Significance
Amorphous silica (SiO2) phases produced by plants are principal mass fluxes in the global silica cycle. The study of silica biomineralization in plants has largely focused on angiosperms, leaving open questions about its early evolution. To address the effect of early plants on the silica cycle, we measured the silica contents of extant members of plant groups known from fossils to have been major components of the terrestrial landscape in the past, as grasses are today. Most of these early-diverging plant lineages accumulate substantial amounts of silica. We compared these observations with the distribution and evolution of plant silica transport proteins, suggesting convergent evolution of silica use. Results presented here outline an extensive evolutionary history of silica biomineralization in plants.
Keywords: phytolith, fern, lycophyte, silicon, aquaporin
Abstract
Biomineralization plays a fundamental role in the global silicon cycle. Grasses are known to mobilize significant quantities of Si in the form of silica biominerals and dominate the terrestrial realm today, but they have relatively recent origins and only rose to taxonomic and ecological prominence within the Cenozoic Era. This raises questions regarding when and how the biological silica cycle evolved. To address these questions, we examined silica abundances of extant members of early-diverging land plant clades, which show that silica biomineralization is widespread across terrestrial plant linages. Particularly high silica abundances are observed in lycophytes and early-diverging ferns. However, silica biomineralization is rare within later-evolving gymnosperms, implying a complex evolutionary history within the seed plants. Electron microscopy and X-ray spectroscopy show that the most common silica-mineralized tissues include the vascular system, epidermal cells, and stomata, which is consistent with the hypothesis that biomineralization in plants is frequently coupled to transpiration. Furthermore, sequence, phylogenetic, and structural analysis of nodulin 26-like intrinsic proteins from diverse plant genomes points to a plastic and ancient capacity for silica accumulation within terrestrial plants. The integration of these two comparative biology approaches demonstrates that silica biomineralization has been an important process for land plants over the course of their >400 My evolutionary history.
In modern ecosystems, land plants play a major role in the silica cycle through the accumulation and synthesis of amorphous biominerals composed of SiO2, known as phytoliths or silica bodies. It is widely appreciated that actively accumulating plants such as grasses are important components of the terrestrial biological pump of silica (1–3). Plant silica also plays a key role in connecting the terrestrial and marine carbon cycles, because silica is an important nutrient for marine silica-biomineralizing primary producers (i.e., diatoms) (1, 2, 4–7). However, both grasses and diatoms evolved in the latter part of the Mesozoic Era (8–10) and rose to ecological dominance within the Cenozoic Era (6, 9, 11–14). Determining precisely when and how the terrestrial–marine silica teleconnections evolved remains an obstacle to reconstructing the history of the silica cycle.
Direct analysis of silica bodies in the fossil record provides limited insight into this problem. When fossiliferous material is macerated, it is often challenging to identify whether residual silica bodies are the result of primary biomineralization or secondary diagenetic processes, and if a living plant origin is suspected, it is often difficult to assign taxonomic identity to the phytolith producer. In addition, with rare exceptions (e.g., ref. 15), lagerstätten that preserve exceptional anatomical detail in fossils, and might therefore be expected to preserve silica bodies, tend to be oversaturated with respect to silica (e.g., ref. 16) or extremely undersaturated with respect to silica (e.g., refs. 17, 18). To account for this, efforts to understand the history of silica biomineralization in terrestrial plants have taken a comparative biology approach (1, 5).
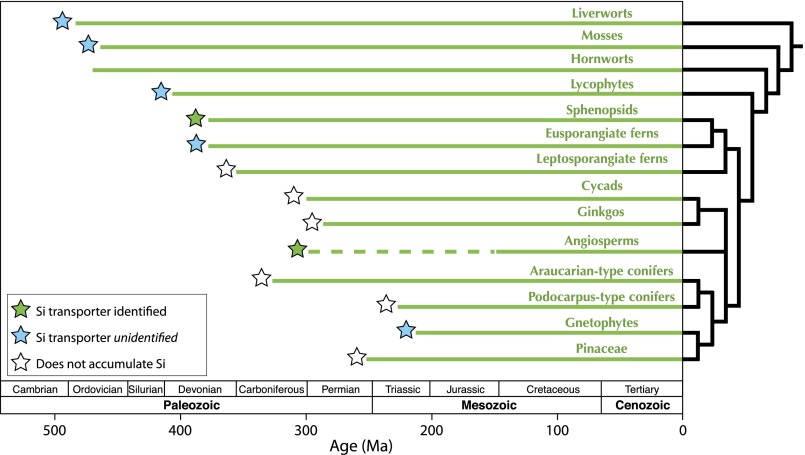
Silica is widely used within plants for structural support and pathogen defense (19–21), but it remains a poorly understood aspect of plant biology. Recent work on the angiosperm Oryza sativa demonstrated that silica accumulation is facilitated by transmembrane proteins expressed in root cells (21–24). Phylogenetic analysis revealed that these silicon transport proteins were derived from a diverse family of modified aquaporins that include arsenite and glycerol transporters (19, 21, 25, 26). A different member of this aquaporin family was recently identified that enables silica uptake in the horsetail Equisetum, an early-diverging fern known to accumulate substantial amounts of silica (27). However, despite a growing number of fully sequenced genomes, angiosperm-type silicon transporters are not found within the gymnosperms or in spore-bearing plants, including plant lineages that are known to contain many weight-percent silica (25, 28) (Fig. 1). A more complete understanding of the distribution and mechanisms of silica accumulation within these early-diverging lineages is a necessary precondition for assessing the evolutionary history of silica biomineralization in terrestrial plants.
