Abstract
Mammary serine protease inhibitor (maspin) is an important tumor suppressor gene whose expression is associated not only with tumor growth inhibition but also with decreased angiogenesis and metastasis. Maspin expression is down-regulated in metastatic tumors by epigenetic mechanisms, including aberrant promoter hypermethylation. We have constructed artificial transcription factors (ATFs) as novel therapeutic effectors able to bind 18-bp sites in the maspin promoter and reactivate maspin expression in cell lines that harbor an epigenetically silenced promoter. In this article, we have investigated the influence of epigenetic modifications on ATF-mediated regulation of maspin by challenging MDA-MB-231 breast cancer cells, comprising a methylated maspin promoter, with different doses of ATFs and chromatin remodeling drugs: the methyltransferase inhibitor 5-aza-2′-deoxycytidine and the histone deacetylase inhibitor suberoylanilide hydroxamic acid. We found that the ATFs synergized with both inhibitors in reactivating endogenous maspin expression. The strongest synergy was observed with the triple treatment ATF-126 + 5-aza-2′-deoxycytidine + suberoylanilide hydroxamic acid, in which the tumor suppressor was reactivated by 600-fold. Furthermore, this combination inhibited tumor cell proliferation by 95%. Our data suggest that ATFs enhance the efficiency of chromatin remodeling drugs in reactivating silenced tumor suppressors. Our results document the power of a novel therapeutic approach that combines both epigenetic and genetic (sequence-specific ATFs) strategies to reactivate specifically silenced regions of the genome and reprogram cellular phenotypes.
Introduction
Tumor suppressor genes play an essential role in controlling unscheduled cell proliferation and they act as gatekeepers that block neoplastic processes in tissues. Due to the pivotal role of tumor suppressor gene inactivation during tumor progression, these genes are primary targets in cancer therapeutics. Inactivation can occur via a variety of mechanisms, such as point mutations, deletions, and epigenetic modifications (1–3). Epigenetic modifications, such as DNA and histone methylation and histone deacetylation, result in a compact chromatin configuration that silences entire DNA regions (1–6). At the promoter level, this compact chromatin topology restricts the physical access of the polymerase II complex to regulatory sequence domains, resulting in inhibition of tumor suppressor transcription (7–9). Unlike genetic alterations, which irreversibly inactivate tumor suppression expression, epigenetic modifications are potentially reversible (9–11).
The reversible nature of epigenetic silencing offers a unique opportunity for therapeutic intervention by reactivating endogenous tumor suppressor genes. Several chromatin remodeling drugs have been developed to release the repressed state of tumor suppressor genes. These drugs act by inhibiting DNA methyltransferases or histone deacetylases (HDAC), resulting in increased promoter accessibility and enhanced tumor suppressor gene transcription (12 – 14). To date, the most widely used chromatin remodeling drug is the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-2′-dC), recently approved for therapeutic treatment (15). Several methyltransferase inhibitors [such as 5-aza-dC and MG98 (16, 17)] and HDAC inhibitors [such as suberoylanilide hydroxamic acid (SAHA; ref. 18), valproic acid (19), and pivaloyloxymethyl butyrate (20, 21)] are presently in phase I and II clinical trials. The small-molecule inhibitors 5-aza-2′-dC and SAHA have been used to reactivate tumor suppressor genes aberrantly methylated in aggressive tumor cells, such as desmocollin 3 (22), gelsolin (23), and mammary serine protease inhibitor (maspin; refs. 13, 22). Moreover, several reports have shown that methyltransferase and HDAC inhibitors are able to synergize to reactivate tumor suppressor expression (24, 25). Nevertheless, potential limitations for the use of these drugs in cancer patients include their toxicity, lack of target specificity, and development of acquired drug resistance (6, 26). Thus, there is a need for the development of novel strategies to increase the targeted efficiency and specificity of current anticancer drugs.
Our laboratory has recently applied a new strategy to specifically reactivate tumor suppressor genes silenced by epigenetic mechanisms in aggressive tumors (27). We have targeted the tumor suppressor gene maspin using three rationally designed artificial transcription factors (ATFs). These ATFs comprise six sequence-specific zinc finger (ZF) domains, designed to recognize unique 18-bp sites in the maspin promoter. The ZFs were linked to the VP64 activator domain, which mediates promoter up-regulation. We found that the capability of the ATFs to up-regulate maspin depended on the cell line analyzed, indicating that the structure of the chromatin can influence ATF-mediated transactivation of maspin. In the aggressive MDA-MB-231 breast cancer cell line, which comprises a methylated and silenced maspin promoter, only one ATF (ATF-126) was able to partially reactivate the endogenous maspin. We hypothesized that the structure of the chromatin (which is found in a more compact configuration in methylated promoters) could act as a partial blockade and restrict ATF-mediated transactivation of maspin. In this article, we have investigated the influence of chromatin structure at the maspin promoter in the context of artificial ATF regulation by challenging MDA-MB-231 cells expressing ATF-126 with different doses of 5-aza-2′-dC and SAHA. We found that ATF synergized with both inhibitors to reactivate maspin expression. The strongest synergy was observed with the triple treatment ATF-126 + 5-aza-2′-dC + SAHA, in which the tumor suppressor was reactivated by 600-fold. Furthermore, this combination inhibited breast tumor cell proliferation by 95%. Our data suggest that ATFs amplify the response of chromatin remodeling drugs in reactivating silenced tumor suppressors. Thus, combinations of low concentrations of chromatin remodeling drugs and sequence-specific ATFs are efficient in reactivating silenced regions of the genome and effectively reprogram cellular phenotypes. This could represent a powerful therapeutic strategy to target a variety of neoplasias through specific reactivation of tumor suppressor genes.
Materials and Methods
Cell Lines
MDA-MB-231 breast carcinoma, MDA-MB-468, MCF-12A, and 293TGagPol cell lines were obtained from the American Type Culture Collection.
