Abstract
Obesity poses an increased risk of developing metabolic syndrome and closely associated nonalcoholic fatty liver disease, including liver cancer. Satiety hormone leptin-deficient (ob/ob) mice, considered paradigmatic of nutritional obesity, develop hepatic steatosis but are less prone to developing liver tumors. Sustained activation of peroxisome proliferator–activated receptor α (PPARα) in ob/ob mouse liver increases fatty acid oxidation (FAO), which contributes to attenuation of obesity but enhances liver cancer risk. To further evaluate the role of PPARα-regulated hepatic FAO and energy burning in the progression of fatty liver disease, we generated PPARα-deficient ob/ob (PPARαΔob/ob) mice. These mice become strikingly more obese compared to ob/ob littermates, with increased white and brown adipose tissue content and severe hepatic steatosis. Hepatic steatosis becomes more severe in fasted PPARαΔob/ob mice as they fail to up-regulate FAO systems. PPARαΔob/ob mice also do not respond to peroxisome proliferative and mitogenic effects of PPARα agonist Wy-14,643. Although PPARαΔob/ob mice are severely obese, there was no significant increase in liver tumor incidence, even when maintained on a diet containing Wy-14,643. We conclude that sustained PPARα activation–related increase in FAO in fatty livers of obese ob/ob mice increases liver cancer risk, whereas deletion of PPARα in ob/ob mice aggravates obesity and hepatic steatosis. However, it does not lead to liver tumor development because of reduction in FAO and energy burning.
Obesity, a disorder involving chronic energy imbalance resulting from excess caloric intake and reduced energy expenditure, was estimated to affect 671 million adults globally in 2013.1,2 Furthermore, in 2013, another 1.4 billion adults were considered to become overweight.1 Overall, the prevalence of overweight with a body mass index of ≥25 to <30 kg/m2, and obesity with a body mass index of ≥30 kg/m2, in adults aged >18 years has increased substantially during the past three decades (1980 to 2013).1 In the United States, obesity rates are among the highest in the world, with approximately 70% of Americans being overweight or obese.2 Because of the established health risks, such as insulin resistance, metabolic syndrome, type 2 diabetes mellitus, atherogenic dyslipidemia, and nonalcoholic fatty liver disease, obesity has become a major global health challenge.1 Nonalcoholic fatty liver disease of obesity begins with simple hepatic steatosis that progresses to nonalcoholic steatohepatitis with inflammation, hepatocellular injury, liver cell proliferation, and fibrous scarring, culminating in end-stage liver disease of cirrhosis and liver cancer.3–6 On the basis of the burgeoning pandemic of nutritional obesity, it is projected that 25 million Americans will likely develop nonalcoholic steatohepatitis by 2025, with approximately 20% progressing to cirrhosis of liver, with an added risk of developing hepatocellular carcinoma.7
In obesity, the unburnt energy is conserved in the form of fat (triacylglycerol), first in adipocytes considered limitless reservoirs of fat and subsequently in liver considered as a surrogate reservoir for fat, when adipose fat stores are nearly saturated.8–10 Liver is a central player in whole body energy homoeostasis by its ability to metabolize glucose and fatty acids, with surplus glucose converted to fat for storage.11 Hepatic steatosis occurs under a variety of conditions, especially when the rate of hepatic fatty acid uptake from plasma and de novo fatty acid synthesis from glucose is greater than the rate of fatty acid oxidation (FAO).12 Excess storage of lipid in liver without inflammation (bland hepatic steatosis) by itself is insufficient to increase liver cancer risk in obesity, as noted in ob/ob mice deficient in satiety hormone leptin.13 Enhancement of FAO in these ob/ob fatty livers by sustained activation of peroxisome proliferator–activated receptor (PPAR)-α leads to nonalcoholic steatohepatitis and enhanced endoplasmic reticulum stress, contributing to a high incidence of liver tumors.14 PPARα, because of its unique ability to regulate FAO in liver, plays a significant role in the pathogenesis of hepatic steatosis and in the development of hepatocellular carcinomas in rats and mice.15,16 PPARα-null mice (PPARα−/−) are unable to up-regulate the expression of FAO-associated genes and, as a consequence, they develop hepatic steatosis but fail to develop liver tumors in response to chronic exposure to peroxisome proliferators.17–19 Accordingly, these observations support the concept that PPARα-regulated increases in FAO contribute to the development of liver tumors in nonalcoholic fatty liver disease.3,4
Herein, we examined the impact of PPARα deficiency on the obesity and fatty liver of leptin-deficient mice. PPARα-deficient mice were crossed with heterozygous leptin-deficient OB/ob mice to generate PPARα-deficient ob/ob (PPARαΔob/ob) mice. Deletion of PPARα in these obese ob/ob mice aggravates obesity because of increases in white and brown fat content and fasting-induced hepatic steatosis because of the failure to increase fatty acid oxidation capacity in the absence of PPARα. Despite severe hepatic steatosis, PPARαΔob/ob mice do not develop liver tumors, because they fail to increase FAO and energy burning necessary for the induction of oxidative and endoplasmic reticulum stresses that play a role in liver tumor development.14,15
Materials and Methods
Animals
Acox1-deficient (Acox1−/−) mice20 and PPARα-deficient (PPARα−/−) mice21 were maintained on a C57BL/6J background as breeding colonies. The generation of Acox1-deficient ob/ob (Acox1Δob/ob) mice was described previously.22 To generate PPARα-deficient ob/ob mice (PPARαΔob/ob), heterozygous leptin-deficient OB/ob mice (The Jackson Laboratory, Bar Harbor, ME) were crossed with PPARα−/− mice to obtain heterozygous PPARα+/−/OB/ob mice, which were further bred to produce PPARα-deficient ob/ob double-mutant mice designated PPARαΔob/ob mice (Supplemental Figure S1).
The mice were genotyped by PCR of genomic DNA isolated from tail tips obtained at the age of 3 weeks. To identify ob/ob mice, primers used were as follows: 5′-TGTCCAAGATGGACCAGACTC-3′ (forward) and 5′-ACTGGTCTGAGGCAGGGAGCA-3′ (reverse). For PPARα-null mice, the primers used were as follows: 5′-CTTGGGTGGAGAGGCTATTC-3′ (forward) and 5′-AGGTGAGATGACAGGAGATC-3′ (reverse). For genotyping Acox1 mice, the primers used were as follows: 5′-TATTCGGCTATGACTGGGCACA-3′ (forward) and 5′-GATGGATACTTTCTCGGCAGGA-3′ (reverse).22
All mice were maintained in polypropylene cages in a temperature-controlled (23°C) environment using a standard photoperiod (12 hours light and 12 hours dark cycle) with lights on at 6 am. Mice were provided rodent chow (Teklad 7904; Harlan-Teklad, Indianapolis, IN) with or without added PPARα activator Wy-14,643 (0.05% or 0.125% w/w) and water ad libitum.
For food intake measurement, three mice were housed individually, and the daily caloric intake was determined over a 7-day period and normalized against the initial body weight (kcal/g body weight per day). These measurements were repeated three times using different batches of mice. To assess hepatocyte proliferation, mice were administered 0.5 mg/mL bromodeoxyuridine (BrdUrd) in drinking water and sacrificed at the end of 3 days. All procedures of animal handling were approved by the Institutional Animal Care and Use Committees of Northwestern University (Chicago, IL; protocol number 2013-3198).
Glucose Tolerance and Insulin Tolerance Tests
Wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice (aged 3 months; five mice in each group) were used for determining glucose tolerance and insulin tolerance essentially as described previously.22 Briefly, glucose (1.5 mg/g body weight in phosphate-buffered saline) was administered i.p. to mice that were maintained without food for 6 hours.22,23 Blood obtained from tail vein at 0, 30, 60, and 120 minutes after administering glucose was used for measuring glucose using a glucose meter (One Touch; LifeScan, Milpitas, CA). For testing insulin tolerance, mice deprived of food for 4 hours were given insulin (0.75 mU/g body weight in phosphate-buffered saline; Sigma, St. Louis, MO) by an i.p. injection. Blood glucose levels were determined at 0, 15, 30, and 60 minutes after insulin injection.
Biochemical Assays
Liver samples were homogenized for extraction of lipid using chloroform/methanol method. Triacylglycerol (Thermo Electron, Louisville, CO) and total cholesterol (Wako Diagnostics, Richmond, VA) levels were determined as described elsewhere.24 Blood obtained from retro-orbital veins was used for triglyceride and total cholesterol determinations.25,26
Histology and Immunohistochemistry
For histological analysis, liver slices were fixed in 4% paraformaldehyde and processed for embedding in paraffin. Paraffin sections (4 μm thick) were cut and stained with hematoxylin and eosin. For visualization of cellular fat, frozen sections of liver (approximately 5 μm thick) were stained with 0.5% Oil Red O solution for 30 minutes in a 60°C oven and then in 85% propylene glycol solution for 5 minutes. After rinsing with distilled water, sections were stained with Gill's hematoxylin for 2 seconds, washed, and mounted with aqueous mounting medium.22,27 Paraffin sections were also used for Sirius Red staining.28 Liver sections were also processed for the localization of catalase, l-PBE (enoyl-Coenzyme A, hydratase; EHHADH), and BrdUrd, as described elsewhere.20,29 BrdUrd nuclear labeling indices were obtained by analyzing immunohistochemically stained liver sections.30 All images were acquired by a light microscope adapted to a high-resolution camera (AxioCam; Carl Zeiss, Oberkochen, Germany) and analyzed by the computer using AxioVisionRel software version 4.8 (Carl Zeiss).