Fig. 1.
Stratigraphic ranges (42) and evolutionary relationships (59–61) between major terrestrial plant lineages. Although the angiosperm macrofossil record only extends to the Early Cretaceous Period (62), a strict interpretation of their position as sister group to all other seed plant clades implies an earlier origin, shown here with a dashed line (60, 63). For the purposes of this article, we define Araucarian-type conifers as Araucariaceae and extinct relatives, and Podocarpus-type conifers as Podocarpaceae and extinct relatives. Filled stars mark clades that accumulate >1 wt % silica (dry matter), color coded for identified and unidentified silicon transport proteins.
Results and Discussion
We measured SiO2 content within and across a diverse set of terrestrial plants (88 different plants from 23 families) collected in southern California, with a focus on lesser-studied lineages with long fossil records. Silica content was assessed gravimetrically on bulk above-ground plant tissues, using a modified dry ashing technique, and the resulting silica bodies were imaged using scanning electron microscopy and microscale energy dispersive spectroscopy (Methods). We combined these results with previously published observations (20–22, 29) to build a coherent picture of silica biomineralization in land plants (Fig. 2).
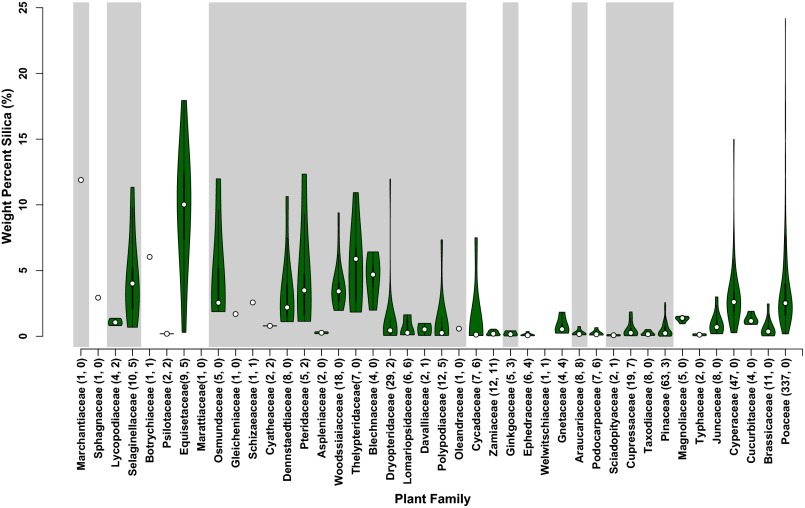
Fig. 2.
Violin plots of silica abundance in terrestrial plant families (white dots are medians; top and bottom of the thick bar mark first and third quartile ranges, respectively; green fill shows kernel density estimates; n = 688). After the family name in parentheses is the number of total analyses within that group, followed by the number of analyses in that group from this study. Families are arranged from left to right in rough order of evolutionary divergence. For clarity, we did not display data from several angiosperm families that are not known to accumulate silica. Conifers, and more recently diverging fern clades (e.g., Polypodiaceae), have the lowest medians, whereas liverworts, mosses, lycophytes, and eusporangiate ferns have higher weight percentage silica (dry matter), with median values that approach or exceed those found in grasses (Poaceae) and sedges (Cyperaceae).
