Abstract
Progress of development is commonly reconstructed from imaging snapshots of chemical or mechanical processes in fixed tissues. As a first step in these reconstructions, snapshots must be spatially registered and ordered in time. Currently, image registration and ordering are often done manually, requiring a significant amount of expertise with a specific system. However, as the sizes of imaging data sets grow, these tasks become increasingly difficult, especially when the images are noisy and the developmental changes being examined are subtle. To address these challenges, we present an automated approach to simultaneously register and temporally order imaging data sets. The approach is based on vector diffusion maps, a manifold learning technique that does not require a priori knowledge of image features or a parametric model of the developmental dynamics. We illustrate this approach by registering and ordering data from imaging studies of pattern formation and morphogenesis in three model systems. We also provide software to aid in the application of our methodology to other experimental data sets.
Keywords: Temporal ordering, Image registration, Vector diffusion maps, Zebrafish, Drosophila
Summary: Learning algorithms allow developmental dynamics to be reconstructed through the automatic registration and ordering of fixed images of developing tissues.
INTRODUCTION
In one of the common approaches to studies of developmental dynamics, a group of embryos is fixed and stained to visualize a particular biochemical or morphological process within a developing tissue. The developmental dynamics must then be reconstructed from multiple embryos, each of which contributes only a snapshot of the relevant process along its developmental trajectory (Jaeger et al., 2004; Fowlkes et al., 2008; Peter and Davidson, 2011). Importantly, the ‘age’ of any given embryo arrested in its development is often only approximately known. Typically, what is known is a certain time window to which a collection of embryos belongs (Castro et al., 2009; Ng et al., 2012; Richardson et al., 2014). Furthermore, images are often collected in different spatial orientations. In order to recover the developmental dynamics from such data sets, snapshots of different embryos must first be spatially aligned or registered, and then ordered in time.
Temporal ordering and registration of images can be done manually when the number of images is small and the differences between them are visually apparent. Fig. 1 shows a caricature of fish development that illustrates the processes of growth and patterning. In this case, temporal ordering can be accomplished by arranging the fish by size, which is monotonic with the developmental progress. Image registration is based on obvious morphological landmarks, such as the positions of the head and the fins. In contrast to this example, real data pose nontrivial challenges for both registration and temporal ordering. In general, the landmarks needed for registration, as well as the attributes that can be used to order the data, are not known a priori. Additional challenges arise from embryo-to-embryo variability, sample size and measurement noise.
Fig. 1.
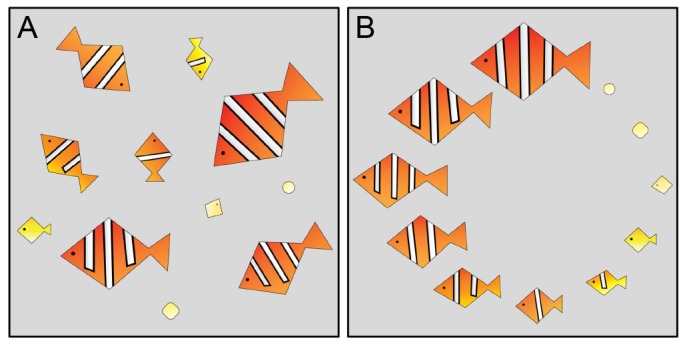
Caricature illustrating the tasks of image registration and temporal ordering. (A) Images of ‘samples’, each in a different orientation and at a different stage of development. (B) Registered and ordered samples. For this caricature, the registration and ordering is straightforward because the data set is small, the landmarks are visually apparent, and the developmental changes are easy to recognize.
We present a robust algorithmic approach to simultaneous registration and temporal ordering. In contrast to a number of previous methodologies (Greenspan et al., 1994; Rowley et al., 1998; Zhao et al., 2003; Zitová and Flusser, 2003; Hajnal and Hill, 2010; Dubuis et al., 2013), ours does not rely on the a priori knowledge of landmarks for registration or markers of developmental progression. The approach is based on vector diffusion maps (Singer and Wu, 2012), a manifold learning algorithm that simultaneously addresses the problems of registration and temporal ordering. This algorithm is one of several nonlinear dimensionality reduction techniques that have been developed over the past decade (Roweis and Saul, 2000; Tenenbaum et al., 2000; Belkin and Niyogi, 2003; Coifman et al., 2005; Coifman and Lafon, 2006) for applications ranging from the analysis of cryo-electron microscopy (cryo-EM) images of individual molecules (Singer et al., 2011; Zhao and Singer, 2014) to face recognition (Lafon et al., 2006) and the classification of CT scans (Fernández et al., 2014).
Here, the vector diffusion maps algorithm is adapted for the analysis of images of tissues in studies of developmental dynamics, with the main objective of revealing stereotypic developmental trajectories from fixed images. To illustrate our approach, we analyze four experimental data sets. The first two data sets come from live imaging studies of Drosophila and zebrafish embryogenesis. In both of these examples, the correct rotational orientation and temporal order are independently known, and these data sets will be used to validate our approach. Our third data set consists of images from fixed Drosophila embryos, for which the correct orientation and order are unknown; here, we will show how the algorithm can help uncover developmental dynamics that are not readily apparent. Our final data set consists of z-stacks of Drosophila wing discs, which we will use to illustrate how our methods can be used to analyze specific types of 3D imaging data. We also show how to compute an average trajectory from a set of registered and ordered fixed images to remove noise due to intersample variability and obtain a smooth description of the underlying developmental dynamics.
RESULTS
Vector diffusion maps for registration and temporal ordering
Vector diffusion maps (Singer and Wu, 2012) is a manifold learning technique developed for data sets that contain two sources of variability: geometric symmetries, such as rotations of the images, which one would like to factor out; and ‘additional’ directions of variability, such as temporal dynamics, which one would like to uncover. Vector diffusion maps combine two algorithms – angular synchronization (Singer, 2011) for image registration, and diffusion maps (Coifman et al., 2005) for extracting intrinsic low-dimensional structure in data – into a single computation. We will use the algorithm to register images of developing tissues with respect to planar rotations, as well as uncover the main direction of variability after removing rotational symmetries. Although, in general, images may contain variations due to rotations, translations and scaling, we will remove the relevant translations and/or scaling via relatively simple image preprocessing, and focus only on factoring out rotations using the vector diffusion maps algorithm. In the case that all relevant symmetries can be removed with straightforward preprocessing, our algorithms can extract the main direction of variability within the imaging data set. We assume that this main direction of remaining variability in these images is parametrized by the developmental time of each embryo. As a consequence, uncovering this direction should reveal the underlying dynamics.
