Abstract
To investigate the relationship between linker histone H1 and protein-protein interactions in the nucleolus, biochemical and proteomics approaches were used to characterize nucleoli purified from cultured human and mouse cells. Mass spectrometry identified 175 proteins in human T-cell nucleolar extracts that bound to sepharose-immobilized H1 in vitro. Gene ontology analysis found significant enrichment for H1 binding proteins with functions related to nucleolar chromatin structure and RNA polymerase I transcription regulation, rRNA processing, and mRNA splicing. Consistent with the affinity binding results, H1 existed in large (400 to >650 kDa) macromolecular complexes in human T cell nucleolar extracts. To complement the biochemical experiments, the effects of in vivo H1 depletion on protein content and structural integrity of the nucleolus were investigated using the H1 triple isoform knock out (H1ΔTKO) mouse embryonic stem cell (mESC) model system. Proteomic profiling of purified wild type mESC nucleoli identified a total of 613 proteins, only ~60% of which were detected in the H1 mutant nucleoli. Within the affected group, spectral counting analysis quantitated 135 specific nucleolar proteins whose levels were significantly altered in H1ΔTKO mESC. Importantly, the functions of the affected proteins in mESC closely overlapped with those of the human T cell nucleolar H1 binding proteins. Immunofluorescence microscopy of intact H1ΔTKO mESC demonstrated both a loss of nucleolar RNA content and altered nucleolar morphology resulting from in vivo H1 depletion. We conclude that H1 organizes and maintains an extensive protein-protein interaction network in the nucleolus required for nucleolar structure and integrity.
Keywords: Mass spectrometry, chromatin structure and gene expression, protein-protein interactions, ribosome biogenesis, messenger RNA splicing
Introduction
The H1 linker histones make up a family of abundant chromatin-associated proteins involved in the packaging of eukaryotic genomes. In mammals there are seven somatic H1 sequence variants or subtypes (H1.0-H1.5 and H1X), and four germ cell-specific variants (H1t, H1T2, HILS1 and H100). The expression, nuclear distribution and posttranslational modification of H1 subtypes vary across cell types, and differ during cell development and differentiation (reviewed in (1-3)). H1 family members bind nucleosomes and are organizational constituents of chromatin in vitro and in vivo (1,4). The nucleosome is formed by wrapping 147 base pairs of DNA ~1.7 times around an octamer of core histones (H2A, H2B, H3, and H4) (5). Chromatin contains arrays of nucleosomes separated by free ‘linker’ DNA. Genomic chromatin of most eukaryotic cells contains ~0.7 H1 per nucleosome (6). Linker histones bind to nucleosomes and linker DNA to stabilize condensed higher order chromatin structures in vitro (7,8), and chromatin decompaction occurs in vivo when linker histones are genetically depleted from cells (9,10). Altogether, the current paradigm holds that linker histones modulate nuclear processes such as transcription, replication, and DNA repair through their interactions with DNA and effects on chromatin condensation (1-3,11).
More recently it has become apparent that linker histones also bind specifically to proteins. More than 100 functionally diverse proteins from mammalian nuclear extracts were found to interact with H1.0 in vitro, and biophysical experiments demonstrated that H1 binds directly to several of these proteins (FACT, U2AF65 and SF2/ASF) (12). Immunoprecipitation of tagged proteins expressed in mouse cells demonstrated interactions between specific H1 variants and DNMT1 or DNMT3B, and in vitro studies with pure proteins showed that the interactions were direct (13). Drosophila H1 co-purified with numerous ribosomal proteins in co-immunoprecipitation experiments, and endogenous ribosomal proteins co-localize with H1 in condensed nuclear chromatin in vivo (14). Purification of the epitope tagged H1.2 isoform from a HeLa derived cell line identified a large complex containing H1.2 and many regulatory proteins, including PARP1, FIR, YB1, PURα, CAPERα, and WDR5 (15). The interaction of H1.2 with these proteins was direct, as was its binding to p53 (15). While compelling, the functional significance of H1-dependent protein-protein interactions remains largely unexplored.
In addition to their high levels in nuclear chromatin, linker histones are components of the nucleolus (16,17). The nucleolus is a non-membranous nuclear organelle that consists of densely packaged ribosomal DNA (rDNA), RNA, and protein lacking a soluble compartment. MS-based analyses of purified nucleoli from various cell types have established that the nucleolar proteome is extensive. Collectively, over 4500 human nucleolar proteins have been identified (18) with the largest single source provided by Jurkat T-cells (16). A primary function of the nucleolus is ribosome biogenesis, which consists of the coupled processes of rDNA transcription, rRNA processing and pre-ribosome assembly. Initially, RNA polymerase I (Pol I)-mediated transcription of the 47S precursor ribosomal RNA (pre-rRNA) occurs from tandemly repeated rDNA loci (19,20). Only a fraction (~50%) of rDNA loci are transcriptionally active; the remaining loci are inactive or silent depending on the cell type (19,21) and cell growth and differentiation conditions (22). Silent rDNA chromatin is thought to resemble inactive nuclear heterochromatin through the recruitment and action of H1 and other chromatin architectural proteins, chromatin remodeling complexes, transcriptional repressors and co-repressors, and epigenetic silencing factors (19,21). In contrast, transcriptionally active rDNA requires the recruitment of RNA polymerase I (Pol I) by the Pol I transcription upstream binding factor (UBF) (22) and subsequent loss of canonical nucleosome structure (23). Once transcribed, the 47S pre-rRNA is covalently modified and cleaved by processing proteins into mature 28S, 18S, and 5.8S rRNAs, which are then assembled with ribosomal proteins into pre-ribosomal particles. It is being increasingly recognized that the nucleolus is plurifunctional, with vital roles in cell cycle control, DNA repair, cell senescence, apoptosis and development that extend beyond ribosome biogenesis (24-26).
The function of linker histones in the nucleolus remains an open question, with most evidence suggesting a role in transcriptional silencing, presumably mediated through effects on rDNA chromatin structure (27). In contrast, we have recently hypothesized that H1 may play a more global role in the nucleolus acting through extensive protein-protein interactions (12). To address our hypothesis, we performed two types of experiments with nucleoli purified from cultured mammalian cells. The first set of studies identified the proteins in human Jurkat T cell nucleolar extracts that bound to sepharose-immobilized H1 and characterized the oligomeric state of nucleolar H1. The second set of studies applied quantitative proteomics to determine the effects of in vivo H1 depletion on the protein composition of the nucleolus using the H1 triple isoform knockout (H1ΔTKO) mouse embryonic stem cell (mESC) model developed previously (10,28). Collectively, our biochemical and proteomic studies suggest that H1 is a nucleolar hub protein responsible for establishing and maintaining an extensive protein-protein interaction network involved in Pol I transcription, precursor rRNA processing, mRNA metabolism, and various metabolic and stress activities.
Results
Identification of H1 binding proteins in purified T-cell nucleoli
To test our hypothesis that H1 interacts with an extensive network of proteins in the nucleolus (12), we first performed biochemical studies of H1 binding proteins in Jurkat T cell nucleoli (Fig. 1a). Jurkat cells were chosen because of their extensively characterized nucleolar proteome (16). Affinity binding experiments were carried out with a soluble DNase I-treated nucleolar extract derived from Jurkat T-cells and sepharose-immobilized H1. Nucleolar H1 binding proteins were identified using liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS).
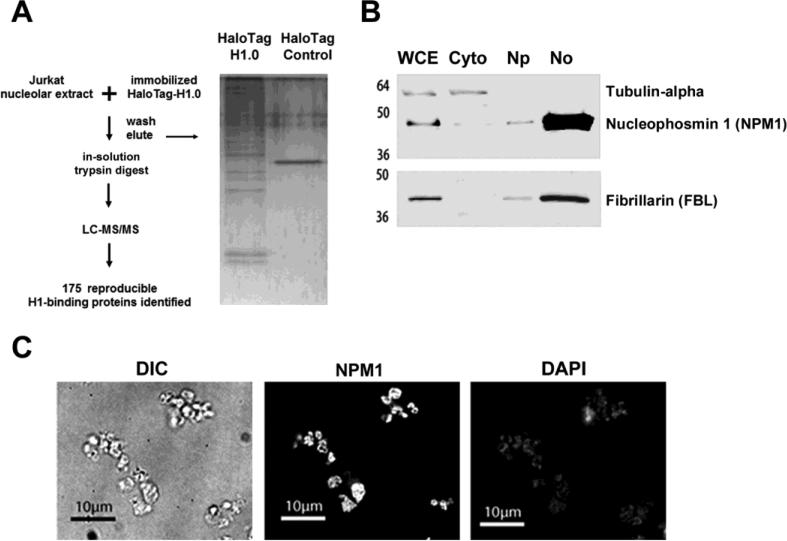
Fig. 1.