Sodium Bisulfite Genomic Sequencing of the Maspin Promoter
Genomic DNA (1.5 μg) was modified with sodium bisulfite using EZ DNA Methylation-Gold kit (Zymo Research). The maspin promoter was amplified from the bisulfite-modified DNA by PCR using primers specific to the bisulfite-modified sequence of the maspin promoter: 5′-TAGGATTTTAAAAAGAAATTTTTTG-3′(forward primer) and 5′-CCCACCTTACTTACCTAAAATCACA-3′(reverse primer). The PCR products were cloned and 10 positive recombinants were sequenced. The methylation status of individual CpG sites was determined by comparison of the sequence obtained with the known maspin sequence.
ATF Retroviral Transduction
The retroviral vector pMX-6ZFs-VP64-IRES-GFP (28) was first cotransfected with a plasmid (pMDG.1) expressing the vesicular stomatitis virus envelope protein into 293TGag-Pol cells to produce retroviral particles. Transfection was done using Lipofectamine as recommended (Invitrogen). The viral supernatant was used to infect the host cell lines, and the infection efficiency was assessed by flow cytometry (FACSCalibur, BD Biosciences) using green fluorescent protein as marker.
Drug Treatments
ATF-transduced cells and control cells (0.25 × 106 untransduced cells, cells transduced with empty retroviral vector, and a control ATF that does not regulate maspin) were seeded in 10-cm tissue culture plates. These samples were treated with different concentrations of 5-aza-2′-dC (0, 0.025, 0.05, 0.125, 0.25, 0.625, 1.25, 2.5, 3.75, 6.25, 12.5, 25, 62.5, and 125 μg/mL; Sigma) or SAHA (0, 0.0133, 0.026, 0.066, 0.132, 0.26, 0.4, 0.66, 1.32, 2.6, 3.97, and 5.3 μg/mL; BioVision) or both inhibitors (5-aza-2′-dC and SAHA) during 48 h in a 37°C, 5% CO2 incubator. Cells were collected, and the RNA was extracted, reverse transcribed, and processed for real-time quantification of maspin.
Real-time PCR Expression Assays
ATF-transduced cells and control cells, drug treated or nontreated, were collected, and the RNA was extracted and 2.5 μg were used for reverse transcription. Quantification of maspin and VP64 activator domain was obtained by real-time quantitative PCR using fluorescent Taqman assays (Applied Biosystems) as described (27). The primers and probes for the VP64 activator domain were the following: 5′-AAGCGACGCATTGGATGAC-3′(forward primer), 5′-GGAACGTCGTACGGGTAGTTAATT3′(reverse primer), and 5′-6FAM-TCGGCTCCGATGCT-MGBNFQ-3′(probe). Real-time PCR data were analyzed using the comparative 2−ΔΔCT method (SDS 2.1 RQ software, Applied Biosystems) and results were expressed as “fold change” in maspin RNA expression normalized to GAPDH and relative to the vehicle-treated control (29).
Proliferation Assays
Proliferation assays were done measuring cell viability determined by a survival assay (XTT, Roche, according to the manufacturer’s instructions). To measure the effect of ATF-126, 5-aza-2′-dC, and SAHA in cell viability, MDA-MB-231 breast cancer cells were transduced with different concentrations of ATF-126 plasmid or/and treated with 5-aza-2′-dC or SAHA (same concentration described earlier). Twenty-four hours after transfection, 3,000 cells per well (5 wells per concentration) were seeded into 96-well format tissue culture plates. Cell viability was measured using the XTT assay by monitoring the absorbance (405 nm) of the cells at 0 and 72 h after transduction and 48 h of drug treatment.
Experimental Drug Dose-Effect Plots
Dose-effect curves and median-effect plots were generated for each set of the real-time and proliferation data samples using the software package PharmToolsPro (McCary Group; ref. 30). The median-effect dose (Dm50) and the slope (m) were calculated from the median-effect plots and introduced in the isobologram equation for the calculation of the CI (28, 31–33). The CI isobologram equation [CI = (D)1/(Dx)1 + (D)2/(Dx)2 + (D)3/(Dx)3] was used for data analysis of three-drug combination (31, 32). CI < 1, CI = 1, and CI > 1 indicate synergy, additive effect, and antagonism, respectively.
Statistical Analysis
Real-time PCR and viability experiments were repeated thrice using three independently processed samples. For each sample, we did triplicate acquisitions. Differences between all treatments were analyzed by the ANOVA test with a critical level of significance set up at P < 0.05, and significant differences between groups of treatments were analyzed with post hoc Turkish test using the software GraphPad Prism v.5.
Results
ATFs Reactivate Maspin in Combination with 5-Aza-2′-dC and SAHA
In a previous report, we have described the construction of three ATFs designed to bind 18-bp sites in the maspin proximal promoter (27). The ATFs were constructed by linkage of six sequence-specific ZF domains with the VP64 transactivator domain (Fig. 1A). Each ZF is a compact 30-amino acid domain composed of a recognition α-helix packed with two antiparallel β-strands via the coordination of a zinc ion. The α-helix of each ZF specifically recognizes 3 bp in the DNA or “recognition triplet.” The main contact positions are residue +6 of the recognition helix, which interacts with the 5′nucleotide position of the triplet, residue +3, interacting with the middle base, and position −1, which makes H-bonding contacts with the 3′nucleotide of the triplet (34).
Figure 1.
ATFs designed to reactivate the maspin promoter. A, schematic representation of a 6ZF-ATF. B, cytosine methylation status of maspin in MDA-MB-231 breast cancer cells. X axis, nucleotide position relative to the transcription start site; Y axis, percentage of methylation along the maspin promoter. The maspin proximal promoter region (−495 to +134) was originally reported by Zhang et al. (53). 5-Methylcytosine levels were obtained by sodium bisulfite genomic sequencing of the maspin promoter from genomic DNA of untransduced MDA-MB-231 cells. Transcription factor–binding sites were included [p53-binding sites (35) and ATF-binding sites (27)]. Red nucleotides indicate the methylated cytosines in the ATF-binding sites.