Immunoblotting
For immunoblotting, 40 μg liver protein samples were subjected to 4% to 20% SDS-PAGE, transferred to nitrocellulose membrane, and blotted using antibodies against PPARα, l-3-hydroxyacyl-CoA dehydrogenase (l-PBE), acyl-CoA oxidase 1, palmitoyl (Acox1), short-chain acyl-CoA dehydrogenase (SCAD), medium-chain acyl-CoA dehydrogenase (MCAD), long-chain acyl-CoA dehydrogenase, peroxisomal 3-ketoacyl-CoA thiolase A (PTL), d-3-hydroxyacyl-CoA dehydratase (d-PBE), and catalase, as described elsewhere.29,31 β-Actin (antibody sc-47778; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used as loading control.
Northern Blotting and Real-Time PCR
Total RNA was isolated from mouse liver with TRIzol reagent (Invitrogen, Life Technologies, Carlsbad, CA). For Northern blotting, RNA was glyoxylated, separated on a 1% agarose gel, transferred to a nylon membrane, hybridized at 65°C in Rapid-hyb buffer (GE Healthcare, Piscataway, NJ), and probed with α-32P–labeled cDNA.29,32 RNAs (28S and 18S) were set as the loading controls. For real-time PCR, total RNA isolated from liver was reverse transcribed to make cDNA using the Superscript III first-strand synthesis system for RT-PCR (Invitrogen Life Technologies). The primers used for the real-time PCR are listed in Table 1. Quantitative expression of genes was checked using SYBR Green (Applied Biosystems, Foster City, CA) in triplicate and normalized with 18S ribosomal RNA. PCR was composed of 1 μL (100 ρmol) of forward and reverse primers and 10 μL of 2× SYBR Green PCR Master Mix and performed by using an ABI 7300 system (Applied Biosystems). The generation of specific PCR products was confirmed by melting curve analysis, and the relative gene expression changes were measured using the comparative Ct method:
Table 1.
Primers for Genotyping, Quantitative PCR, and Northern Blot Analysis
| Primer name | Forward primer | Reverse primer |
|---|---|---|
| PPARα ko geno | 5′-CTTGGGTGGAGAGGCTATTC-3′ | 5′-AGGTGAGATGACAGGAGATC-3′ |
| PPARα wt geno | 5′-CCATCCAGATGACACCTTCC-3′ | 5′-TCTCTTGCAACAGTGGGTGC-3′ |
| ob/ob geno | 5′-TGTCCAAGATGGACCAGACTC-3′ | 5′-ACTGGTCTGAGGCAGGGAGCA-3′ |
| Neo geno | 5′-TATTCGGCTATGACTGGGCACA-3′ | 5′-GATGGATACTTTCTCGGCAGGA-3′ |
| ACOX1 geno | 5′-CCGCAAGCCATCCGACATTC-3′ | 5′-ATTCAGTGGGTCAGGCGACTGC-3′ |
| PPARα | 5′-GGGCTCCGAGGGCTCTGTCA-3′ | 5′-TGCAGCTCCGATCACACTTGTCG-3′ |
| Acox1 | 5′-GCCAAGGCGACCTGAGTGAGC-3′ | 5′-ACCGCAAGCCATCCGACATTC-3′ |
| l-PBE/Ehhadh | 5′-GGTCGTTGGAGTTCCTGTTGCT-3′ | 5′-TGGGCAAGCTTGGGACTGGC-3′ |
| SCAD | 5′-GCTGAGTGGTGCAGGCTTG-3′ | 5′-CCATTGGTGAAAGGGGTGATC-3′ |
| MCAD | 5′-GGATGACGGAGCCAATG-3′ | 5′-GGGTGTCGGCTTCCACAATG-3′ |
| LCAD | 5′-GACGGCGGGCAAGTGTATC-3′ | 5′-GCAGGCGATCGAGCTTCAC-3′ |
| Cyp4a1 | 5′-AGGATGAGGGAGAGCTGGAAAAGAT-3′ | 5′-GACTCCACTGGCTGTGGTGTCATG-3′ |
| Cyp4a3 | 5′-GCAGAAGGCCAGGAAGAGACAC-3′ | 5′-CCAGAGCATAGAAAATCCAGGAAATT-3′ |
| PPARγ | 5′-CCACAGTTGATTTCTCCAGCATTTC-3′ | 5′-CAGGTTCTACTTTGATCGCACTTTG-3′ |
| PGC1α | 5′-CTCCATGCCTGACGGCACCC-3′ | 5′-GCAGGGACGTCTTTGTGGCT-3′ |
| Sirt1 | 5′-GGATGATATGACGCTGTGGC-3′ | 5′-AGAGACGGCTGGAACTGTCC-3′ |
| FGF21 | 5′-TGGGGGTCTACCAAGCATAC-3′ | 5′-AAGGCTCTACCATGCTCAGG-3′ |
| PEPCK | 5′-TGAACTGACAGACTCGCCCT-3′ | 5′-GTCTTCCCACAGGCACTAGG-3′ |
| PTL | 5′-TCTCCAGGACGTGAGGCTAAA-3′ | 5′-CGCTCAGAAATTGGGCGATG-3′ |
| Nrf2 | 5′-GATGGACTTGGAGTTGCCAC-3′ | 5′-GTTTGGGAATGTGGGCAACC-3′ |
| P8 | 5′-CTCCCTCTCCAGAACCTCACT-3′ | 5′-ACCAAGAGAGAAGCTGCTGC-3′ |
| 18S | 5′-AAACGGCTACCACATCCAAG-3′ | 5′-CCTCCAATGGATCCTCGTTA-3′ |
ACOX, acyl-Coenzyme A oxidase; FGF, fibroblast growth factor; LCAD, long-chain acyl-CoA dehydrogenase; l-PBE, l-3-hydroxyacyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase; PGC, peroxisome proliferator-activated receptor gamma, coactivator; PPAR, peroxisome proliferator–activated receptor; PTL, 3-ketoacyl-CoA thiolase; SCAD, short-chain acyl-CoA dehydrogenase.
Statistical Analysis
Data were analyzed by one-way analysis of variance using SPSS software version 11.5 (SPSS, Chicago, IL). P < 0.05 was considered significant.
Results
Obese ob/ob Mice Lacking PPARα Become Super Obese
Because of subfertility of homozygous ob/ob mice, heterozygous OB/ob mice were mated with PPARα−/− (PPARα−/− with OB/OB) mice to generate double-heterozygous PPARα+/− OB/ob mice. These mice, when intercrossed, yielded PPARαΔob/ob mice used in these studies (Supplemental Figure S1A). PCR amplification of genomic DNA from double-nullizygous PPARαΔob/ob mice yielded 55- and 100-bp bands for ob/ob and a 280-bp band for PPARα−/− (Supplemental Figure S1B). DNA from WT mice showed one 143-bp band and one 155-bp band for PPARα+/+ and OB/OB, separately (Supplemental Figure S1B).
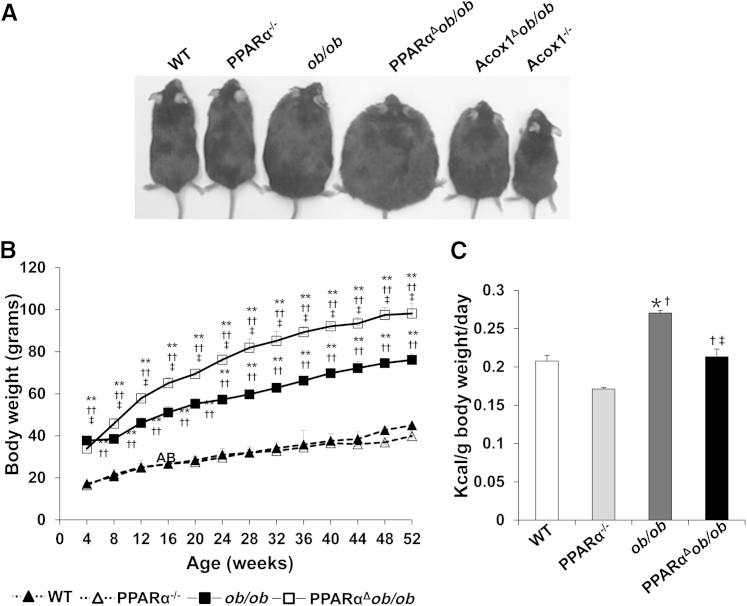
PPARα mediates the pleiotropic responses induced in liver by peroxisome proliferators.16,21,33 These include hepatomegaly, peroxisome proliferation, and transcriptional activation of genes encoding peroxisomal, mitochondrial, and microsomal FAO enzymes.16,34 Previous studies demonstrated that Acox1 deletion disrupts metabolism in WT mice, and ob/ob mice are smaller and leaner than both WT and ob/ob mice.20,22 In the absence of Acox1, the unmetabolized substrates of this enzyme serve as endogenous PPARα activators.16,32 Sustained activation of PPARα by its endogenous ligands in Acox1Δob/ob mice (Figure 1A) increases hepatic fatty acid oxidation and attenuates obesity.22 In contrast, deletion of PPARα in the ob/ob background (PPARαΔob/ob) resulted in a further increase in somatic growth compared to their ob/ob littermates (Figure 1, A and B). From weaning until approximately the age of 6 weeks, somatic growth of PPARαΔob/ob mice did not significantly differ from ob/ob mice. However, by the age of 24 weeks, PPARαΔob/ob mice gained more weight than ob/ob mice and were distinctly more obese (super obese) than ob/ob mice (Figure 1, A and B). There was no significant difference in food intake of WT and PPARα−/− mice, but food consumption in ob/ob mice was slightly more than that of PPARαΔob/ob mice. These findings suggest that increased obesity observed in PPARαΔob/ob mice is not due to increased food intake but related to decreased energy expenditure associated with PPARα deletion (Figure 1C).
