Abstract
The regulatory factors governing adult mesenchymal stem cells (MSCs) physiology and their tumorigenic potential are still largely unknown, which substantially delays the identification of effective therapeutic approaches for the treatment of aggressive and lethal form of MSC-derived mesenchymal tumors, such as undifferentiated sarcomas. Here we have developed a novel platform to screen and quickly identify genes and pathways responsible for adult MSCs transformation, modeled undifferentiated sarcoma in vivo, and, ultimately, tested the efficacy of targeting the identified oncopathways. Importantly, by taking advantage of this new platform, we demonstrate the key role of an aberrant LRF-DLK1-SOX9 pathway in the pathogenesis of undifferentiated sarcoma with important therapeutic implications.
Keywords: LRF, Pokemon, Mesenchymal Stem Cell, Dlk1, Sarcomas
INTRODUCTION
Tumorigenesis and stem cell differentiation are frequently tied together, in the way that, genetic and functional loss of genes responsible for cell commitment and differentiation are selected during the tumorigenic process, since undifferentiated cells are often endowed with longer life spans, higher survival and proliferative potential, and are often resistant to treatment (1).
Adult mesenchymal stem cells (MSCs) are known for their ability to self-renew as well as differentiate into cells of varying mesenchymal lineages, such as chondrocytes, osteoblasts and adipocytes (2, 3). Importantly, mounting evidence now implicates adult MSCs as the cell of origin of human undifferentiated sarcomas, one of the most aggressive and lethal soft tissue tumors (4–8). Undifferentiated sarcomas are generally treated by surgical resection (whenever possible), radiotherapy, and chemotherapy; all options, however, that minimally change the very poor overall survival in patients. Limited genetic and molecular analyses, along with the absence of faithful in vivo models enabling pre-clinical testing of targeting specific tumorigenic pathways, are together the main factors impeding the development of new and more effective therapeutic options. While specific forms of sarcoma (i.e. osteosarcoma) have been successfully modeled in genetically engineered mice (9–12), current protocols to model undifferentiated sarcomas are generally based on transplantation of human tumor cell lines in immune compromised mice (13), in vitro expanded, and spontaneously transformed heterogeneous mouse primary mesenchymal cells (4, 14, 15), or in vitro-genetically modified human bone marrow MSCs (16, 17), whose purity and stemness features have recently been debated (3, 18). Although these studies have proved helpful in uncovering aspects of sarcomagenesis, such protocols fail to mirror the real onset and progression of undifferentiated sarcomas since they cannot control key factors such as the type and number of the genetic alterations driving the tumorigenic process, or cell of origin (19). In turn, these approaches will hardly allow the discovery of key genetic drivers involved in the onset and progression of undifferentiated sarcomas, and, most importantly, of potential druggable targets with clinical relevance. To fill this void, we have developed a novel ex vivo/in vivo genetic platform that will allow the discovery of genetic drivers responsible for adult MSC transformation and the generation, in vivo, of undifferentiated sarcomas.
RESULTS
Optimized culture conditions to prevent MSCs spontaneous transformation lead to the development of a new genetic platform to model sarcomagenesis
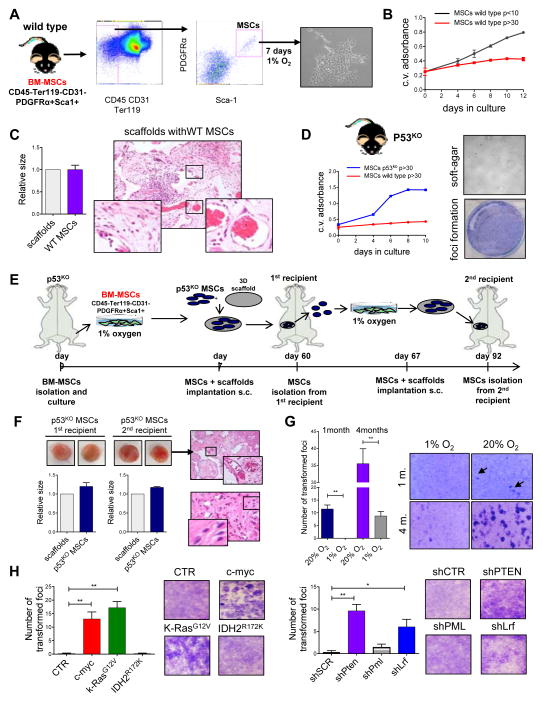
In order to model undifferentiated sarcomas, we selectively isolated from the bone marrow of mice a cell population highly enriched for adult MSCs (20, 21) (BM-MSCs: CD45−CD31−Ter119−Sca1+PDGFRα+, Fig. 1A), grew them in vitro, in conditions that maintain their stemness properties, and then studied the genetic drivers leading to their transformation. We have recently described that mimicking in vitro the hypoxic conditions characterizing the natural environment of MSCs within the bone, favors the expansion of adult BM-MSCs, while maintaining their stem features (21). This analysis led us to discover that, unexpectedly and in contrast with what has been previously reported for mesenchymal cells cultured in regular oxygen concentrations (20% oxygen) (4, 14, 15, 22), primary adult BM-MSCs cultured in hypoxic conditions (1% oxygen) did not undergo spontaneous in vitro transformation; on the contrary they showed progressive reduction in the proliferation rate during the culture (Fig. 1B). Moreover, once seeded into scaffolds and implanted subcutaneously in mice, MSCs remained vital even after months, showing abilities to recruit blood vessels within the scaffold, but not to form tumors, or to show marks of neoplastic transformation (Fig. 1C).
Figure 1.
New genetic platform to study genes responsible for sarcomagenesis. (A) MSCs were isolated from the bone marrow of p53KO mice as CD31−CD45−Ter119−Sca1+PDGFRα+, and cultured at 1% of oxygen. After 7 days in culture cells formed visible CFU-F colonies. (B) Growth curve with wild type MSCs maintained in culture for less then 10 passages or more than 30 passages before the day 0 of the proliferation assay. Results are shown as one representative experiment out of 2 independent biological replicates. (C) Scaffolds seeded with wild type MSCs isolated from bone marrow and grown 7 days in hypoxic conditions. The chart on the left show the size of the collected scaffolds, while the pictures on the right are section of the scaffolds showing blood vessels, and not transformed mesenchymal cells. n=6 mice implanted with scaffolds. (D) Growth curve of wild type or p53KO MSCs isolated from bone marrow and maintained for more than 30 passages in hypoxic conditions is shown on the left, while foci formation assay (lower panel) and growth in anchorage-independent manner (soft agar, upper panel) performed with p53KO MSCs are shown on the right. Results are shown as one representative experiment out of 3 independent biological replicates. (E) Schematic overview of the experimental design. The tumorigenic potential of MSCs cultured in hypoxic conditions was assessed seeding cells into scaffolds and implanting them subcutaneously into mice. Two serial implantations have been performed. (F) Representative pictures of scaffolds seeded with p53KO MSCs, and implanted subcutaneously in mice are shown on the left, while H&E staining of scaffold sections are shown on the right. Charts below the pictures of the scaffolds represent the size of the scaffolds collected from mice (from 1st recipient on the left, as well from the 2nd recipient on the right), compared to the size of the scaffold before its implantation. The upper right picture shows the blood vessels recruited by cells within the scaffold. The lower right pictures shows non-transformed mesenchymal cells seeded within the scaffold. (G) Focus formation assay with p53KO MSCs cultured for one month or 4 months at 20% of oxygen or 1% of oxygen. Quantification of transformed foci is shown on the left while representative pictures are shown on the right. Results are shown as one representative experiment out of 2 independent biological replicates. (H) Foci formation assay performed with p53KO MSCs transduced in vitro with vectors expressing c-myc, K-RasG12V and IDH2R172K, or silenced with shRNA for PML, PTEN and LRF The carts shown on the left represent the quantification of the observed foci of transformation. n=3 independent biological experiments.
Loss of p53 has been firmly implicated in the pathogenesis of undifferentiated sarcomas in human (23). We therefore assessed the impact of p53 inactivation in our model system. Differently to wild type MSCs, primary p53KO adult MSCs maintained in vitro in hypoxic conditions were characterized by high proliferation rate even after numerous passages, as evidences of a status of immortalization (Fig. 1D). Surprisingly, however, p53KO MSCs did not show signs of neoplastic transformation in hypoxic growth conditions in vitro, such as the ability to form foci of transformation in the dedicated assay or sizable colonies in soft agar (Fig. 1D). In order to test their tumorigenic potential in vivo, we next seeded p53KO MSCs into scaffolds (24) and transplanted them subcutaneously in syngeneic C57BL/6, or nude mice (1rst recipients). Two months after the implantation, the scaffolds were collected, cells within them were expanded in hypoxic conditions, and were then used for a second round of implantation (2nd recipients) (Fig. 1E). Similarly to wild type MSCs, p53KO MSCs remained vital within scaffolds. They recruited blood vessels, and they did not show any signs of neoplastic transformation in both 1rst and 2nd recipients, which resulted in the inability to generate tumors in serially transplanted animals (Fig. 1F).