Experimental design and isolation of nucleoli from cultured T cells. (a) Schematic representation of H1-affinity binding experiments and proteomics workflow (left). Purified nucleoli from Jurkat T-cells were used as the protein source for H1-binding experiments. Proteins that bound HaloTag-H1 or Halo Tag control beads were eluted, digested with trypsin protease, and identified by LC-MS/MS analysis. A portion of the eluate was resolved by SDS-PAGE and stained with silver (right). (b) Immunoblots of Jurkat T-cell subcellular fractions (5 μg protein per lane) probed with the indicated antibodies (right). The fractions (top) and molecular weight standards (left) are indicated. Whole cell extract (WCE), cytoplasm (Cyto), nucleoplasm (Np) and nucleolar (No). (c) Nucleolar fractions from Jurkat T-cells were fixed in 4% paraformaldehyde and analyzed by differential interference contrast (DIC) microscopy or indirect immunofluorescence with either α-nucleophosmin antibody or stained with 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI).
T-cell nucleoli were purified as described (16). Briefly, ~20×106 Jurkat cells were lysed in hypotonic buffer and nuclei were isolated by centrifugation through a 0.25 M sucrose buffer layer. Nuclei were disrupted by sonication and nucleoli were isolated by centrifugation through a 0.35 M sucrose layer. Equivalent total protein from either whole cell or subcellular fractions containing the cytoplasm, nucleoplasm and nucleoli were resolved by SDS-PAGE and probed for the cytoplasmic protein α-tubulin, and nucleolar proteins nucleophosmin 1 (NPM1/B23) and fibrillarin (FBL). Relative to the signal in the whole cell extract, strong enrichment for both NPM1 and FBL was observed in the nucleolar fraction, while α-tubulin was found exclusively in the cytoplasmic fraction (Fig. 1b). The nucleolar fraction was then examined by differential interference contrast (DIC) and high-resolution immunofluorescence (IF) microscopy (Fig. 1c). DIC imaging revealed the presence of a nearly uniform population of 2-3 μM diameter particles. The particles visualized by DIC microscopy stained intensely for NPM1, while staining by 4',6-diamidino-2-phenylindole (DAPI) was relatively low and diffuse as determined by IF, indicating that the isolated nucleoli were devoid of contamination by either whole nuclei or large chromosomal DNA fragments.
The purified T-cell nucleoli were the source of proteins used for affinity binding experiments. Nucleolar proteins were extracted from isolated nucleoli under native conditions and incubated with sepharose-immobilized H1.0 (HaloTag-H1.0) or HaloTag control resin using the HaloTag system described previously (12). Unbound proteins were removed by extensive washes with a modified RIPA buffer containing 200 mM NaCl and bound proteins were eluted in denaturing buffer and precipitated. The eluted proteins were digested in-solution with trypsin, and analyzed by LC-MS/MS. Compound lists of MS/MS spectra from two biological replicates of HaloTag-H1 and two HaloTag control experiments were generated using Xcalibur 2.2 software (Thermo Scientific) and searched against the Human Uniprot protein database concatenated to a reverse database using the Mascot database search engine (version 2.3) and the Sorcerer™-SEQUEST® version 3.5. For each sample, search results were imported and pooled using probabilistic protein identification algorithms (29) employed in Scaffold software (30) (Version 4, Proteome Software). Peptide and protein probability thresholds of 95% and 99%, respectively, were applied and a minimum of two unique peptides were required for protein identification. LC-MS/MS identified 175 nucleolar proteins that were observed in duplicate HaloTag-H1 samples and absent from the HaloTag controls (Supplementary Table 1). Approximately 25% of the nucleolar H1 binding proteins overlapped with the nuclear H1-binding proteins previously identified by our group (12) (Supplementary Fig. 1).
To categorize the molecular functions of the candidate H1 binding proteins we first queried gene ontology (GO) databases using Scaffold 4 software. A high percentage of H1 binding proteins bind nucleic acids; 28% of the proteins bind RNA and 25% bind DNA or chromatin (data not shown). Next, the DAVID Bioinformatics Resources gene ontology tool (http://david.abcc.ncifcrf.gov, (31)) was used to determine gene groups that were significantly enriched relative to the Homo sapiens database for various biological processes (Fig. 2). The most significant GO biological process terms with the lowest p-values were selected from each cluster of related functions. Proteins with functions associated with chromatin disassembly and nucleosome organization were among the most highly enriched, including histone H2B, H3.3, H1.0 , H1.3, the core histone variants H2AX, H2A.Z, the multifunctional nucleolar protein NPM1, the histone chaperones NAP1L1, NAP1L, SET, DEK, the SWI/SNF complex subunit SMARCC1, and replication factor MCM2. Other chromatin-associated proteins that bound H1.0 in our experiments included PARP1, HDAC1, and macro-H2A.1, all of which are components of silenced rDNA chromatin (32-34). Constituents of active rDNA chromatin that also bound H1 included UBF (22,27), nucleolin (35), and DNA topoisomerase I (TOP1) (36). Additional identified H1 binding proteins that act in a chromatin environment included CBX1, 3, 5 (HP1α, HP1β, HP1γ), transcription elongation factors SPT5 and SPT6, PAF1 complex subunits (PAF1 and LEO1), DNA repair proteins (XRCC6/Ku70, XRCC5/Ku80), and DNA topoisomerase II-alpha (TOP2A).
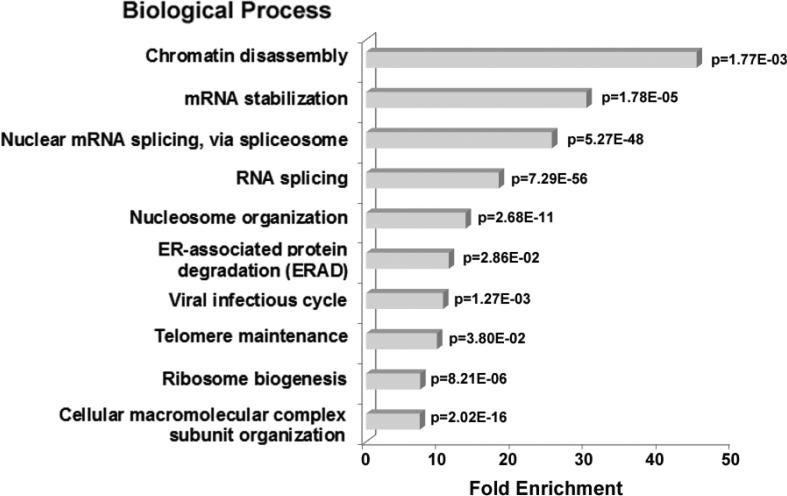
Fig. 2.
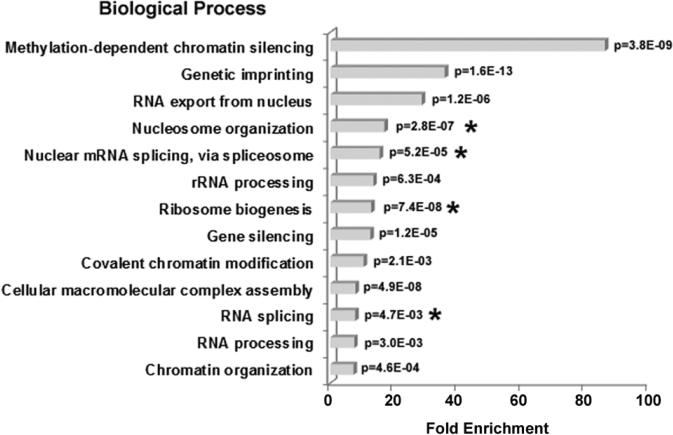
GO biological process enrichment in nucleolar H1 binding proteins identified by LC-MS/MS analysis. The bar graph represents the fold enrichment of GO biological processes terms in H1 binding proteins relative to the Human gene database using the DAVID Bioinformatics Resources functional annotation clustering algorithm (31). The most highly enriched functions were chosen from a cluster of similar terms. Biological processes with <4-fold enrichment are not shown. A p-value for each term is indicated.
H1 binding proteins with functions associated with RNA metabolism made up the other highly enriched categories in the DAVID analysis (Fig. 2). 47S pre-rRNA processing is an essential step in ribosome biogenesis (20,37,38). Pre-rRNA processing proteins that bound immobilized H1 included NolC1/Nopp140, the Box H/ACA snoRNP NHP2, Box C/D snoRNPs (NOP56, NOP58), and U3 snoRNPs (U355K, MPP10). Pre-mRNA processing proteins were also overrepresented in our dataset. Although splicing of pre-mRNA has never been directly connected to ribosome biogenesis or any other biological process in the nucleolus, numerous proteomic profiling studies have identified pre-mRNA processing proteins as components of purified nucleoli (16,17,26,38,39). Interestingly, one third of the 175 H1-bound proteins identified by our experiments function in pre-mRNA processing, including subunits of the spliceosomal small nuclear ribonucleoprotein (snRNP) complexes U1, U2, U4/U6, and U5, many heterogeneous nuclear ribonucleoproteins (hnRNPs), serine/arginine-rich (SR) splicing factors, poly(A)-binding protein and YBX1. Other functional categories enriched ~10-fold in the GO analysis were ERAD, viral infectious cycle, telomere maintenance and macromolecular complex subunit organization. Together, our biochemical experiments indicate that H1 binds either directly or indirectly to at least 175 nucleolar proteins whose biological functions span the known functions of the nucleolus and are particularly enriched with respect to rDNA chromatin structure and pre-rRNA and mRNA metabolism (Fig. 2).