Figure 1.
PPARαΔob/ob mice become super obese. A: Physical appearance of chow-fed 16-month-old wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. Acox1Δob/ob and Acox1−/− mice that manifest PPARα activation by endogenous ligands are included for comparison. B: Age-related body weight changes of WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice, aged 4 to 52 weeks. C: For food consumption estimation, three mice in each group were housed in separate cages, and daily food intake (kcal) per body weight was determined by measuring 7-day consumption. ∗P < 0.05, ∗∗P < 0.01 versus WT; †P < 0.05, ††P < 0.01 versus PPARα−/−; ‡P < 0.05 versus ob/ob.
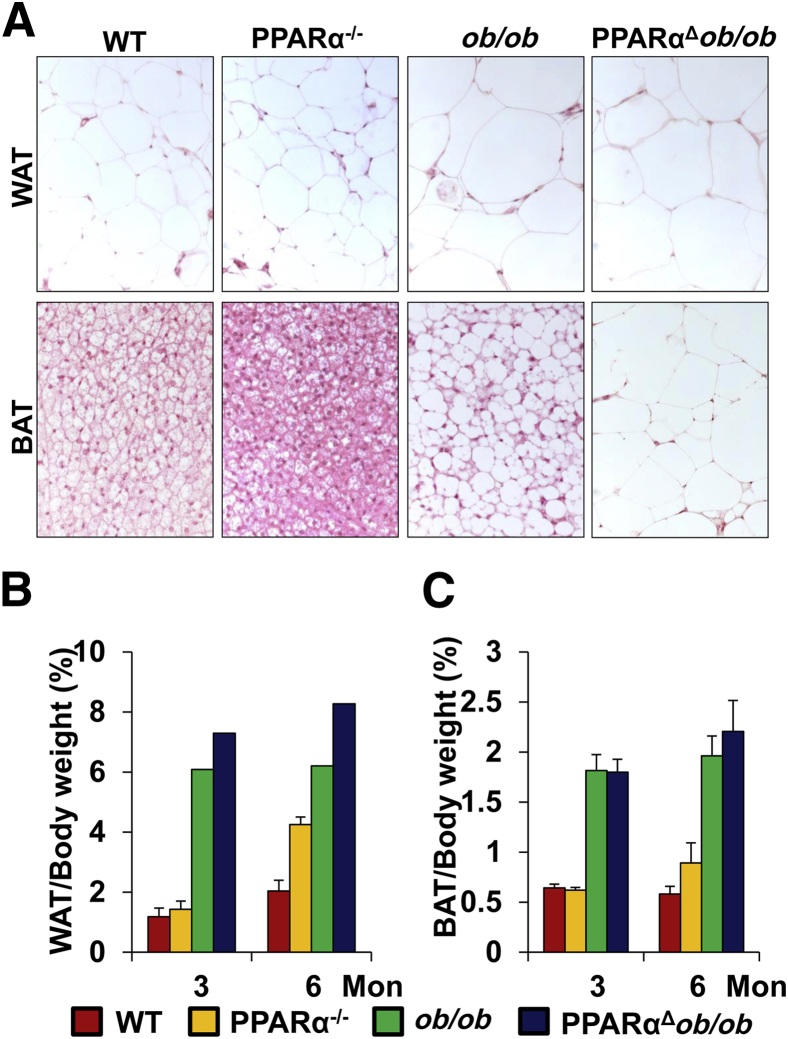
Examination of 3- and 6-month-old PPARαΔob/ob and ob/ob mice revealed a consistent increase in the amount of inguinal white adipose tissue (WAT) and interscapular brown adipose tissue (BAT) in both ob/ob and PPARαΔob/ob mice, but the increase was more prominent in double nulls (Supplemental Table S1). At 6 months of age, the WAT in WT mice was 0.56 g compared to 4.22 g in ob/ob mice and 5.87 g in PPARαΔob/ob mice. In 6-month-old WT mouse, BAT was 0.14 g compared to 1.19 g in ob/ob mice and 1.37 g in PPARαΔob/ob mice (Supplemental Table S1).
Histological examination of WAT revealed no significant difference in the adipocyte size of PPARαΔob/ob mice when compared with ob/ob mice (Figure 2A). In both ob/ob and PPARαΔob/ob mice, BAT whitened with several large lipid droplets, and this change was considerably greater in PPARαΔob/ob mice (Figure 2A). At 6 months of age, the WAT and BAT body weight ratios were increased in PPARαΔob/ob mice (Figure 2, B and C). Leptin-deficient ob/ob mice exhibit hyperphagia and obesity, along with hyperglycemia and hyperinsulinemia.35 PPARα knockout mice appear normal, but the circulating glucose is lower, with increased serum insulin level.23 PPARαΔob/ob mice exhibited higher serum glucose level and lower insulin level compared with WT and ob/ob mice (Supplemental Figure S2, A and B). PPARα deficiency improved insulin and glucose tolerance in obese mice.
Figure 2.
Fat accumulation in the inguinal white adipose tissue (WAT) and scapular brown adipose tissue (BAT). A: Histological features of WAT and BAT of 3-month-old wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. B and C: Fat:body weight ratios.
PPARα Deficiency Aggravates Hepatic Steatosis in ob/ob Mice
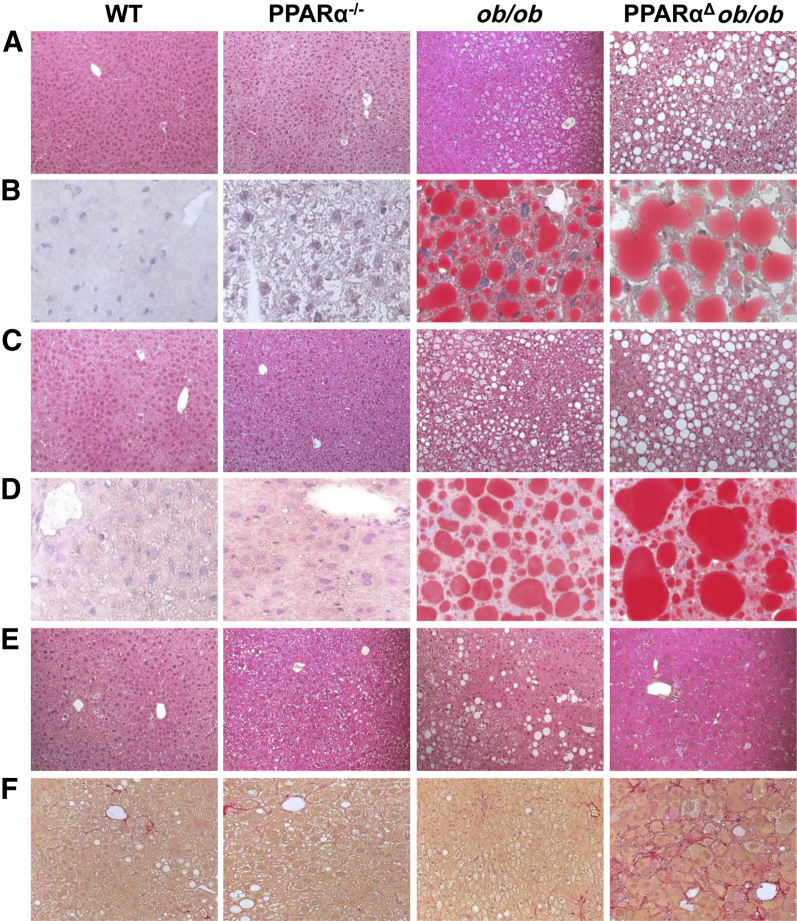
PPARα deficiency in ob/ob mice results in an increased accumulation of fat in liver as compared to that in ob/ob livers (Figure 3). At 3 and 6 months of age, liver/body weight ratios were higher in PPARαΔob/ob mice than in ob/ob mice (Supplemental Table S1), and on histological examination, including Oil Red O staining for fat, these livers revealed prominent macrovesicular steatosis (Figure 3). Overall, in PPARαΔob/ob mouse hepatocytes, fat droplets appeared much larger at 3 and 6 months of age (Figure 3). Hepatic triglyceride and cholesterol content was higher in 6-month-old PPARαΔob/ob mice as compared to WT, PPARα−/−, and ob/ob mice (Supplemental Figure S2, C and D).