Previous published data reported spontaneous transformation of murine MSCs cultured in regular oxygen conditions after several passages (14, 15). We therefore analyzed the spontaneous transformation of p53KO MSC populations culturing them for 1 month or 4 months in low (1%) or high (20%) oxygen tension, and then performed a “focus formation assay”. As shown in Figure 1G, cells cultured for 1 month at 1% of oxygen were not able to generate transformed foci; while, on the contrary, cells kept at 20% of oxygen formed several foci of transformation, which increased in number and size during the culture. Importantly, we also noticed that MSC cultures kept at 20% of oxygen showed a significant increase in the number of cells characterized by several (n>5) nuclear dots of γH2AX in comparison to the same cells kept at 1% of oxygen (Supplementary Fig. S1A), thus defining a condition of increased DNA damage linked to the 20% oxygen condition, primary cause of genomic instability in replicating cells (25).
Overall, these data led us to hypothesize that loss of p53 functions in human MSCs may be necessary but not sufficient to trigger sarcomagenesis. In addition, in vitro hypoxic growth conditions, by maintaining genomic stability of primary adult p53-null MSCs and by preventing their spontaneous neoplastic transformation, might represent the cornerstone for the development of a tightly controlled genetic platform aimed at identifying specific genetic alterations that, in combination with p53 loss, could dictate adult MSCs transformation and development of undifferentiated sarcomas. To test this hypothesis, we decided to challenge our platform with oncogenic stresses previously implicated in sarcomagenesis, and assess their capacity to transform p53-null MSCs. Specifically, in p53-null MSCs maintained in hypoxic conditions we over-expressed c-myc (26), K-RasG12V (27) and IDH2R172K (13), while we knocked-down Pten (28). The expression of c-myc and K-RasG12V, as well as the loss of Pten (but not the expression of IDH2R172K) were indeed able to trigger p53-null MSCs transformation in vitro, and represented proofs of principle for the validity of our approach (Fig. 1H and Supplementary Fig. S1B–G).
Identification of novel genes involved in sarcomagenesis through the MSC-derived platform
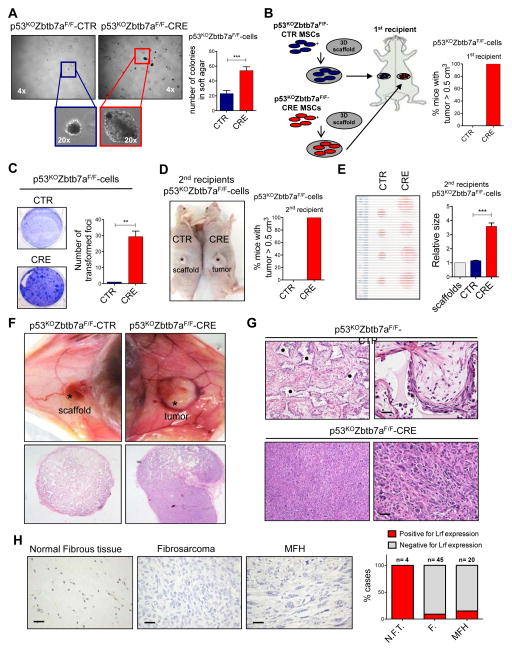
We next tested whether this new platform would be useful in identifying new genes implicated in sarcomagenesis. Since undifferentiated sarcomas have been suggested to originate through the combined deregulation in adult MSCs of genes involved in cellular proliferation/apoptosis (such as p53), and genes implicated in the regulation of stem cell differentiation (29, 30), we decided to couple two known regulators of stem cells maintenance and differentiation with the loss of p53. The regulators enrolled in the platform were PML (31) and LRF/Pokemon [encoded by the gene ZBTB7A (Zinc finger and BTB domain-containing protein 7A)], which is emerging as one of the master regulators of differentiation for both hematopoietic and non-hematopoietic stem/progenitor cells, and has been attributed oncogenic or tumor suppressive functions depending on the specific cellular context (32). While Pml knockdown in p53-null MSCs did not trigger neoplastic transformation, adult p53-null MSCs knocked down for Lrf, showed features of neoplastic transformation in vitro (Fig. 1H and Supplementary Fig. S1F–G), suggesting a possible role for LRF as suppressor of sarcomagenesis. In order to fully determine this new role of Lrf/Pokemon in sarcomagenesis, we generated a cohort of p53KOZbtb7aF/F mice, and derived adult MSCs as previously described (Fig. 1A). Zbtb7a was knocked out in vitro by transducing p53KOZbtb7aF/F MSCs with lentiviral vectors containing CRE cDNA, or an empty vector as control (hereafter referred to as p53KOZbtb7aF/F-CRE or p53KOZbtb7aF/F-CTR for brevity) (Supplementary Fig. S2A). Although p53KOZbtb7aF/F-CRE and p53KOZbtb7aF/F-CTR cells showed similar anchorage-dependent growth rate (Supplementary Fig. S2B), the anchorage-independent colony-forming capacity of p53KOZbtb7aF/F-CRE cells, as well as the capacity to form transformation foci in vitro, was significantly higher than that of p53KOZbtb7aF/F-CTR cells (Fig. 2A and data not shown). Off-target effects of the CRE infection were ruled out upon transduction of p53KOZbtb7a+/+ cells with the same lentiviral vectors containing CRE cDNA, or an empty vector as control used in p53KOZbtb7aF/F cells, and then performing a focus formation assay (Supplementary Fig. S2C).
Figure 2.
Lrf loss in MSCs leads to formation of mesenchymal tumors. (A) Soft-agar assay for detecting anchorage independent cell growth of p53KOZbtb7aF/F-CTR and p53KOZbtb7aF/F-CRE MSCs. Representative pictures of the colonies are shown on the left, while the quantification on the right is shown as average of 3 biological independent replicates ± SEM. (B) Schematic overview of the experimental design is shown on the left, while the percentage of mice with a tumor bigger that 0.5 cm3 is shown on the right. p53KOZbtb7aF/F-CTR cells were not transformed, and only scaffolds within mesenchymal cells were recovered after transplantation. (n=2 CTR, n=2 CRE). (C) Detection of transformation status of p53KOZbtb7aF/F-CTR or p53KOZbtb7aF/F-CRE cells. Pictures of the foci are shown on the left, while the quantification of the transformed foci is shown on the right. The quantification on the right is shown as pooled from 3 independent experiments, mean ± SEM. (D) Tumors generated by p53KOZbtb7aF/F-CTR or p53KOZbtb7aF/F-CRE cells transplanted within scaffolds into second recipient mice. Representative pictures of mice are shown on the left, while the percentage of mice with a tumor bigger that 0.5 cm3 (scaffold size used as control) is shown on the right. (n=4 CTR, n=7 CRE) (E) Sizes of tumors generated by p53KOZbtb7aF/F-CTR or p53KOZbtb7aF/F-CRE cells transplanted within scaffolds into second recipient mice. Pictures of the collected tumors are shown on the left, while the relative size of tumors is shown on the right (scaffold size used as control). (F–G) Tumors generated by p53KOZbtb7aF/F-CRE cells compared p53KOZbtb7aF/F-CTR cells collected from 1st recipients, seeded into scaffolds and the transplanted into 2nd recipients. H&E staining showing the morphology of collected tumors or mesenchymal cells on top of the scaffold. p53KOZbtb7aF/F-CTR cells were not able to generate tumors in vivo and only scaffolds within mesenchymal cells were recovered after transplantation, while p53KOZbtb7aF/F-CRE cells originated undifferentiated sarcomas. (* indicates the scaffold. Scale bars: 20 μm). (H) LRF expression in human undifferentiated sarcomas. (M.F.H. malignant fibrous hystocytoma; F. fibrosarcomas; N.F.T. normal fibrous tissue; scale bars 30μm).