The observed pattern of silica abundance among extant plants (Figs. 1 and 2) implies a protracted evolutionary history of silica biomineralization and indicates that many plant groups with long fossil records precipitate substantial amounts of silica. Accumulation of silica is widespread among diverse land plant families, and variance within groups is also high. Consistent with previous work, plants with high silica concentrations include members of the monocots, specifically grasses and sedges (20, 29). However, we also observed that many members of early-diverging lineages (e.g., Sellaginellacae, Equisetaceae, Marattiacaeae, and Osmundaceae; toward the left of Fig. 2) contain as much or greater amounts of silica than the grasses and sedges (20). The only groups that show consistently low silica abundances are found in the gymnosperms, including the conifers, ginkgo, and many cycads. Exceptions are Gnetum gnemon and Cycas revoluta, which have greater than 1% dry weight silica. Beyond gnetophytes and cycads, however, there is a general paucity of silica in gymnosperms, suggesting this form of biomineralization is not an important feature of their biology (23, 29, 30).
In addition, the evolution of seed plants must then require either multiple gains or losses of silica biomineralization. The hypothesis that some lineages of seed plants (Araucarian-type conifers, Podocarpus-type conifers, Pinaceae) have lost biomineralization capacity is possible (20, 21); however, the observation that two gnetophytes, Gnetum and Ephedra, each accumulate silica and contain silicified cell walls (Figs. 2 and 3) complicates this scenario. This distribution of silica abundance either implies a secondary gain of biomineralization within the gnetophytes, and a loss in the last common ancestor of all gymnosperms or several independent losses of Si accumulation within gymnosperms. We use this hypothetical framework to evaluate evolution of the molecular mechanisms of silica accumulation in terrestrial plants.
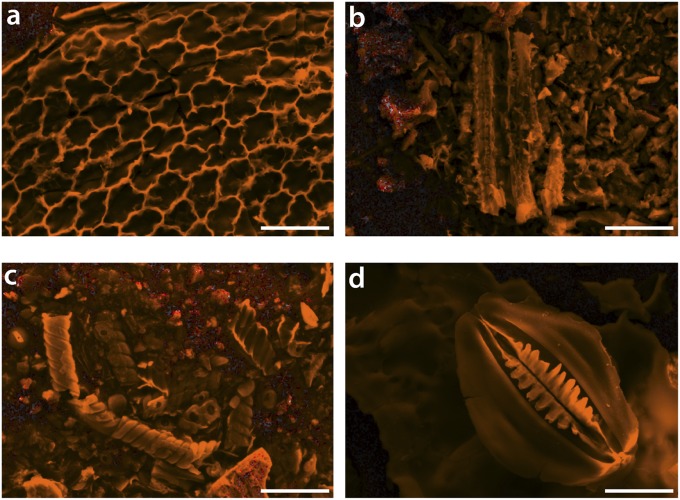
Fig. 3.
Secondary electron images of silica bodies (grayscale) overlaid with Si maps from energy-dispersive X-ray spectroscopy (orange). (Scale bars, 25 μm.) (A) Selaginella sp., (B) Gnetum gnemon, (C) Ephedra californicum, and (D) Equisetum hyemale. Some distinct mineralized plant tissues can be recognized: in A, epidermal cell walls are silicified; in C, possible silicified vascular tissue; and in D, a silicified stomatal complex. It is noteworthy that these tissues are all near the sites of transpiration.
The most well characterized means by which plants accumulate silicic acid from soil water is via transmembrane proteins with selective pores that belong to a plant-specific subfamily of the aquaporins termed nodulin 26-like intrinsic proteins (NIPs) (25, 31–33). Our observations from electron microscopy and spectroscopy confirmed the presence of silicified cell wall structures in diverse taxa, including Equisetum, Selaginella, and Gnetum species (Fig. 3). Where we can resolve anatomical structures in the SiO2 residues, the most heavily biomineralized structures are parts of the vascular system, epidermal cells, and stomata. This is consistent with the hypothesis of a transpiration-driven transport process in these plant groups, in which silicic acid is assimilated by roots and is subsequently deposited as silica bodies throughout the plant via distillation (21, 23).
The NIPs can be subdivided into three groups on the basis of phylogenetic relationships (31, 34) (Fig. 4A). Amino acid residues that surround the narrowest portion of the pore confer a selectivity filter responsible for the exclusion of larger molecules, termed the aromatic/arginine filter (Ar/R filter or gate) (25, 35). NIPI and NIPII groups are thought to be responsible for the movement of a range of solutes, including arsenite and glycerol (36). Of the three major lineages of the NIP proteins, selective transport of orthosilicic acid has been demonstrated in members of the NIPIII (Lsi1) and the NIPII (Fig. 4A, arrows) groups, where the presence of a relatively large aperture at the Ar/R filter is thought to permit the passage of silicic acid compared with smaller constrictions in other NIPs that would only allow smaller solutes passage (21, 22, 24, 27, 37).