Angular synchronization uses pairwise alignment information to register a set of images in a globally consistent way. A schematic illustration of angular synchronization is shown in Fig. 2A, where each image is represented as a vector, and the goal is to align the entire set of vectors given pairwise alignment measurements. We first compute the angles needed to align pairs of vectors (or images), which in general requires no notion of a template function (Ahuja et al., 2007; Sonday et al., 2013). In this work, we aligned pairs of images with respect to rotations by exhaustively searching over a discretized space of rotation angles to minimize the Euclidean distance between the pixels. However, pairwise alignments can also be computed by aligning appropriate image landmarks or features (Dryden and Mardia, 1998). When the data are noisy, these pairwise measurements may be inaccurate, and so we utilize all pairwise measurements to align the set of images robustly. Using the alignment angles between all pairs of vectors, angular synchronization finds the set of rotation angles (one angle for each vector) that is most consistent with all pairwise measurements (see algorithms in the supplementary material); this is illustrated in Fig. 2B. In this schematic, registration via angular synchronization is trivial, as the pairwise measurements contain no noise. However, the algorithm can register data sets even when many of the pairwise measurements are inaccurate (Singer, 2011).
Fig. 2.
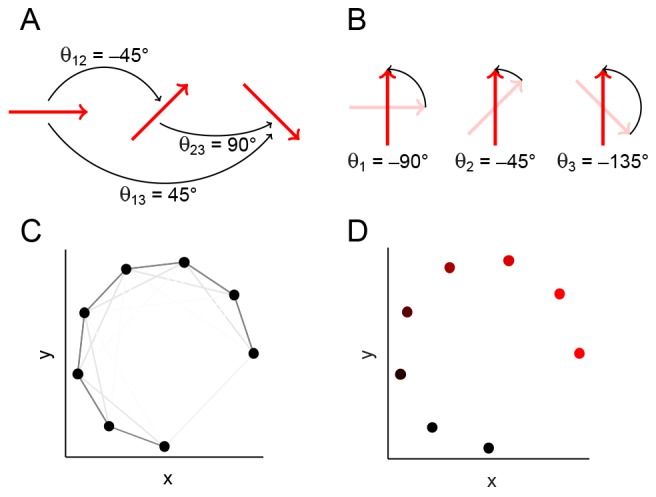
Schematic illustrating angular synchronization and diffusion maps. (A) A set of vectors, each in a different orientation. The pairwise alignment angles are indicated. (B) The vectors from A, each rotated about their midpoint so that the set is globally aligned. Note that the chosen rotation angles are consistent with the pairwise alignments in A: the difference between a pair of angles in B is the same as the pairwise angle in A. (C) Data points (black) that lie on a 1D nonlinear curve in two dimensions. Each pair of points is connected by an edge, and the edge weight is related to the Euclidean distance between the points through a Gaussian kernel (see algorithms in the supplementary material), so that pairs of data points that are close are connected by darker (‘stronger’) edges. (D) The data in C, colored by the first (non-trivial) eigenvector from the diffusion map computational procedure. The color intensity is monotonic with the perceived curve arc length, thus parametrizing the curve.
After removing variability due to rotations, the developmental dynamics may be revealed by ordering the data along the 1D curve that parametrizes most of the remaining variability in the data. Such a curve can be discovered using diffusion maps (Coifman et al., 2005), a nonlinear dimensionality reduction technique that reveals a parametrization of data that lies on a low-dimensional manifold in high-dimensional space. The idea is illustrated in Fig. 2C, where the data are 2D points that lie on a 1D (nonlinear) curve. We use local information about the data to find a parametrization that respects the underlying manifold geometry, so that points that are close in high-dimensional space (e.g. images which look similar) are close in our parametrization. This idea of locality is denoted by the weight of the edges in Fig. 2C: data points that are close are connected by dark edges and, clearly, the dark edges are more ‘informative’ about the low-dimensional structure of the data. The color in Fig. 2D depicts the 1D parametrization or ordering of the data that we can detect visually. A detailed example of using vector diffusion maps to register and order synthetic data is given in supplementary material Fig. S2, and a step-by-step tutorial of the diffusion maps implementation is provided with the software. In our working examples, each data point will be of much higher dimension (e.g. a pixelated image or 3D voxel data), and so we cannot extract this low-dimensional structure visually. Instead, we will use diffusion maps, which automatically uncovers a parametrization of our high-dimensional data from the eigenvectors of the appropriate matrix (see algorithms in the supplementary material). Furthermore, the corresponding eigenvalues will allow us to test our assumption that our data approximately lie on a 1D manifold (see supplementary material Figs S3-S6).
Method validation using live imaging
Drosophila gastrulation
To validate the proposed approach, we first applied our algorithm to a data set where the true temporal order and rotational orientation of the images were known a priori. This data set was obtained through live imaging near the posterior pole of a vertically oriented Drosophila embryo during the 20 min spanning the late stages of cellularization through early gastrulation. During this time window, the ventral furrow is formed where the ventral side buckles towards the center of the embryo, internalizing the future muscle cells and forming a characteristic ‘omega’ shape. Germband extension then causes cells from the ventral side to move towards the posterior pole of the embryo, and then wrap around to the dorsal side (Leptin, 2005). At the end of this process, cells that were originally on the ventral and posterior side of the embryo find themselves on the dorsal side, causing a similar omega to appear on the dorsal side.
