressing Ucp1; whereas RFP will pulse-label cells that express Ucp1 upon tamoxifen-induced CRE deletion. The study presented evidence that a subpopulation of adipocytes within white adipose depots switch between appearances and gene expression patterns of both white and thermogenic adipocytes. It is possible that a bona fide “transdifferentiation” happens between different types of adipocytes. However, it is worth pointing out that UCP1 is a functional marker whose expression level fluctuates significantly as the activity of thermogenic adipocytes is regulated. Hence, it is possible that the GFP and RFP label the same subpopulation of adipocytes at various states of thermogenic activation. Further studies with better-defined cell identity markers will provide alternative approaches to thoroughly investigating these important questions.
Cell surface marker-based approaches
Early work to identify differentially expressed markers in different adipose depots and different types of fat cells not only provided some hints about their potential functional differences but also generated candidate cell surface markers to separate thermogenic preadipocytes from the stromal vascular fraction (SVF) with fluorescence-activated cell sorting (FACS). Through gene expression analysis of murine clonal cell lines, CD137 and TMEM26 were identified as beige selective cell surface proteins. It was shown that the CD137-high subpopulation of the inguinal SVF has higher expression of thermogenic genes compared to the CD137-low subpopulation from the same depot, suggesting that the observations in immortalized cell lines also apply to primary beige and white fat cells [17]. Compared to sorting with established markers with decades of refinement (e.g., cell surface markers for hematopoietic stem cells), which clearly separate positive and negative subpopulations, better sorting strategies for thermogenic fat cells are still yet to be achieved. Further optimization with combined use of multiple markers will be attained as more information about these cells becomes available. To compare functions of subpopulations of adipose precursors, it is essential to ensure the SVF undergoes minimal stress throughout the procedure so that all subpopulations can achieve equal and robust differentiation.
In a different approach to discover markers, gene expression was correlated with either expression of Adipoq or Ucp1, leading to the identification of ASC-1 as a marker for white adipocytes and PAT2 and P2RX5 as specific markers for thermogenic fat cells [41]. A distinguishing feature of this study is that these markers are also present in human white and thermogenic fat cells, offering possibilities of prospective isolation of human thermogenic fat cells. These markers will enable us to thoroughly investigate the distinct functions of thermogenic fat in humans and potentially lead to clinical applications such as autologous transplantation of thermogenic precursor cells back into individuals who undergo liposuction procedures to counteract obesity.
Using several cell surface markers that have been reported to be important for stem cell functions in other tissues, a strategy was optimized to isolate the “CD24+” subpopulation (Lin−:CD29+:CD34+:Sca-1+:CD24+), which was shown to be highly adipogenic in vitro and in vivo [42]. It was further demonstrated that this CD24+ subpopulation consists of an early stage of progenitors that will further commit into adipose cell fate as they lose the expression of CD24 [9]. Both CD24+ and CD24− cells express platelet-derived growth factor receptor α (PDGFRα), which has been shown to be expressed in adipogenic cells within the skeletal muscle [43, 44]. Using CD34, Sca-1, and PDGFRα, a subpopulation was separated from the abdominal adipose depot, and it was proposed that these cells can differentiate into either white or thermogenic fat depending on different environmental cues [45]. This observation suggests that the PDGFRα+ subpopulation within the visceral depot may represent an earlier stage of progenitors before the cells commit into diverged fates of being either white or thermogenic adipocytes.
PDGFRα+ and Ebf2-regulated GFP+ cells were sorted, and it was demonstrated that EBF2, an important regulator of brown fat cell fate [46], may also participate in functional regulation of beige adipocytes [47]. This approach combines the use of cell surface markers and genetic labeling with functional markers and may represent a promising strategy to investigate thermogenic fat function until better defined exclusive markers become available.