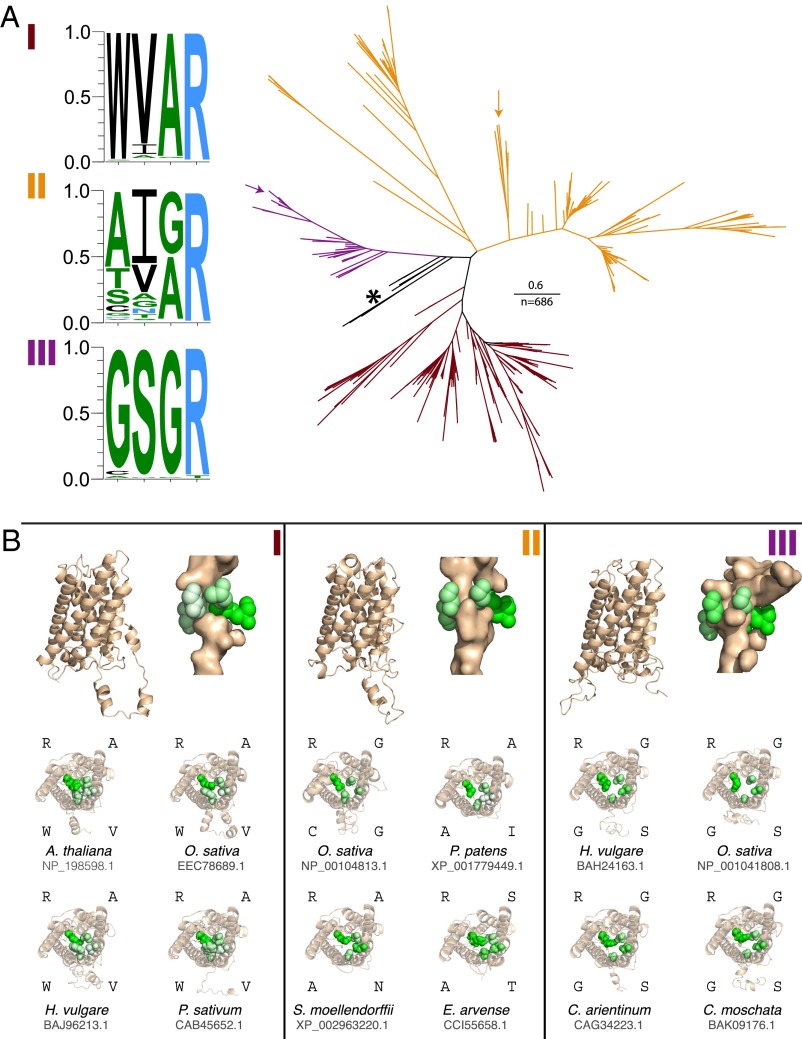
Fig. 4.
Phylogeny and predicted structures of NIP clades. (A) Phylogenic tree of major NIP clades with NIPI (maroon), NIPII (orange), and NIPIII (purple). The frequency of amino acid occurrence at the Ar/R filter are displayed for the NIPI, NIPII, and NIPIII groups as hydrophilic = RKDENQ (blue), neutral = SGHTAP (green), and hydrophobic = YVMCLFIW (black). The maximum of each scale is 1.0, or 100% probability. Unclassified NIP groups with conserved Ar/R residues FAAR are indicated by an asterisk. Verified silica transporters include the NIPIIIs and some members of the NIPIIs, shown here with arrows. (B) Structural models of four representatives of each of the NIP subgroups. Pore-lining residues are identified with PoreWalker. Residues of the Ar/R filter are colored by hydrophobicity on a green scale (white: hydrophobic; green: hydrophilic). A longitudinal transmembrane view of three Orzya sativa representatives of the NIPs (NIPI, NIPII, and NIPIII) are depicted at the top of each panel. Below is the same pore type as above, with three additional ribbon diagrams rotated to show a transverse view of the pore and four Ar/R gate residues.