Finally, in order to test their tumorigenic potential in vivo we seeded p53KOZbtb7aF/F-CRE or p53KOZbtb7aF/F-CTR MSCs into the scaffolds and implanted them into mice following the same protocol described in figure 1E. Two months after implantation, we observed that p53KOZbtb7aF/F-CTR cells were not able to generate tumor in mice (as expected, only non-transformed mesenchymal cells were recovered from the scaffolds), while the p53KOZbtb7aF/F-CRE cells underwent neoplastic transformation and generated tumors in all the transplanted mice (Fig. 2B). Cells were then recovered from the scaffolds, characterized for Lrf expression (Supplementary Fig. S2D), and further tested for their neoplastic potential in vitro, and then in vivo in a second round of transplantation. Once again, p53KOZbtb7aF/F-CRE and p53KOZbtb7aF/F-CTR cells recovered from the scaffold showed similar rates of anchorage-dependent proliferation (Supplementary Fig. S2E), but only the p53KOZbtb7aF/F-CRE cells displayed the capacity to form transformed foci (Fig. 2C). Importantly, once re-transplanted in secondary recipients (2nd recipients), p53KOZbtb7aF/F-CRE cells originated tumors larger than 5 mm in less than two weeks (7/7 mice; 2 nude mice and 5 C57Bl/6), while none of the 4 mice (2 nude mice and 2 C57Bl/6) re-transplanted with p53KOZbtb7aF/F-CTR cells originated tumors (Fig. 2D–G). Histopathological analysis of scaffolds recovered from mice revealed that p53KOZbtb7aF/F-CRE cells originated undifferentiated sarcomas, which were able to egress the scaffolds and to invade tissues nearby. On the contrary, only non-transformed mesenchymal cells were present in the scaffolds implanted with p53KOZbtb7aF/F-CTR cells (Fig. 2F–G).
Since our platform identified Lrf as a tumor suppressor gene in triggering the transformation of MSCs, we investigated the expression of LRF through immunohistochemistry analyses on a comprehensive human tissue microarray (TMA) containing n=45 undifferentiated mesenchymal tumors (fibrosarcomas (F.) and n=20 malignant fibrous hystocytomas, (M.F.H)), compared to n=4 normal fibrous tissues (N.F.T.) (Fig. 2H). All normal fibrous tissues (4/4) as well as human adult MSCs (Supplementary Fig. S2F) were characterized by LRF expression, while, in sharp contrast, malignant undifferentiated sarcomas turned out strongly negative, (41/45 cases of fibrosarcomas and 19/20 case of malignant fibrous hystocytomas).
Overall, these results demonstrate that our platform may turn out as an important tool to identify new oncogenes and tumor suppressors involved in the development of human sarcomas.
LRF is essential for MSCs commitment and differentiation
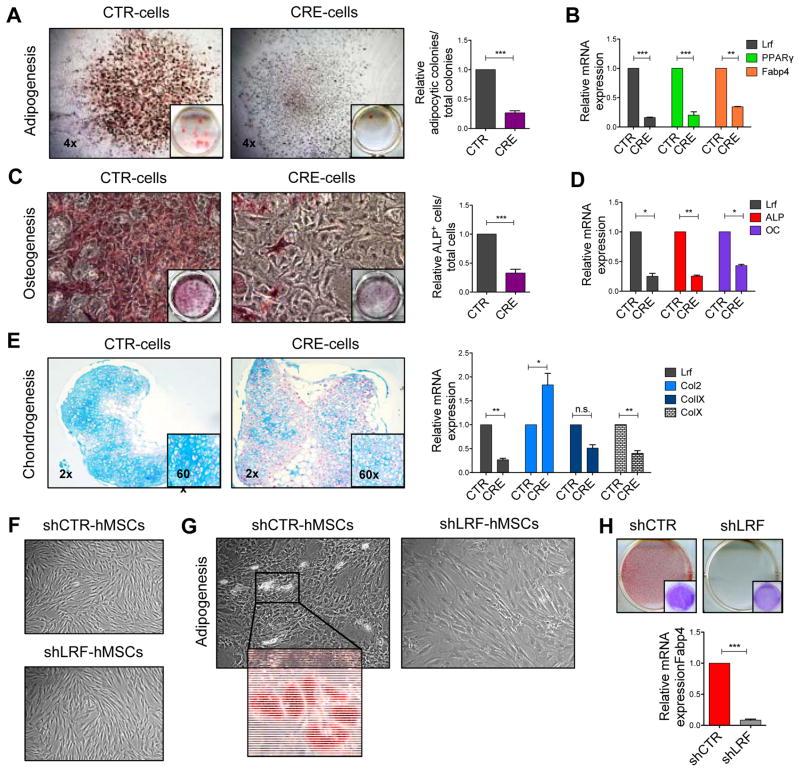
We next investigated the biological processes through which LRF loss could trigger sarcomagenesis. LRF/Pokemon has been described in the stemness maintenance/differentiation in different cell lineages (32), however, nothing is known about its role in adult MSCs. For this reason, we first investigated the possibility that Lrf could trigger sarcomagenesis by blocking the differentiation capacity of MSCs. Accordingly, we derived MSCs from Zbtb7a-floxed mice (Zbtb7aF/F) and investigated their commitment/differentiation ability towards osteoblasts, chondrocytes, and adipocytes upon Lrf knock out in vitro through transduction with CRE-lentiviral vector (from now referred as CRE-cells or CTR-cells). Despite the profound reduction of Lrf expression (Supplementary Fig. S3A), CRE-cells remained similar to CTR-cells in terms of morphology, size and number of the CFU-F colonies generated (Supplementary Fig. S3B), as well as to numbers of cells undergoing to senescence (Supplementary Fig. S3C) or to apoptosis (Supplementary Fig. S3D–E). However, as shown in Fig. 3A–E, CRE-cells displayed differentiation defects when compared to CTR-cells. CRE-cells generated only 30% of the Oil-Red-O positive CFU-F colonies generated by CTR-cells in response to adipogenic induction (Fig. 3A), and accordingly, the expression of Pparγ and Fabp4 during differentiation was significantly lower in CRE-cells compared to CTR-cells (Fig. 3B). Similarly, CRE-cells treated with osteogenic stimulating factors originated less Alp+-osteoblasts than CTR-cells (Fig. 3C) and, CRE-cells expressed lower levels of Alp and osteocalcin (Oc) mRNAs compared to controls (Fig. 3D). Finally, regarding chondrogenesis, CRE-cells originated chondrocyte micro-masses surrounded by less extracellular matrix (as shown by Toluidine blue staining) than CTR-cells. Interestingly, CRE-cells expressed significantly higher amounts of Col2 in comparison to CTR-cells, although the expression of markers of terminal differentiation such as ColIX and ColX was significantly reduced suggesting that chondrocyte progenitors are favored in their initial commitment, yet they fail to reach a mature terminal differentiation (Fig. 3E).
Figure 3.
Lrf is a key factor for MSCs commitment and differentiation. (A) MSCs differentiation into mature adipocytes. Adipocytic colonies stained with Oil-Red-O are shown on the left, while the relative quantification of adipocytic colonies (positive for Oil-Red-O) among the total number of colonies (CFU-F) is shown on the right. The quantification on the right is shown as average of 5 independent experiments ± SEM. (B) Lrf, Pparγ and Fabp4 relative mRNA expression in CTR-cells and CRE-cells during adipogenesis. Results are shown as one representative experiments out of 2 independent biological replicates. (C) MSCs differentiation toward mature osteoblasts. Representative pictures of Alp+ cells are shown one the left while the relative number of Alp+ cells is shown on the right. The quantification on the right is the average of 3 independent experiments ± SEM. (D) Lrf, Alp and Oc relative mRNA expression in CTR-cells and CRE-cells during osteogenesis. Results are shown as one representative experiments out of 2 independent biological replicates. (E) MSCs differentiation toward chondrocytes. Representative images of chondrocytes originated from CTR-cells and CRE-cells stained with Toluidine blue are shown on the left (n=2 independent biological replicates). Relative mRNA expression levels of Lrf, Col2, ColIX and ColX are shown on the right. Results are shown as average of 2 independent biological replicates ± SEM. (F) Morphology of human MSCs transduced with shCTR or shLRF and cultured at 1% of oxygen. (G) Human MSCs transduced with shCTR or shLRF and induced to adipogenesis, at day 7 of the differentiation process. Magnification shows in detail fully differentiated adipocytes. (H) Human MSCs differentiation into adipocytes, at day 15 of the differentiation process. Adipocytes stained with Oil-Red-O are shown in the top panel, while the relative mRNA expression of FABP4 is shown in the chart.