Figure 3.
PPARαΔob/ob mice exhibit severe hepatic steatosis. PPARα deficiency aggravates hepatic steatosis in ob/ob mice (see ob/ob versus PPARαΔob/ob). Liver sections of 3-month-old (A and B), 6-month-old (C and D), 16-month-old (E) wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice maintained on a chow diet were stained with hematoxylin and eosin (A, C, and E) and Oil Red O (B and D). F: Sirius Red staining of liver sections of 16-month-old mice. PPARαΔob/ob mice reveal a discernible increase in Sirius Red–positive fibrous septa.
Aging Attenuates Hepatic Steatosis in PPARαΔob/ob Mice and Induces Hepatic Oval Cell Proliferation
There was a reduction in hepatic steatosis in both ob/ob and PPARαΔob/ob mice aged 16 months or older (Figure 3). In PPARαΔob/ob mice, liver displayed cellular heterogeneity with mild pericellular fibrosis and emerging clusters of oval cells (Figure 3). Severe hepatic steatosis in double-knockout mice increased hepatocyte apoptosis and hepatocyte regeneration independent of PPARα to replace the dead liver cells (data not shown). Oval cells in PPARαΔob/ob mouse liver, occurring either singly or in clusters, stained positively on immunohistochemical analysis (Supplemental Figure S3A) for oval cell markers A6, CD45, CK19, and epithelial cell adhesion molecule.36,37 Quantitative PCR analysis revealed robust expression of these genes in aged PPARαΔob/ob mouse livers, but albumin levels were low and α-fetoprotein was high (Supplemental Figure S3B).
Severe Fatty Liver in Fasted PPARαΔob/ob Mice
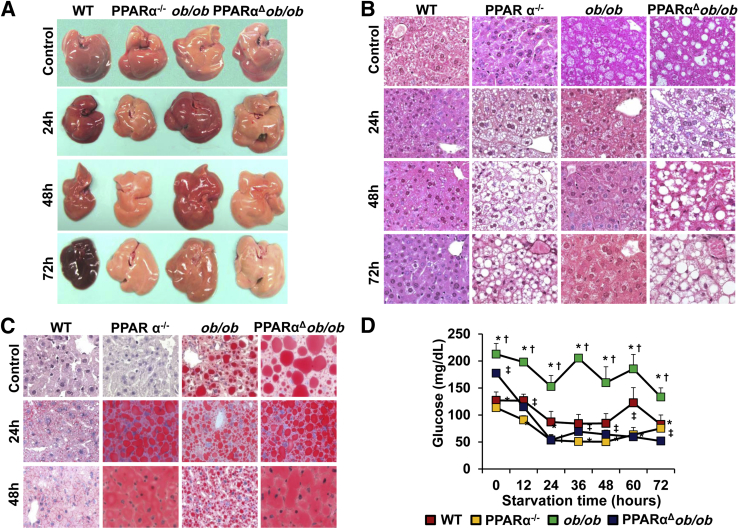
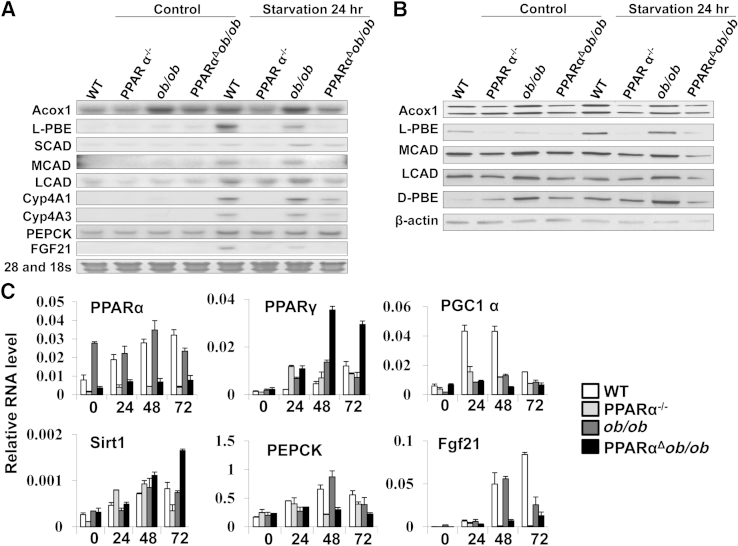
During periods of starvation, triglycerides stored in adipose tissue are hydrolyzed to free fatty acids and mobilized into plasma to reach liver. In liver, they are oxidized by the mitochondrial β-oxidation system and, to a lesser extent, by the peroxisomal β-oxidation, as well as by CYP4A-catalyzed microsomal ω-oxidation pathways to generate ketone bodies that include acetoacetate, 3-hydroxybutyrate (alias β-hydroxybutyrate), and acetone.38 These serve as fuel for nonhepatic peripheral tissues.38 Because PPARα is vital for the transcriptional regulation of fatty acid–metabolizing enzymes in liver, the deficiency of this transcription factor in starvation leads to hepatic steatosis.39,40 To ascertain the combined effects of decreased energy burning due to PPARα deficiency and increased food intake due to lack of leptin during starvation, WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice were fasted for 24, 48, and 72 hours (Figure 4). After 24 hours starvation, PPARα−/− and PPARαΔob/ob mouse livers were paler and larger, suggestive of fatty change when compared to fasted WT and ob/ob livers (Figure 4A). The difference in pallor was exaggerated with prolonged fasting (Figure 4A). In contrast, pallor in ob/ob mouse liver was reduced after starvation (Figure 4A).
Figure 4.
PPARα deficiency increases hepatic steatosis after starvation and reduces plasma glucose significantly. A: Representative gross images of livers of fed and 24-, 48-, and 72-hour fasted wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. B: Hematoxylin and eosin staining of livers of WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice fed and fasted for 24, 48, and 72 hours. C: Oil Red O–stained liver sections confirm fat accumulation in PPARα−/− and PPARαΔob/ob mice, but attenuation of steatosis in ob/ob mice. D: Plasma glucose levels are higher in ob/ob mice compared to other genotypes. ∗P < 0.05 versus WT, †P < 0.05 versus PPARα−/−, and ‡P < 0.05 versus ob/ob.
Hematoxylin and eosin and Oil O Red staining results confirmed severe hepatic steatosis in PPARα−/− and PPARαΔob/ob mice after starvation (Figure 4, B and C). Hepatic steatosis observed in fed ob/ob mice diminished due to fasting-induced activation of PPARα regulated fatty acid oxidation in liver (Figure 4, B and C). No fat droplets are seen in WT and ob/ob mouse livers starved for 48 and 72 hours (Figure 4, B and C). The glycogen content in livers of all animals was reduced after fasting (data not shown). Serum glucose levels of PPARα−/− and PPARαΔob/ob mice were reduced to approximately 47% and 30.3% of basal level after 24 hours starvation, respectively, and no further reduction was noted with prolonged fasting (Figure 4D).
Northern and Western blot analyses of liver samples obtained from WT and ob/ob mice starved for 24 hours revealed increases in the mRNA and/or protein levels of Acox1, l-PBE (EHHADH), MCAD, long-chain acyl-CoA dehydrogenase, CYP4A1, CYP4A3, and fibroblast growth factor 21 after 24 hours of fasting (Figure 5, A and B). Quantitative PCR data revealed increases in PPARα level in livers of fasted WT and ob/ob mice (Figure 5). Increased mRNA content of peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC1α) was noted in 24- and 48-hour fasted WT livers. Sirt1 level increased in PPARαΔob/ob mouse liver at 48 and 72 hours starvation, and phosphoenolpyruvate carboxykinase level increased in normal and ob/ob mouse livers at 48 hours of fasting (Figure 5C).
Figure 5.
Constitutive and 24-hour starvation inducible levels of fatty acid oxidation gene expression in livers of wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. Representative Northern (A) and Western (B) blots to assess the gene expression changes in liver. C: Quantitative PCR data of key enzymes linked to acute fasting. Acox, acyl-Coenzyme A oxidase; d-PBE, d-3-hydroxyacyl-CoA dehydratase; FGF, fibroblast growth factor; LCAD, long-chain acyl-CoA dehydrogenase; l-PBE, l-3-hydroxyacyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase; PGC, peroxisome proliferator-activated receptor gamma, coactivator; SCAD, short-chain acyl-CoA dehydrogenase.