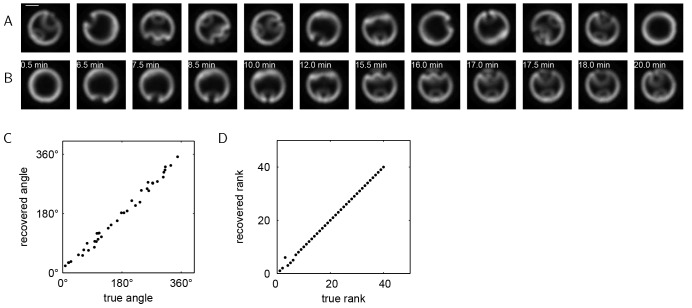
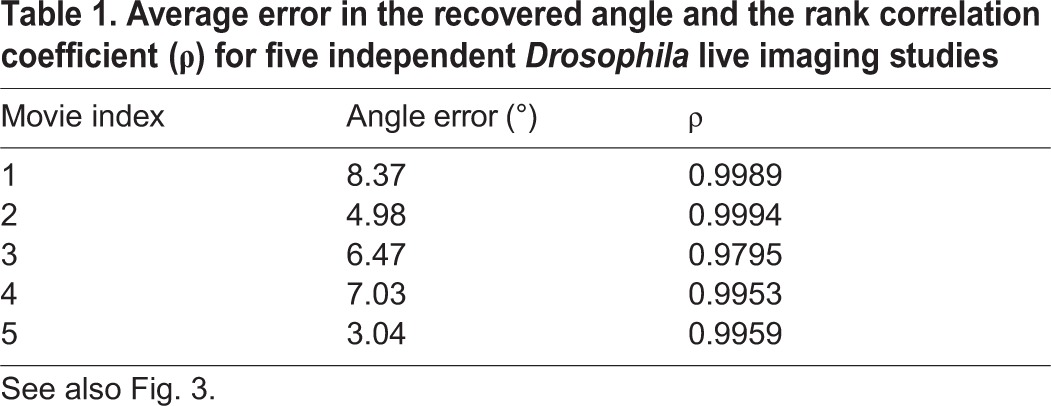
Fig. 3A shows selected images from this live imaging data set, which contains 40 consecutive frames taken at 30 s intervals at a fixed position within a single embryo. Each image shows an optical cross-section near the posterior pole of a vertically oriented developing Drosophila embryo, with the nuclei labeled by Histone-RFP. Each frame was arbitrarily rotated, and the order of the frames was scrambled. The task is now to register these images and order them in time to reconstruct the developmental trajectory.
Fig. 3.
Method validation using live imaging of Drosophila embryos. (A) Selected images from a live imaging study of a Drosophila embryo during gastrulation. Scale bar: 50 μm. Each frame is in an arbitrary rotational orientation and the order of the frames has been shuffled. (B) Images from A registered and ordered by vector diffusion maps. The dorsal side of each embryo now appears at the top of each image and the ventral side appears at the bottom. The real time of each frame is also indicated, where t=0 corresponds to the initiation of ventral furrow formation. (C) The correlation between the recovered rotation angle (using vector diffusion maps) and the true rotation angle. The average absolute error in the recovered angles is 8.37°. (D) The correlation between the recovered rank (using vector diffusion maps) and the true rank. The rank correlation coefficient is 0.9989 (see also Table 1).
We used vector diffusion maps to register and order the images. Fig. 3B shows the images from Fig. 3A now registered and ordered; the real time for each frame is also indicated. With a small number of exceptions, the recovered ordering is consistent with the real-time dynamics. Fig. 3C,D show the correlations between the recovered and true angles and rank orders, respectively, for the entire data set. Both the angles and the ranks are recovered with a high degree of accuracy. We note that determining which end of the trajectory corresponds to early in the developmental progression is a post-processing task that requires some a priori information.
To assess the robustness of the proposed methodology, we repeated this procedure with four additional data sets extracted from independent live imaging studies spanning the same developmental time period. The results are shown in Table 1. The errors in the recovered angles are all less than 10°, and the rank correlation coefficients are consistently greater than 95%, indicating that our methodology can reproducibly order data of this type.
Table 1.
Average error in the recovered angle and the rank correlation coefficient (ρ) for five independent Drosophila live imaging studies
Zebrafish epiboly
As another validation for the proposed methodology, we applied our algorithm to a time-lapse movie of zebrafish embryogenesis. We used publicly available live imaging data of zebrafish embryogenesis [https://zfin.org/zf_info/movies/Zebrafish.mov (Karlstrom and Kane, 1996)]. Taken with a differential interference contrast (DIC) microscope, the movie records the first 17 h of zebrafish development, from the single-cell stage to 16-somite stage. We selected 120 consecutive frames from this movie that capture 5.5 h of epiboly (3.5-9 h after fertilization). In this experiment, embryos were immobilized for imaging so that the position and orientation remained fixed (Kane et al., 1996). At the start of the time window, cells have divided 10-11 times and have accumulated in a cell mass above the yolk. The cell mass is then compressed and the animal-vegetal axis of the embryo (vertical axis in Fig. 4) shortens to form a spherical embryo by the end of the fourth hour of development. Then, the yolk syncytial layer, which forms the boundary between the yolk and the cell mass, moves upward, forming a dome-shaped structure. During this stage, the cells rearrange to form a uniform layer about four cells thick. With time, this cell layer then spreads across the yolk and expands toward the vegetal pole. At the end of epiboly, the blastoderm completely engulfs the yolk.
Fig. 4.
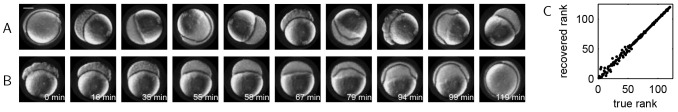
Method validation using live imaging of a zebrafish embryo. (A) Selected images from a movie of zebrafish epiboly. Scale bar: 200 μm. Each frame is in an arbitrary rotational orientation and the order of the frames has been shuffled. (B) Images from A after registration and ordering using vector diffusion maps. The real time of each frame is also indicated. (C) Correlation between the rank recovered using vector diffusion maps and the true rank. The rank correlation coefficient is 0.9954. The larger errors in the recovered ranks towards the beginning of the trajectory are due to slow cell movement within that time window.