To evaluate the distribution and evolution of silica transport biochemistry in land plants, we constructed a phylogenetic tree and built structural models of key members from all three NIP subgroups (Methods). The phylogenetic analyses recover the expected salient relationships between the NIP subgroups with ∼25 times more sequence data then previous reports (25, 31, 33). Results show a complex pattern of functional evolution. Two of the three NIP subgroups have highly conserved residues at Ar/R gate positions (Fig. 4A). NIPI is predominantly WVAR (Fig. 4A, maroon). Nearly all NIPIII members display GSGR (Fig. 4A, purple), with the exceptions of a CSGR-bearing homolog in the Cucurbitaceae (Cucumis melo and Cucumis sativus), in which silicon transporters were bred out for rind softening (25, 38), and also in the string bean Phaseolus vulgaris, which has a single NIPIII homolog with ASGR, and in Eucalyptus grandis, which contains a homolog coding for GSPT at the Ar/R gate position. In contrast, NIPII is highly diverse (Fig. 4A, orange). The earliest diverging NIPs are found in the moss Physcomitrella patens, the lycophyte Selaginella moellendorffii, and the fern Adiantum capillus-verneris (Fig. 4A, asterisk). Sister to these are bacterial NIP-like major intrinsic proteins (MIPs) (39) represented here by sequences from Ktedonobacter racemifer and Nitrolancea hollandica, both members of the Chloroflexi. Both these bacterial NIP-like MIPs and the early diverging plant NIPs display the Ar/R gate residues FAAR (or NNAR, in the case of the Selaginella moellendorffii homolog XP_002986711.1). Proteins with this motif have not been studied in vivo, but the prevalence of FAAR residues suggests that the last common ancestor to the plant NIPs may have had conserved function. NIPIIIs form a clade derived from NIPs with the FAAR motif and are only found in angiosperms. Based upon their conserved Ar/R filter, NIPIIIs facilitate silicic acid uptake (21, 25) (Fig. 4). NIPIs form a diverse clade, but with conserved pore residues, and presumably function as water, glycerol, and lactic acid transporters (25). In contrast, NIPIIs are not only diverse but also show extreme sequence diversity at the Ar/R gate. Our structural models (Fig. 4B) are consistent with the hypothesis that they may transport a range of larger molecules (25). Included in the NIPIIs are a group of recently identified, highly efficient (twice the silicic acid conductance of Lsi1) silicic acid transporters with the previously unreported Ar/R gate residues STAR from the horsetail Equisetum arvense, demonstrating that porins facilitating silicic acid transport have evolved at least twice in plants (27). Notably, the model structure of the ANAR porins from Selaginella moellendorffii has similar size and chemistry to both the STAR porin found in Equisetum arvense and the NIPIIIs (Fig. 4B).
A reasonable evolutionary scenario that satisfies both biochemical and empirical silicic acid abundance data begins with the evolution of NIP-like proteins with an Ar/R conformation of FAAR in bacteria from an ancestral aquaporin, followed by horizontal gene transfer into early terrestrial plants, resulting in the FAAR NIPs found in mosses. The ancestral NIPs subsequently diversified into the NIPI and NIPII clades found throughout land plants, including the functional diversity of pore residues found in the NIPIIs, at least some of which enable selective silicic acid uptake (STAR porin). Despite many fully sequenced genomes, NIPIIs are rare in gymnosperms. It is possible that silica biomineralization was lost in the last common ancestor of seed plants, and angiosperm NIPIIIs constitute a secondary gain of function (25, 39), with gnetophyte silica biomineralization currently unresolved, awaiting further molecular data. The NIP phylogeny implies an adaptive radiation of metalloid (including silicic acid) transport early within land plants (25) and is consistent with our observations of silica biomineralization in early-diverging lineages (34).
Summary
In order of appearance, major players in the terrestrial silica cycle include some bryophytes (liverworts), lycophytes, and early-diverging vascular plants (horsetails, eusporangiate ferns), followed by gnetophytes and grasses. Terrestrial plant lineages with roots in the Paleozoic Era, including lycophytes, horsetails, and ferns, accumulate silica at abundances comparable to or exceeding many siliceous angiosperm lineages. Combining our results with stratigraphic ranges of silica-biomineralizing plants from the fossil record, we hypothesize that a terrestrial silica cycle must have developed no later than the time of the Rhynie Chert, which contains fossilized stem group bryophytes and vascular plants (411–407 Ma, early Devonian Period) (40–42). As a consequence, plants may have had a significant effect on the terrestrial silica cycle throughout the Middle and Late Paleozoic Era, with much of the fluxes cycled through lycophytes, horsetails, and early-diverging lineages of ferns that dominated terrestrial ecosystems at this time. A decrease in continental accumulation of silica may have followed throughout the Mesozoic Era as a consequence of the radiation of conifers, perhaps with some modest silica accumulation in gnetophytes and cycads. Finally, large-scale changes in the terrestrial silica cycle likely occurred with the rise of grasslands late in the Cenozoic Era (11–13, 43).
Methods
Bulk Plant Silica Analysis Using Dry Ashing.