PPARα Ligand Does Not Induce Liver Tumors in PPARαΔob/ob Mice
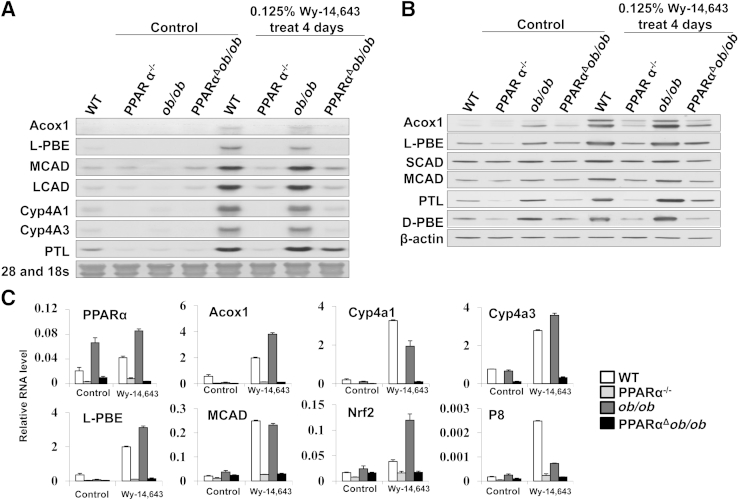
Previous studies have shown that increased FAO and energy combustion in liver leads to liver tumor development.15 Sustained activation of PPARα in ob/ob mice attenuates obesity by increasing hepatic FAO but increases the risk for liver tumor development, in part, related to excess energy combustion.22 To study the influence of PPARα deficiency in PPARαΔob/ob mice, we first evaluated the effects of short-term administration of Wy-14643, a PPARα ligand (Figure 6). We found, as expected, that PPARαΔob/ob mice fail to respond to PPARα ligand Wy-14,643 administered in powdered chow at 0.125% for 4 days (Figures 6 and 7).
Figure 6.
Constitutive and Wy-14,643–inducible levels of fatty acid oxidation gene expression in livers of wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. Representative Northern (A) and Western (B) blots to assess the gene expression changes in liver. C: Quantitative PCR data of key enzymes linked to fatty acid oxidation. Acox, acyl-Coenzyme A oxidase; d-PBE, d-3-hydroxyacyl-CoA dehydratase; LCAD, long-chain acyl-CoA dehydrogenase; l-PBE, l-3-hydroxyacyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; PTL, 3-ketoacyl-CoA thiolase; SCAD, short-chain acyl-CoA dehydrogenase.
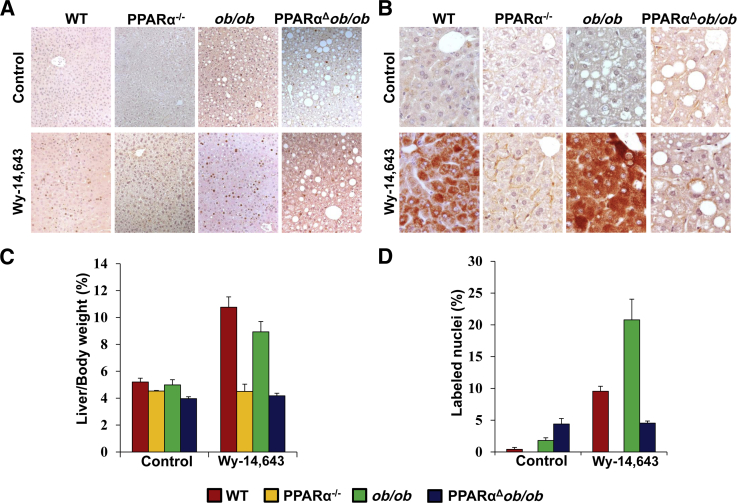
Figure 7.
Liver cell proliferation and induction of l-3-hydroxyacyl-CoA dehydrogenase (l-PBE) resulting from PPARα activation by dietary Wy-14,643 for 4 days. Bromodeoxyuridine (BrdUrd) labeling (A) and l-PBE immunohistochemical staining (B) of livers of 2-month-old wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice fed with chow diet or 0.125% Wy-14,643 for 4 days. Liver/body weight ratio (C) and hepatocyte proliferation (D) assessed by BrdUrd labeling ratio by calculating BrdUrd-positive hepatocyte nuclei.
Induction of PPARα-regulated genes in liver, in particular those involved in FAO, was not observed in PPARαΔob/ob mice (Figure 6). We also evaluated BrdUrd incorporation in hepatocyte nuclei and expression level of l-PBE, the second enzyme of the peroxisomal β-oxidation, by immunohistochemical analyses (Figure 7, A and B). The liver/body weight ratio increased in WT and ob/ob mice fed Wy-14,643 for 4 days, but no increase was evident in PPARαΔob/ob mice (Figure 7C). BrdUrd labeling indices in liver increased variably in WT and ob/ob mice given Wy-14,643 (Figure 7, A and D). It would appear that the increased nuclear labeling in these mice is PPARα linked, and the small, but perceptible, increase in control and Wy-14,643–treated PPARαΔob/ob mice is PPARα independent.
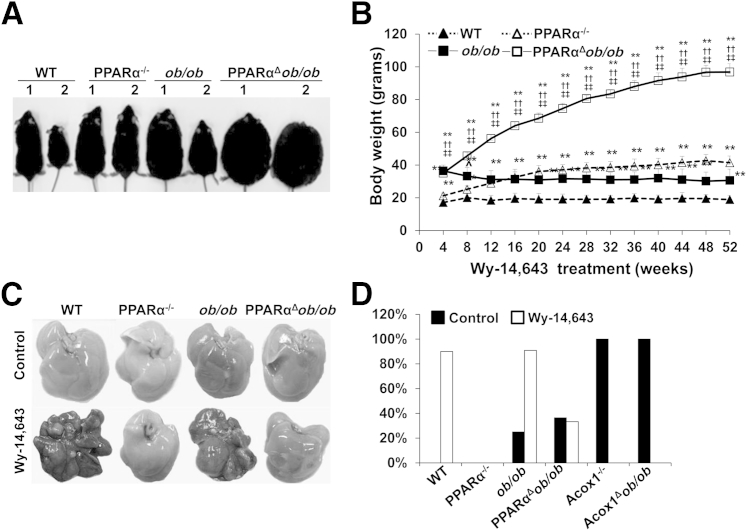
Sustained activation of PPARα by both synthetic and endogenous ligands results in the development of hepatocellular carcinomas in mice.15,22,32 Herein, we examined the role of chronic exposure to PPARα ligand Wy-14,643 in PPARαΔob/ob mice (Figure 8). In WT and ob/ob mice chronically fed a Wy-14,643–containing diet, body weights were lower than the chow-fed controls, whereas absence of PPARα, in PPARα−/− mice and PPARαΔob/ob mice, exerted no significant change in body weights (Figure 8, A and B). All mice that survived were sacrificed at 52 weeks of age, and liver tumor incidence was obtained (Figure 8, C and D). Although all ob/ob mice survived at 1 year, the survival probability was reduced during this period in PPARαΔob/ob mice. Liver tumors were seen in 9 of 10 WT, and 10 of 11 ob/ob, mice fed PPARα ligand, but the incidence of liver tumors in ob/ob mice maintained on normal chow was low (3/12). PPARαΔob/ob mice fed a normal or Wy-14,643–containing diet had tumor incidence similar to that of chow-fed ob/ob mice. No liver tumors were detected in PPARα−/− mice maintained on a control (0/10) or Wy-14,643–containing diet (0/10). We included data obtained from Acox1−/− and Acox1Δob/ob mice on a control diet. Both these groups with sustained PPARα activation due to endogenous ligands developed a high incidence of liver tumors. All tumors were well to moderately differentiated hepatocellular carcinomas.
Figure 8.
PPARαΔob/ob mice fail to respond to PPARα ligand Wy-14,643 induced body weight reduction and liver tumorigenesis. A: Physical appearance of chow-fed (1) or 0.05% Wy-14,643–fed (2) for 14 months (aged 16 months) wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. B: Body weight changes of mice treated with 0.05% Wy-14,643 for 8 to 52 weeks. C: Liver appearance of WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice fed normal chow (control) or treated with 0.05% Wy-14,643 for 14 months. Liver tumors are seen in Wy-14,643–fed WT and ob/ob mice. D: Incidence of liver tumors. PPARα activation by Wy-14,643 induces liver tumors in ob/ob mice, but absence of PPARα in ob/ob mice attenuates liver tumorigenesis. All Acox1-null mice and Acox1Δob/ob mice develop liver tumors because of sustained PPARα activation by endogenous ligands. ∗∗P < 0.01 versus WT, ††P < 0.01 versus PPARα−/−, and ‡‡P < 0.01 versus ob/ob.