As in the example of Drosophila embryo live imaging, the 2D frames were randomly rotated and shuffled (Fig. 4A). We then used vector diffusion maps to register and order the frames. The results are shown in Fig. 4B. The recovered rotations and order are consistent with the expected developmental dynamics, as shown in the correlations between the recovered and true ranks (Fig. 4C). Quantitatively, the rank correlation coefficient for this data set is 0.9954, and the average error in the recovered angle is 4.14°. Some errors in ordering images of the early embryo result from slow cell movement during the early developmental stage when cells divide and accumulate above the yolk. During epiboly, cell movement is more dynamic and the recovered ordering is more consistent with the real dynamics.
In summary, we have shown that our approach to temporal ordering performs very well on imaging data of two different developmental processes (Drosophila gastrulation and zebrafish epiboly) taken with two different imaging methods (fluorescent microscopy and DIC), where the true temporal order is known a priori. Provided that significant dynamics exist within the data set and that the developmental trajectory is well sampled, the developmental dynamics can be recovered.
Data sets with intersample variability
Fixed images of Drosophila gastrulation
We have analyzed how our algorithm performs on two model data sets where all images come from a single embryo. In practice, we are interested in cases in which each image comes from a different embryo, and the largest source of noise in the considered data set arises from embryo-to-embryo variability. To demonstrate that our methods are robust to such variations, we constructed a synthetic timecourse data set by selecting a random image from one of five Drosophila live imaging data sets (those data sets used in Fig. 3) at each time point. The resulting data set is spatially unregistered, scrambled in time, and reflects embryo-to-embryo variability. The median rank correlation coefficient when ordering such a synthetic timecourse using our methodology was 0.77, indicating that the algorithm can recover the temporal order even under noisy conditions.
We then applied our approach to a data set where the true rotational orientation and temporal order were not known a priori. Fig. 5A shows selected images from a set of 120 images of developing Drosophila embryos that cover a 30 min interval spanning late cellularization through gastrulation. This data set is more complex than the live imaging data sets in that it contains significantly more images, each of which provides information about tissue morphology and the spatial distribution of two regulatory proteins. Each image shows an optical cross-section of the posterior view of a different embryo at a different rotational orientation and fixed at a different (and unknown) developmental time. The nuclei (gray) were labeled with the DNA stain DAPI. Embryos were stained with an antibody that recognizes Twist (Twi, green), a transcription factor that specifies the cells of the future muscle tissue. Another signal is provided by the phosphorylated form of the extracellular signal regulated kinase [dpERK (ERK is also known as Rolled in Drosophila), red], an enzyme that, in this context, specifies a subset of neuronal cells (Lim et al., 2013).
Fig. 5.
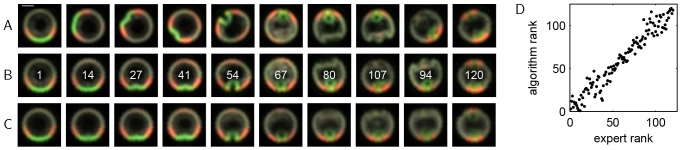
Analysis of images of fixed Drosophila embryos. (A) Images of Drosophila embryos stained for nuclei (gray), Twi (green) and dpERK (red). Scale bar: ∼50 μm (images have been rescaled to remove slight interembryo size variations). Each image is of a different embryo arrested at a different developmental time and in a different rotational orientation. (B) Data from A, registered and ordered using vector diffusion maps. The ‘expert rank’ for each image is indicated. (C) A representative developmental trajectory obtained from local averaging of the entire set of registered and ordered images (see smooth trajectories from registered and ordered images in the supplementary material). (D) Correlation between the image ranks calculated from the vector diffusion maps algorithm and the ranks obtained from ordering by an expert. The rank correlation coefficient is 0.9716.
Fig. 5B shows the selected images in Fig. 5A now registered and ordered using vector diffusion maps. Registered and ordered images of individual embryos can then be used to construct a representative average trajectory. Each snapshot in the average trajectory is the (weighted) average of a group of successive images from the registered and ordered data set (see smooth trajectories from registered and ordered images in the supplementary material). Averaging successive images removes some of the interembryo variability, so that sequential snapshots of this averaged trajectory, as shown in Fig. 5C, serve as a summary of the stereotypic developmental dynamics.
From this average trajectory, we can now easily see the developmental progression consistent with the known dynamics: dpERK first appears as two lateral peaks at the ventrolateral side of the embryo, and a third dpERK peak then appears at the dorsal side of the embryo. During mesoderm invagination, the two ventrolateral dpERK peaks merge together, eventually forming, together with Twi, the omega shape. The dorsal dpERK peak then disappears during germband extension as cells from the ventral side wrap around to the dorsal side. At the end of this process, similar omegas formed by Twi and dpERK appear on the dorsal side of the embryo; these patterns are most readily seen in the last image of Fig. 5C. Thus, vector diffusion maps can accomplish the tasks presented in the caricature in Fig. 1, even in the absence of information about image landmarks and without a priori knowledge of developmental features.
To evaluate the quality of our registration and ordering, we can use prior knowledge about the developmental system. The Twi signal is known to form a single peak at the ventralmost point of the embryo. We found that the standard deviation in the location of this peak in the set of registered images was ∼8°, indicating that the algorithm successfully aligns the ventralmost points of the images. Because the developmental time of each embryo cannot be easily estimated, we have few options for evaluating the quality of our temporal ordering. We compared the ordering obtained from vector diffusion maps with the ordering provided by a trained embryologist who is knowledgeable about the developmental progression and the important image features. The ranks from the ordering provided by the embryologist, which we will refer to as the ‘expert rank’, are indicated for the images in Fig. 5B, and the rank correlation (Fig. 5D) shows that our ordering is consistent with the expert ordering.
Fixed z-stacks of Drosophila wing discs
In this section we show that the approach can readily be applied to 3D data. We restrict ourselves to the case in which an obvious fixed axis exists, so that only rotations of the 3D data around this axis need be taken into account. This does not constitute an inherent limitation for vector diffusion maps. Although, for simplicity, here we will not discuss the general case, incorporating general 3D symmetries is possible (Arie-Nachimson et al., 2012; Cucuringu et al., 2013; Wang and Singer, 2013).