Our previous results show that the efficiency of maspin activation by ATFs depended on the particular cell line analyzed, indicating that the structure of the chromatin may influence the ATF-mediated regulation of the endogenous promoter. To investigate the influence of promoter topology in ATF regulation, we focused our studies on the MDA-MB-231 cell line, an aggressive cell estrogen receptor–negative breast cancer cell line that comprises a methylated and silenced maspin promoter (35). First, we verified the methylation status of the maspin promoter in the MDA-MB-231 background by doing sodium bisulfate sequencing of the maspin proximal promoter and we mapped the ATF-binding sites in this sequence. As shown in Fig. 1B, the ATF-126–binding site contained two methylated cytosines, whereas the ATF-97–binding site comprised one methylated cytosine. In contrast, both ATF-452–binding and the two p53-binding sites in the maspin promoter mapped in methylation-free regions. The same methylation pattern was found in other aggressive cancer cell lines comprising a silenced maspin promoter (data not shown). The ATF-binding sites were not found mutated or deleted in all 10 genomic clones processed by sequencing. This agrees with previous reports (35–37), which showed that maspin gene is not found mutated or deleted in tumor cells, but its promoter is silenced by epigenetic mechanisms. Consistent with this epigenetic silencing, we found that MDA-MB-231 cells have no detectable maspin protein as assessed by Western blotting (Fig. 2C). When retrovirally transduced in MDA-MB-231 cells, only the ATF-126 was able to strongly reactivate the promoter (70-fold relative to controls, in the absence of drugs), whereas ATF-97 and ATF-452 alone had a much weaker activity (27).
Figure 2.
ATFs synergize with chromatin remodeling drugs to reactivate maspin expression. A, real-time quantification of maspin in untransduced MDA-MB-231 cells, cells retrovirally transduced with an empty retroviral vector (control), cells transduced with ATFs, and cells transduced with a control retrovirus lacking the ZF domains (pMXVP64SS). These samples were treated with 5-aza-2′-dC (1.0 μg/mL) or SAHA (0.5 μg/mL) or both inhibitors (same concentrations) using complete cell culture medium for the dilution of the drugs during 48 h in a 37°C, 5% CO2 incubator. Cells were collected, and the RNA was extracted, reverse transcribed, and processed for real-time maspin quantification. Real-time PCR data were analyzed using the comparative 2−ΔΔCT method and expressed as fold change in maspin mRNA expression normalized to GAPDH and relative to the vehicle-treated control (52). Differences between treatments were analyzed using ANOVA test and the post hoc Turkish test; critical level of significance was set up at P < 0.05. B, real-time expression analysis of maspin mRNA levels in the breast cancer cell lines MCF-12A, MDA-MB-468, and MDA-MB-231. MDA-MB-231 cells were transduced with a control empty retroviral vector, with ATF-126, and with ATF-126 in the presence of 5-aza-2′-dC (1.0 μg/mL) and SAHA (0.5 μg/mL). MCF-12A was used as a normalizing control. C, Western blot for the detection of maspin in the MCF-12A, MDA-MB-468, and MDA-MB-231 cell lines. MDA-MB-231 cells were transduced with control vector or ATF-126 and treated with a combination of 5-aza-2′-dC/SAHA (0.5 and 1 μg/mL, respectively).
To investigate the influence of methylation and chromatin structure on ATF regulation, we challenged ATF-expressing cells with 5-aza-2′-dC and SAHA (Fig. 2). These drugs are known to induce a more relaxed promoter topology, which facilitates the access of transcription factors and DNA polymerase complex (7, 8, 38). 5-Aza-2′-dC causes inhibition of DNA methyltransferase activity. The DNA methyltransferase is bound irreversible to the DNA through the 5-aza-2′-dC residues, which results in a depletion of soluble DNA methyltransferase protein levels. The lack of DNA methyltransferase availability leads to a DNA replication with global demethylation (22, 39–41). SAHA interacts with HDAC enzymes at the catalytic site inhibiting their activity. This process leads to histone acetylation, which opens the chromatin structure, increasing transcriptional activity (42, 43). We hypothesized that remodeling the chromatin in the MDA-MB-231 cell line toward a more open configuration facilitated by 5-aza-2′-dC or/and SAHA enhances the efficiency of ATF regulation of maspin. To test this hypothesis, we first retrovirally transduced ATF-97, ATF-126, and ATF-452 into MDA-MB-231 cells. Additionally, cells were transduced with a control empty retroviral vector (control) and with a retroviral vector lacking the ZF domains (pMXVP64SS). These samples were treated with 5-aza-2′-dC (5 μg/mL) or SAHA (0.5 μg/mL) or both inhibitors (5 and 0.5 μg/mL) and processed by real-time PCR for quantification of maspin mRNA levels. These concentrations were chosen in the range of the median-effect dose [Dm50, the concentration of inhibitor giving rise to 50% of maximum maspin mRNA up-regulation (31, 32, 44) calculated for these drugs (Fig. 3A–C)]. Maspin mRNA levels were calculated as a “fold change in mRNA expression” relative to the vehicle-treated MDA-MB-231 cell line as explained in Materials and Methods. Previously, we have found that in the absence of inhibitor only ATF-126 was able to strongly up-regulate maspin compared with control cells (cells transduced with an empty retroviral vector), whereas ATF-97 and ATF-452 had a much weaker effect (27). To compare differences between treatments and to evaluate synergisms, we used an ANOVA test with a critical level of significance set up at P < 0.05. Significant differences between groups of treatments were analyzed with post hoc Turkish test.
Figure 3.