Samples were collected in and around Southern California. Source locations include: the Caltech grounds; The Huntington Library, Art Collections, and Botanical Gardens; and Rancho Santa Ana Botanic Garden; the private collection of Loran M. Whitelock; and commercial sources. Plant material (∼1 g wet weight leaf, sporophyte, or photosynthetic surface sample) was rinsed and dried and then combusted at 500 °C (44). The sample ashes were subsequently washed in 10% (vol/vol) HCl at 70 °C, incubated again in 15% (vol/vol) H2O2 at 70 °C, and dehydrated in ethanol. Final SiO2 masses are presented as percentage initial sample dry weight. Typical uncertainty of the dry ashing method is less than 0.1 wt % SiO2 (45, 46), which is much smaller than the natural variation between tissues of a single plant (e.g., ref. 30). Previously published silica abundance data whose primary sources could be verified (e.g., ref. 29) were combined with our results. The complete list of all silica abundances are reported as SiO2 wt % in Dataset S1.
Imaging and Elemental Mapping of Silica Bodies.
A representative subset of washed ash powders was selected for imaging and elemental analysis via electron microscopy and energy dispersive spectroscopy. Samples were pressed gently on to a carbon tape-coated SEM stub and either carbon or palladium sputter-coated and then imaged with a Zeiss 1550 VP Field Emission Scanning Electron Microscope to observe microstructures. Chemistry was also confirmed by creating spectral element maps with an Oxford INCA Energy 300 X-ray Energy Dispersive Spectrometer system.
NIP Phylogeny and Structure Prediction.
Sequences were collected from the National Center for Biotechnology Information (NCBI) nr/nt database using the NIP homolog XP_002986711.1 as a query. A thousand sequences were retrieved and aligned and manipulated with Jalview (47) and CLUSTALO (48), with a full distance matrix for each iteration and 10 iterations. The alignment was manually trimmed to obtain an alignment block, and a tree was constructed with Fasttree (49). This first tree was used to identify the NIP group. NIP sequences were then collected along with three closely related bacterial homologs, leaving 686 unique NIP protein sequences (Dataset S2). These were then realigned with CLUSTALW (50), using default gap extension and opening penalties, and the Gonnet substitution matrix. Prottest (51) was then used to identify an appropriate evolutionary model for tree construction. Fasttree was again used to construct a tree, and this tree was then used as a starting tree for optimization by PhyML (52). The tree (Dataset S3) was constructed with JTT+I+G, 8 rate substitution categories, the best of NNIs and SPRs, and aBayes was used to evaluate branch supports. From this phylogeny of NIP proteins, each lineage (I, II, and III) was analyzed for sequence conservation, using WebLogo3 (53). Residues in the NIP Ar/R filter region were then selected to display diversity at these positions. A representative subset of NIPI, NIPII, and NIPIII sequences was selected for structure prediction to visualize pore geometries (Dataset S4). Models of NIP homologs were generated through sequence submission to the iterative threading assembly refinement (I-TASSER) server (54–56). The top model based on C-score was selected for further analysis (57). One of each NIP type was analyzed, using PoreWalker (58) to identify pore-lining residues from the modeled structures and observe constriction at the Ar/R gate. All structures were visualized using PyMol.
Supplementary Material
Acknowledgments
We thank George Rossman and Victoria Orphan for laboratory equipment, Chi Ma for assistance with electron microscopy and spectroscopy, and Sean Lahmeyer (The Huntington Library, Art Collections, and Botanical Gardens), Loran M. Whitelock (Eagle Rock, CA), and Lucinda McDade (Rancho Santa Ana Botanic Garden) for aid in sample collection. This project was partially supported by an OK Earl Postdoctoral Scholarship (Caltech) and a San Andreas Visiting Fellowship to The Huntington Library, Art Collections, and Botanical Gardens (J.P.W.). S.E.M. was partially supported by the Agouron Institute as a Geobiology Fellow. W.W.F. acknowledges support from the Agouron Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500289112/-/DCSupplemental.