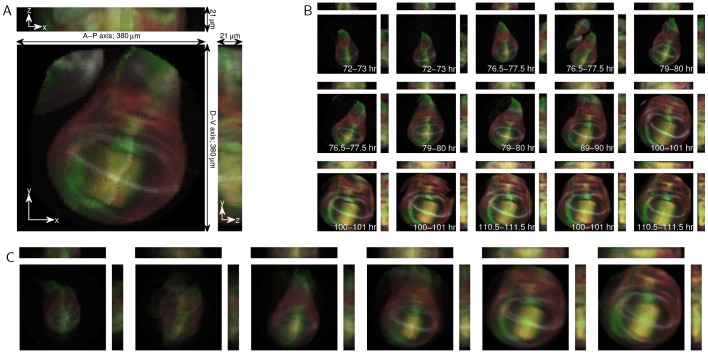
To demonstrate this approach, we used an existing 3D data set of fixed Drosophila wing imaginal discs (Hamaratoglu et al., 2011). Imaginal discs are groups of progenitor cells in fly larva that will transform into specific organs during metamorphosis. The wing disc is an imaginal disc that gives rise to the wing, thorax and hinge. The data set is composed of 46 fixed wing discs with developmental times that range from 72 to 112 h after fertilization. Each disc contains 21 z-slices taken at 1 μm intervals. The discs were dissected from larvae expressing the Dad-GFP reporter construct (green) and stained with antibodies that recognize Spalt (red), Wingless (gray) and Patched (gray), the factors that play important roles in disc patterning and growth (Fig. 6A).
Fig. 6.
Analysis of 3D Drosophila wing disc z-stacks. (A) Maximum projections of an example 3D Drosophila wing disc z-stack. The anterior-posterior (A-P) and dorsal-ventral (D-V) axes are indicated. Discs express the Dad-GFP reporter construct (green) and are stained for Spalt (red), Wingless (gray) and Patched (gray). Projections along the x, y and z axes are shown. (B) Example 3D images, ordered using diffusion maps. The time cohort, as assessed by an expert, is indicated for each image, and the rank correlation coefficient between the diffusion maps ordering and the expert timing is 0.9427. (C) The average developmental trajectory for the registered and ordered images.
In the wing disc, the anterior-posterior and dorsal-ventral axes are significantly longer than the third principal axis (see Fig. 6A). Therefore, we need not consider registration in all three dimensions, and can instead focus on registering the wing discs with respect to rotations only in the x-y plane. To register the data, we first aligned the maximum intensity projections using angular synchronization. We then used these rotations to register the full 3D data in the x-y plane. Because the maximum intensity projections are 2D images, this step is no more computationally intensive than in the previous examples. Such an approach is possible when there are distinct major and minor axes within a 3D sample, which reduces the rotational degrees of freedom.
We then used diffusion maps to order the registered 3D data. Fig. 6B shows selected images from the data set ordered by diffusion maps. In the original data set, each disc was assigned to one of six time classes (72-73, 76.5-77.5, 79-80, 89-90, 100-101 and 110.5-111.5 h after fertilization, Fig. 6B) by an expert. In the ordered set, the size of the wing disc grows, and the intensity of the Dad-GFP signal increases as a function of time. The rank correlation coefficient based on the time class is 0.9436. The registration errors are primarily due to some wing discs having extra tissue attached to them (such as the image in Fig. 6A and the fourth image in Fig. 6B). Even with such obstructions, we can accurately order the images and extract a stereotypical developmental trajectory, as shown in Fig. 6C, by averaging (see smooth trajectories from registered and ordered images in the supplementary material). We can now clearly see the growth of the wing disc, even though averaging somewhat blurs some finer scale structures.
Computational requirements
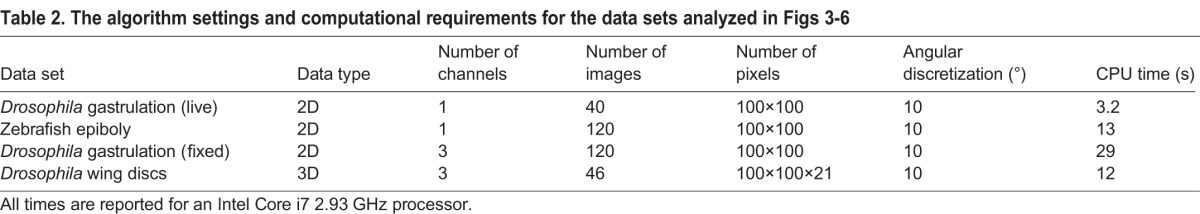
The computational costs for our methodology are outlined in Fig. 7 and Table 2. The computational time is a function of the number of images in the data set, the number of pixels in each point, and the angular resolution to compute the pairwise rotations (see registering images in the supplementary material). Furthermore, the computation of the pairwise rotational alignments, which accounts for the majority of the computational time, can be easily parallelized, and only a subsample of the pairwise alignments need be computed for larger data sets for accurate recovery of the underlying rotations (Singer, 2011). Because the computational cost increases with the image resolution, we chose to subsample all of our data sets to 100×100 pixels. This resolution allowed us to rapidly analyze our data sets while retaining all of the relevant developmental features. However, as can be seen from the computational costs in Fig. 7 and Table 2, it is feasible to use our algorithms to analyze higher-resolution images.
Fig. 7.
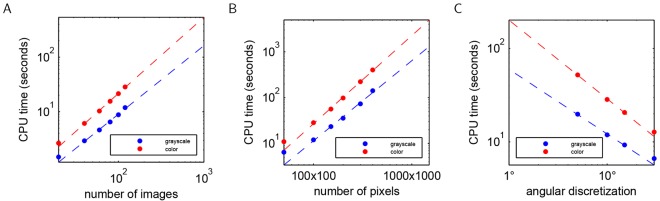
Computational requirements for the presented methodology. (A) CPU time as a function of the number of images in the data set (for 100×100 pixel images and 10° angular discretization). Empirically, the CPU time scales as ∼n1.33 in number of images. (B) CPU time as a function of the number of pixels in the images (for 120 images and 10° angular discretization). Empirically, the CPU time scales as ∼n1.83 in the number of pixels. (C) CPU time as a function of the number of rotations (for 120 images of 100×100 pixels). Empirically, the CPU time scales as ∼n−0.77 in the angular discretization.
Table 2.