Finally, to corroborate our findings in human MSCs (hMSCs), LRF was knocked down through specific shRNAs taking advantage of commercially available hMSCs cultured under the same conditions used for mouse MSCs. The ability of sh-LRF and sh-CTR hMSCs to originate mature adipocytes was evaluated as previously described for mouse MSCs. Both shLRF and shCTR transduced hMSCs behaved comparably in culture, without displaying features of apoptosis or senescence (Figure 3F). However, while shCTR-hMSCs showed initial signs of adipocytic differentiation starting from day 7 (Figure 3G) and were fully differentiated at day 15 (Figure 3H), shLRF-hMSCs remained completely undifferentiated at day 7 (Figure 3G), and generated only few mature cells at day 15. Accordingly, expression levels of FABP4 were reduced in shLRF-hMSCs compared to shCTR-hMSCs (Figure 3H). Analysis of human MSCs therefore confirms the results obtained in mouse MSCs, as well as the evolutionary conserved critical role for LRF in determining MSCs cell fate decisions.
Lrf promotes MSCs commitment through the transcriptional repression of Dlk1
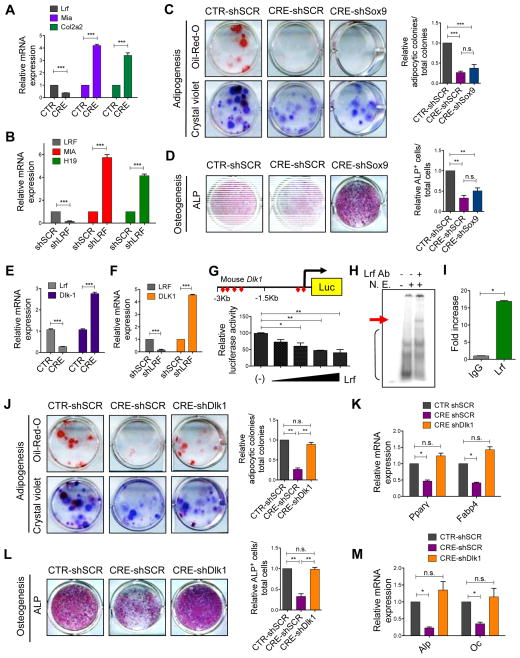
Based on these data showing that LRF/Pokemon governs the commitment capacity of MSCs, and having excluded a possible role of two well characterized pathways regulated by LRF/Pokemon such as ARF (33) and NOTCH1 (34) in this process (data not shown), we aimed to understand which pathways are regulated by Lrf in MSCs, in order to identify possible new players involved in the genesis of undifferentiated sarcoma and, in turn, potential targets for therapy. Considering that oncogenic and tumor suppressive pathways are often wired to regulate cell lineage commitment and differentiation in physiological conditions (35), we focused our attention on DLK1 (Delta-like-1)/SOX9 (SRY (sex determining region Y)-box 9) pathway, that is know to be critical in mesenchymal lineages determination (36–40). In particular, we focused on SOX9, as it has been shown to be involved in the differentiation process of MSCs (41), in addition to being recently described as functionally antagonized by Lrf in prostate tumorigenesis (42, 43). CTR-cells and CRE-cells were first analyzed by RT-qPCR for the expression of Sox9 transcriptional target genes (44, 45); both genes were over-expressed in CRE-cells as compared to CTR-cells (Fig. 4A). Similarly, the knock-down of LRF in hMSCs resulted in the up-regulation of SOX9-activity (Figure 4B). Next, in order to address the critical question of whether Lrf mediates MSC commitment through Sox9, CRE-cells were knocked down for Sox9 (Supplementary Fig. S4A), and induced to differentiate. CTR-shSCR-cells, CRE-shSCR-cells and CRE-shSox9-cells originated a comparable number of colonies as detected by crystal violet, while down-regulation of Sox9 levels and activity did not rescue the capacity of CRE-cells to differentiate into mature adipocytes (Fig. 4C and Supplementary Fig. S4B–C). Similarly, down-regulation of Sox9 in CRE-cells negligibly rescued the ability of MSCs to differentiate into mature osteoblasts (Fig. 4D).
Figure 4.
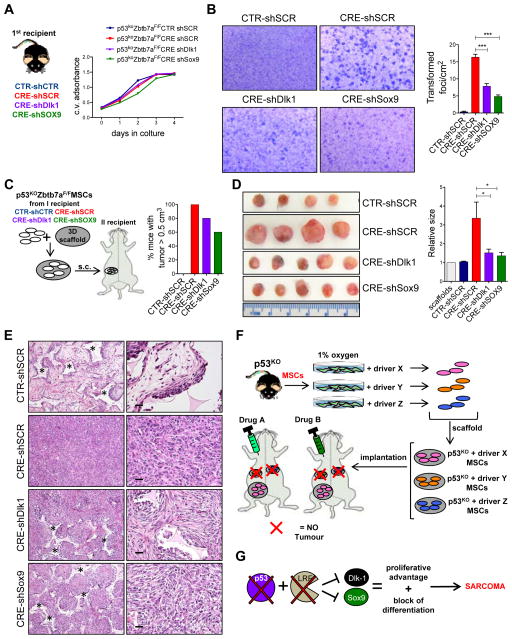
Lrf regulates the differentiation process of MSCs through the repression of Dlk1. (A) Relative mRNA expression levels of Lrf, Mia and Col2a2 in CTR-cells and CRE-cells. Results are shown as one representative experiments out of 2 independent biological replicates. (B) Relative mRNA expression levels of LRF, H19 and MIA in human MSCs transduced with shCTR or shLRF. (C) Adipogenesis potential of CTR-shSCR-cells, CRE-shSCR-cells and CRE-shSox9-cells. Adipocytic colonies stained with Oil-Red-O (upper panels) or crystal violet (lower panels) are shown on the left, while the relative quantification of adipocytic colonies is shown on the right. The quantification on the right is presented as average of 3 independent experiments ± SEM. (D) Osteogenesis potential of CTR-shSCR-cells, CRE-shSCR-cells and CRE-shSox9-cells. Representative pictures of Alp staining are shown on the left, while the relative numbers of Alp+ cells are shown on the right. The quantification on the right is presented as average of 3 independent experiments ± SEM. (E) Lrf and Dlk1 relative mRNA expression in CTR-cells and CRE-cells. Results are shown as average of 5 biological independent replicates ± SEM. (F) Relative mRNA expression levels of LRF and DLK1 in human MSCs transduced with shCTR or shLRF. (G) Luciferase assay for detecting the activity of Lrf on Dlk1 promoter. Results are shown as average of 3 biological independent replicates ± SEM. (H) EMSA assay showing a shift in presence of a specific probe representative for binding site #4 and a super-shift in presence of anti-Lrf antibody. (I) Chromatin immunoprecipitation of Lrf and Dlk1 promoter region. Results are shown as average of 3 biological independent replicates ± SEM. (J) Adipogenesis potential of CTR-shSCR-cells, CRE-shSCR-cells and CRE-shDlk1-cells. Adipocytic colonies stained with Oil-Red-O (upper panels) or crystal violet (lower panels) are shown on the left while the relative quantification of adipocytic colonies is shown on the right. The quantification on the right is presented as average of 3 biological independent replicates ± SEM. (K) Relative mRNA expression levels of Pparγ and Fabp4 in adipocytes derived from CTR-shSCR-cells, CRE-shSCR-cells and CRE-shDlk1-cells. Results are shown as average of 3 biological independent replicates ± SEM. (L) Osteogenesis potential of CTR-shSCR-cells, CRE-shSCR-cells and CRE-shDlk1-cells. Representative pictures of Alp staining are shown on the left, while the relative numbers of Alp+ cells is shown on the right. The quantification on the right is presented as average of 3 biological independent replicates ± SEM. (M) Alp and Oc relative mRNA expression in osteoblasts derived from CTR-shSCR-cells, CRE-shSCR-cells and CRE-shDlk1-cells. Results are shown as average of 3 biological independent replicates ± SEM.
Another critical regulator of MSCs commitment is DLK1 (46–50). We therefore decided to investigate the possibility that Lrf could regulate Dlk1 activity, and through it the commitment of MSCs. Intriguingly, CRE-cells showed a significant increase in Dlk1 expression compared to CTR-cells (Fig. 4E). Similarly, knock-down of LRF resulted in the up-regulation of DLK1 in hMSCs (Figure 4F). To determine whether Lrf could directly regulate Dlk1 expression, we cloned the Dlk1 promoter sequence into a luciferase construct and performed in vitro transactivation assays. This analysis revealed that Lrf efficiently repressed the basal promoter activity in a dose-dependent manner (Fig. 4G), and, among the six putative Lrf consensus regions within the promoter of Dlk1, we discovered that only the two consensus sequences (indicated as 4 and 5 in Supplementary Fig. S4D), closest to the transcriptional starting site, were necessary for Lrf repression of Dlk1 transcription. Additionally, the ability of Lrf to bind Dlk1 promoter was confirmed performing electrophoretic mobility shift (EMSA) assays (Fig. 4H), and chromatin-immunoprecipitation (ChIP) (Fig. 4I) in 3T3L1 mouse mesenchymal cell line. In order to functionally validate the Lrf/Dlk1 axis in MSC commitment, we knocked down Dlk1 in CRE-cells, and induced the generated cells to differentiate toward adipocytes and osteoblasts. As expected, CRE-shSCR-cells failed to differentiate compared to CTR-shSCR-cells; but, critically, the concomitant inactivation of Dlk1 (CRE-shDlk1-cells) rescued their defects of adipogenesis (Fig. 4J–K and Supplementary Fig. S4E–F), and osteogenesis (Fig. 4L–M).