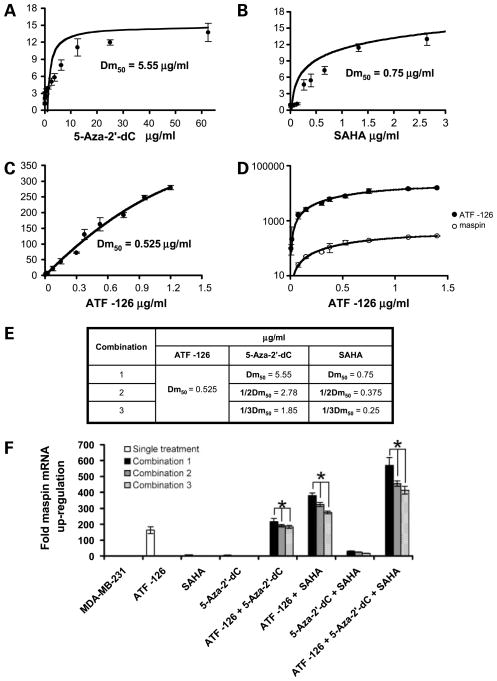
ATFs synergize with low concentrations of chromatin remodeling drugs to reactivate maspin mRNA expression. A and B, dose-effect plots assessing changes in maspin mRNA levels in cells treated with different concentrations of 5-aza-2′-dC and SAHA. Fold maspin mRNA levels were evaluated by real-time PCR using as a normalized control vehicle-treated cells. C, dose-effect plots assessing changes in maspin mRNA levels in cells transduced with different concentrations of ATF-126. D, mRNA expression changes of ATF directly correlate with changes on maspin mRNA levels, as evaluated by real-time PCR using primers specific for the ATF and maspin, respectively. Changes in mRNA expression of the ATF were generated, varying the amount of ATF-encoded DNA in the retroviral transduction. E, concentrations of ATF, 5-aza-2′-dC, and SAHA used in each combination tested. F, real-time expression analysis of maspin mRNA expression in ATF-transduced cells treated with specific combinations of 5-aza-2′-dC, SAHA, and both inhibitors, as indicated in E. MDA-MB-231 cells were transduced with ATF-126 (0.525 μg/mL; the Dm50 for maspin expression) and treated with three different concentrations (Dm50, 1/2Dm50, and 1/3Dm50) of either 5-aza-2′-dC, SAHA, or both compounds. Cells were collected, and the RNA was extracted, reverse transcribed, and processed for real-time quantification of maspin. Using fold change in maspin, mRNA expression was calculated using the comparative 2−ΔΔC T method as described above (52). Differences between treatments were analyzed using ANOVA test and the post hoc Turkish test; critical level of significance was set up at P < 0.05.
As shown in Fig. 2A, particular ATFs synergized with chromatin remodeling drugs in reactivating maspin expression. ATF-452, which had a poor activity in up-regulating the promoter (3.2-fold), was not able to synergize with 5-aza-2′-dC, SAHA, or both inhibitions in up-regulating maspin expression. ATF-97 up-regulated maspin by 14-fold and synergized with both inhibitors when used separately: 5-aza-2′-dC (63-fold maspin up-regulation) and SAHA (156-fold). However, the triple treatment ATF-126 + 5-aza-2′-dC + SAHA did not significantly further improved regulation. ATF-126–transduced cells up-regulated maspin by 70-fold. This ATF synergized with both inhibitors, 5-aza-2′-dC (161-fold) and SAHA (376-fold), in reactivating maspin. In contrast with the other ATFs, the triple treatment ATF-126 + 5-aza-2′-dC + SAHA exhibited synergy, up-regulating maspin mRNA levels by 600-fold. This stimulatory effect leads to an 8.26-fold change in maspin mRNA expression relative to a breast cancer cell line carrying a nonmethylated promoter (the MDA-MB-468 cell line) and to ~40% of the expression levels observed in nontransformed breast epithelial cell lines, such as MCF-12A (Fig. 2B and C). In contrast with breast cancer cells, nontransformed breast epithelial cells express very high levels of maspin (37). Our data suggest that other epigenetic marks, in addition to methylation and histone acetylation, might contribute to maspin silencing in tumor cells.
Because the three ATFs target distinct 18-bp sites along the maspin promoter, their particular responses to the inhibitors could reflect local differences in methylation and/or acetylation levels in the chromatin. Overall, these experiments suggested that modifications of the chromatin leading to a more compact promoter topology could partially block or impair ATF regulation, probably by affecting ATF binding.
ATF-126 Synergizes with Low Concentrations of 5-Aza-2′dC and SAHA in Reactivating Maspin Expression
We subsequently focused our studies on ATF-126 because among all the ATFs analyzed it exhibited the strongest response in reactivating maspin in combination with chromatin remodeling drugs. High concentration or persistent exposure of tumor cells with chromatin remodeling drugs can potentially result in high toxicity (38, 45). Thus, novel approaches to reactivate tumor suppression expression while minimizing the exposure of tumor cells to the drugs are desired. We next investigated if synergy between the ATF and the chromatin remodeling drugs was maintained when low concentrations of inhibitors (below their Dm50) were used. The Dm50 was calculated for each treatment, 5-aza-2′-dC, SAHA, and ATF-126, using dose-effect plots in the MDA-MB-231 breast cancer cell line (Fig. 3A–D). In these experiments, MDA-MB-231 cells were treated with different concentrations of 5-aza-2′-dC (0.025–125 μg/mL; Fig. 3A) or SAHA (0.07–5.3 μg/mL; Fig. 3B) during 48 h and maspin mRNA expression levels were monitored by real-time PCR assays. For ATF-126, cells were transduced with increasing concentrations of ATF-encoded DNA (0–1.3 μg/mL; Fig. 3C), and 72 h after transduction, cells were processed by real-time PCR to detect ATF-126 and maspin mRNA levels. The concentration of ATF-126 DNA used in the transfection correlated with ATF-126 mRNA levels detected in MDA-MB-231–transduced cells, as assessed by real-time PCR using ATF-specific primers (Fig. 3D). ATF-126 reactivated maspin in a concentration-dependent manner, reaching a maximum effect of 300-fold maspin mRNA relative to control cells, whereas 5-aza-2′-dC and SAHA induced a maximum of 13-fold maspin up-regulation relative to control cells. The dose-response plots were used to calculate the Dm50 for each single treatment. For ATF-126, the Dm50 (0.525 μg/mL) was calculated as 50% maspin up-regulation at 72 h after transduction. For the inhibitors, the Dm50 was calculated as 50% maspin up-regulation after 48 h of drug treatment, being 5.55 and 0.75 μg/mL for 5-aza-2′dC and SAHA, respectively (Fig. 3A and B).