The algorithm settings and computational requirements for the data sets analyzed in Figs 3-6
The requisite user intervention and parameter tuning required for our method is relatively minor. As a first step, images must be preprocessed so that the Euclidean distance between the pixels is informative. Our software provides several preprocessing options (such as blurring, rescaling and mean-centering), as well as some guidance for what options to select depending on the system of interest. Two algorithmic parameters – the angular discretization to compute the pairwise alignments, and the diffusion maps kernel scale that determines which data points are ‘close’ (see Fig. 2 and algorithms in the supplementary material) – must also be defined. We also provide some guidance on selecting these parameters, and we find that the results are robust to both of these parameters. Overall, the tasks of image preprocessing and parameter selection are relatively simple compared with the manual registration and ordering of images, and so this methodology is promising for much larger imaging data sets that are impractical to evaluate manually.
DISCUSSION
Temporal ordering of large-scale data has been performed in the context of molecular profiling studies, in which data points are vectors describing the expression levels of different mRNAs (Gupta and Bar-Joseph, 2008; Anavy et al., 2014; Trapnell et al., 2014). At the same time, temporal ordering of imaging data sets was undertaken with a significant amount of manual intervention and using registered images as a starting point (Fowlkes et al., 2008; Surkova et al., 2008; Yuan et al., 2014), or using some a priori knowledge of the developmental processes under examination (Dubuis et al., 2013). In contrast to most of the existing registration approaches, which rely on a knowledge of appropriate landmarks in the images (Dryden and Mardia, 1998), such as the eyes in face-recognition applications (Zhao et al., 2003), algorithms based on angular synchronization can register images even in the absence of such information, making them relevant for a wide variety of applications.
Angular synchronization and vector diffusion maps have been used to reconstruct molecular shapes from cryo-electron microscopy images (Singer et al., 2011; Singer and Wu, 2012; Zhao and Singer, 2014). Because of high levels of instrument noise in these data, thousands of images were needed for successful shape reconstruction. Based on the results presented here, we expect that much smaller data sets will be sufficient for successful reconstruction of developmental trajectories from snapshots of fixed tissues. In general, the size of the data set required for accurate registration and ordering is a function of the instrument noise, interembryo variability, and the complexity of the developmental dynamics.
The benefits of our approach to image data mining are twofold. First, the algorithm can accomplish the tasks of registration and ordering in a single step. Furthermore, because our methodology is nonlinear, it can successfully order data sets that contain complex dynamics (see supplementary material Table S1 for a comparison of ordering using linear principal component analysis versus vector diffusion maps for the data sets presented in this paper). We expect nonlinear techniques to be necessary for larger data sets that span a wider dynamic range. The main utility of our proposed methodology lies in the analysis of data sets containing hundreds of images from systems that have not been well studied. For such data sets, manual ordering of the images can be nontrivial, and our algorithms can clearly accelerate uncovering of the underlying developmental dynamics.
We acknowledge that our methods, although general, do have limitations. The first is that we require enough data to sufficiently sample the developmental trajectory. Therefore, for very small and/or very noisy data sets, our algorithms might fail. Second, the pertinent image features need to be large compared with the noise and the image resolution. In all of our examples, the relevant expression patterns and morphological structures span several pixels and are large compared with both the instrument noise and embryo-to-embryo variability, making the Euclidean distance between pixels a good measure of image similarity.
Vector diffusion maps allow us to automatically register images, which is an essential task for many applications. Simultaneously, the algorithm provides us with parameters to describe each image. In the examples presented here, we have focused on ordering the images in time using the first vector diffusion maps coordinate. In general, we can recover several coordinates that concisely and comprehensively describe the data set. This parametrization can then be used for typical data analysis tasks, such as outlier detection and model fitting. Furthermore, images taken from different viewing directions can be analyzed, as the vector diffusion maps parametrization will organize the images according to the viewing angle (Singer et al., 2011). Another direction for future work is related to the joint analysis of data sets provided by different imaging approaches, such as merging live imaging data of tissue morphogenesis with snapshots of cell signaling and gene expression from fixed embryos (Rübel et al., 2010; Krzic et al., 2012; Dsilva et al., 2013; Ichikawa et al., 2014). It would also be interesting to explore the connections between our proposed approach and recently developed methods for the ordering and classification of face images (Kemelmacher-Shlizerman et al., 2011, 2014). Given the rapidly increasing volumes of imaging data from studies of multiple developmental systems, we expect that the dimensionality reduction approaches discussed in this work will prove increasingly useful for biologists and motivate future applications and algorithmic advances.
MATERIALS AND METHODS
Drosophila embryo experiments
Oregon-R was used as the wild-type Drosophila strain. Embryos were collected and fixed at 22°C. Monoclonal rabbit anti-dpERK (1:100, Cell Signaling, cat. 4370) and rat anti-Twist (1:500; a gift from Eric Wieschaus, Princeton University) were used to stain proteins of interest; DAPI (1:10,000, Vector Laboratories) was used to visualize nuclei, and Alexa Fluor secondary antibodies (1:500, Invitrogen) were used. The Histone-RFP strain (Bloomington Stock Center) was used to obtain time-lapse movies of gastrulating embryos at 22°C. Live embryos were loaded to the microfluidic device with PBST (PBS with 0.02% Triton X-100) to keep them oxidized, and fixed embryos were loaded with 90% glycerol.
Drosophila embryo microscopy
A Nikon A1-RS scanning confocal microscope and Nikon 60× Plan-Apo oil objective were used to image Drosophila embryos. Embryos were collected, stained, and imaged together under the same microscope settings. End-on imaging was performed using the microfluidics device described previously (Chung et al., 2011). Images were collected at the focal plane ∼90 μm from the posterior pole of the embryo (see supplementary material Fig. S1).
Image preprocessing
Images were subsampled, normalized, blurred and centered prior to diffusion maps analysis to remove any variations due to the experimental and imaging framework. Details of the specific preprocessing operations applied to each imaging data set are given in the image preprocessing section in the supplementary material.
Software and imaging data
All algorithms and analysis were implemented in MATLAB (R2013b, MathWorks) and are described further in the methods in the supplementary material. Software, including documentation and tutorials, along with the full imaging data sets used in this paper are available at genomics.princeton.edu/stas/publications.html under ‘Code for the temporal ordering and registration of images in studies of developmental dynamics’.