Lrf acts as oncosuppressor of mesenchymal tumorigenesis by controlling the activity of Dlk1 and Sox9
Both DLK1 and SOX9 have been described as oncogenes in several tumor types (36–39). Since we have demonstrated that Lrf negatively controls both the expression of Dlk1 and the activity of Sox9 in primary non-transformed MSCs, and that, in a p53-null MSCs, Lrf acts as a tumor suppressor gene, we next used our platform in order to investigate the possibility that Dlk1 and Sox9 deregulation might be responsible for transformation of MSCs and sarcomagenesis after Lrf loss. p53KOZbtb7aF/F-CRE cells or p53KOZbtb7aF/F-CTR cells collected from scaffolds (1st recipient), were grown in hypoxia for 7 days and then transduced with shRNA in order to silence Dlk1 or Sox9. Four different cellular types were then obtained: p53KOZbtb7aF/F-CTR-shSCR, p53KOZbtb7aF/F-CRE-shSCR, p53KOZbtb7aF/F-CRE-shDlk1 and p53KOZbtb7aF/F-CRE-shSox9 (in the figures indicated only as CTR-shSCR, CRE-shSCR, CRE-shDlk1 and CRE-shSox9 for brevity) (Fig. 5A and Supplementary Fig. S5A–B). Cells were then used both in vitro and in vivo experiments to test their tumorigenic potential. In an anchorage-dependent proliferation assay, all the cell types showed similar division rates (Fig. 5A), but their capacity to form transformed foci in the culture was significantly different. While p53KOZbtb7aF/F-CRE-shSCR cells were extremely prone to form foci of transformation, down regulation of Dlk1 (p53KOZbtb7aF/F-CRE-shDlk1) or Sox9 (p53KOZbtb7aF/F-CRE-shSox9) in p53KOZbtb7aF/F-CRE cells significantly limited their oncogenic potential in vitro (Fig. 5B and Supplementary Fig. S5C). In order to analyze in vivo the possible implications of Dlk1 and Sox9 hyperactivity in the tumorigenic process induced by the absence of Lrf, we transplanted scaffolds containing CTR-shSCR, CRE-shSCR, CRE-shDlk1 or CRE-shSox9 cells subcutaneously in the flank of recipient mice, and then evaluated their tumorigenic potential. One month after implantation, 4/4 mice implanted with scaffolds containing p53KOZbtb7aF/F-CRE-shSCR cells, 4/5 mice implanted with scaffolds containing p53KOZbtb7aF/F-CRE-shDlk1 cells and 3/5 mice implanted with scaffolds containing p53KOZbtb7aF/F-CRE-shSox9 cells developed visible tumors, while no tumors were observed in mice implanted with scaffolds containing p53KOZbtb7aF/F-CTR-shSCR cells (0/4) (Fig. 5C). Although 80% of mice implanted with p53KOZbtb7aF/F-CRE-shDlk1 and 60% of mice implanted with p53KOZbtb7aF/F-CRE-shSox9 MSCs developed tumors, these lesions were significantly smaller than the p53KOZbtb7aF/F-CRE-shSCR tumors (Fig. 5D), and only a few transformed multinucleated cells were present within the scaffolds containing p53KOZbtb7aF/F-CRE-shDlk1 cells or p53KOZbtb7aF/F-CRE-shSox9 cells (Fig. 5E), revealing that both these two oncogenes participate to undifferentiated sarcoma formation from MSCs.
Figure 5.
Lrf plays a role as oncosuppressor gene in mesenchymal tumor through Dlk1 and Sox9. (A) Schematic overview of the experimental design is depicted on the left, while the growth curve of p53KOZbtb7aF/F-CTR-shSCR, p53KOZbtb7aF/F-CRE-shSCR, p53KOZbtb7aF/F-CRE-shDlk1 and p53KOZbtb7aF/F-CRE-shSox9 cells is shown on the right. Data show one representative experiment out of 3 independent biological replicates. (B) Detection of transformation status of p53KOZbtb7aF/F-CTR-shSCR, p53KOZbtb7aF/F-CRE-shSCR, p53KOZbtb7aF/F-CRE-shDlk1 and p53KOZbtb7aF/F-CRE-shSox9 cells. Pictures of the transformed foci are shown on the left, while their quantification on the right is presented as average of 3 biological independent replicates ± SEM. (C) Experimental design for in vivo sarcomagenesis is shown in the left panel, while the percentage of mice with tumor is shown on the right. (D) Representative pictures of tumors generated by p53KOZbtb7aF/F-CTR-shSCR, p53KOZbtb7aF/F-CRE-shSCR, p53KOZbtb7aF/F-CRE-shDlk1 and p53KOZbtb7aF/F-CRE-shSox9 cells implanted within scaffolds into recipient mice (n=4 CTR-shSCR, n=4 CRE-shSCR, n=5 CRE-shDlk1, n=5 CRE-shSox9) are shown on the left while the relative size of tumors is shown on the right (scaffold size used as control). (E) Representative H&E staining showing the morphology of collected tumors or mesenchymal cells on top of the scaffold. p53KOZbtb7aF/F-CTR cells were not able to generate tumors in vivo and only scaffolds within mesenchymal cells were recovered after transplantation, while p53KOZbtb7aF/F-CRE cells, p53KOZbtb7aF/F-CRE-shDlk1 and p53KOZbtb7aF/F-CRE-shSox9 cells originated undifferentiated sarcomas. (* indicates the scaffold. Scale bars: 20 μm). (F) Schematic overview of the genetic platform for discovering new oncopathways involved within the sarcomagenesis process, and new targeted-therapies (G) Schematic overview of Lrf involvement as tumor suppressor gene in undifferentiated sarcomas. Lrf/Dlk1 and Lrf/Sox9 pathways are examples of results obtained with the application of the genetic platform.
DISCUSSION
Adult MSCs have been proposed as the cell of origin of human undifferentiated sarcomas, one of the most aggressive and lethal soft tissue tumors (4–8). However, a comprehensive analysis of the molecular mechanisms dictating the onset and progression of undifferentiated sarcomas is still missing, consequently, limiting the generation of pre-clinical models faithfully recapitulating the human disease, and more importantly, the development of new and effective therapeutic options for this lethal tumor. A major current hurdle in studying and modeling the pathogenesis of sarcoma is represented by the need to deliver the appropriate genetic perturbation(s) to a specific putative cell of origin (19). Here, we employed primary adult mouse bone marrow MSCs sorted according to the expression of specific markers, and tested for stemness potential. By modifying their culturing condition, we have developed a new experimental ex vivo/in vivo platform to deconstruct the molecular genetics underlying the pathogenesis of undifferentiated sarcomas. Comparable findings were obtained using commercially available unsorted human MSC. It must be noted however that recent reports question the use of unsorted human MSCs collected from bone marrow and solely selected on their ability to adhere to culture dish for their inherent heterogeneity (3). Our approach nevertheless rests on the discovery that culturing mouse and human MSCs in hypoxic conditions prevents their spontaneous propensity to transform. As we recently reported, MSCs reside in bone marrow areas characterized by low oxygen concentration. Compared to other stromal cells within the endosteal bone marrow, MSCs express higher levels of HIF-1α and HIF-2α transcripts, showing a distinctive hypoxic profile (21). Therefore, the enforced switch from an anaerobic to an aerobic environment due to culturing in a 20% oxygen condition could determine excessive levels of reactive oxygen species (ROS) and, in turn, increasing amount of unrepaired DNA damage, prelude to genetic instability and neoplastic transformation. By preventing the spontaneous transformation of wild type as well as p53-null MSCs, this new approach will turn out to be very useful to: i) quickly asses the relevance of specific genetic alterations within the tumorigenesis process of MSCs; ii) characterize oncogenic or oncosuppressive functions and molecular pathways controlled by the newly identified genetic alterations; iii) to evaluate, pre-clinically, both in vitro and in vivo (in mice implanted with scaffolds), the efficacy of novel targeted therapies towards the eradication of this lethal disease (Fig. 5F). The relevance of this new approach is based on its ability to control the cell of origin of the disease, its simplicity and velocity. Furthermore, it can easily be coupled with the use of shRNA/over-expressing vectors/libraries in vitro, or the new developing system Crispr/Cas9 for genome editing of both coding and non-coding genes to be tested individually or in combination. Importantly, we demonstrate the utility of this discovery platform. Specifically, we have defined a role for LRF as tumor suppressor gene in adult MSCs and identified LRF as a key factor in the control of the early steps of adult MSC commitment. Additionally, we have characterized two evolutionary conserved oncosuppressive mechanisms regulated by LRF in adult MSCs: its ability to transcriptionally repress DLK1 expression, and to inhibit SOX9 activity (Fig. 5G). DLK1 is a trans-membrane protein that, once cleaved by enzyme TACE/ADAM17, releases the soluble factor FA-1 (fetal antigen-1) (51). Thus, the use of neutralizing antibodies against DLK1 (52), alone or in combination with inhibitors of SOX9 down-stream factors may offer a window of opportunity for the development of novel therapeutic strategies for this lethal form of cancer.