References
- 1.Epstein E. The anomaly of silicon in plant biology. Proc Natl Acad Sci USA. 1994;91(1):11–17. doi: 10.1073/pnas.91.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conley DJ. Terrestrial ecosystems and the global biogeochemical silica cycle. Global Biogeochem Cycles. 2002;16(4):68. [Google Scholar]
- 3.Carey JC, Fulweiler RW. The terrestrial silica pump. PLoS ONE. 2012;7(12):e52932. doi: 10.1371/journal.pone.0052932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raven JA. The transport and function of silicon in plants. Biol Rev Camb Philos Soc. 1983;58(2):179–207. [Google Scholar]
- 5.Raven JA. Cycling silicon – the role of accumulation in plants. New Phytol. 2003;158(3):419–421. doi: 10.1046/j.1469-8137.2003.00778.x. [DOI] [PubMed] [Google Scholar]
- 6.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305(5682):354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 7.Frings P, et al. Lack of steady-state in the global biogeochemical si cycle: Emerging evidence from lake Si sequestration. Biogeochemistry. 2014;117(2-3):255–277. [Google Scholar]
- 8.Harper HE, Knoll AH. Silica, diatoms, and cenozoic radiolarian evolution. Geology. 1975;3(4):175–177. [Google Scholar]
- 9.Edwards EJ, et al. C4 Grasses Consortium The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science. 2010;328(5978):587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 10.Philippe H, et al. Comparison of molecular and paleontological data in diatoms suggests a major gap in the fossil record. J Evol Biol. 1994;7(2):247–265. [Google Scholar]
- 11.Stromberg CAE. Using phytolith assemblages to reconstruct the origin and spread of grass-dominated habitats in the great plains of North America during the Late Eocene to Early Miocene. Palaeogeogr Palaeoclimatol Palaeoecol. 2004;207(3-4):239–275. [Google Scholar]
- 12.Strömberg CAE. Decoupled taxonomic radiation and ecological expansion of open-habitat grasses in the Cenozoic of North America. Proc Natl Acad Sci USA. 2005;102(34):11980–11984. doi: 10.1073/pnas.0505700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stromberg CAE, Feranec RS. The evolution of grass-dominated ecosystems during the late Cenozoic. Palaeogeogr Palaeoclimatol Palaeoecol. 2004;207(3-4):199–201. [Google Scholar]
- 14.Katz ME, Finkel ZV, Grzebyk D, Knoll AH, Falkowski PG. Evolutionary trajectories and biogeochemical impacts of marine eukaryotic phytoplankton. Annu Rev Ecol Evol Syst. 2004;35:523–556. [Google Scholar]
- 15.Prasad V, Strömberg CAE, Alimohammadian H, Sahni A. Dinosaur coprolites and the early evolution of grasses and grazers. Science. 2005;310(5751):1177–1180. doi: 10.1126/science.1118806. [DOI] [PubMed] [Google Scholar]
- 16.Kidston R, Lang WH. On Old Red Sandstone plants showing structure, from the Rhynie Chert bed, Aberdeenshire. Part i. Rhynia gwynne-vaughani kidston & lang. Trans R Soc Edinb. 1917;51(24):761–784. [Google Scholar]
- 17.Scott AC, Mattey DP, Howard R. New data on the formation of Carboniferous coal balls. Rev Palaeobot Palynol. 1996;93(1-4):317–331. [Google Scholar]
- 18.Hatcher PG, Lyons PC, Thompson CL, Brown FW, Maciel GE. Organic matter in a coal ball: Peat or coal? Science. 1982;217(4562):831–833. doi: 10.1126/science.217.4562.831. [DOI] [PubMed] [Google Scholar]
- 19.Cooke J, Leishman MR. Is plant ecology more siliceous than we realise? Trends Plant Sci. 2011;16(2):61–68. doi: 10.1016/j.tplants.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Ma JF, Takahashi E. 2002. Soil, Fertilizer, and Plant Silicon Research in Japan. (Elsevier, Amsterdam)
- 21.Ma JF, Yamaji N. Functions and transport of silicon in plants. Cell Mol Life Sci. 2008;65(19):3049–3057. doi: 10.1007/s00018-008-7580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma JF, et al. A silicon transporter in rice. Nature. 2006;440(7084):688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- 23.Mitani N, Ma JF. Uptake system of silicon in different plant species. J Exp Bot. 2005;56(414):1255–1261. doi: 10.1093/jxb/eri121. [DOI] [PubMed] [Google Scholar]
- 24.Mitani N, Yamaji N, Ma JF. Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflugers Arch. 2008;456(4):679–686. doi: 10.1007/s00424-007-0408-y. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Zhu Z. Functional divergence of the NIP III subgroup proteins involved altered selective constraints and positive selection. BMC Plant Biol. 2010;10(1):256. doi: 10.1186/1471-2229-10-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma JF, et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA. 2008;105(29):9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grégoire C, et al. Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 2012;72(2):320–330. doi: 10.1111/j.1365-313X.2012.05082.x. [DOI] [PubMed] [Google Scholar]