To study the synergy between ATF-126 and the chromatin remodeling drugs in reactivating maspin, we designed different drug combination experiments where the ATF-126 was used at its Dm50 and the inhibitors were used at their respective Dm50 (combination 1), 1/2Dm50 (combination 2), and 1/3Dm50 (combination 3; Fig. 3F). As a control, MDA-MB-231 cells were subjected to single treatments (transduced with ATF-126 for 72 h or exposed to 5-aza-2′-dC or SAHA for 48 h) and processed by real-time PCR to evaluate maspin mRNA levels. We additionally evaluated maspin mRNA levels for double and triple treatments using specific combinations of control cells, ATF-126–transduced cells, 5-aza-2′-dC, and SAHA (Fig. 3E). In all the combinations tested, we found that ATF-126 was able to synergize with 5-aza-2′-dC, SAHA, and both inhibitors to reactivate maspin. We found that the most effective combination in activating maspin was the triple treatment ATF-126 + 5-aza-2′-dC + SAHA, with a 600-fold maspin up-regulation, when all the compounds were combined at their Dm50 (drug combination 1). Furthermore, ATF-126 synergized with 5-aza-2′-dC and SAHA in reactivating maspin expression by 413-fold even using 1/3Dm50 of inhibitors (drug combination 3). No statistical difference was observed between combination 2 (when 5-aza-2′-dC and SAHA were used at their 1/2Dm50) and combination 3 (when 5-aza-2′-dC and SAHA were used at their 1/3Dm50). Overall, these results indicate that ATF-126 strongly synergized with a combination of methyltransferase and HDAC inhibitors to reactivate maspin expression and this synergism was maintained when low concentrations of inhibitors (below their Dm50) were used.
ATF-126 Synergizes with 5-Aza-2′dC and SAHA to Inhibit Tumor Cell Viability
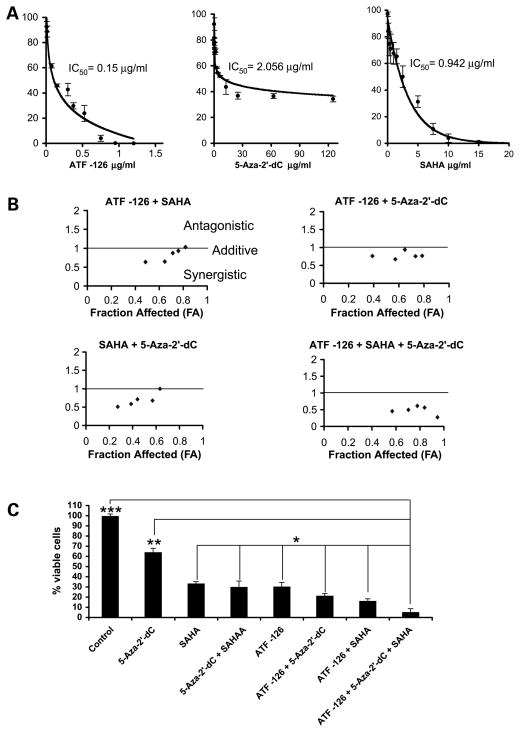
Several reports showed that induction of maspin mRNA expression results in inhibition of tumor cell proliferation by enhancement of apoptosis. We next investigated if ATF-126 could also synergize with 5-aza-2′-dC and SAHA in inducing inhibition of tumor cell growth. MDA-MB-231 cells were transduced with a control vector or with ATF-126, and 72 h after transduction, these cells were treated with 5-aza-2′-dC, SAHA, or both inhibitors during 48 h. Tumor cell viability was evaluated using survival assays {2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt (XTT) assays}. The dose-effect plots for ATF-126, 5-aza-2′-dC, and SAHA (Fig. 4A) were used to calculate the inhibitory concentration (IC50, the concentration of ATF-126 or inhibitor giving rise to 50% of inhibition of tumor cell growth at 72 h after transduction or after 48 h of drug treatment). The IC50 values were 0.15, 2.056, and 0.942 μg/mL for ATF-126, 5-aza-2′-dC, and SAHA, respectively. To evaluate synergisms, we applied a standard combinatorial method, which uses the isobologram equation to calculate the combinatorial index (CI). The interaction between drugs is defined as synergistic if CI < 1, antagonistic if CI > 1, and additive if CI = 1 (Fig. 4B). Control-transduced or ATF-126–transduced cells were challenged with different concentrations of 5-aza-2′-dC, SAHA, or both inhibitors, as shown in Table 1. We used concentrations of ATF/5-aza-2′-dC/SAHA in the range of the IC50 value, which resulted on 30% to 80% of tumor cell growth inhibition. As shown in Fig. 4B, ATF-126 synergized with 5-aza-2′-dC, SAHA, and both inhibitors (5-aza-2′-dC + SAHA) for the majority of the drug combinations tested. The synergistic effect for the double treatments ATF-126 + 5-aza-2′-dC, ATF-126 + SAHA, and 5-aza-2′-dC + SAHA was higher when low concentrations were used in each combination. For the triple treatment ATF-126 + 5-aza-2′-dC + SAHA, we observed a synergistic effect with all the combinations tested. The lowest (most synergistic) CI was achieved by the combination with the highest dose of ATF-126 and inhibitors, which results in 95% of inhibition of tumor cell viability compared with vehicle-treated control cells (Fig. 4C; doses are indicated in Table 1).
Figure 4.