Epigenetic regulation of thermogenic fat cells
The role of epigenetic regulation in adipose tissue function has been increasingly appreciated in recent years [48]. Studies in both 3T3-L1 immortalized white fat and human adipose stromal cells showed that many fundamental aspects of transcriptional networks for adipogenesis act in concert with epigenetic regulation [48]. Given that environmental signaling acutely controls thermogenic functions, it is particularly interesting to study how development and function of thermogenic fat are regulated through chromatin structure changes. Several epigenetic regulators have been shown to be closely involved in brown and/or beige fat functions. Jhdma2a, an H3K9-specific demethylase, has been shown to be essential for the adrenergic-stimulated UCP1 expression in brown fat. Loss of Jhdma2a in mice leads to obesity with both regular and high-fat diets, although this phenotype may be caused in part by defective β-oxidation in skeletal muscle in the absence of Jhdma2a [49]. Euchromatic histone-lysine N-methyltrasferse 1 (EHMT1) is another key epigenetic regulator involved in brown fat function. Studies with fat-specific EHMT1-KO mice display impaired development of brown fat and increased obesity [50]. Further mechanistic investigations revealed that this brown adipose tissue (BAT)-enriched methyltransferase controls brown fat cell fate through interactions with PRDM16 [50]. Nicotinamide N-methyltransferase (NNMT) regulates histone methylation, and loss of function of NNMT in white adipose tissue and in the liver increases energy expenditure, counteracting diet-induced obesity through a mechanism independent of classical brown fat activity [51]. In vitro experiments with an immortalized fat cell line demonstrated that at least some of the systemic effects are regulated through cell-autonomous mechanisms in the adipocytes [51]. Future studies will determine whether and how different types of adipocytes are distinctively regulated on the epigenetic level and lead to a better understanding of the interplay between various genetic, behavioral, and environmental causes of obesity.
Thermogenic fat and its therapeutic potential in humans
Six years after initial reports of thermogenic fat cells in human adults, an overwhelming amount of data on gene expression, histology, and function of these adipocytes leaves little room for doubt regarding the presence of these cells in humans [3–5, 52–54]. Studies are now emerging that address the potential significance of their impact on systemic metabolism.
Using the cell-type specific markers identified through analysis with murine clonal brown, beige, and white cell lines provided evidence that UCP1+ cells resident within the human supraclavicular region are more similar to murine beige fat cells instead of brown fat cells [17]. Several later reports independently confirmed the presence of beige-like thermogenic fat cells in human adults [55, 56]. A recent study showed that some of the UCP1+ fat cells in the neck area express genes enriched in murine interscapular brown fat [57]. Another report proposed that UCP1+ cells in the human supraclavicular region consist of both classical brown and beige adipocytes [58]. It is worth noting that clonal cell lines were later independently generated from the biopsies used in this study [58] and unbiased genome-wide expression analyses revealed these thermogenic cells possess molecular signatures much more similar to murine beige adipocytes than brown adipocytes [59]. Given the heterogeneity in the human genetic makeup and our still limited understanding of the distinction between brown and beige fat, the debate regarding the molecular identities of human thermogenic fat is still ongoing.
Several concerns and caveats should be taken into consideration when designing future studies on human thermogenic fat. When the sample size is small (not on the epidemiological scale), analysis of differences between so-called “BAT” and “WAT” should be conducted with biopsies from the same person to offset individual variance. Because thermogenic gene expression (e.g., UCP1) may vary significantly depending on the activation states of thermogenic fat cells (particularly so for beige fat), all the biopsies for one experiment should be collected under exactly the same conditions. It has been shown that the activity of human thermogenic fat varies significantly as environmental temperatures change, both upon acute cold exposure and as seasons change [60–62].
In addition to their molecular identity, another essential question regarding human thermogenic fat is whether the amount of this type of fat cell present in an average human constitutes a significant influence over systemic metabolism. It was speculated that one year of maximal activation of thermogenic fat could convert chemical energy into heat equivalent to the energy stored in 8 pounds of white fat [63, 64]. New studies have shown that activated human thermogenic fat significantly improves whole body glucose homeostasis and insulin sensitivity [65, 66]. Given that thermogenic fat activities induced by cold exposure alone can lead to reduced body weight [67], it is reasonable to expect that ongoing endeavors to develop thermogenesis-enhancing therapeutics for weight loss will succeed in the foreseeable future.