METHODS
Mice
Transgenic mice Zbtb7afl/fl were generated as described (34); p53KO mice were purchased from The Jackson Lab, and generated as described (53). All the experimental animals were kept in C57Bl/6J pure background. In some specific experiments immunodeficent mice were used (The Jackson Lab B6.Cg-Foxn1nu/J). Animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Cell Lines and human MSCs
3T3-L1 cells were purchased from ATCC (ATCC-CL-173) and cultured following the vendor direction. Cell lines were tested for mycoplasma (MycoAlert, Lonza), but not further authenticated. Human MSC were purchased from Lonza (PT-2501). According to the vendor cells are positive for CD105, CD166, CD29, and CD44, and test negative for CD14, CD34 and CD45. Cells were cultured and induced to differentiate following the vendor directions, but not further authenticated. Cells were maintained in culture with mesenchymal stem cell basal medium (Lonza, PT-3238) supplemented with growth factors (Lonza, PT-3001). During adipogenesis cells were cultured in adipogenic induction medium (Lonza, PT-3102B), followed by adipogenic maintenance medium (Lonza, PT-3102A), according to the vendor instructions.
Mesenchymal Stem Cells maintenance
MSCs were derived from C57BL/6 wild type, LrfF/F, and p53koLrfF/F mice. Long bones were collected, crushed and digested with collagenase II (1mg/ml) for 1 hour shaking at 37°C. Recovered cells were stained and FACS-sorted as: CD45−CD31−Ter119−Sca1+PDGFRα+, and cultured at the density of 1000 cells in a T25 flask. MSCs were cultured using complete MesenCult medium (STEMCELL Technologies) and maintained in humidified chamber with 5% CO2 and 1% O2, half medium was changed every 3 days. After 7 days in culture at 1% O2 cells formed visible CFU-F colonies, after this point cells were split once reached 80% of confluence.
Mesenchymal Stem Cells differentiation
MSCs were cultured using complete MesenCult medium (STEMCELL Technologies) and maintained in humidified chamber with 5% CO2 and 1% O2. For inducing the differentiation, the MesenCult medium was changed with specific medium for each differentiation (see below). During the differentiation process cells were maintained in regular oxygen concentration, 5% CO2 and 37°C. For adipocytes differentiation MSCs colonies were treated with StemXVivo™ Osteogenic/Adipogenic medium (R&D Systems, CCM007) plus adipogenic supplements (R&D Systems, CCM011). Medium was changed every other day for 7 days. Mature adipocytes were identified with Oil-Red-O (Sigma) following manufacturer procedure. Briefly, Oil-Red-O stock solution was prepared solubilizing Oil-red-O powder in isopropanol (0,35g/100ml isopropanol), stirring it over night. Cells were washed once in PBS and then and fixed with 10% formalin (Sigma) for 30 minutes at RT. The working Oil-Red-O solution was prepared mixing 3 parts of the stock solution with 2 parts of ddH2O. Fixed cells were washed once to remove the formalin with water and then treated 5 minutes with 60% isopropanol; then they were treated for 5 minutes with the working solution. After the treatment cells were washed with water to eliminate Oil-Red-O precipitates. For Osteocytes differentiation CFU-F forming MSCs were collected and re-plated at a density of 20.000 cells/well in a 12-wells plate. Once reached 40% confluence, cells were treated with StemXVivo Osteogenic/Adipogenic medium (R&D Systems, CCM007) together with osteogenic supplement (R&D Systems, CCM009). Medium was changed every three days for 20 days. Mature osteoblasts were stained with Leukocyte Alkaline Phosphatase kit (Sigma Cat. 85L3R) according to the manufacturer procedures. For chondrocytes differentiation 150.000 MSCs were pelleted in StemXVivo Chondrogenic medium (R&D Systems, CCM005) with chondrogenic Supplement (R&D Systems, CCM006). Tubes were then incubated at 37° C and 5% CO2 with loosen cap. Medium was changed every 3–4 days for 20 days. Micro-masses were then collected, embedded into paraffin, sectioned and stained with Toluidine Blue.
Flow cytometry
Cells were analyzed using LRSII (BD, Pharmingen) and sorted using FACS-ARIA II (BD, Pharmingen). The following antibodies were used: anti-CD45 FITC, anti-CD31 FITC, anti-Ter119 FITC, anti-Sca1 Pacific Blue, anti-PDGFRα PE (all purchased from Biolegend); Annexin-V PE (BD, pharmingen).
Immunofluorescence
Cells were fixed with 4% PFA for 10 minutes, washed with PBS and permeabilized with PBS, Triton-X100 0.2% for 10 minutes. Blocking before antibodies has been performed in PBS, Triton-X100 0.2% and 10% FBS for 30 minutes. Primary antibody p-histone H2AX (Cell Signaling) was incubated over night in blocking buffer, and an anti-Rabbit-488 was used as secondary antibody.
Immunohistochemistry and human tumor samples
A TMA (Tissue Microarray) of human fibrous tissue, fibromas, fibrosarcomas and Malignant fibrous histiocytomas was purchased from US Biomax, Inc. (SO2084). IHC was performed on 5-mm paraffin sections with the avidin-biotin-peroxidase method. The following primary antibody was used: LRF (Bethyl lab, cat # A300-549) 1:400 for human staining. Antigen retrieval was performed using citrate buffer. Samples were evaluated assessing the overall positivity of the neoplastic tissue and comparing it with normal fibrous tissue.
Luciferase assay
One day before transfection, cells were plated into 24-well plate at a density of 70–80%. The cells were transfected with the plasmids DNA (pGl3-Luc-Dlk1 promoter and pcDNA3-Lrf) and lipofectamine 2000 (Invitrogen) for 24 hours according to the manufacturer’s recommendation. 48 hours post-transfection, cells were lysed and analyzed for luciferase activity using the Dual-luciferase assay system (Promega). pRL-SV40-Renillar was used a control for transfection efficiency.
EMSA
Electrophoretic Mobility Shift Assay
For EMSA, 3T3L1 cells were resuspended in lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5% Nonidet P-40, 150 mM NaCl, 1 mM dithiothreitol, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitors). After 20 min on ice, extracts were centrifuged at 16000 × g for 20 min at 4 °C to remove cell debris. Protein concentration in supernatants was determined using Bio-Rad protein assay. A 26-mer DNA oligonucleotide containing the Dlk1 promoter sequence with putative Lrf binding site (5′-GGCTCGTCGGAGGGCTTCGGCTTTTC-3′) was end-labeled with 32P ((10 μM oligonucleotide, 1 μl kinase buffer 10 × (40 mM, Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM dithiothreitol), 2 μl ATP (ATP (γ-32P) (5,000 Ci mmol 1)) and 0.5 μl of the kinase (10 units μl 1)) and annealed to the complementary strand. For binding reactions, 30 μg of whole cell extract were added to gel shift buffer (20 mm HEPES pH 8, 25 mm KCl, 0.1 mm EDTA, 2 mm MgCl2, 0.5 mm dithiothreitol, 0.025% Nonidet P-40, 2 mm spermidine, 10% glycerol, 0.1 mg/ml acetylated bovine serum albumin, 120 ng of double-stranded poly(d[I-C])) containing the labeled oligonucleotide in a final volume of 30 μl. Reactions were incubated for 30 min at room temperature, and electrophoresed on a non-denaturing 4% polyacrylamide gel before autoradiography. Supershift was obtained by adding 500ng of anti-LRF antibody (13E9) to the binding reaction.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described (54) with the Magna ChIP G Chromatin Immunoprecipitation Kit (Millipore) using a hamster monoclonal antibody against Pokemon (LRF) (33). Primer sequences used for PCR were as follows: Dlk1_4,5_Fwd: cccagggacaggcagtaaggtt, Dlk1_4,5_Rev: ccaaacgcacaccacgaagat. 3T3L1 cells were crossed linked with formaldehyde for 5 minutes and terminated with 0.125M glycine. Cells were sonicated to generate chromatin with average size of 500bp. Monocloncal anti-LRF antibody were first incubated with a hamster bridging antibody (Jackson ImmunoResearch Laboratories), followed by addition of the 3T3L1 chromatin. Immunoprecipitated chromatin was assayed by quantitative PCR using the Dlk1-specific primers.