ATF-126 synergizes with 5-aza-2′-dC and SAHA in inhibiting tumor cell viability. A, ATF-126, 5-aza-2′-dC, and SAHA induce inhibition of tumor cell viability in a dose-dependent manner. Dose-effect curves for cells transduced with different concentrations of the DNA of ATF-126 or treated with different concentrations of 5-aza-2′-dC and SAHA (Table 1).3 The effects of the ATFs and the chromatin remodeling drugs in inhibiting tumor cell viability were measured by the ability of metabolic active cells to reduce the tetrazolium salt XTT to orange-colored compounds of formazan. Dose-effect curves and median-effect plots were generated for each set of samples using the software package PharmToolsPro (28). B, CI for cells transduced with ATF-126 and treated with 5-aza-2′-dC (ATF-126 + 5-aza-2′-dC), SAHA (ATF-126 + SAHA), and both inhibitors (ATF-126 + 5-aza-2′-dC + SAHA). Nontransduced cells were treated with both inhibitors (5-aza-2′-dC + SAHA). CI was calculated from the median-effect plots (31) to measure the synergistic action between ATF-126, 5-aza-2′-dC, and SAHA in the MDA-MB-231 breast cancer cell line. CI < 1 defines a synergistic interaction, and CI > 1 defines an antagonistic drug interaction. The straight line at CI = 1 represents additive effects. C, inhibition of tumor cell viability on ATF-126 transduction and/or treatment with chromatin remodeling drugs. For single treatments, MDA-MB-231 cells were transduced with ATF-126 (0.525 μg/mL) or treated with 5-aza-2′-dC (3.75 μg/mL) and SAHA (1.32 μg/mL) for 48 h at 37°C and 5% CO2. The same concentrations were used for the following combinations: 5-aza-2′-dC + SAHA, ATF-126 + 5-aza-2′-dC, ATF-126 + SAHA, and ATF-126 + 5-aza-2′-dC + SAHA. Cell viability was measured using the XTT assay, as described above. The data were analyzed using an ANOVA test and a post hoc Turkish test, as described in Materials and Methods. The asterisks indicate that the triple treatment decreased significantly tumor cell viability compared with all the other treatments tested. *, P = 0.05; **, P = 0.01; ***, P = 0.001.
Table 1.
CI values for single, double, and triple combinations of ATF-126, 5-aza-2′-dC, and SAHA
| ATF/Drug (μg/mL)
|
fa | CI | ||
|---|---|---|---|---|
| ATF-126 | SAHA | 5-aza-2′-dC | ||
| 0.075 | 0.13215 | 0.484 | 0.633 | |
| 0.150 | 0.264 | 0.645 | 0.647 | |
| 0.300 | 0.396 | 0.713 | 0.874 | |
| 0.375 | 0.661 | 0.758 | 0.930 | |
| 0.525 | 1.322 | 0.819 | 1.028 | |
| 0.075 | 0.250 | 0.389 | 0.763 | |
| 0.150 | 0.625 | 0.571 | 0.673 | |
| 0.300 | 1.250 | 0.648 | 0.939 | |
| 0.375 | 2.500 | 0.733 | 0.755 | |
| 0.525 | 3.750 | 0.786 | 0.767 | |
| 0.132 | 0.250 | 0.272 | 0.507 | |
| 0.264 | 0.625 | 0.386 | 0.583 | |
| 0.396 | 1.250 | 0.441 | 0.712 | |
| 0.661 | 2.500 | 0.570 | 0.675 | |
| 1.322 | 3.750 | 0.634 | 1.000 | |
| 0.075 | 0.132 | 0.250 | 0.569 | 0.456 |
| 0.150 | 0.264 | 0.625 | 0.704 | 0.497 |
| 0.300 | 0.396 | 1.250 | 0.779 | 0.613 |
| 0.375 | 0.661 | 2.500 | 0.839 | 0.561 |
| 0.525 | 1.322 | 3.750 | 0.947 | 0.275 |
NOTE: Experimental dose combinations of ATF-126, SAHA, and 5-aza-2′-dC are indicated. MDA-MB-231 cells were transduced with ATF-126 and treated with 5-aza-2′-CC or SAHA or both for 48 h. Cell viability was measured by using an XTT assay.
Abbreviation: fa, fraction of cells affected by the treatment (no viable cells).
As shown in Fig. 4C, the triple treatment ATF-126 + 5-aza-2′-dC + SAHA was significantly the most efficient in decreasing tumor cell viability compared with all the other treatments. The double combinations ATF-126 + SAHA and ATF-126 + 5-aza-2′-dC reduced significantly cell viability compared with all the single treatments (ATF-126, 5-aza-2′-dC, and SAHA and vehicle; P = 0.05). However, our data in Fig. 4C did not reveal statistical differences between the following treatments: SAHA, 5-aza-2′-dC + SAHA, and ATF-126. The apparent discrepancy between the RNA and the viability data could be explained by the fact that these experiments measure different outcomes. Unlike real-time, which specifically measures maspin mRNA levels, cell viability is a complex phenotype involving many different gene products, including tumor suppressors such as maspin. The higher effect of SAHA and 5-aza-2′-dC observed in viability assays might suggest that these compounds reactivate many tumor suppressor genes, not just maspin (46, 47). Although off-target effects are possible with ATFs, these proteins have been engineered to reactivate specifically maspin and are not expected to regulate other tumor suppressor genes. We are presently investigating putative off-target effects of ATF-126.
To verify that the effect of the ATFs and inhibitors was specific for tumor cells and not normal epithelial cells, we did the same viability assays in a nontransformed breast epithelial cell line, the MCF-12A. In contrast with the MDA-MB-231 cell line, none of the combinations of ATFs and inhibitor was able to significantly up-regulate maspin expression nor decrease cell viability as assessed by the ANOVA and post hoc Turkish tests (Supplementary Fig. S1).3
Discussion
In this article, we have investigated the influence of promoter structure in the regulation of the tumor suppressor gene maspin by ATFs. We have focused our analysis on the highly invasive, metastatic breast cancer cell line MDA-MB-231, which comprises a maspin promoter silenced by methylation and transcriptional repression (35), and on ATF-126, the strongest maspin regulator in this cell line (27). We have challenged MDA-MB-231 cells expressing ATF-126 with different doses of the methyltransferase inhibitor 5-aza-2′-dC and the HDAC1 inhibitor SAHA. These drugs interfere with repressive mechanisms, which maintain inaccessible chromatin structure: aberrant cytosine methylation and recruitment of HDAC complexes. Consequently, these inhibitors are able to relax the chromatin, facilitating access to the polymerase II transcriptional machinery (7 – 9). We hypothesized that disruption of the epigenetic silencing mediated by methyltransferase and HDAC inhibitors coupled to ATFs would result in an enhanced up-regulation of silenced genes. Our work shows that ATFs synergized with chromatin remodeling drugs to reactivate endogenous maspin expression. We found that maspin reactivation in response to the inhibitors depended on the ATF-binding site analyzed. It could be that, in the endogenous gene, these sites map in regions of the promoter that contain different levels of histone/methylcytosine modifications. It is also possible that other endogenous factors, such as additional epigenetic marks in the nucleosome, the positioning of the nucleosomes, and CpG-binding proteins, could affect ATF binding and regulation. The strongest synergy was observed with a triple treatment (ATF-126 + 5-aza-2′-dC + SAHA), in which the tumor suppression was reactivated by 600-fold. Consistent with the tumor-suppressive functions of maspin, we found that this triple drug combination was also the most effective in inhibiting breast tumor cell proliferation in vitro.