Concluding remarks
The recent explosion of interest in thermogenic fat cells has led to significant advances in our understanding of the development and regulation of these potentially metabolically beneficial adipocytes. Yet, many important questions remain contentious, reminding us that science is a process of trial and error. Recent history has witnessed a similar trajectory of progress with regulatory T cells, which were originally identified in the 1970s. It took the field four decades to arrive at a better understanding of what these immune cells do and how they are regulated [68, 69]. In addition to further characterization of “inducible thermogenesis”, thorough investigation of beige-selective transcriptional regulation, hormonal response, and other distinct functions will provide mechanistic insights and enable the development of the necessary research tools to delineate the detailed developmental paths of different types of adipocytes, reveal their unique regulation, and functionally distinguish their individual impact on systemic metabolism.
Highlights.
Thermogenic fat cells can be distinguished by their developmental origins.
Studies with genetic models and FACS reveal regulations of thermogenic adipocytes.
Activated thermogenic fat in humans helps to improve systemic metabolic health.
Acknowledgments
We apologize to those whose work is not cited owing to space limitations. Work in the Wu laboratory is supported by grants K01DK094824 and R03DK100698 from the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health, a Pilot and Feasibility Grant from the Michigan Diabetes Research Center (NIH Grant 2P30-DK020572) and a Young Investigator Grant RGY0082/14 from the Human Frontier Science Program.
Nonstandard abbreviations
- WAT
white adipose tissue
- PET/CT
positron emission tomography – computed tomography
- FACS
fluorescence-activated cell sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ogden CL, et al. Prevalence of childhood and adult obesity in the United States, 2011–2012. Jama. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Fact sheet on obesity and overweight. 2014. [Google Scholar]
- 3.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 5.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 6.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nature medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 8.Townsend KL, Tseng YH. Brown fat fuel utilization and thermogenesis. Trends in endocrinology and metabolism: TEM. 2014;25:168–177. doi: 10.1016/j.tem.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nature cell biology. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry R, et al. Weighing in on adipocyte precursors. Cell metabolism. 2014;19:8–20. doi: 10.1016/j.cmet.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran KV, et al. The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell metabolism. 2012;15:222–229. doi: 10.1016/j.cmet.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crossno JT, Jr, et al. Rosiglitazone promotes development of a novel adipocyte population from bone marrow-derived circulating progenitor cells. The Journal of clinical investigation. 2006;116:3220–3228. doi: 10.1172/JCI28510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sera Y, et al. Hematopoietic stem cell origin of adipocytes. Experimental hematology. 2009;37:1108–1120. 1120 e1101–1104. doi: 10.1016/j.exphem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 15.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JZ, et al. A smooth muscle-like origin for beige adipocytes. Cell metabolism. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukui Y, et al. A new thiazolidinedione, NC-2100, which is a weak PPAR-gamma activator, exhibits potent antidiabetic effects and induces uncoupling protein 1 in white adipose tissue of KKAy obese mice. Diabetes. 2000;49:759–767. doi: 10.2337/diabetes.49.5.759. [DOI] [PubMed] [Google Scholar]
- 21.Laplante M, et al. PPAR-gamma activation mediates adipose depot-specific effects on gene expression and lipoprotein lipase activity: mechanisms for modulation of postprandial lipemia and differential adipose accretion. Diabetes. 2003;52:291–299. doi: 10.2337/diabetes.52.2.291. [DOI] [PubMed] [Google Scholar]
- 22.Wilson-Fritch L, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Molecular and cellular biology. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. The Journal of clinical investigation. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vernochet C, et al. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Molecular and cellular biology. 2009;29:4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Gurmaches J, Guertin DA. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nature communications. 2014;5:4099. doi: 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms MJ, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell metabolism. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of clinical investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen P, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra C, et al. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. The Journal of clinical investigation. 2001;108:1205–1213. doi: 10.1172/JCI13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassmann A, et al. Tamoxifen-inducible Cre-mediated recombination in adipocytes. Genesis. 2010;48:618–625. doi: 10.1002/dvg.20665. [DOI] [PubMed] [Google Scholar]