Discussion
PPARα and Energy Burning
Activation of lipid-sensing nuclear receptor PPARα in liver promotes the uptake, use, and catabolism of fatty acids by transcriptional up-regulation of genes involved in these physiological processes.8,16,33,41 In liver, PPARα plays a central role in the regulation of energy burning by peroxisomal and mitochondrial fatty acid β-oxidation and microsomal ω-oxidation.8,15 PPARα is activated by a plethora of synthetic exogenous ligands that include fibrate and other classes of drugs used in the treatment of hyperlipidemia and also by industrial plasticizers, insecticides, herbicides, and certain organic solvents that are collectively referred to as peroxisome proliferators.15–17
Metabolic Pathways that Generate and Degrade Endogenous PPARα Ligands Exist to Modulate Lipid Metabolism: Acox1 in the Metabolism of Endogenous PPARα Activators
The existence of endogenous biological molecules that activate PPARα in vivo first became evident in mice with the germline deletion of Acox1, the first and rate-limiting enzyme of the peroxisomal fatty acid β-oxidation pathway.20,32 Acox1 deficiency results in the generation of a lean mouse with a complex hepatic phenotype manifesting as steatohepatitis, hepatocellular regeneration with massive spontaneous proliferation of peroxisomes, and concomitant perpetually heightened transcriptional activation of PPARα-regulated genes.20,32 The Acox1-null mice also develop a high incidence of hepatocellular carcinomas.32 These effects are attributed to Acox1 substrates, such as long-chain and very-long-chain fatty acids, fatty acyl CoAs, and other biological molecules that remain unmetabolized in the absence of Acox1; some of these substrates then function as endogenous activators of PPARα.32 Thus, Acox1 was the first enzyme demonstrated to be involved in the metabolism of endogenous PPARα ligands.32 Subsequent studies with other mice with defects in enzymes of FAO established the concept that metabolic pathways that generate and degrade endogenous PPARα ligands exist to modulate lipid metabolism. The sustained overexpression of PPARα-regulated genes involved in FAO in Acox1−/− mice contributes to increased energy burning; consequently, these mice develop lean body mass.20,32
We have previously reported that Acox1Δob/ob mice attenuates obesity and hepatic steatosis, along with an increase in insulin sensitivity.22 As expected, in these Acox1Δob/ob mice, unmetabolized substrates of Acox1 function as endogenous ligands of PPARα to cause sustained transcriptional activation of PPARα and enhanced FAO.16,20,32,42 It is, thus, clear that excess energy burning diminishes the development of obesity in ob/ob mice, with the genetic model of obesity representing excess energy consumption.22,35 Activation of PPARα in Acox1Δob/ob mice increased the expression of genes associated with inflammation and endoplasmic reticulum stress systems contributing to liver tumor development.22
Insights from the PPARαΔob/ob Mice
As a corollary, to establish that the failure to induce PPARα-regulated hepatic fatty acid oxidation and energy burning in ob/ob mice impedes the progression of fatty liver disease and development of liver tumors, we generated PPARαΔob/ob mice. In contrast to Acox1Δob/ob mice that consumed more energy due to lack of leptin but also burnt excess energy in view of the heightened PPARα-regulated fatty acid oxidation, PPARαΔob/ob mice, as expected, also consumed more energy due to leptin deficiency but burnt less energy due to PPARα deficiency. PPARαΔob/ob mice gained more body weight from the age of 24 weeks when compared to ob/ob mice, although they consumed less food per gram weight than ob/ob mice, suggesting a reduction in energy expenditure associated with PPARα deficiency. The excess energy is conserved in WAT in ob/ob and PPARαΔob/ob mice and accounts for increase in fat weight and adipocyte size.8–10 As the process progresses, whitening of BAT ensues. Deletion of PPARα in ob/ob mice aggravated fat accumulation in adipocytes, resulting in hypertrophic fat cells.
Leptin encoded by ob gene regulates appetite, energy homeostasis,41,43,44 and immune function.41–45 Its expression level in the adipocytes is correlated with the lipid content and the corresponding individual adipocyte size.45 Disruption of ob gene results in excess food intake with reduced energy expenditure, which accelerates fat accumulation in adipose tissue and liver and influences whitening of BAT.46–48 As a result, ob/ob mice become obese with increased body weight and several fold increase in fat content compared to WT mice, and become hyperinsulinemic and hyperglycemic.46–48 Because PPARα modulates energy homeostasis, its deletion contributes to the failure to activate fatty acid oxidation systems, which results in an accelerated accumulation of excess energy, first in the adipose tissue and then in the liver. Accordingly, as shown herein, deficiency of PPARα aggravates obesity and hepatic steatosis in ob/ob mice and influences insulin and glucose tolerance. In this context, some PPARα ligands have been shown to reduce hyperinsulinemia and hyperglycemia and increase insulin sensitivity of mice fed with a high-fat diet.46,49 These observations suggest that PPARα deficiency leads to impaired fatty acid oxidation, which aggravates fat accumulation and influences metabolic syndrome in ob/ob mice. In this regard, adenoviral-mediated hyperleptinemia induces PPARα transcription and activation of its target genes and reduces hepatic triacylglycerol content.47 Because adenoviral-mediated induction is transient, the effects of hyperleptinemia reversed as the expression level of PPARα decreased, and fat was regained 2 months after the level of leptin return to normal.47,48,50,51 Moreover, PPARα expression can be programmed by neonatal leptin administration in rat, which reverses the phenotypic effects of maternal undernutrition.52,53 Leptin can induce liver-specific promoter activity via a STAT3/Sp1 mechanism.54 These data suggest that leptin deficiency in PPARα−/− mice attenuates PPARα activity, to further reduce FAO.
Interestingly, aging attenuated hepatic steatosis in both ob/ob mice and PPARαΔob/ob mice. Small cells with oval nuclei arose in both the periphery of the portal tracts and within the hepatic lobular part in PPARαΔob/ob mouse livers. These cells stained positively for hepatic oval cell markers A6, CD45, CK19, and epithelial cell adhesion molecule. Similar cells were not evident in the livers of WT, PPARα−/−, and ob/ob mice. Hepatic oval cells, regarded as stem/progenitor cells in adult livers, can be activated under Dipin, partial hepatectomy, and chronic liver injury conditions.55–59 After chronic injury or impaired proliferation of hepatocytes, facultative adult oval cells proliferate and differentiate into hepatocytes and epithelial cells. Chemical hepatotoxic substances and continuous metabolic stress can thus be considered as potential oval cell activators. In the livers of PPARαΔob/ob mice, severe hepatic steatosis, because of excess fat overload and damaged FAO, leads to hepatocyte apoptosis, with new hepatocyte proliferation independent of PPARα to replace the dead cells.
During acute fasting, metabolic substrates are switched from carbohydrate to fatty acids, which leads to the release of large quantities of free fatty acids from adipose tissue into circulation, reaching the liver. In the liver, fatty acids are metabolized to generate ketone bodies to serve as fuels for other tissues. In WT and ob/ob mice, the intact PPARα senses the influx of fatty acids resulting from starvation to up-regulate enzymes involved in FAO.39,40 We noted a marked increase in Cyp4a1 and Cyp4a3 mRNA levels. During fasting, PGC1α, Sirt1, and fibroblast growth factor 21 were also induced in WT and ob/ob mice. Fasting stimulated an increase in fatty acid mobilization, and an increase in FAO resulted in the reduction of hepatic steatosis in ob/ob mice, which have intact PPARα. Deletion of PPARα in ob/ob mice resulted in severe hepatic steatosis and reduction in plasma glucose level after 24 hours of starvation. FAO enzymes were not induced because of PPARα deficiency. In PPARα-deleted ob/ob mice, PPARγ and Sirt1 expression was induced after starvation, but the relevance is unclear. Furthermore, PPARα−/− mice and PPARαΔob/ob mice fail to respond to Wy-14,643, a potent synthetic PPARα activator. In the absence of PPARα, there is no induction of fatty acid oxidation, peroxisome proliferation, and liver tumorigenesis in PPARα−/− mice and PPARαΔob/ob mice. Previous work has shown that fat accumulation in the livers of ob/ob mice alone will not lead to liver tumors, but sustained activation of PPARα-modulated energy expenditure by endogenous PPARα ligands in ob/ob mice resulted in liver tumor development.22 On the other hand, PPARα deficiency in ob/ob mice further aggravates hepatic steatosis, but does not increase the liver tumor risk, suggesting that fat accumulation in liver is not the direct reason for hepatocellular carcinoma.
In summary, these observations further support the notion that PPARα-regulated signaling plays an important role in the progression of liver diseases in obese mice. PPARα deficiency aggravates obesity-associated hepatic steatosis without increasing liver cancer risk in ob/ob mice, whereas heightened activation of PPARα in ob/ob fatty livers increases the risk of liver cancer.
Footnotes
Supported by NIH grants DK083163 and DK097240 (J.K.R.), R21A1094296 (B.T.), DK60015 and DK58614 (B.P.), and DK60635 (Y.S.K.) and the China Scholarship Council grant CSC 2011630169 (Q.G.).
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosures: None declared.
Supplemental Data
Generation of PPARαΔob/ob mice. A: PPARα−/− mice were mated with heterozygous OB/ob mice to generate PPARα+/−/OB/ob mice, which, when intercrossed, yielded PPARαΔob/ob mice. B: Genotyping for PPARα−/− and ob/ob. Genomic DNA was extracted from tail tips of mice, which served as template for PCR amplification. When separated on 2% agarose gel, PPARα+/+ shows a 143-bp band, PPARα+/− shows 143- and 280-bp bands, and PPARα−/− shows only a 280-bp band. Genotyping of OB/OB (wild-type 155-bp band), OB/ob (heterozygous 55-, 100-, and 155-bp bands), and ob/ob (leptin null 55- and 100-bp bands) mice.