Size exclusion chromatography of endogenous nucleolar H1
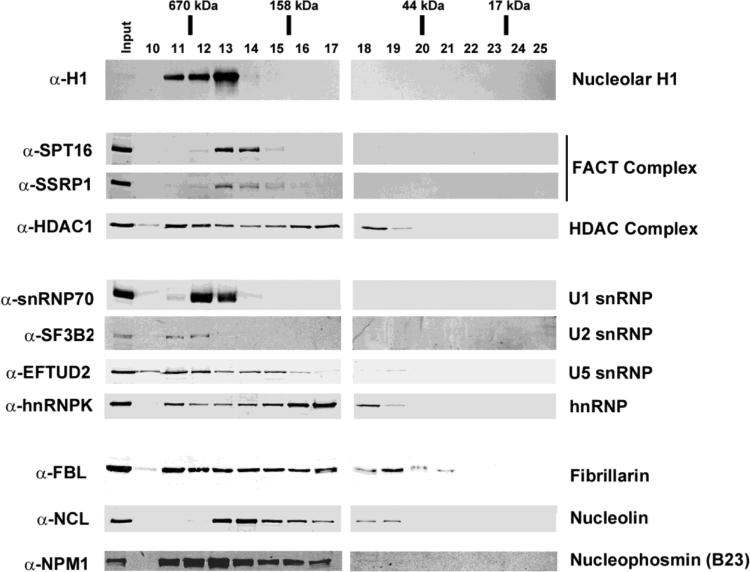
To determine the oligomeric state of endogenous nucleolar H1 and a subset of H1 binding proteins we performed size exclusion chromatography (SEC) of Jurkat T-cell nucleolar extracts under native conditions. Nucleolar proteins were solubilized as in the affinity binding studies and fractionated using a Superdex S200 16/60 column. Fractions 10-25 were TCA precipitated, resuspended in sample buffer, and resolved by SDS-PAGE for immunoblot analysis (Fig. 3). Individual SEC fractions were probed for H1 along with nine other H1 binding proteins identified in the affinity binding experiments (FACT, HDAC1, FBL, nucleolin, NPM1, snRNP70, SF3B2, EFTUD2, and hnRNPK). The observed molecular weight (MW) range for each protein was extrapolated from a standard curve (Table 1 and data not shown). Bacterially expressed H1.0 (22 kDa) eluted in fractions 19-22 in the absence of nucleolar extract, corresponding to an apparent MW of a monomer (21-55 kDa) (data not shown). In distinct contrast, >95% of the endogenous H1 in nucleolar extracts eluted in fractions 11-13, corresponding to a MW distribution of ~378 to >670 kDa. This result indicates that endogenous nucleolar H1 is assembled into large macromolecular complexes. Importantly, each of the nine of the H1 binding proteins examined were present all or in part in the H1-containing fractions (fractions 11-13), suggesting that endogenous nucleolar H1 is complexed with many of the proteins identified in the affinity binding experiments.
Fig. 3.
Size exclusion chromatography (SEC) analysis of purified nucleoli. Immunoblot analysis of nucleolar extract from Jurkat T-cells resolved by SEC under native buffer conditions. Purified nucleoli were sonicated and treated with DNAse I in high salt (0.5 M NaCl) prior to loading on a S200 Sepharose column pre-equilibrated in low salt (200 mM NaCl). Individual fractions (top) were resolved by SDS-PAGE, transferred to PVDF membrane and probed with the indicated primary antibodies (left). The apparent MW range for each protein or protein complex (right) was extrapolated from a standard curve derived from MW standards (Table I).
Table 1.
Molecular Weight of Hi-binding Proteins; the apparent MW for each protein determined by SEC analysis and the predicted MW determined for the amino acid sequences of each protein.
| Protein name | Apparent MW (kDa) | Predicted MW (kDa) |
|---|---|---|
| Linker Histone H1.0 | 21-55 | 21 |
| Nucleolar linker Hisone H1 (all subtypes) (H1) * | 378 to >670 | 22 |
| FACT complex subunit SPT16 * | 199-520 | 120 |
| FACT complex subunit SSRP1 * | 199-520 | 81 |
| Histone deacetylase 1 (HDAC 1) * | 55 to >670 | 55 |
| U1 small nuclear ribonucleoprotein 70 kDa (snRNP 70) * | 378 to >670 | 52 |
| Splicing factor 3B subunit 2 (SF3B2) * | 520 to >670 | 100 |
| 116 kDa U5 small nuclear ribonucleoprotein component (EFTUD2) * | 145 to >670 | 109 |
| Heterogeneous nuclear ribonucleoprotein K (hnRNPK) * | 55 to >670 | 51 |
| rRNA 2′-O-methyltransferase fibrillarin (FBL) | 40 to >670 | 34 |
| Nucleolin (NCL) * | 55 to 378 | 77 |
| Nucleophosmin (NPM1) * | 105 to >670 | 33 |
Effect of in vivo H1 depletion on nucleolar protein composition and abundance
Our biochemical analyses indicated that H1 binds numerous proteins in T cell nucleolar extracts that collectively are involved in many aspects of nucleolar function (Supplementary Table 1, Fig. 2). Assuming that these in vitro interactions are biologically relevant, we hypothesized that H1 depletion in vivo would disrupt key protein-protein interactions and alter the composition of the nucleolar proteome. To address our hypothesis, we took advantage of the H1ΔTKO mESC model system (28). H1ΔTKO mESC harbor null mutations in H1c, H1d and H1e (H1.2, H1.3 and H1.4 in an unified nomenclature (40)), and exhibit a ~50% reduction in the total H1/nucleosome ratio compared to WT mESC (28). This level of H1 depletion results in several specific phenotypes, including defects in transcription (10), DNA methylation (13), and chromosome condensation (10). For our experiments, the protein composition of purified wild type (WT) and H1ΔTKO mESC nucleoli was determined by LC-MS/MS, and the relative abundances of individual proteins in the WT and mutant nucleoli were quantified by spectral counting (SpC) analysis (Fig. 4a).
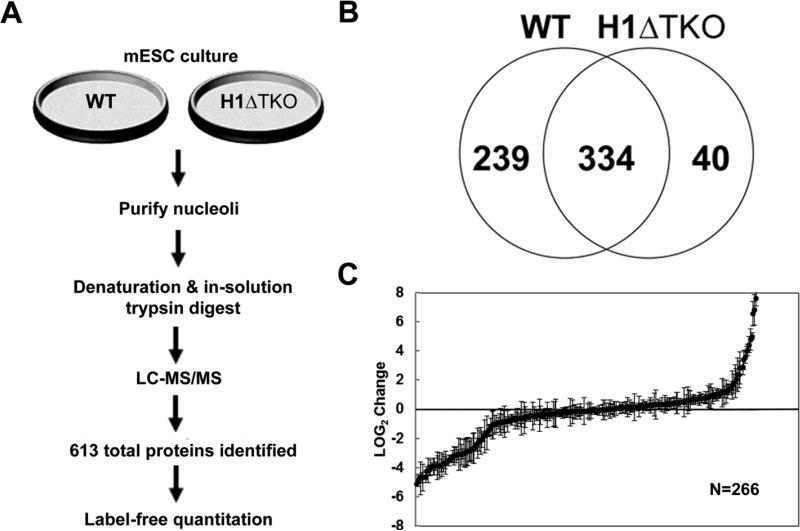
Fig. 4.
Comparative proteomic profile of nucleoli from WT and H1ΔTKO mESC. (a) Schematic representation of sample preparation from either WT or H1ΔTKO mESC and proteomics workflow. Purified nucleoli were treated with nuclease, denatured and digested with trypsin for LC-MS/MS analysis. Protein identification results for biological replicates were compiled for quantitative spectral counting analysis. (b) Venn diagram showing the overlap of proteins identified in WT and H1ΔTKO mESC. (c) Scatter plot representing the normalized LOG2 values of H1ΔTKO over WT control. Three biological repeats were performed for each strain and proteins identified in at least two out of the three samples with a sum of >10 spectra and standard deviations below the average standard deviation of the experiment were included in the analysis. Negative LOG2 values indicate reduced protein levels whereas positive LOG2 values represent elevated protein levels in H1ΔTKO relative to WT mESC.