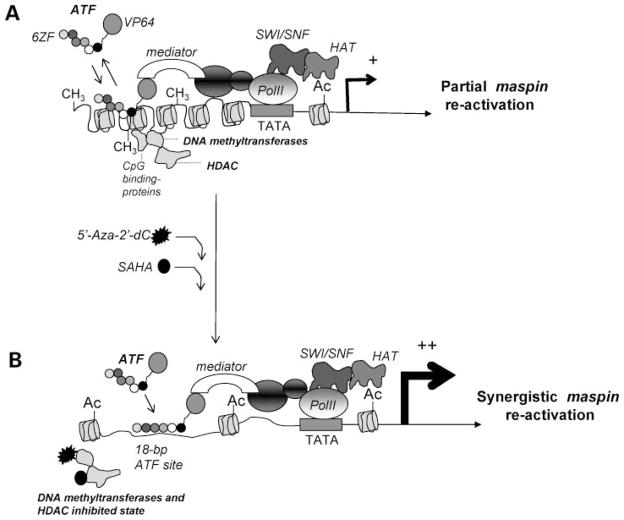
A plausible model explaining this synergy is shown in Fig. 5. In a context of a silenced promoter, methylated CpG islands are associated with methyl-binding proteins, methyltransferases, and HDAC, which maintain the promoter in a compact configuration, inaccessible to the transcriptional machinery (4–6). Likewise, it is possible that in the context of a repressed maspin promoter the ATF-binding sites are not optimally accessible to the ATFs. In the ATF, the ZF domains are linked to the strong transactivator domain VP64, which recruits the mediator protein and other polymerase II–associated proteins (including chromatin remodeling enzymes and histone acetyltransferases), resulting in a partial maspin reactivation. The synergy between the ATF and the chromatin remodeling drugs could be explained by drug-induced enhanced accessibility of the ATFs for their target sites in the maspin promoter.
Figure 5.
A putative model explaining the synergy between the ATF and the chromatin remodeling drugs in reactivating a methylated maspin promoter. A, the binding of ATF to the methylated promoter triggers a partial reactivation of the maspin gene. B, synergistic interaction between the ATFs and chromatin remodeling drugs. On treatment with chromatin remodeling drugs, changes in the chromatin structure facilitate the landing of ATF on the maspin promoter, which enhances the maspin reactivation.
Methyltransferase and HDAC inhibitors interfere with two enzymatic mechanisms of repression: 5-aza-2′-dC inhibits DNA methyltransferase Dnmt1 enzyme (39, 41, 49), whereas SAHA promotes histone acetylation and weakens the histone-DNA interactions (50). Synergy between methyltransferase and HDAC agents in reactivating silenced tumor suppressors has been previously reported by many groups (14, 22, 25, 51, 52). Our results further show that ATF expression highly amplifies the gene reactivation effect of chromatin remodeling drugs with different mechanisms of action. In contrast with chromatin remodeling drugs, which potentially alter many genes in the genome, the ATF is used as a sequence-specific regulator of tumor suppression expression. Our results agree with a report showing that overexpression of the p53 transcription factor in the p53-deficient MDA-MB-231 cell line leads to a synergy with 5-aza-2′-dC in reactivation of the tumor suppressor maspin (13). We have found that, like natural transcription factors, ATFs can strongly synergize with both methyltransferase and HDAC inhibitors. Because ATFs can be designed for virtually any sequence in the human genome, the strategy presented in this article can be potentially applied for the reactivation of any epigenetically silenced promoter. Current ATF technology can generate ATF binding designed sequences with high specificity and selectivity in both in vitro binding assays and in reporter transactivation assays. Only a subset of ATFs designed against a given target promoter results in successful endogenous regulation (27), indicating that subtle aspects of the architecture of endogenous promoters may be key determinants for ATF-mediated regulation. Chromatin modifications could limit the binding of the ATFs in vivo by restricting ATF target site accessibility. This idea is supported by our observations, which show a gain of ATF-mediated regulation of silenced promoters only in the presence of chromatin remodeling drugs with ATFs having poor or no activity in the absence of remodeling-inducing compounds.
Importantly, we found that strong synergy between ATF/chromatin remodeling drugs was maintained in a concentration range of inhibitors below their IC50. Although more experiments need to be done to evaluate the applicability of our findings to experiments using tumor models in vivo, our work shows proof of concept of an exciting strategic approach in therapeutics, which uses ATFs to amplify the apoptotic response of anticancer agents with locus-targeted gene activation while minimizing the exposure/concentration of the drugs.
Acknowledgments
We thank Dr. Lee Graves for the critical reading of the manuscript.
Grant support: NIH/National Cancer Institute grant 1R01CA125273-01, American Lung Association and LUNGevity Foundation grant LD-17098-N, V-Foundation Award, and Department of Defense Idea Award BC051475 (P. Blancafort) and National Cancer Institute grant 1 R21 CA116079-01 (P.M. Lizardi).
Footnotes
Supplementary material for this article is available at Molecular Cancer Therapeutics (http://mct.aacrjournals.org/).