Glucose and insulin tolerance and hepatic triacylglycerol (TG) and cholesterol content. A: Glucose tolerance test (GTT). Mice aged 3 months were fasted for 6 hours, and then given one 1.5 g/kg injection of glucose i.p. Glucose levels were measured at 0, 30, 60, and 120 minutes after injection. B: Insulin tolerance test (ITT) was performed in 3-month-old mice. After 4 hours of fasting, mice were injected with 0.75 mU/kg insulin i.p., and plasma glucose was measured at 0, 15, 30, and 60 minutes. C and D: Total hepatic lipid extracted from 100-mg liver samples was analyzed for liver TG (C) and liver cholesterol (D) Data are shown as means ± SEM. n = 5 (A and B); n = 6 (C and D). ∗P < 0.05, ∗∗P < 0.01 versus wild type; †P < 0.05, ††P < 0.01 versus PPARα−/−; ‡P < 0.05, ‡‡P < 0.01 versus ob/ob.
PPARαΔob/ob mice mobilize hepatic oval cells. A: Immunohistochemical staining of A6, CD45, CK19, and epithelial cell adhesion molecule (EpCAM) in serially cut liver sections obtained from 16-month-old wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. In the livers of PPARαΔob/ob mice, small A6-, CD45-, CK19-, and EpCAM-expressing oval cells are visualized. B: Quantitative PCR analysis to assess hepatic oval cell marker gene expression in the livers of 16-month-old WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice. AFP, alpha-fetoprotein; Alb, albumin.
References
- 1.Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen B.T., Powell L.M. The impact of restaurant consumption among US adults: effects on energy and nutrient intakes. Public Health Nutr. 2014;17:2445–2452. doi: 10.1017/S1368980014001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy J.K. Nonalcoholic steatosis and steatohepatitis, III: peroxisomal beta-oxidation, PPAR alpha, and steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1333–G1339. doi: 10.1152/ajpgi.2001.281.6.G1333. [DOI] [PubMed] [Google Scholar]
- 4.Reddy J.K., Rao M.S. Lipid metabolism and liver inflammation, II: fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 5.Park E.J., Lee J.H., Yu G.Y., He G., Ali S.R., Holzer R.G., Osterreicher C.H., Takahashi H., Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J.C., Horton J.D., Hobbs H.H. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agopian V.G., Kaldas F.M., Hong J.C., Whittaker M., Holt C., Rana A., Zarrinpar A., Petrowsky H., Farmer D., Yersiz H., Xia V., Hiatt J.R., Busuttil R.W. Liver transplantation for nonalcoholic steatohepatitis: the new epidemic. Ann Surg. 2012;256:624–633. doi: 10.1097/SLA.0b013e31826b4b7e. [DOI] [PubMed] [Google Scholar]
- 8.Evans R.M., Barish G.D., Wang Y.X. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Mei M., Yang S., Li Q. Roles of chronic low-grade inflammation in the development of ectopic fat deposition. Mediators Inflamm. 2014;2014:418185. doi: 10.1155/2014/418185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevich-Panigrahi T., Panigrahi S., Wiechec E., Los M. Obesity: pathophysiology and clinical management. Curr Med Chem. 2009;16:506–521. doi: 10.2174/092986709787315568. [DOI] [PubMed] [Google Scholar]
- 11.Tacke F., Luedde T., Trautwein C. Inflammatory pathways in liver homeostasis and liver injury. Clin Rev Allergy Immunol. 2009;36:4–12. doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 12.Fabbrini E., Sullivan S., Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faggioni R., Jones-Carson J., Reed D.A., Dinarello C.A., Feingold K.R., Grunfeld C., Fantuzzi G. Leptin-deficient (ob/ob) mice are protected from T cell-mediated hepatotoxicity: role of tumor necrosis factor alpha and IL-18. Proc Natl Acad Sci U S A. 2000;97:2367–2372. doi: 10.1073/pnas.040561297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J., Viswakarma N., Yu S., Jia Y., Bai L., Vluggens A., Cherkaoui-Malki M., Khan M., Singh I., Yang G., Rao M.S., Borensztajn J., Reddy J.K. Progressive endoplasmic reticulum stress contributes to hepatocarcinogenesis in fatty acyl-CoA oxidase 1-deficient mice. Am J Pathol. 2011;179:703–713. doi: 10.1016/j.ajpath.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra P., Reddy J.K. Peroxisome proliferator-activated receptor-alpha activation and excess energy burning in hepatocarcinogenesis. Biochimie. 2014;98:63–74. doi: 10.1016/j.biochi.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Pyper S.R., Viswakarma N., Yu S., Reddy J.K. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal. 2010;8:e002. doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez F.J., Shah Y.M. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Hays T., Rusyn I., Burns A.M., Kennett M.J., Ward J.M., Gonzalez F.J., Peters J.M. Role of peroxisome proliferator-activated receptor-alpha (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis. 2005;26:219–227. doi: 10.1093/carcin/bgh285. [DOI] [PubMed] [Google Scholar]
- 19.Palkar P.S., Anderson C.R., Ferry C.H., Gonzalez F.J., Peters J.M. Effect of prenatal peroxisome proliferator-activated receptor alpha (PPARalpha) agonism on postnatal development. Toxicology. 2010;276:79–84. doi: 10.1016/j.tox.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan C.Y., Pan J., Chu R., Lee D., Kluckman K.D., Usuda N., Singh I., Yeldandi A.V., Rao M.S., Maeda N., Reddy J.K. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J Biol Chem. 1996;271:24698–24710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.S., Pineau T., Drago J., Lee E.J., Owens J.W., Kroetz D.L., Fernandez-Salguero P.M., Westphal H., Gonzalez F.J. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J., Jia Y., Fu T., Viswakarma N., Bai L., Rao M.S., Zhu Y., Borensztajn J., Reddy J.K. Sustained activation of PPARalpha by endogenous ligands increases hepatic fatty acid oxidation and prevents obesity in ob/ob mice. FASEB J. 2012;26:628–638. doi: 10.1096/fj.11-194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalloyer F., Vandewalle B., Percevault F., Torpier G., Kerr-Conte J., Oosterveer M., Paumelle R., Fruchart J.C., Kuipers F., Pattou F., Fievet C., Staels B. Peroxisome proliferator-activated receptor alpha improves pancreatic adaptation to insulin resistance in obese mice and reduces lipotoxicity in human islets. Diabetes. 2006;55:1605–1613. doi: 10.2337/db06-0016. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y., Zhang X., Chen L., Wu J., Dang H., Wei M., Fan Y., Zhang Y., Zhu Y., Wang N., Breyer M.D., Guan Y. Expression profiling of hepatic genes associated with lipid metabolism in nephrotic rats. Am J Physiol Renal Physiol. 2008;295:F662–F671. doi: 10.1152/ajprenal.00046.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu T., Mukhopadhyay D., Davidson N.O., Borensztajn J. The peroxisome proliferator-activated receptor alpha (PPARalpha) agonist ciprofibrate inhibits apolipoprotein B mRNA editing in low density lipoprotein receptor-deficient mice: effects on plasma lipoproteins and the development of atherosclerotic lesions. J Biol Chem. 2004;279:28662–28669. doi: 10.1074/jbc.M403271200. [DOI] [PubMed] [Google Scholar]
- 26.Fu T., Borensztajn J. Simvastatin causes the formation of cholesterol-rich remnants in mice lacking apoE. Biochem Biophys Res Commun. 2006;341:1172–1176. doi: 10.1016/j.bbrc.2006.01.071. [DOI] [PubMed] [Google Scholar]
- 27.Bai L., Jia Y., Viswakarma N., Huang J., Vluggens A., Wolins N.E., Jafari N., Rao M.S., Borensztajn J., Yang G., Reddy J.K. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miura K., Yang L., van Rooijen N., Brenner D.A., Ohnishi H., Seki E. Toll-like receptor 2 and palmitic acid cooperatively contribute to the development of nonalcoholic steatohepatitis through inflammasome activation in mice. Hepatology. 2013;57:577–589. doi: 10.1002/hep.26081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Y., Qi C., Kashireddi P., Surapureddi S., Zhu Y.J., Rao M.S., Le Roith D., Chambon P., Gonzalez F.J., Reddy J.K. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 30.Viswakarma N., Jia Y., Bai L., Gao Q., Lin B., Zhang X., Misra P., Rana A., Jain S., Gonzalez F.J., Zhu Y.J., Thimmapaya B., Reddy J.K. The Med1 subunit of the mediator complex induces liver cell proliferation and is phosphorylated by AMP kinase. J Biol Chem. 2013;288:27898–27911. doi: 10.1074/jbc.M113.486696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S., Matsusue K., Kashireddy P., Cao W.Q., Yeldandi V., Yeldandi A.V., Rao M.S., Gonzalez F.J., Reddy J.K. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 32.Fan C.Y., Pan J., Usuda N., Yeldandi A.V., Rao M.S., Reddy J.K. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase: implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem. 1998;273:15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 33.Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 34.Reddy J.K., Goel S.K., Nemali M.R., Carrino J.J., Laffler T.G., Reddy M.K., Sperbeck S.J., Osumi T., Hashimoto T., Lalwani N.D., Rao M.S. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc Natl Acad Sci U S A. 1986;83:1747–1751. doi: 10.1073/pnas.83.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halaas J.L., Gajiwala K.S., Maffei M., Cohen S.L., Chait B.T., Rabinowitz D., Lallone R.L., Burley S.K., Friedman J.M. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 36.Nikoozad Z., Ghorbanian M.T., Rezaei A. Comparison of the liver function and hepatic specific genes expression in cultured mesenchymal stem cells and hepatocytes. Iran J Basic Med Sci. 2014;17:27–33. [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen B.E., Goff J.P., Greenberger J.S., Michalopoulos G.K. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology. 1998;27:433–445. doi: 10.1002/hep.510270218. [DOI] [PubMed] [Google Scholar]