Nucleoli from mESC were purified as described for Jurkat T-cells (16), and examined by DIC and high-resolution IF microscopy (data not shown). The WT nucleolar fractions contained 2-3 μM diameter particles that stained robustly for NPM1 and weakly for DAPI, indicating the presence of a highly enriched population of nucleoli lacking chromosomal DNA contamination. Equivalent protein amounts from triplicate samples of WT and H1ΔTKO mESC nucleoli were treated with DNase, TCA precipitated, digested in solution with trypsin, and analyzed by LC-MS/MS. A total of 613 different proteins were identified from the WT and H1ΔTKO mESC nucleoli (Supplementary Table 2); 334 proteins (54%) were found in both WT and H1ΔTKO samples, 40 proteins (7%) were exclusively found in H1ΔTKO, and, significantly, 239 proteins (39%) were only detected in the WT sample (Fig. 4b). These data suggest that disruption of H1 homeostasis in vivo greatly affected the protein composition of nucleoli and resulted in a large reduction in protein number.
Next, the relative abundance of individual proteins identified in WT and H1ΔTKO nucleoli was determined by SpC analysis. SpC analysis is a label-free quantitative method that uses the normalized sum of MS/MS spectra assigned to peptides from a given protein as a measure of abundance (41). Raw spectral counts from three biological replicates of WT and H1ΔTKO were normalized in Scaffold 4 software by applying a scaling factor such that the total spectral counts for each biological replicate were equivalent. Spectral counts of shared peptides were distributed equally among all proteins to which they were assigned. The resulting list of proteins affected by H1 depletion was further filtered by the following criteria: proteins must be present in a minimum of two out of three biological replicates for a given group and the total normalized spectral counts for a given group must be >10. Out of the 613 nucleolar proteins identified, 441 proteins met the filtering criteria and were included in the statistical analysis. The relative fold-change for each protein was calculated by dividing the average SpC for H1ΔTKO by the corresponding value obtained for WT, and a student's t-test was applied to calculate p-values (Supplementary Table 3). The LOG2 fold-change was calculated and plotted on a scatter diagram for proteins with a standard deviation less than or equal to the average standard deviation for each dataset (Fig. 4c). Of the 441 proteins analyzed, the fold-change values of 135 proteins were significantly different in the H1ΔTKO mESC dataset compared to WT (p<0.05). Of these, 46 proteins (34%) exhibited a fold change of ≥1.5 and 77 proteins (57%) exhibited a fold change of ≤0.067. Among the affected proteins were the three remaining H1 isoforms expressed in H1ΔTKO mESC including H1.0, H1.1, and H1.5 which demonstrated a ~12-fold, ~7-fold and ~3-fold increase in the mutant compared to WT, respectively (Supplementary Table 3). These findings are consistent with H1.0, H1.1, and H1.5 expression in H1ΔTKO embryos which were previously reported as having a relative increase of ~10-fold, ~2-fold and ~2.5-fold, respectively (28). Therefore, it is possible that H1 isoform-specific interactions (involving H1.0, H1.1, and H1.5) increase in H1ΔTKO mESC, while H1 isoform-independent interactions decrease due to the net decrease in total H1 (by ~ 50%). Consequently, all of the significantly affected proteins identified by SpC analysis were grouped together for functional clustering analysis using the DAVID Bioinformatics Resources gene ontology tool (http://david.abcc.ncifcrf.gov, (31)), regardless of the direction of the observed change.
The fold-enrichment of the biological processes associated with the affected proteins was determined relative to the Mus musculus database used by DAVID (Fig. 5). The biological processes with the lowest p-values were selected from each cluster of related functions. Proteins with functions involving epigenetic regulation of gene silencing and nucleosome organization were among the most highly enriched. De novo DNMTs (DNMT3a, DNMT3b) and Polycomb Group proteins (RNF2, EED) are epigenetic regulators that were affected in H1ΔTKO mESC. Other chromatin-associated proteins included histone deacetylase 1 (HDAC 1), SIN3a, H2AX tyrosine-142 kinase (Baz1b), chromatin remodeling factors (SMARCA4, BPTF), core histone variants (H3.3 and H2A.X) and linker histone H1 itself. Thirty-three out of the 135 (24%) of the affected proteins were associated with ribosome biogenesis, including many pre-rRNA processing proteins (NOL9, BOP1, DIS3, FBL, PDCD11, RPS14, UTP18, UTP20, WDR36). Eighteen out of 135 (13%) affected proteins have functions related to pre-mRNA splicing, including subunits of the U1 (snRNP70), U2 (SF3B2), U4/U6 (PRPF4) and U5 (PRPF8) spliceosomal snRNP subcomplexes, and several hnRNP proteins (hnRNPK, hnRNPC, hnRNPM and PCBP1). Together these results are consistent with the affinity binding studies in human nucleoli (Fig. 2), and suggest that H1 organizes a network of proteins with functions involving rDNA chromatin structure, rRNA processing and mRNA metabolism in the nucleolus.
Fig. 5.
GO Biological process enrichment in proteins altered in H1ΔTKO mESC. The bar graph shows the fold enrichment of GO biological processes terms in the H1-depleted dataset relative to the Mus musculus database using the DAVID Bioinformatics Resources functional annotation clustering algorithm (31). The most highly enriched functions were chosen from a cluster of similar terms. A p-value for each term is indicated. Asterisks (*) indicated terms shared with the H1-binding proteins identified in T-cells (Fig. 2.).
Validation of SpC analysis
We have interpreted the SpC results as being due to direct effects on specific nucleolar H1-protein interactions. These included H1 knockouts (H1.2, H1.3 and H1.4) as well as the remaining H1 isoforms that were overexpressed in H1ΔTKO mESC (H1.0, H1.1 and H1.5). However, because H1 affects the expression of specific RNA polymerase II (Pol II)-directed genes (1-3,10), it is possible that the changes in protein composition observed in H1ΔTKO mESC were due to variations in the gene expression of affected proteins though Pol II-mediated mechanisms. In order to distinguish between these two possibilities and validate our SpC analysis, the relative amount of affected proteins in both whole cells and nucleoli of WT and H1ΔTKO mESC was determined by immunoblot analysis (Fig. 6a-c).
Fig. 6.
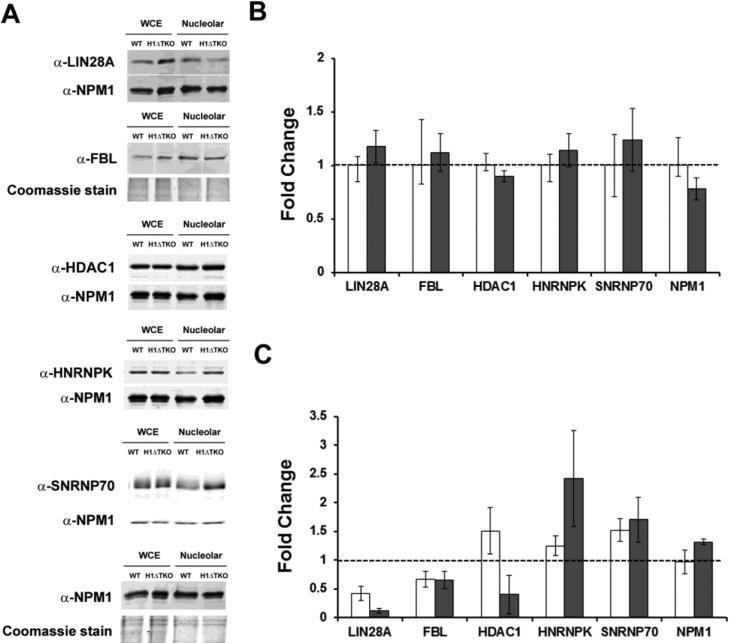
Expression of H1ΔTKO mESC proteins and validation of SpC analysis. (a) Immunoblot analysis of extracts from either WT or H1ΔTKO mESC was used to determine the relative abundance of proteins identified by SpC analysis in either the whole cell (b) or in the nucleolus (c). Five μg of total protein from each sample was resolved by SDS-PAGE and probed for the indicated proteins (left). As a loading control, the same immunoblot was either probed with α-NPM1 or the gel was stained with coomassie. (b) The fold-change expression in mESC whole cell extracts was determined by dividing the normalized signal for H1ΔTKO (closed bars) by WT (open bars) set at 1.0 for each of the indicated proteins. (c) The fold-change expression of candidate proteins in nucleolar extracts was determined by dividing the normalized values for H1ΔTKO by WT for each of the indicted proteins. Bar graphs represent the values obtained from either immunoblot analysis (open bars) or SpC analysis (closed bars). Values are the averages ± the standard deviations of triplicate samples and normalized to the loading control present in each lane.