- 28.Anderberg HI, Kjellbom P, Johanson U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front Plant Sci. 2012;3:33. doi: 10.3389/fpls.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodson MJ, White PJ, Mead A, Broadley MR. Phylogenetic variation in the silicon composition of plants. Ann Bot (Lond) 2005;96(6):1027–1046. doi: 10.1093/aob/mci255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carnelli AL, Madella M, Theurillat J-P. Biogenic silica production in selected alpine plant species and plant communities. Ann Bot (Lond) 2001;87(4):425–434. [Google Scholar]
- 31.Zardoya R. Phylogeny and evolution of the major intrinsic protein family. Biol Cell. 2005;97(6):397–414. doi: 10.1042/BC20040134. [DOI] [PubMed] [Google Scholar]
- 32.Johanson U, Gustavsson S. A new subfamily of major intrinsic proteins in plants. Mol Biol Evol. 2002;19(4):456–461. doi: 10.1093/oxfordjournals.molbev.a004101. [DOI] [PubMed] [Google Scholar]
- 33.Abascal F, Irisarri I, Zardoya R. Diversity and evolution of membrane intrinsic proteins. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840(5):1468–1481. doi: 10.1016/j.bbagen.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Zardoya R, Ding X, Kitagawa Y, Chrispeels MJ. Origin of plant glycerol transporters by horizontal gene transfer and functional recruitment. Proc Natl Acad Sci USA. 2002;99(23):14893–14896. doi: 10.1073/pnas.192573799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290(5491):481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 36.Wallace IS, Roberts DM. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry. 2005;44(51):16826–16834. doi: 10.1021/bi0511888. [DOI] [PubMed] [Google Scholar]
- 37.Mitani-Ueno N, Yamaji N, Zhao F-J, Ma JF. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot. 2011;62(12):4391–4398. doi: 10.1093/jxb/err158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piperno DR, Holst I, Wessel-Beaver L, Andres TC. Evidence for the control of phytolith formation in Cucurbita fruits by the hard rind (Hr) genetic locus: Archaeological and ecological implications. Proc Natl Acad Sci USA. 2002;99(16):10923–10928. doi: 10.1073/pnas.152275499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danielson JAH, Johanson U. 2010. Phylogeny of major intrinsic proteins. MIPs and Their Role in the Exchange of Metalloids, eds Jahn TP, Bienert GP (Landes Bioscience and Springer Science+Business Media, New York), pp 19-29.
- 40.Mark DF, et al. 40Ar/39Ar dating of hydrothermal activity, biota and gold mineralization in the Rhynie hot-spring system, Aberdeenshire, Scotland. Geochim Cosmochim Acta. 2011;75(2):555–569. [Google Scholar]
- 41.Kidston R, Lang WH. On Old Red Sandstone plants showing structure, from the Rhynie chert bed, Aberdeenshire. Part iii. Asteroxlon mackiei, kidston and lang. Trans R Soc Edinb. 1920;52(26):643–680. [Google Scholar]
- 42.Taylor TN, Taylor EL, Krings M. Paleobotany: The biology and evolution of fossil plants. 2nd Ed Academic Press; Burlington, MA: 2008. [Google Scholar]
- 43.Derry LA, Kurtz AC, Ziegler K, Chadwick OA. Biological control of terrestrial silica cycling and export fluxes to watersheds. Nature. 2005;433(7027):728–731. doi: 10.1038/nature03299. [DOI] [PubMed] [Google Scholar]
- 44.Parr JF, Lentfer CJ, Boyd WE. A comparative analysis of wet and dry ashing techniques for the extraction of phytoliths from plant material. J Archaeol Sci. 2001;28(8):875–886. [Google Scholar]
- 45.Ali MW, Zoltai SC, Radford FG. A comparison of dry and wet ashing methods for the elemental analysis of peat. Can J Soil Sci. 1988;68(2):443–447. [Google Scholar]
- 46.Jorhem L. Dry ashing, sources of error, and performance evaluation in aas. Mikrochim Acta. 1995;119(3-4):211–218. [Google Scholar]
- 47.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larkin MA, et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 51.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pellegrini-Calace M, Maiwald T, Thornton JM. PoreWalker: A novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLOS Comput Biol. 2009;5(7):e1000440. doi: 10.1371/journal.pcbi.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doyle JA. Seed ferns and the origin of angiosperms. J Torrey Bot Soc. 2006;133(1):169–209. [Google Scholar]
- 60.Mathews S. Phylogenetic relationships among seed plants: Persistent questions and the limits of molecular data. Am J Bot. 2009;96(1):228–236. doi: 10.3732/ajb.0800178. [DOI] [PubMed] [Google Scholar]
- 61.Qiu Y-L, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103(42):15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun G, et al. Archaefructaceae, a new basal angiosperm family. Science. 2002;296(5569):899–904. doi: 10.1126/science.1069439. [DOI] [PubMed] [Google Scholar]
- 63.Frohlich MW, Chase MW. After a dozen years of progress the origin of angiosperms is still a great mystery. Nature. 2007;450(7173):1184–1189. doi: 10.1038/nature06393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.