- 38.van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J Inherit Metab Dis. 1991;14:407–420. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- 39.Kersten S., Seydoux J., Peters J.M., Gonzalez F.J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto T., Cook W.S., Qi C., Yeldandi A.V., Reddy J.K., Rao M.S. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 41.Park H.K., Ahima R.S. Leptin signaling. F1000Prime Rep. 2014;6:73. doi: 10.12703/P6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy J.K., Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y.T., Shimabukuro M., Koyama K., Lee Y., Wang M.Y., Trieu F., Newgard C.B., Unger R.H. Induction by leptin of uncoupling protein-2 and enzymes of fatty acid oxidation. Proc Natl Acad Sci U S A. 1997;94:6386–6390. doi: 10.1073/pnas.94.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimabukuro M., Koyama K., Chen G., Wang M.Y., Trieu F., Lee Y., Newgard C.B., Unger R.H. Direct antidiabetic effect of leptin through triglyceride depletion of tissues. Proc Natl Acad Sci U S A. 1997;94:4637–4641. doi: 10.1073/pnas.94.9.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maffei M., Fei H., Lee G.H., Dani C., Leroy P., Zhang Y., Proenca R., Negrel R., Ailhaud G., Friedman J.M. Increased expression in adipocytes of ob RNA in mice with lesions of the hypothalamus and with mutations at the db locus. Proc Natl Acad Sci U S A. 1995;92:6957–6960. doi: 10.1073/pnas.92.15.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerre-Millo M., Gervois P., Raspe E., Madsen L., Poulain P., Derudas B., Herbert J.M., Winegar D.A., Willson T.M., Fruchart J.C., Berge R.K., Staels B. Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J Biol Chem. 2000;275:16638–16642. doi: 10.1074/jbc.275.22.16638. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y., Yu X., Gonzales F., Mangelsdorf D.J., Wang M.Y., Richardson C., Witters L.A., Unger R.H. PPAR alpha is necessary for the lipopenic action of hyperleptinemia on white adipose and liver tissue. Proc Natl Acad Sci U S A. 2002;99:11848–11853. doi: 10.1073/pnas.182420899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orci L., Cook W.S., Ravazzola M., Wang M.Y., Park B.H., Montesano R., Unger R.H. Rapid transformation of white adipocytes into fat-oxidizing machines. Proc Natl Acad Sci U S A. 2004;101:2058–2063. doi: 10.1073/pnas.0308258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J.M., Doyle P.J., Iglesias M.A., Watson D.G., Cooney G.J., Kraegen E.W. Peroxisome proliferator-activated receptor (PPAR)-alpha activation lowers muscle lipids and improves insulin sensitivity in high fat-fed rats: comparison with PPAR-gamma activation. Diabetes. 2001;50:411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- 50.Wang M.Y., Lee Y., Unger R.H. Novel form of lipolysis induced by leptin. J Biol Chem. 1999;274:17541–17544. doi: 10.1074/jbc.274.25.17541. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y.T., Wang Z.W., Higa M., Newgard C.B., Unger R.H. Reversing adipocyte differentiation: implications for treatment of obesity. Proc Natl Acad Sci U S A. 1999;96:2391–2395. doi: 10.1073/pnas.96.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gluckman P.D., Lillycrop K.A., Vickers M.H., Pleasants A.B., Phillips E.S., Beedle A.S., Burdge G.C., Hanson M.A. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci U S A. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vickers M.H., Gluckman P.D., Coveny A.H., Hofman P.L., Cutfield W.S., Gertler A., Breier B.H., Harris M. Neonatal leptin treatment reverses developmental programming. Endocrinology. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- 54.Garratt E.S., Vickers M.H., Gluckman P.D., Hanson M.A., Burdge G.C., Lillycrop K.A. Tissue-specific 5' heterogeneity of PPARalpha transcripts and their differential regulation by leptin. PLoS One. 2013;8:e67483. doi: 10.1371/journal.pone.0067483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox I.J., Roy-Chowdhury J. Hepatocyte transplantation. J Hepatol. 2004;40:878–886. doi: 10.1016/j.jhep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 56.Jochheim-Richter A., Rudrich U., Koczan D., Hillemann T., Tewes S., Petry M., Kispert A., Sharma A.D., Attaran F., Manns M.P., Ott M. Gene expression analysis identifies novel genes participating in early murine liver development and adult liver regeneration. Differentiation. 2006;74:167–173. doi: 10.1111/j.1432-0436.2006.00066.x. [DOI] [PubMed] [Google Scholar]
- 57.Golding M., Sarraf C.E., Lalani E.N., Anilkumar T.V., Edwards R.J., Nagy P., Thorgeirsson S.S., Alison M.R. Oval cell differentiation into hepatocytes in the acetylaminofluorene-treated regenerating rat liver. Hepatology. 1995;22:1243–1253. doi: 10.1016/0270-9139(95)90635-5. [DOI] [PubMed] [Google Scholar]
- 58.Petersen B.E., Zajac V.F., Michalopoulos G.K. Hepatic oval cell activation in response to injury following chemically induced periportal or pericentral damage in rats. Hepatology. 1998;27:1030–1038. doi: 10.1002/hep.510270419. [DOI] [PubMed] [Google Scholar]
- 59.Davies R.A., Knight B., Tian Y.W., Yeoh G.C., Olynyk J.K. Hepatic oval cell response to the choline-deficient, ethionine supplemented model of murine liver injury is attenuated by the administration of a cyclo-oxygenase 2 inhibitor. Carcinogenesis. 2006;27:1607–1616. doi: 10.1093/carcin/bgi365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of PPARαΔob/ob mice. A: PPARα−/− mice were mated with heterozygous OB/ob mice to generate PPARα+/−/OB/ob mice, which, when intercrossed, yielded PPARαΔob/ob mice. B: Genotyping for PPARα−/− and ob/ob. Genomic DNA was extracted from tail tips of mice, which served as template for PCR amplification. When separated on 2% agarose gel, PPARα+/+ shows a 143-bp band, PPARα+/− shows 143- and 280-bp bands, and PPARα−/− shows only a 280-bp band. Genotyping of OB/OB (wild-type 155-bp band), OB/ob (heterozygous 55-, 100-, and 155-bp bands), and ob/ob (leptin null 55- and 100-bp bands) mice.
Glucose and insulin tolerance and hepatic triacylglycerol (TG) and cholesterol content. A: Glucose tolerance test (GTT). Mice aged 3 months were fasted for 6 hours, and then given one 1.5 g/kg injection of glucose i.p. Glucose levels were measured at 0, 30, 60, and 120 minutes after injection. B: Insulin tolerance test (ITT) was performed in 3-month-old mice. After 4 hours of fasting, mice were injected with 0.75 mU/kg insulin i.p., and plasma glucose was measured at 0, 15, 30, and 60 minutes. C and D: Total hepatic lipid extracted from 100-mg liver samples was analyzed for liver TG (C) and liver cholesterol (D) Data are shown as means ± SEM. n = 5 (A and B); n = 6 (C and D). ∗P < 0.05, ∗∗P < 0.01 versus wild type; †P < 0.05, ††P < 0.01 versus PPARα−/−; ‡P < 0.05, ‡‡P < 0.01 versus ob/ob.
PPARαΔob/ob mice mobilize hepatic oval cells. A: Immunohistochemical staining of A6, CD45, CK19, and epithelial cell adhesion molecule (EpCAM) in serially cut liver sections obtained from 16-month-old wild-type (WT), PPARα−/−, ob/ob, and PPARαΔob/ob mice. In the livers of PPARαΔob/ob mice, small A6-, CD45-, CK19-, and EpCAM-expressing oval cells are visualized. B: Quantitative PCR analysis to assess hepatic oval cell marker gene expression in the livers of 16-month-old WT, PPARα−/−, ob/ob, and PPARαΔob/ob mice. AFP, alpha-fetoprotein; Alb, albumin.