Proteins from whole cell extracts derived from WT and H1ΔTKO mESC were resolved by SDS-PAGE and detected using primary antibodies raised against several of the affected proteins in H1ΔTKO mESC identified by SpC analysis (p-value <0.05) (Fig. 6a). The fold-change expression for LIN28A, FBL, HDAC1, hnRNPK, snRNP70, and NPM1 was calculated by dividing the normalized signals obtained for H1ΔTKO by the corresponding WT values (Fig. 6b). Because NPM1 expression levels were nearly equivalent in WT and H1ΔTKO, the α-NPM1 antibody was used as a loading standard for protein normalization (with exception for FBL due to similar MW and co-migration with NPM1). Quantitative immunoblot analysis of WT and H1ΔTKO whole cell extracts found no significant differences in the normalized protein levels of LIN28A, FBL, HDAC1, hnRNPK, snRNP70 and NPM1 suggesting that their cellular expression is largely unaffected by the H1ΔTKO mutation.
Next, similar comparisons were made using the nucleolar fractions derived from WT and H1ΔTKO mESC. The normalized fold-change values for each protein determined by immunoblot analysis were plotted with the values obtained by SpC analysis (Fig. 6c). The fold-change values determined for LIN28A and FBL were consistent with the values obtained by SpC analysis (0.42-fold and 0.11-fold, respectively for LIN28A and 0.66-fold and 0.65-fold, respectively, for FBL). The fold-change values determined for hnRNPK and snRNP70 were both elevated in H1ΔTKO and were consistent with the values determined by SpC analysis (1.24-fold and 2.42-fold, respectively, for hnRNPK and 1.52-fold and 1.70-fold, respectively, for snRNP70). HDAC1 on the other hand exhibited a 1.5-fold increase by immunoblot analysis and a 0.4-fold decrease by SpC analysis. The observed disagreement between the immunoblot and SpC analyses of HDAC1 levels is likely attributable to the detection of identical peptides derived from alternative HDAC1 isoforms or HDAC family members by LC-MS/MS, while amino acids 467-482 were exclusively detected by the α-HDAC1 antibody used in the immunoblot analysis. In summary, the protein expression levels measured by immunoblot are generally consistent with the SpC analysis of purified nucleoli. Importantly, comparison of the protein levels in the whole cells of H1ΔTKO and WT mESC suggest that a general defect in cellular abundance is unlikely, and instead represents a nucleolar-specific defect.
Altered nucleolar morphology and RNA content in intact H1ΔTKO mESC
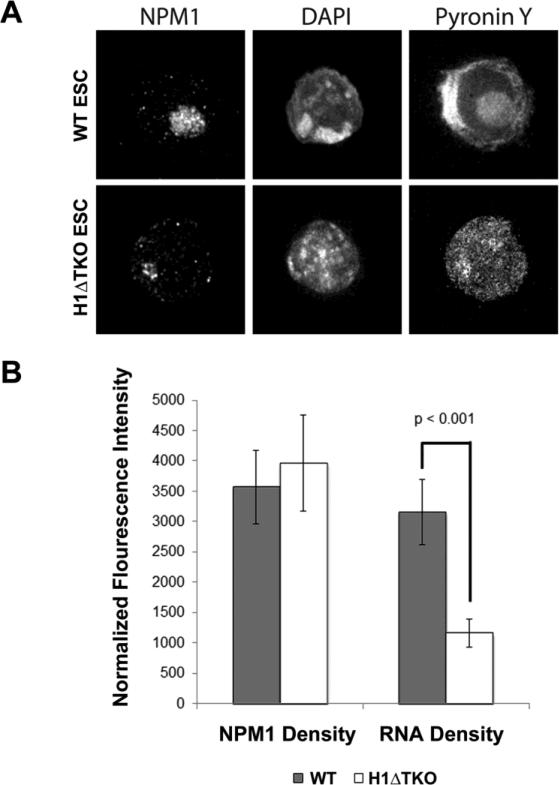
Given the enrichment of DNA- and RNA-binding proteins that bound H1 in vitro and the pronounced differences in protein composition between the WT and H1ΔTKO mESC nucleoli, we speculated that the nucleoli from H1ΔTKO mESC would have altered morphology and exhibit reduced RNA levels. Therefore, we assayed nucleolar morphology and RNA content by IF microscopy (Fig. 7a and b). WT and H1ΔTKO mESC were stained with α-NPM1 antibody, DAPI to block double-stranded DNA (dsDNA), and Pyronin Y to stain all remaining nucleic acid (primarily RNA). The nucleolar size determined by Pyronin Y and NPM1 overlap was reduced by ~20% in H1ΔTKO mESC compared to WT (Fig. 7a and data not shown). The fluorescence signal intensity for NPM1 and Pyronin Y were measured for WT and H1ΔTKO mESC and normalized to the defined nucleolar area. Consistent with both our immunoblot and SpC analyses, NPM1 signal density was not significantly altered in H1ΔTKO mESC compared to WT. However, Pyronin Y density decreased nearly 2.5-fold in the H1 mutant nucleoli.
Fig. 7.
Immunofluorescence microscopy of WT and H1ΔTKO mESC. (a) High-resolution immunofluorescence images of mESC stained with NPM1 antibody, DAPI and Pyronin Y. B, The fluorescence intensity for NPM1 and Pyronin Y was plotted for WT (closed bar) and H1ΔTKO (closed bar) mESC. Fluorescence signal was normalized to nucleolar area in each cell. Error bars represent the standard deviation among >25 cells. Pyronin Y (RNA) signal density deceases in H1ΔTKO mESC (student t-test p<0.001).
Discussion
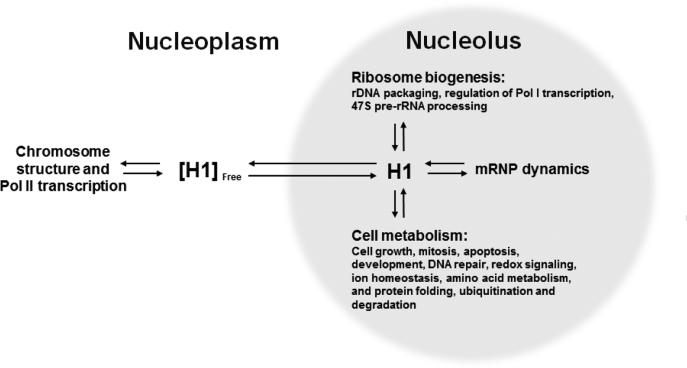
The current paradigm of linker histone function maintains that H1 is a chromatin architectural protein and effector of Pol II genes in the nucleus (6,11,42)). H1 is generally thought to act through protein-DNA interactions to promote chromatin condensation and regulate transcription (1-3). H1 has also been described as an adaptor protein for transcription factors at Pol II-regulated genes (43). Here, biochemical and proteomic studies have identified a large number of nucleolar proteins that either bound immobilized H1 in vitro or were significantly perturbed in nucleoli upon H1 depletion in H1ΔTKO mESC in vivo. These results support an important new role for H1-mediated protein-protein interactions in the nucleolus, and suggest that H1 organizes an extensive protein-protein interaction network (PPIN) connecting rDNA chromatin structure and Pol I transcription to ribosome biogenesis, mRNA-associated ribonucleoprotein (mRNP) dynamics, and various aspects of cell metabolism (Fig. 8). The nucleolus is a steady-state supramolecular assemblage whose protein constituents are in constant flux with the nucleoplasm (44,45). We therefore hypothesize that H1 controls nucleolar protein dynamics and prolongs the residency of target proteins through specific H1-protein interactions, i.e., H1 acts as a protein hub in the nucleolus (46). Like H1 itself, many of the H1 binding proteins are predicted to have one or more large regions of intrinsic disorder by the FoldIndex program (47), with the disorder being most prevalent in the splicing and chromatin-associated proteins. A potential connection between intrinsic disorder and hub proteins previously has been noted (48). SEC analysis of endogenous nucleolar H1 (Fig. 3) suggests that H1 hub activity is linked to the assembly of large multi-protein complexes. The proposed role for H1 is analogous to other multifunctional nucleolar hub proteins such as nucleolin and NPM1, (46,49,50), both of which are H1 binding proteins (Supplemental Table 1). Thus, a direct prediction of our hypothesis is that altered H1 expression in vivo would disrupt key nucleolar PPINs and lead to significant changes in nucleolar protein composition and morphology. Indeed, quantitative proteomic profiling and IF microscopy of H1ΔTKO mESC nucleoli revealed this to be the case. Altogether, our results are consistent with the view of H1 as a highly connected nodule in a network of steady-state protein-protein and protein-nucleic acid interactions in the nucleolus.
Fig. 8.