Viral vectors
Lentiviral vector expressing CRE, shCTR and shLRF, and retroviral vectors containing c-myc, and K-RasG12V were obtained from Addgene. Viral vectors of control as well as containing shRNA against Lrf, Pml, Pten, Dlk1 and Sox9 were obtained from open biosystem. Vectors containing IDH2R172K were cloned in our lab. All the viral particles were produced in transfecting 293T cells with packaging vectors of second generation.
Anchorage-independent cell growth
Soft agar colony formation assay was carried out seeding 1 × 104 MSCs in DMEM containing 0.4% low-melting agarose and 10% FCS. The cells were then plated in six wells plates previously coated with DMEM containing 1% of low-melting agarose and 10% FCS. The number of colonies was scored 3 weeks later, and quantification has been done using ImageJ.
Focus formation assay
MSCs were seeded at a concentration of 1x105 cell per well in a 6-wells plate and cultured for 10 days in complete medium (DMEM + 10% FBS) at 37°C. Once the formed visible foci cells were fixed with 10% formalin for 30 minutes, and the stained with crystal violet.
In vivo sarcomagenesis
As previously described, 3D scaffolds made with reticulated polycarbonate polyurethane urea matrix (Synthecon) were used (24). Briefly, scaffolds (5mm x 2mm) were put into the wells of a 96-wells plated, and seeded with MSCs at a concentration of 1x105 cells/scaffold. Wells were completely filled with 200μl of complete MesenCult medium, and left in culture (in regular oxygen concentration) for at least 6 hours. Scaffolds were washed with PBS in order to eliminate not attached cells, and implanted sub-cutaneoulsy into mice flanks. When recovered scaffolds/tumors were fixed in formalin and embedded into paraffin, sectioned and stained then with hematoxylin and eosin. Animal experiments were performed in accordance with the guidelines of the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Pathological analysis of the tumors has been performed at Dana-Farber Rodent Histopathology Core.
Western blot
For western blot, cell lysates were prepared with RIPA buffer. The following antibodies were used: hamster anti-Lrf clone 13E9, rabbit polyclonal anti-cPARP (Cell Signaling Cat. 9532), mouse polyclonal anti-β-actin (Sigma-aldrich), and mouse monoclonal anti-HSP90 (BD Biosciences).
Senescence detection
MSCs were cultured in hypoxia, before and for 4 days after the transduction. After the transduction cells were collected and seeded 10 x 105 cells/well in a 12-well-plate. Cells were then treated with SA-β-galactosidase over night. Senescence was detected using senescence detection kit (Calbiochem) following the manufacturer protocol.
Plasmids
The entire Dlk1 mouse promoter sequence was obtained by PCR from a BAC clone. FW (5′ ccgagctcgggagtgccatttcatttaa 3′) and RV (5′ ccgctagcaaagccagcaggagcaagag 3′) primers were used. The fragment was cloned into pGL3-Luc enhancer (Promega) in SacI and NheI sites. All the mutants were obtained by using the Quickchange mutagenesis kit (Stratagene) and confirmed by sequencing. The mutated version of this plasmid was generated by utilizing the Dlk1 3′UTR as template and modifying the putative Lrf binding sites using the QuikChange II XL Site-Directed Mutagenesis Kit.
The mutagenic primers used were:
site Mutant 1AB forward 5′-ggaagggaaaatggagtctagagacggggagagactcacctcactagtctggg-3′
reverse 5′-ccttcccttttacctcagatctctgcccctctctgagtggagtgatcagaccc-3′;
site Mutant 2 forward 5′-gggctcacctcactagtctagagttccttgtactctatgtgcccc-3′
reverse 5′-cccgagtggagtgatcagatctcaaggaacatgagatacacgggg-3′;
site Mutant 3 forward 5′-ggagcccttatctcaggaatctagccccaagatcctctc-3′
reverse 5′-cctcgggaatagagtccttagatcggggttctaggagag-3′;
site Mutant 4 forward 5′-tgtgccgaaaggtgtgtttgggttagagattcgtggggcaagtgc-3′
reverse 5′-acacggctttccacacaaacccaatctctaagcaccccgttcacg-3′;
site Mutant 5 forward 5′-caatggcaaggctcgtcgaaagacctcggcttttcgtggtggt-3′
reverse 5′-gttaccgttccgagcagctttctggagccgaaaagcaccacca-3′.
RNA extraction and RT-PCR
Cellular RNA was extracted using Quick-RNA MicroPrep (Zymo Research, R1050) and then retro-transcribed to cDNA using iScript cDNA Synthesis kit (Bio-Rad, 170-8890). All the analyzed mRNA was detected using TaqMan FAM-conjugated probes (Applied Biosystems). Each target was run in triplicate and expression level was normalized to mouse β2-microglobulin. Details are provided in Supplemental Experimental Procedures.
Taq-Man RT-PCR probes
All the probes were purchased from Applied Biosystems.
β2-microglobulin: Mm00437764_m1
Sox9: Mm00448840_m1
Mia: Mm00444563_m1
Col2: Mm01309565_m1
Col9: Mm00483836_m1
Col10: Mm00487041_m1
c/ebpδ: Mm00786711_s1
Runx2: Mm00501584_m1
Fabp4: Mm00445878_m1
PPARγ: Mm01184322_m1
ALP: Mm00475834_m1
Osteocalcin (OC): Mm00649782_gH
Zbtb7a: Mm00657132_m1 and ZBTB7A: Hs00792219_m1
Dlk1: Mm00494477_m1
Hes1: Mm01342805_m1
Hey1: Mm00468865_m1
p21: Mm04205640_g1
MIA: Hs00197954_m1
H19: Hs00262142_g1
LRF: Hs00252415_s1
DLK1: Hs00171584_m1
FABP4: Hs01086177_m1
Statistic
Data were analyzed using unpaired t-test (GraphPad Prism, GraphPad Software, Inc.). Values of P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001 (t-test).
Supplementary Material
SIGNIFICANCE.
The paucity of therapeutic options for the treatment of sarcoma calls for a rapid and effective preclinical assessment of new therapeutic modalities. We have here developed a new platform to deconstruct the molecular genetics underlying the pathogenesis of sarcoma and to evaluate in vivo the efficacy of novel targeted therapies.
Acknowledgments
We thank Thomas Garvey for insightful editing and all members of the Pandolfi lab for critical discussion. We thank Dennis M. Bonal for the IHC staining.
GRANT SUPPORT
Jlenia Guarnerio has been granted leave of absence from the University Vita-Salute San Raffaele, Milan and she was funded partially by a fellowship from the University Vita-Salute San Raffaele, and partially by a grant from the Fondazione Cariplo to Rosa Bernardi and Pier Paolo Pandolfi. Andrea Lunardi has been supported in part by a fellowship from the Istituto Toscano Tumori (ITT, Italy). Riccardo Taulli has been granted leave of absence from the Department of Oncology, University of Turin School of Medicine. This work has been supported through the NIH grant (R01 CA102142-7) to Pier Paolo Pandolfi.
Footnotes
The authors disclose no potential conflicts of interest.