Supplementary Material
Acknowledgements
We thank Fisun Hamaratoglu and Markus Affolter for providing the wing disc data; and Angela DePace, Granton Jindal, Adam Finkelstein, Thomas Funkhouser and John Storey for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
C.J.D. was supported by the Department of Energy Computational Science Graduate Fellowship (CSGF) [DE-FG02-97ER25308] and the National Science Foundation Graduate Research Fellowship [DGE 1148900]. B.L. and S.Y.S. were supported by the National Institutes of Health [R01GM086537]. H.L. was supported by the National Science Foundation Grant Emerging Frontiers in Research and Innovation (EFRI) [1136913]. A.S. was supported by the Air Force Office of Scientific Research [FA9550-12-1-0317]. I.G.K. was supported by the National Science Foundation (CS&E Program). Deposited in PMC for release after 12 months.
Author contributions
Scientific approaches were developed by C.J.D., B.L., H.L., A.S., I.G.K. and S.Y.S. Experiments were performed by B.L. Data analysis was performed by C.J.D. and B.L. The manuscript was prepared and edited by C.J.D., B.L., H.L., A.S., I.G.K. and S.Y.S.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.119396/-/DC1
References
- Ahuja S., Kevrekidis I. G. and Rowley C. W. (2007). Template-based stabilization of relative equilibria in systems with continuous symmetry. J. Nonlinear Sci. 17, 109-143 10.1007/s00332-005-0801-7 [DOI] [Google Scholar]
- Anavy L., Levin M., Khair S., Nakanishi N., Fernandez-Valverde S. L., Degnan B. M. and Yanai I. (2014). BLIND ordering of large-scale transcriptomic developmental timecourses. Development 141, 1161-1166 10.1242/dev.105288 [DOI] [PubMed] [Google Scholar]
- Arie-Nachimson M., Kovalsky S. Z., Kemelmacher-Shlizerman I., Singer A. and Basri R. (2012). Global motion estimation from point matches. In Second International Conference on 3D Imaging, Modeling, Processing, Visualization and Transmission (3DIMPVT) 2012, pp. 81-88 IEEE. [Google Scholar]
- Belkin M. and Niyogi P. (2003). Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 15, 1373-1396 10.1162/089976603321780317 [DOI] [Google Scholar]
- Castro C., Luengo-Oroz M. A., Desnoulez S., Duloquin L., Fernandez-de-Manuel L., Montagna S., Ledesma-Carbayo M. J., Bourgine P., Peyrieras N. and Santos A. (2009). An automatic quantification and registration strategy to create a gene expression atlas of zebrafish embryogenesis. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009, 1469-1472 10.1109/IEMBS.2009.5332436 [DOI] [PubMed] [Google Scholar]
- Chung K., Kim Y., Kanodia J. S., Gong E., Shvartsman S. Y. and Lu H. (2011). A microfluidic array for large-scale ordering and orientation of embryos. Nat. Methods 8, 171-176 10.1038/nmeth.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman R. R. and Lafon S. (2006). Geometric harmonics: a novel tool for multiscale out-of-sample extension of empirical functions. Appl. Comput. Harmon. Anal. 21, 31-52 10.1016/j.acha.2005.07.005 [DOI] [Google Scholar]
- Coifman R. R., Lafon S., Lee A. B., Maggioni M., Nadler B., Warner F. and Zucker S. W. (2005). Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proc. Natl. Acad. Sci. USA 102, 7426-7431 10.1073/pnas.0500334102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucuringu M., Singer A. and Cowburn D. (2013). Eigenvector synchronization, graph rigidity and the molecule problem. Inf. Inference 1, 21-67 10.1093/imaiai/ias002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden I. L. and Mardia K. V. (1998). Statistical Shape Analysis. John Wiley. [Google Scholar]
- Dsilva C. J., Talmon R., Rabin N., Coifman R. R. and Kevrekidis I. G. (2013). Nonlinear intrinsic variables and state reconstruction in multiscale simulations. J. Chem. Phys. 139, 184109 10.1063/1.4828457 [DOI] [PubMed] [Google Scholar]
- Dubuis J. O., Samanta R. and Gregor T. (2013). Accurate measurements of dynamics and reproducibility in small genetic networks. Mol. Syst. Biol. 9, 639 10.1038/msb.2012.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Á., Rabin N., Coifman R. R. and Eckstein J. (2014). Diffusion methods for aligning medical datasets: location prediction in CT scan images. Med. Image Anal. 18, 425-432 10.1016/j.media.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Fowlkes C. C., Hendriks C. L. L., Keränen S. V. E., Weber G. H., Rübel O., Huang M.-Y., Chatoor S., DePace A. H., Simirenko L., Henriquez C. et al. (2008). A quantitative spatiotemporal atlas of gene expression in the drosophila blastoderm. Cell 133, 364-374 10.1016/j.cell.2008.01.053 [DOI] [PubMed] [Google Scholar]
- Greenspan H., Belongie S., Goodman R. and Perona P. (1994). Rotation invariant texture recognition using a steerable pyramid. In Proceedings of the 12th IAPR International Conference on Pattern Recognition, Vol. 2, pp. 162-167. [Google Scholar]
- Gupta A. and Bar-Joseph Z. (2008). Extracting dynamics from static cancer expression data. IEEE/ACM Trans. Comput. Biol. Bioinform. 5, 172-182 10.1109/TCBB.2007.70233 [DOI] [PubMed] [Google Scholar]
- Hajnal J. V. and Hill D. L. (2010). Medical Image Registration. CRC Press. [Google Scholar]
- Hamaratoglu F., de Lachapelle A. M., Pyrowolakis G., Bergmann S. and Affolter M. (2011). Dpp signaling activity requires Pentagone to scale with tissue size in the growing Drosophila wing imaginal disc. PLoS Biol. 9, e1001182 10.1371/journal.pbio.1001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T., Nakazato K., Keller P. J., Kajiura-Kobayashi H., Stelzer E. H. K., Mochizuki A. and Nonaka S. (2014). Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3D tracking tools. Nat. Protoc. 9, 575-585 10.1038/nprot.2014.035 [DOI] [PubMed] [Google Scholar]