Model for a nucleolar H1 protein-protein interaction network (PPIN). H1 transitions between the nucleoplasm and the nucleolus to regulate the dynamics of proteins associated with the H1 protein-protein interaction network (PPIN). For nuclear genes, a major role for H1 is to modulate Pol II transcription. In the nucleolus, the H1 PPIN functions in various aspects of ribosome biogenesis, pre-mRNA splicing (or mRNP dynamics), and an assortment of other functions related to cellular health and disease. Through these interactions, H1 functions as a hub protein that directly impacts the nuclear steady-state equilibrium and accumulation of proteins and RNP complexes in the nucleolus.
The nucleolar organizer regions (NORs) are tandemly repeated rDNA sequences located on chromosomes 13, 14, 15, 21 and 22 in humans (51) that spatially cluster together to nucleate the formation of the nucleolus (19,21,26). Active rRNA gene repeats are transcribed by Pol I to produce the 47S pre-rRNA, which is subsequently modified and processed into the mature rRNAs found in the ribosome (20). However, approximately half of rRNA genes are transcriptionally active and the remaining loci are silenced, i.e., they cannot be transcribed by Pol I (19,21,22). Previously, H1 has been shown to associate with rDNA in vivo (52) and has been proposed to help silence rDNA (22,53). Unexpectedly, we identified both positive and negative regulators of Pol I transcription associated with H1. UBF establishes and maintains transcriptionally active rDNA loci (22,27), and other positive Pol I regulators including nucleolin (35), TOPI (36), and transcription elongation factors PAF1 (54) and FACT (55) were identified by LC-MS/MS analysis in our affinity binding studies. Through these interactions, H1 may modulate Pol I activity and control the balance between transcriptionally active and silent rDNA repeats. The silent rDNA repeats are maintained in a repressive chromatin structure characterized by the assembly into regular nucleosomal arrays, high levels of DNA methylation, histone H4 hypoacetylation, histone H3 methylation, and a specific set of proteins including DNMTs (33), PARP1 (34), HDAC1 (56), macro-H2A.1 (32), and H1 (22). We found that HP1 (CBX1, -3, -5), PARP1, Macro-H2A.1, and HDAC1 repressor proteins bound immobilized H1 in vitro (Supplementary Table 1). Moreover, H1, HDAC1, PARP1, and DNMTs (DNMT3a, DNMT3b) exhibited altered levels in ΔH1TKO mESC nucleoli (Supplementary Table 3). Based on these associations, we favor a mechanism in which H1 functions dynamically to mediate the assembly and/or disassembly of a specific heterochromatic nucleoprotein complex minimally consisting of the chromatin fiber, HP1, PARP1, MarcroH2A1, DNMTs and H1 itself. Finally, we note that H1 also bound casein kinase II (CKII) α and β subunits in vitro (Supplementary Table 1). CKII is a multifunctional nucleolar serine/threonine kinase that phosphorylates Pol I (57), UBF (58), FACT (59), nucleolin (60,61), NPM1 (62,63), and TOP 1 (64). CKII is also a component of the pre-rRNA processing machinery (37) (discussed below). Taken together, our results suggest that a major module of the H1 PPIN is dedicated to the assembly, maintenance, and regulation of both active and silent chromatin structure at NORs.
A large proportion of nucleolar proteins identified in our studies function downstream of Pol I transcription and are required for 47S pre-rRNA processing. Co-transcriptional processing of pre-rRNA has been demonstrated in yeast (65,66) and has been proposed for mammals (67). Our results reveal a novel connection between rDNA packaging, Pol I transcription, and 47S pre-rRNA processing through the H1 PPIN. Numerous protein components of either the small-subunit (SSU) processome including subunits of the U3 snoRNP, UTPA/t-UTP, UTP-B, UTP-C, Mpp10, and CKII subcomplexes or components of the exosome were identified in our datasets. These included proteins that either bound immobilized H1.0 in vitro in at least one biological replicate (Supplemental Table 1 and data not shown) or proteins affected in H1ΔTKO mESC nucleoli (Supplementary Table 3). The SSU processome is part of the 90S pre-ribosomal particle formed around the newly synthesized 47S pre-rRNA, and is required for early RNA cleavage and modification (20,38). The exosome consists of 3’-5’ exonucleases which act directly on the 90S pre-ribosomal particle to generate the pre-40S and pre-60S particles (20,38). Importantly, proteins with functions involving the maturation or export of the pre-40S or pre-60S ribosomal particles were absent from our datasets further indicating that the H1 network is limited to the early steps of ribosome biogenesis.
Our biochemical and proteomic data also connect H1 to nuclear pre-mRNA processing proteins. Approximately 30% of the proteins that bound H1 in vitro and ~10% of the affected proteins in H1ΔTKO mESC nucleoli are pre-mRNA splicing proteins. The nucleolus coordinates various protein and RNA networks to generate a diverse array of RNP particles (24,26,68). Collectively, evidence from eukaryotes implicate the nucleolus in signal-recognition particle (SRP) biogenesis (69); telomerase RNP dynamics (70); tRNA processing by RNase P (71); small interfering RNA (siRNA) and microRNA (miRNA) dynamics in plants (72,73), and let-7 miRNA regulation in mammals (74,75); noncoding RNA (ncRNA) synthesis and protein sequestration through the nucleolar detention pathway (NoDP) (76); snRNP biogenesis including U6 snRNA processing (77-79); and mRNA export, surveillance or biogenesis (24,26,68). Although the question as to whether pre-mRNA itself is a component of H1 binding complexes is not addressed experimentally here, results from our SEC analysis of nucleolar extracts indicate that H1 associates with either the entire spliceosome complex or with multiple sub-complexes as they accumulate in the nucleolus. Moreover, it is likely that native nucleolar complexes that either bound H1 in vitro or co-fractionated with H1 by SEC are associated with their respective cognate RNAs.
The notion of the ‘plurifunctional nucleolus’ is continually reinforced by growing experimental evidence that links the nucleolus to cellular growth, proliferation, development, stress response and aging (24-26,80,81). GO analysis of the entire H1 PPIN (Supplementary Table 1 and 3) hints at a potential role for H1 in cellular health and disease. Specifically, we found low enrichment (< 4.0-fold using DAVID) for biological processes related to cell growth, mitosis, apoptosis, development, DNA repair, redox signaling, ion homeostasis, amino acid metabolism, and protein folding, ubiquitylation and degradation (data not shown). These underrepresented biological processes further widen the scope of the H1 PPIN beyond ribosome biology and reveal novel associations between H1 and metabolic proteins that transition through the nucleolus.
Methods and Materials
Cell culture
Human Jurkat cells, clone E6-1 (ATCC TIB-152) were maintained in an incubator at 37°C, 5% CO2 in RPMI-1640 (ATCC) medium supplemented with 10% fetal bovine serum (FBS). mESC (derived from WW6 cell line) were cultured in DMEM with L-glutamine (Gibco) supplemented with 10% ESC-screened FBS (Hyclone), 2-mercaptoethanol (55 μM), MEM non-essential amino acids and 104 U/mL LIF (Chemicon), and grown on mouse embryonic fibroblast (MEF (CF-1)) (ATCC SCRC-1040) feeder layers pre-treated with 10 μg/mL mitomycin C (Sigma-Aldrich) (as described (10)). To eliminate MEF feeder cells from downstream applications, confluent cultures were diluted 1:3 and plated on gelatin-coated culture dishes (lacking feeder cells) and incubated at 37°C, 5% CO2 for 48 h. Cells were harvested with trypsin and residual MEF feeder cells were allowed to re-adhere for 45 min before mESC suspensions were collected for biochemical and proteomic studies.
Isolation of nucleoli and protein extraction
Jurkat and mESC nucleoli were isolated using a sucrose layering method as described (16,17). Whole cells were lysed in hypotonic buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT) and layered over S1 buffer (0.25 M sucrose, 10 mM MgCl2). Nuclei were pelleted and resuspended in S2 buffer (0.35 M sucrose, 0.5 mM MgCl2), sonicated on ice using a Misonix sonicator (5 times 10s bursts, 10% amplitude), and layered over S3 buffer (0.88 M sucrose, 0.5 mM MgCl2). Nucleoli were pelleted, washed in S2 buffer, and stored at −70°C. Soluble nucleolar extract was prepared as described (82) by disruption in a high salt modified RIPA buffer (50 mM Tris pH7.6, 500 mM NaCl, 1% IGEPAL, 0.5% deoxycholate (DOC), protease inhibitor (PI) cocktail), sonicated on ice using a Misonix sonicator (3 times 10s bursts, 3% amplitude), and treated with 20 U DNase I (Roche) at 4°C for 30 min. The supernatant containing the soluble fraction was recovered by centrifugation at 22,000 rcf for 20 min at 4°C. The extract was then adjusted to 150 mM NaCl and a portion (0.5mg protein) was used directly for either H1 pull-downs or for chromatography experiments. All protein concentrations were determined by either BCA (Pierce) or Bio-RAD protein assay according to the manufacturer's directions. For proteomic profiling experiments, pelleted nucleoli (50 μg of total protein) were resuspended in S2, digested in 5 U/mL Benzonase (Novagen), and precipitated (6% TCA, 0.02% DOC). Pellets were then washed twice with cold acetone and stored at −20°C. Protein pellets were then resuspended in 8 M Urea prior to a 5 min sonication. ProteaseMAX surfactant trypsin enhancer (Promega) was added to a final concentration of 0.11% and supplemented with 29 mM Ambic. Disulfide bonds were then reduced with DTT and alkylated with iodoacetamide (IAA). Proteins were digested for 3 h at 37°C using Trypsin Gold (Promega). The digestion was halted by the addition of 0.5% TFA and samples were dried in a vacuum evaporator. The resulting peptides were purified using a C18 TopTip (Glygen, Columbia, MD), dried, and then resuspended in 3% acetonitrile/0.1% formic acid prior to MS analysis.