- 33.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RK, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell metabolism. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ZV, et al. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology. 2010;151:2933–2939. doi: 10.1210/en.2010-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang QA, et al. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature medicine. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang QA, et al. Improved methodologies for the study of adipose biology: insights gained and opportunities ahead. J Lipid Res. 2014;55:605–624. doi: 10.1194/jlr.R046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinert RB, et al. Tamoxifen-Induced Cre-loxP Recombination Is Prolonged in Pancreatic Islets of Adult Mice. PloS one. 2012;7:e33529. doi: 10.1371/journal.pone.0033529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenwald M, et al. Bi-directional interconversion of brite and white adipocytes. Nature cell biology. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 41.Ussar S, et al. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Science translational medicine. 2014;6:247ra103. doi: 10.1126/scitranslmed.3008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodeheffer MS, et al. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 43.Joe AW, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uezumi A, et al. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature cell biology. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 45.Lee YH, et al. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell metabolism. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell metabolism. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proceedings of the National Academy of Sciences of the United States of America; 2014. pp. 14466–14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tateishi K, et al. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohno H, et al. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manieri M, et al. Morphological and immunohistochemical features of brown adipocytes and preadipocytes in a case of human hibernoma. Nutr Metab Cardiovasc Dis. 2010;20:567–574. doi: 10.1016/j.numecd.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 53.Zingaretti MC, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- 54.Frontini A, et al. White-to-brown transdifferentiation of omental adipocytes in patients affected by pheochromocytoma. Biochim Biophys Acta. 2013;1831:950–959. doi: 10.1016/j.bbalip.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Sharp LZ, et al. Human BAT Possesses Molecular Signatures That Resemble Beige/Brite Cells. PloS one. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nature medicine. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 57.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature medicine. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell metabolism. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 59.Shinoda k, Luijten IHN, Hasegawa Y, Hong Haemin, Sonne Si B, Kim Miae, Xue Ruidan, Chondronikona Maria, Cypess Aaron M, Tseng Yu-Hua, Nedergaard Jan, Sidossis Labros S, Kajimura Shingo. genetic and functional characterization of clonally-derived adult human brown adipocytes. Nature Medicine. 2015 doi: 10.1038/nm.3819. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouellet V, et al. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J Clin Endocrinol Metab. 2011;96:192–199. doi: 10.1210/jc.2010-0989. [DOI] [PubMed] [Google Scholar]
- 62.van der Lans AA, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of clinical investigation. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Enerback S. Human brown adipose tissue. Cell metabolism. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Ma SW, Foster DO. Uptake of glucose and release of fatty acids and glycerol by rat brown adipose tissue in vivo. Canadian journal of physiology and pharmacology. 1986;64:609–614. doi: 10.1139/y86-101. [DOI] [PubMed] [Google Scholar]
- 65.Chondronikola M, et al. Brown Adipose Tissue Improves Whole Body Glucose Homeostasis and Insulin Sensitivity in Humans. Diabetes. 2014;63:4089–4099. doi: 10.2337/db14-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee P, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 2014;63:3686–3698. doi: 10.2337/db14-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneshiro T, et al. Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of clinical investigation. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Josefowicz SZ, et al. Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakaguchi S, et al. Regulatory T cells - a brief history and perspective. European journal of immunology. 2007;37(Suppl 1):S116–123. doi: 10.1002/eji.200737593. [DOI] [PubMed] [Google Scholar]
- 70.Villanueva CJ, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell metabolism. 2011;13:413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]