- Jaeger J., Surkova S., Blagov M., Janssens H., Kosman D., Kozlov K. N., Manu, Myasnikova E., Vanario-Alonso C. E., Samsonova M. et al. (2004). Dynamic control of positional information in the early Drosophila embryo. Nature 430, 368-371 10.1038/nature02678 [DOI] [PubMed] [Google Scholar]
- Kane D. A., Hammerschmidt M., Mullins M. C., Maischein H.-M., Brand M., van Eeden F., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C.-P. et al. (1996). The zebrafish epiboly mutants. Development 123, 47-55. [DOI] [PubMed] [Google Scholar]
- Karlstrom R. O. and Kane D. A. (1996). A flipbook of zebrafish embryogenesis. Development 123, 461-462. [DOI] [PubMed] [Google Scholar]
- Kemelmacher-Shlizerman I., Shechtman E., Garg R. and Seitz S. M. (2011). Exploring photobios. ACM Trans. Graph. 30, 61 10.1145/2010324.1964956 [DOI] [Google Scholar]
- Kemelmacher-Shlizerman I., Suwajanakorn S. and Seitz S. M. (2014). Illumination-aware age progression. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2014, pp. 3334-3341. [Google Scholar]
- Krzic U., Gunther S., Saunders T. E., Streichan S. J. and Hufnagel L. (2012). Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730-733 10.1038/nmeth.2064 [DOI] [PubMed] [Google Scholar]
- Lafon S., Keller Y. and Coifman R. R. (2006). Data fusion and multicue data matching by diffusion maps. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1784-1797 10.1109/TPAMI.2006.223 [DOI] [PubMed] [Google Scholar]
- Leptin M. (2005). Gastrulation movements: the logic and the nuts and bolts. Dev. Cell 8, 305-320 10.1016/j.devcel.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Lim B., Samper N., Lu H., Rushlow C., Jiménez G. and Shvartsman S. Y. (2013). Kinetics of gene derepression by ERK signaling. Proc. Natl. Acad. Sci. USA 110, 10330-10335 10.1073/pnas.1303635110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L. L., Sunkin S. M., Feng D., Lau C., Dang C. and Hawrylycz M. J. (2012). Large-scale neuroinformatics for in situ hybridization data in the mouse brain. Int. Rev. Neurobiol. 104, 159-182 10.1016/B978-0-12-398323-7.00007-0 [DOI] [PubMed] [Google Scholar]
- Peter I. S. and Davidson E. H. (2011). A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635-639 10.1038/nature10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L., Stevenson P., Venkataraman S., Yang Y., Burton N., Rao J., Christiansen J. H., Baldock R. A. and Davidson D. R. (2014). EMAGE: electronic mouse atlas of gene expression. In Mouse Molecular Embryology (ed. Lewandoski M.), pp. 61-79 Springer; 10.1007/978-1-60327-292-6_5 [DOI] [PubMed] [Google Scholar]
- Roweis S. T. and Saul L. K. (2000). Nonlinear dimensionality reduction by locally linear embedding. Science 290, 2323-2326 10.1126/science.290.5500.2323 [DOI] [PubMed] [Google Scholar]
- Rowley H. A., Baluja S. and Kanade T. (1998). Rotation invariant neural network-based face detection. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition 1998, pp. 38-44. [Google Scholar]
- Rübel O., Ahern S., Bethel E. W., Biggin M. D., Childs H., Cormier-Michel E., DePace A., Eisen M. B., Fowlkes C. C., Geddes C. G. R. et al. (2010). Coupling visualization and data analysis for knowledge discovery from multi-dimensional scientific data. Procedia Comput. Sci. 1, 1757-1764 10.1016/j.procs.2010.04.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A. (2011). Angular synchronization by eigenvectors and semidefinite programming. Appl. Comput. Harmon. Anal. 30, 20-36 10.1016/j.acha.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A. and Wu H.-T. (2012). Vector diffusion maps and the connection Laplacian. Commun. Pure Appl. Math. 65, 1067-1144 10.1002/cpa.21395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Zhao Z., Shkolnisky Y. and Hadani R. (2011). Viewing angle classification of cryo-electron microscopy images using eigenvectors. SIAM J. Imaging Sci. 4, 723-759 10.1137/090778390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonday B., Singer A. and Kevrekidis I. G. (2013). Noisy dynamic simulations in the presence of symmetry: data alignment and model reduction. Comput. Math. Appl. 65, 1535-1557 10.1016/j.camwa.2013.01.024 [DOI] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu, Myasnikova E., Samsonova A. A., Spirov A., Vanario-Alonso C. E., Samsonova M. and Reinitz J. (2008). Characterization of the Drosophila segment determination morphome. Dev. Biol. 313, 844-862 10.1016/j.ydbio.2007.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum J. B., De Silva V. and Langford J. C. (2000). A global geometric framework for nonlinear dimensionality reduction. Science 290, 2319-2323 10.1126/science.290.5500.2319 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S. and Rinn J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381-386 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. and Singer A. (2013). Exact and stable recovery of rotations for robust synchronization. Info. Inference 2, 145-193 10.1093/imaiai/iat005 [DOI] [Google Scholar]
- Yuan L., Pan C., Ji S., McCutchan M., Zhou Z.-H., Newfeld S. J., Kumar S. and Ye J. (2014). Automated annotation of developmental stages of Drosophila embryos in images containing spatial patterns of expression. Bioinformatics 30, 266-273 10.1093/bioinformatics/btt648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. and Singer A. (2014). Rotationally invariant image representation for viewing direction classification in cryo-EM. J. Struct. Biol. 186, 153-166 10.1016/j.jsb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Chellappa R., Phillips P. J. and Rosenfeld A. (2003). Face recognition: a literature survey. ACM Comput. Surveys (CSUR) 35, 399-458 10.1145/954339.954342 [DOI] [Google Scholar]
- Zitová B. and Flusser J. (2003). Image registration methods: a survey. Image Vis. Comput. 21, 977-1000 10.1016/S0262-8856(03)00137-9 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.