H1 pull-downs and size exclusion chromatography (SEC)
H1 pull-down experiments, and protein expression and purification were performed as previously described (12) using purified recombinant Halo Tag or HaloTag-H1.0 fusion protein as bait and a soluble nucleolar extract as prey. Briefly, Halo Tag control or HaloTag-H1.0 (~7 nmol each) were pre-bound to Halo resin in Halo Buffer (10 mM Tris-Cl pH 7.6, 150 mM NaCl, 0.05% IGEPAL, 0.5 mM DTT, 0.5 mM EDTA, PI cocktail) at 4°C for 16 h then washed with Halo buffer containing 1 M NaCl to remove unbound proteins. The resin was then equilibrated in a low salt modified RIPA buffer (150 mM NaCl) and incubated with 0.5 mg of extract at 4°C for 16 h. Unbound proteins were removed by washing the resin in a modified RIPA buffer (200 mM NaCl) prior to protein elution with 1% SDS, 1M NaCl, and followed by TCA precipitation. For LC-MS/MS analysis, eluted samples were denatured, reduced, modified and digested with trypsin as described above. For SEC analysis, nucleolar extract (0.5 mg protein) or gel filtration standards (Bio-Rad) were loaded onto a Superdex 200 16/60 column equilibrated in SEC buffer (50 mM Tris pH7.6, 200 mM NaCl, 0.001% IGEPAL, 0.05% DOC, PI cocktail). 4 mL fractions were collected, TCA precipitated and resuspended in Laemmli Sample Buffer for subsequent immunoblot analysis.
LC-MS/MS analysis
Tryptic peptides were purified and concentrated using an on-line enrichment column (Thermo Scientific 5 μm, 100 μm ID × 2 cm C18 column). Subsequent chromatographic separation was performed on a reverse phase nanospray column (Thermo Scientific EASYnano-LC, 3 μm, 75 μm ID × 100mm C18 column) using a 90 minute linear gradient from 10%-30% buffer B (100% ACN, 0.1% formic acid) at a flow rate of 400 nanoliters/min. Peptides were eluted directly into the mass spectrometer (Thermo Scientific Orbitrap Velos) and spectra were collected over a m/z range of 400-2000 Da using a dynamic exclusion limit of 2 MS/MS spectra of a given peptide mass (exclusion duration of 90 s). The instrument was operated in Orbitrap-LTQ mode where precursor measurements were acquired in the orbitrap (60,000 resolution) and MS/MS spectra (top 20) were acquired in the LTQ ion trap. Compound lists of the resulting spectra were generated using Xcalibur 2.2 software (Thermo Scientific) with an S/N threshold of 1.5 and 1 scan/group. For quality control, instrument functionality and stability was monitored using the Mass QC software (Proteome Software) (83). Quality control samples were injected at least once every 24 h during the analysis, and the data was analyzed using the Mass QC software. Values for all metrics were within normal limits throughout the duration of the experiment indicating instrument stability and data robustness.
Analysis of proteomic data
MS/MS spectra were searched against the Human or Mouse Uniprot protein database concatenated to a reverse database (Version 03/06/2013, 175,312 sequence entries or version 05/16/2013, 101,600 sequence entries) using the Mascot database search engine (version 2.3) and the Sorcerer™-SEQUEST® version 3.5. A 0.00% or 0.04% peptide FDR (false discovery rate), respectively, was calculated by Scaffold based on hits to the reverse database (84). Search parameters were as follows: monoisotopic mass, parent ion mass tolerance of 20 ppm, fragment ion mass tolerance of 0.8 Da, fully tryptic peptides with 1 missed cleavage, variable modification of oxidation of M and fixed modification of carbamidomethylation of C. Search results for each independently analyzed sample were imported and combined using probabilistic protein identification algorithms (29) implemented in Scaffold software (30)(Version 4, Proteome Software, Portland, OR). Data filters were applied such that only spectra with a 95% peptide probability, a 99% protein probability, and a minimum of two unique peptides per protein were considered for the analysis. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Manual validation of MS/MS spectra was performed for all protein identifications above the probability thresholds that were based on only two unique peptides. Criteria for manual validation included the following: 1) minimum of 80% coverage of theoretical y or b ions (at least 5 in consecutive order); 2) absence of prominent unassigned peaks greater than 5% of the maximum intensity; and 3) indicative residue specific fragmentation, such as intense ions N-terminal to proline and immediately C-terminal to aspartate and glutamate (used as additional parameters of confirmation.)
Immunoblot analysis and antibodies
Antibodies used in this study included anti-TUBA1B (11224-1-AP), HNRNPK (11426-1-AP), NPM1 (60096-1-Ig, 10306-1-AP), EFTUD2 (10208-1-AP), FBL (16021-1-AP), and SUPT16H (20551-1-AP) (purchased from Proteintech Group, Inc.), and anti-nucleolin (sc8031), HDAC1 (sc81598), SSRP1 (sc74536), SNRNP70 (sc390899), SF3B2 (sc101133) (purchased from Santa Cruz Biotechnology, Inc.), and anti-Lin28A (ab63740: Abcam). For immunoblot analysis, proteins were resolved on a 15% or 12% SDS polyacrylamide gel and transferred to PVDF membrane (Immobilon-FL, Millipore). Blots were incubated with primary antibody diluted 1:1000 in 3% non-fat dried milk, TBS buffer overnight at 4°C. Blots were then probed using fluorescent conjugated secondary antibodies (LI-COR) according to the manufacturer's instructions and visualized using the LI-COR Odyssey scanner and software. For H1 detection, blots were probed with anti-H1 (sc8030 and sc393358) together with anti-H10 (sc377468) (Santa Cruz Biotechnology, Inc.) diluted 1:1000 in TBS with 3% non-fat dried milk and detected with alkaline phosphatase conjugated secondary antibody (Santa Cruz Biotechnology, Inc.) according to the manufacturer's instructions.
Immunofluorescence and cell imaging
Nucleoli were fixed in solution with 4% para-formaldehyde (PFA) (Electron Microscopy Sciences) in phosphate buffered saline (PBS) for 20 min at room temperature prior to incubation on acid washed and poly L-lysine coated glass coverslips for 16 h at 4°C. Coverslips were washed with PBS with 0.1% TritonX-100 (Sigma) and blocked in PBS with 10% boiled donkey serum (BDS) (Jackson ImmunoResearch) for 1 h at room temperature, and probed with α-NPM1 primary antibody (diluted 1:100 in PBS with 5% BDS) for 16 h at 4°C. Coverslips were then washed (PBS with 0.1% TritonX-100) and stained with fluorescently labeled secondary antibody (Jackson ImmunoResearch) (diluted 1:300 in PBS with 5% BDS) for 45 min at room temperature. For 4', 6-diamidino-2-phenylindole (DAPI) staining, coverslips were washed (PBS with 0.1% TritonX-100), stained with DAPI (2 ng/mL in PBS), and mounted on microscope slides in 90% glycerol and 0.5% N-propyl gallate. For whole cell imaging, cells were cultured on 10 mm Microwell (0.16-0.19 mm) petri dishes (MatTek, P35G-1.5-10-C), permeablized with 1% TritonX-100 in PBS for 5 min and fixed with 4% PFA in PBS for 20 min. Cells were then probed with α-NPM1 antibody and stained with DAPI using the methods described above. mESC were also stained with Pyronin Y (10μg/mL) and mounted to microscope slides with ProLong Gold Antifade (Life Technologies). Samples were imaged on a DeltaVision PersonalDV Imaging System (Applied Precision) equipped with a Photometrics CoolSnap HQ2 camera (Roper Scientific) and a 60X/1.42NA Planapochromat DIC oil immersion lens (Olympus). Z-stacks at 0.2 μm intervals were deconvolved using SoftWorx Suite (Applied Precision) and fluorescence intensities were measured in the SoftWorx suite on maximum projections of Z-stacks.