AUTHOR’s CONTRIBUTIONS
Conception and design: J. Guarnerio, L. Riccardi A. Lunardi, P.P. Pandolfi
Development of methodology: J. Guarnerio
Analysis and interpretation of data: j. Guarnerio, L. Riccardi, R. Taulli, R.T. Bronson, A. Lunardi, P.P. Pandolfi
Writing, review, and/or revision of the manuscript: J. Guarnerio, A. Lunardi, P.P. Pandolfi
Administrative, technical, or material support: T. Maeda, L. Riccardi, G. Wang, R. M. Hobbs, M. S. Song, P. Sportoletti, R. Bernardi
Provided histology and immunohistochemistry: J. Guarnerio, R.T. Bronson, M. Castillo-Martin, C. Cordon-Cardo
References
- 1.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 2.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8(9):726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 3.Bianco P, Cao X, Frenette PS, Mao JJ, Robey PG, Simmons PJ, et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19(1):35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolar J, Nauta AJ, Osborn MJ, Panoskaltsis Mortari A, McElmurry RT, et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007;25(2):371–379. doi: 10.1634/stemcells.2005-0620. [DOI] [PubMed] [Google Scholar]
- 5.Choi J, Curtis SJ, Roy DM, Flesken-Nikitin A, Nikitin AY. Local mesenchymal stem/progenitor cells are a preferential target for initiation of adult soft tissue sarcomas associated with p53 and Rb deficiency. Am J Pathol. 2010;177(5):2645–2658. doi: 10.2353/ajpath.2010.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubio R, Garcia-Castro J, Gutierrez-Aranda I, Paramio J, Santos M, Catalina P, et al. Deficiency in p53 but not retinoblastoma induces the transformation of mesenchymal stem cells in vitro and initiates leiomyosarcoma in vivo. Cancer Res. 2010;70(10):4185–4194. doi: 10.1158/0008-5472.CAN-09-4640. [DOI] [PubMed] [Google Scholar]
- 7.Mohseny AB, Hogendoorn PC. Concise review: mesenchymal tumors: when stem cells go mad. Stem Cells. 2011;29(3):397–403. doi: 10.1002/stem.596. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez R, Rubio R, Menendez P. Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res. 2012;22(1):62–77. doi: 10.1038/cr.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin PP, Pandey MK, Jin F, Raymond AK, Akiyama H, Lozano G. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogenesis. 2009;30(10):1789–1795. doi: 10.1093/carcin/bgp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22(12):1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466(7310):1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao J, Jiang MM, Jiang L, Salvo JS, Zeng HC, Dawson B, et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26(3):390–401. doi: 10.1016/j.ccr.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27(18):1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura M, Miura Y, Padilla-Nash HM, Molinolo AA, Fu B, Patel V, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells. 2006;24(4):1095–1103. doi: 10.1634/stemcells.2005-0403. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Huso DL, Harrington J, Kellner J, Jeong DK, Turney J, et al. Outgrowth of a transformed cell population derived from normal human BM mesenchymal stem cell culture. Cytotherapy. 2005;7(6):509–519. doi: 10.1080/14653240500363216. [DOI] [PubMed] [Google Scholar]
- 16.Funes JM, Quintero M, Henderson S, Martinez D, Qureshi U, Westwood C, et al. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc Natl Acad Sci U S A. 2007;104(15):6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez R, Tornin J, Suarez C, Astudillo A, Rubio R, Yauk C, et al. Expression of FUS-CHOP fusion protein in immortalized/transformed human mesenchymal stem cells drives mixoid liposarcoma formation. Stem Cells. 2013;31(10):2061–2072. doi: 10.1002/stem.1472. [DOI] [PubMed] [Google Scholar]
- 18.Frenette PS, Pinho S, Lucas D, Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu Rev Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- 19.Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA, et al. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117(11):3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa S, Mabuchi Y, Kubota Y, Nagai Y, Niibe K, Hiratsu E, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med. 2009;206(11):2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarnerio J, Coltella N, Ala U, Tonon G, Pandolfi PP, Bernardi R. Bone Marrow Endosteal Mesenchymal Progenitors Depend on HIF Factors for Maintenance and Regulation of Hematopoiesis. Stem Cell Reports. 2014;2(6):794–809. doi: 10.1016/j.stemcr.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou YF, Bosch-Marce M, Okuyama H, Krishnamachary B, Kimura H, Zhang L, et al. Spontaneous transformation of cultured mouse bone marrow-derived stromal cells. Cancer Res. 2006;66(22):10849–10854. doi: 10.1158/0008-5472.CAN-06-2146. [DOI] [PubMed] [Google Scholar]
- 23.Xiao W, Mohseny AB, Hogendoorn PC, Cleton-Jansen AM. Mesenchymal stem cell transformation and sarcoma genesis. Clin Sarcoma Res. 2013;3(1):10. doi: 10.1186/2045-3329-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaiselbuh SR, Edelman M, Lipton JM, Liu JM. Ectopic human mesenchymal stem cell-coated scaffolds in NOD/SCID mice: an in vivo model of the leukemia niche. Tissue Eng Part C Methods. 2010;16(6):1523–1531. doi: 10.1089/ten.tec.2010.0179. [DOI] [PubMed] [Google Scholar]
- 25.Fenech M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today. 2002;7(22):1128–1137. doi: 10.1016/s1359-6446(02)02502-3. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu T, Ishikawa T, Sugihara E, Kuninaka S, Miyamoto T, Mabuchi Y, et al. c-MYC overexpression with loss of Ink4a/Arf transforms bone marrow stromal cells into osteosarcoma accompanied by loss of adipogenesis. Oncogene. 2010;29(42):5687–5699. doi: 10.1038/onc.2010.312. [DOI] [PubMed] [Google Scholar]
- 27.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13(8):992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 28.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42(8):715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren YX, Finckenstein FG, Abdueva DA, Shahbazian V, Chung B, Weinberg KI, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68(16):6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez R, Rubio R, Gutierrez-Aranda I, Melen GJ, Elosua C, Garcia-Castro J, et al. FUS-CHOP fusion protein expression coupled to p53 deficiency induces liposarcoma in mouse but not in human adipose-derived mesenchymal stem/stromal cells. Stem Cells. 2011;29(2):179–192. doi: 10.1002/stem.571. [DOI] [PubMed] [Google Scholar]
- 31.Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP. Role of LRF/Pokemon in lineage fate decisions. Blood. 2013;121(15):2845–2853. doi: 10.1182/blood-2012-11-292037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433(7023):278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 34.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316(5826):860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3(12):895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 36.Espina AG, Mendez-Vidal C, Moreno-Mateos MA, Saez C, Romero-Franco A, Japon MA, et al. Induction of Dlk1 by PTTG1 inhibits adipocyte differentiation and correlates with malignant transformation. Mol Biol Cell. 2009;20(14):3353–3362. doi: 10.1091/mbc.E08-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res. 2010;16(17):4363–4373. doi: 10.1158/1078-0432.CCR-10-0138. [DOI] [PubMed] [Google Scholar]
- 38.Yanai H, Nakamura K, Hijioka S, Kamei A, Ikari T, Ishikawa Y, et al. Dlk-1, a cell surface antigen on foetal hepatic stem/progenitor cells, is expressed in hepatocellular, colon, pancreas and breast carcinomas at a high frequency. J Biochem. 2010;148(1):85–92. doi: 10.1093/jb/mvq034. [DOI] [PubMed] [Google Scholar]
- 39.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72(5):1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang GL. A. Lrf suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Gen. 2013 doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 2009;9(3):287–302. doi: 10.1016/j.cmet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Lunardi A, Zhang J, Chen Z, Ala U, Webster KA, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013;45(7):739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu CJ, Zhang Y, Xu K, Parsons D, Alfonso D, Di Cesare PE. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front Biosci. 2007;12:3899–3910. doi: 10.2741/2359. [DOI] [PubMed] [Google Scholar]
- 44.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14(5):757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 45.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17(4):2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73(4):725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 47.Smas CM, Chen L, Zhao L, Latasa MJ, Sul HS. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J Biol Chem. 1999;274(18):12632–12641. doi: 10.1074/jbc.274.18.12632. [DOI] [PubMed] [Google Scholar]
- 48.Kim KA, Kim JH, Wang Y, Sul HS. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007;27(6):2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nueda ML, Garcia-Ramirez JJ, Laborda J, Baladron V. dlk1 specifically interacts with insulin-like growth factor binding protein 1 to modulate adipogenesis of 3T3-L1 cells. J Mol Biol. 2008;379(3):428–442. doi: 10.1016/j.jmb.2008.03.070. [DOI] [PubMed] [Google Scholar]
- 50.Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, Kassem M. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19(5):841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Sul HS. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor alpha converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol Cell Biol. 2006;26(14):5421–5435. doi: 10.1128/MCB.02437-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu NL, Asahina K, Wang J, Ueno A, Lazaro R, Miyaoka Y, et al. Hepatic stellate cell-derived delta-like homolog 1 (DLK1) protein in liver regeneration. J Biol Chem. 2012;287(13):10355–10367. doi: 10.1074/jbc.M111.312751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 54.Maeda T, Ito K, Merghoub T, Poliseno L, Hobbs RM, Wang G, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17(4):527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.