References
- 1.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;2:163–8. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;4:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 3.Bourdon JC. p53 and its isoforms in cancer. Br J Cancer. 2007;3:277–82. doi: 10.1038/sj.bjc.6603886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;6:1187–97. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome—components and functional correlates. Genes Dev. 2006;20:3215–31. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;3:255–75. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 8.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;1:S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 10.Bastian PJ, Yegnasubramanian S, Palapattu GS, et al. Molecular biomarker in prostate cancer: the role of CpG island hypermethylation. Eur Urol. 2004;6:698–708. doi: 10.1016/j.eururo.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Nuyt AM, Szyf M. Developmental programming through epigenetic changes. Circ Res. 2007;4:452–5. doi: 10.1161/01.RES.0000260292.95612.ac. [DOI] [PubMed] [Google Scholar]
- 12.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;1:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 13.Oshiro MM, Watts GS, Wozniak RJ, et al. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene. 2003;22:3624–34. doi: 10.1038/sj.onc.1206545. [DOI] [PubMed] [Google Scholar]
- 14.Hellebrekers DM, Griffioen AW, van Engeland M. Dual targeting of epigenetic therapy in cancer. Biochim Biophys Acta. 2007;1:76–91. doi: 10.1016/j.bbcan.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Samlowski WE, Leachman SA, Wade M, et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: inhibition of promoter-specific and global genomic DNA methylation. J Clin Oncol. 2005;17:3897. doi: 10.1200/JCO.2005.06.118. [DOI] [PubMed] [Google Scholar]
- 16.Winquist E, Knox J, Ayoub JP, et al. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;2:159–67. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- 17.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;20:1498–506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 18.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;1:65–76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 19.Münster P, Marchion D, Bicaku E, et al. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J Clin Oncol. 2007;15:1979–85. doi: 10.1200/JCO.2006.08.6165. [DOI] [PubMed] [Google Scholar]
- 20.Patnaik A, Rowinsky EK, Villalona MA. A phase I study of pivaloyloxymethyl butyrate, a prodrug of the differentiating agent butyric acid, in patients with advanced solid malignancies. Clin Cancer Res. 2002;7:2142–8. [PubMed] [Google Scholar]
- 21.Reid T, Valone F, Lipera W, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;3:381–6. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 dimethylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;1:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 23.Primeau M, Gagnon J, Momparler RL. Synergistic antineoplastic action of DNA methylation inhibitor 5-AZA-2′-deoxycytidine and histone deacetylase inhibitor depsipeptide on human breast carcinoma cells. Int J Cancer. 2003;2:177–84. doi: 10.1002/ijc.10789. [DOI] [PubMed] [Google Scholar]
- 24.Zhu WG, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anti-Canc Agents. 2003;3:187–99. doi: 10.2174/1568011033482440. [DOI] [PubMed] [Google Scholar]
- 25.Hurtubise A, Momparler RL. Effect of histone deacetylase inhibitor LAQ824 on antineoplastic action of 5-aza-2′-deoxycytidine (decitabine) on human breast carcinoma cells. Cancer Chemother Pharmacol. 2006;5:618–25. doi: 10.1007/s00280-006-0225-6. [DOI] [PubMed] [Google Scholar]
- 26.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;25:11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltran A, Parikh S, Liu Y, et al. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene. 2007;19:2791–8. doi: 10.1038/sj.onc.1210072. [DOI] [PubMed] [Google Scholar]
- 28.Blancafort P, Magnenat L, Barbas CF., III Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–74. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;4:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Tallarida RJ. Drug synergism and dose-effect data analysis. Florida: CRC Press; 2000. pp. 15–71. [Google Scholar]
- 31.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 32.Chou TC. Assessment of synergistic and antagonistic effects of chemotherapeutic agents in vitro. Contrib Gynecol Obstet. 1994;19:91–107. [PubMed] [Google Scholar]
- 33.Berenbaum MC. What is synergy? Pharmacol Rev. 1989;2:93–141. [PubMed] [Google Scholar]
- 34.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2. 1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 35.Futscher BW, Oshiro MM, Wozniak RJ, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Med. 2002;31:175–9. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 36.Zou Z, Anisowicz A, Hendrix MJ, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;5146:526–9. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 37.Domann FE, Rice JC, Hendrix MJC, Futscher BW. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer. 2000;85:805–10. doi: 10.1002/(sici)1097-0215(20000315)85:6<805::aid-ijc12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Baylin SB, Schuebel KE. Genomic biology: the epigenomic era opens. Nature. 2007;7153:553–60. doi: 10.1038/448548a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;4:2041–8. [PubMed] [Google Scholar]
- 40.Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;2:151–8. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. The DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyl-transferases 1 and 3B. Mol Cell Biol. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;15:1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 43.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;1:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 44.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;3:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 45.Brock MV, Herman JG, Baylin SB. Cancer as a manifestation of aberrant chromatin structure. Cancer J. 2007;1:3–8. doi: 10.1097/PPO.0b013e31803c5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmond JC, Raynaud j, Tung E, et al. Discovery of epigenetically silenced genes in acute myeloid leukemias. Leukemia. 2007;21:1026–34. doi: 10.1038/sj.leu.2404611. [DOI] [PubMed] [Google Scholar]
- 47.Kumagai T, Wakimoto N, Yin D, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;3:656–65. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 48.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;5:1241–6. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;35:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 50.Rundall BK, Denlinger CE, Jones DR. Suberoylanilide hydroxamic acid combined with gemcitabine enhances apoptosis in non-small cell lung cancer. Surgery. 2005;2:360–7. doi: 10.1016/j.surg.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Shaker S, Bernstein M, Momparler LF, Momparler RL. Preclinical evaluation of antineoplastic activity of inhibitors of DNA methylation (5-aza-2′-deoxycytidine) and histone deacetylation (trichostatin A, depsipeptide) in combination against myeloid leukemic cells. Leuk Res. 2005;5:437–44. doi: 10.1016/s0145-2126(02)00222-9. [DOI] [PubMed] [Google Scholar]
- 52.Becker JC, Ugurel S, Bröcker EB, Schrama D, Houben R. New therapeutic approaches for solid tumors: histone deacetylase, methyltransferase and proteasome inhibitors. J Dtsch Dermatol Ges. 2006;2:108–13. doi: 10.1111/j.1610-0387.2006.05920.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang M, Magit D, Sager R. Expression of maspin in prostate cells is regulated by a positive ets element and a negative hormonal responsive element site recognized by the androgen receptor. Proc Natl Acad Sci U S A. 1997;94:5673–8. doi: 10.1073/pnas.94.11.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]