Supplementary Material
Highlights.
Linker histone H1 is a DNA binding protein that regulates chromatin structure.
H1 binds to over 170 proteins derived from purified nucleoli from Jurkat T-cells.
Nucleoli from H1-depleted cells have altered protein and RNA composition.
H1 has a key function in the nucleolus acting through protein-protein interactions.
Acknowledgements
We thank the Proteomics and Metabolomics Facility at CSU for data acquisition and analytical support. This work was supported by the National Institutes of Health Grants GM045916 (to J. C. H.), GM088371 (to J. G. D.) and CA079057 (to A. S.), and was also supported by a grant from Colorado State University, College of Veterinary Medicine and Biomedical Sciences, College Research Council (to Q. A. W.).
Abbreviations
- DAPI
4',6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DNMTs
DNA methyltransferases
- dsDNA
double-stranded DNA
- ERAD
endoplasmic reticulum (ER)-associated protein degradation
- FDR
false discovery rate
- GO
gene ontology
- H1ΔTKO
H1 triple isoform knockout
- IAA
iodoacetamide
- IF
immunofluorescence
- MEF
mouse embryonic fibroblast
- mESC
mouse embryonic stem cell
- miRNA
microRNA
- mRNP
messenger RNA (mRNA)-associated ribonucleoprotein
- MS
mass spectrometry
- MW
molecular weight
- ncRNA
noncoding RNA
- NoDP
nucleolar detention pathway
- NORs
nucleolar organizer regions
- PBS
phosphate buffered saline
- PcG
Polycomb group
- PFA
paraformaldehyde
- Pol I
RNA polymerase I
- Pol II
RNA polymerase II
- PPIN
protein-protein interaction network
- pre-mRNA
precursor messenger RNA
- pre-RNA
precursor ribosomal RNA
- rDNA
ribosomal DNA
- RIPA
radio-immunoprecipitation assay
- SEC
size exclusion chromatography
- siRNA
small interfering RNA
- snRNP
small nuclear ribonucleoprotein
- SpC
spectral counting
- SR
serine/arginine-rich
- SRP
signal recognition particle
- SSU
small-subunit
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Kowalski A, Palyga J. Linker histone subtypes and their allelic variants. Cell biology international. 2012;36:981–996. doi: 10.1042/CBI20120133. [DOI] [PubMed] [Google Scholar]
- 3.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biological chemistry. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 4.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 7.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 8.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Szerlong HJ, Hansen JC. Nucleosome distribution and linker DNA: connecting nuclear function to dynamic chromatin structure. Biochem Cell Biol. 2011;89:24–34. doi: 10.1139/O10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalashnikova AA, Winkler DD, McBryant SJ, Henderson RK, Herman JA, DeLuca JG, Luger K, Prenni JE, Hansen JC. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013;41:4026–4035. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci U S A. 2013;110:1708–1713. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarboui MA, Wynne K, Elia G, Hall WW, Gautier VW. Proteomic profiling of the human T-cell nucleolus. Molecular immunology. 2011;49:441–452. doi: 10.1016/j.molimm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Current biology : CB. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database--2008 update. Nucleic Acids Res. 2009;37:D181–184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annual review of cell and developmental biology. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 20.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and molecular life sciences : CMLS. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartova E, Horakova AH, Uhlirova R, Raska I, Galiova G, Orlova D, Kozubek S. Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2010;58:391–403. doi: 10.1369/jhc.2009.955435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001;29:3241–3247. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature reviews. Molecular cell biology. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 26.Shaw P, Brown J. Nucleoli: composition, function, and dynamics. Plant physiology. 2012;158:44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics : official journal of the DNA Methylation Society. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 30.Searle BC, Turner M, Nesvizhskii AI. Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. Journal of proteome research. 2008;7:245–253. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Cong R, Das S, Douet J, Wong J, Buschbeck M, Mongelard F, Bouvet P. macroH2A1 histone variant represses rDNA transcription. Nucleic Acids Res. 2014;42:181–192. doi: 10.1093/nar/gkt863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grummt I, Langst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 35.Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grierson PM, Acharya S, Groden J. Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutation research. 2013;743-744:89–96. doi: 10.1016/j.mrfmmm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phipps KR, Charette J, Baserga SJ. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley interdisciplinary reviews. RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coute Y, Burgess JA, Diaz JJ, Chichester C, Lisacek F, Greco A, Sanchez JC. Deciphering the human nucleolar proteome. Mass spectrometry reviews. 2006;25:215–234. doi: 10.1002/mas.20067. [DOI] [PubMed] [Google Scholar]
- 39.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Molecular biology of the cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics & chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 42.McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- 43.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell research. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 45.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochemistry and cell biology. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 46.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 48.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. The FEBS journal. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 49.Hernandez-Verdun D, Roussel P, Gebrane-Younes J. Emerging concepts of nucleolar assembly. J Cell Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- 50.Nalabothula N, Indig FE, Carrier F. The Nucleolus Takes Control of Protein Trafficking Under Cellular Stress. Molecular and cellular pharmacology. 2010;2:203–212. [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vujatovic O, Zaragoza K, Vaquero A, Reina O, Bernues J, Azorin F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. FACT facilitates chromatin transcription by RNA polymerases I and III. Embo J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. Embo J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bierhoff H, Dundr M, Michels AA, Grummt I. Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol Cell Biol. 2008;28:4988–4998. doi: 10.1128/MCB.00492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg HG, Grummt I. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. Embo J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Keller DM, Scott JD, Lu H. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J Biol Chem. 2005;280:11869–11875. doi: 10.1074/jbc.M413944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider HR, Reichert GH, Issinger OG. Enhanced casein kinase II activity during mouse embryogenesis. Identification of a 110-kDa phosphoprotein as the major phosphorylation product in mouse embryos and Krebs II mouse ascites tumor cells. Eur J Biochem. 1986;161:733–738. doi: 10.1111/j.1432-1033.1986.tb10501.x. [DOI] [PubMed] [Google Scholar]
- 61.Li D, Dobrowolska G, Krebs EG. The physical association of casein kinase 2 with nucleolin. J Biol Chem. 1996;271:15662–15668. doi: 10.1074/jbc.271.26.15662. [DOI] [PubMed] [Google Scholar]
- 62.Chan PK, Aldrich M, Cook RG, Busch H. Amino acid sequence of protein B23 phosphorylation site. J Biol Chem. 1986;261:1868–1872. [PubMed] [Google Scholar]
- 63.Szebeni A, Hingorani K, Negi S, Olson MO. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein b23. J Biol Chem. 2003;278:9107–9115. doi: 10.1074/jbc.M204411200. [DOI] [PubMed] [Google Scholar]
- 64.Bandyopadhyay K, Li P, Gjerset RA. CK2-mediated hyperphosphorylation of topoisomerase I targets serine 506, enhances topoisomerase I-DNA binding, and increases cellular camptothecin sensitivity. PLoS One. 2012;7:e50427. doi: 10.1371/journal.pone.0050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 66.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown JW, Shaw PJ. The role of the plant nucleolus in pre-mRNA processing. Current topics in microbiology and immunology. 2008;326:291–311. doi: 10.1007/978-3-540-76776-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci U S A. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Etheridge KT, Banik SS, Armbruster BN, Zhu Y, Terns RM, Terns MP, Counter CM. The nucleolar localization domain of the catalytic subunit of human telomerase. J Biol Chem. 2002;277:24764–24770. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- 71.Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110(Pt 7):829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 72.Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 73.Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogt EJ, Meglicki M, Hartung KI, Borsuk E, Behr R. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development. 2012;139:4514–4523. doi: 10.1242/dev.083279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 77.Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2'-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerbi SA, Lange TS. All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli; Identification of the nucleolar localization element of U6 snRNA. Molecular biology of the cell. 2002;13:3123–3137. doi: 10.1091/mbc.01-12-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lange TS, Gerbi SA. Transient nucleolar localization Of U6 small nuclear RNA in Xenopus Laevis oocytes. Molecular biology of the cell. 2000;11:2419–2428. doi: 10.1091/mbc.11.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki A, Kogo R, Kawahara K, Sasaki M, Nishio M, Maehama T, Sasaki T, Mimori K, Mori M. A new PICTure of nucleolar stress. Cancer science. 2012;103:632–637. doi: 10.1111/j.1349-7006.2012.02219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chamousset D, Mamane S, Boisvert FM, Trinkle-Mulcahy L. Efficient extraction of nucleolar proteins for interactome analyses. Proteomics. 2010;10:3045–3050. doi: 10.1002/pmic.201000162. [DOI] [PubMed] [Google Scholar]
- 83.Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, Billheimer DD, Blackman RK, Bunk DM, Cardasis HL, et al. Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Molecular & cellular proteomics : MCP. 2010;9:225–241. doi: 10.1074/mcp.M900223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. Journal of proteome research. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.