Abstract
Although phospholipase B (PLB) enzymes have been described in eukaryotes from yeasts to mammals, their biological functions are poorly understood. Here we describe the characterization of plb1, one of five genes predicted to encode PLB homologs in the fission yeast, Schizosaccharomyces pombe. The plb1 gene is dispensable under normal growth conditions but required for viability in high-osmolarity media and for normal osmotic stress-induced gene expression. Unlike mutants defective in function for the stress-activated MAP kinase Spc1, plb1Δ cells are not hypersensitive to oxidative or temperature stresses, nor do they undergo a G2-specific arrest in response to osmotic stress. In addition to defects in osmotic stress response, plb1Δ cells exhibit a cold-sensitive defect in nutrient-mediated mating repression, a phenotype reminiscent of mutants in the cyclic AMP (cAMP) pathway. We show that, like plb1Δ cells, mutants in the cAMP pathway are defective for growth in high-osmolarity media, demonstrating a previously unrecognized role for the cAMP pathway in osmotic stress response. Furthermore, we show that gain-of function in the cAMP pathway can rescue the osmosensitive growth defect of plb1Δ cells, suggesting that the cAMP pathway is a potential downstream target of the actions of Plb1 in S. pombe.
Keywords: Schizosaccharomyces pombe, Osmotic stress response, Phospholipase B, cAMP pathway
Introduction
Phospholipases comprise a large group of structurally and functionally diverse enzymes that hydrolyze one or more ester linkages in glycerophospholipids (Ansell and Hawthorne 1964). In addition to playing roles in membrane phospholipid metabolism and dietary lipid digestion, the lipolytic activities of phospholipases can lead to the release of bioactive molecules that regulate diverse biological processes, including cell cycle progression, apoptosis, cytoskeletal organization, neurological function, and inflammatory responses (Colley et al. 1997; James and Downes 1997; Gomez-Cambronero and Keire 1998; Pei and Williamson 1998; Wang et al. 2000; Capper and Marshall 2001; Cockcroft 2001; Nomura et al. 2001). Phospholipases are classified according to the specificity of the ester linkage that is cleaved, although the nomenclature can at times be confusing because many phospholipases have multiple enzymatic activities (Ansell and Hawthorne 1964; Loo et al. 1997; Wang and Dennis 1999, Ghannoum 2000). Among this group of multi-functional phospholipases, phospholipase B (PLB) enzymes are perhaps the most poorly understood in terms of their biological functions. Strictly speaking, PLB is the designation given to phospholipases that catalyze the hydrolysis of fatty acids from both the sn-1 and sn-2 positions of glycerophospholipids (Ansell and Hawthorne 1964). However, all enzymatically characterized PLB proteins have been shown to possess lysophospholipase activity, which cleaves fatty acids from lysophospholipids (Gassama-Diagne et al. 1989; Lee et al. 1994), and PLB enzymes isolated from fungi possess a transacylase activity that catalyzes the synthesis of phospholipids from lysophospholipids (Saito et al. 1991; Lee et al. 1994; Ghannoum 2000).
PLB enzyme activities have been described in eukaryotes from fungi to mammals (Gassama-Diagne et al. 1989; Masuda et al. 1991; Saito et al. 1991; Lee et al. 1994; Takemori et al. 1998). Fungi possess a class of highly homologous PLB enzymes that exhibit little sequence similarity to PLB proteins identified thus far in other eukaryotes (Ghannoum 2000). Fungal PLB proteins exhibit a limited degree of sequence homology to members of the cytosolic phospholipase A2 (cPLA2) family of proteins (Sharp et al. 1994), several of which have been shown to possess the lysophospholipase and transacylase activities common to fungal PLB enzymes (Leslie 1991; Reynolds et al. 1993; Hanel and Gelb 1995; Loo et al. 1997). The physiological functions of fungal PLB enzymes are largely unknown. Three closely related members of the fungal PLB family, encoded by the PLB1, PLB2, and PLB3 genes, have been described in the budding yeast, Saccharomyces cerevisiae (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999). Simultaneous disruption of all three PLB genes in S. cerevisiae abolishes detectable levels of PLB and lysophospholipase activities, but does not result in any obvious phenotypic defects under normal growth conditions (Merkel et al. 1999). A fourth, more distantly related PLB-like protein, Spo1, is required for meiosis and sporulation in S. cerevisiae (Tevzadze et al. 1996). In the pathogenic dimorphic fungus Candida albicans, as well as the pathogenic yeast Cryptococcus neoformans, PLB enzymes have been shown to function as virulence factors (Ibrahim et al. 1995; Cox et al. 2001; Mukherjee et al. 2001). However, as in budding yeast, the roles of these enzymes, if any, in cell physiology during the noninvasive growth phases of these fungi have not been determined. A homolog of yeast PLBs has also been described in the filamentous fungus Penicillium notatum, but its physiological functions are likewise unknown (Masuda et al. 1991; Saito et al. 1991).
Although PLB genes have yet to be described in the fission yeast Schizosaccharomyces pombe, the genome sequence of this organism contains at least five genes predicted to encode members of the fungal PLB family of proteins. In this paper, we describe the characterization of one of these genes, which we have named plb1. Our results demonstrate that the plb1 gene product is required for osmotic stress response and for nutrient-mediated repression of sexual differentiation in S. pombe. We provide evidence that the functions of the Plb1 may be mediated, at least in part, by the cAMP pathway and in doing so establish a new role for the cAMP pathway in osmotic stress response in S. pombe.
Materials and methods
Yeast strains, DNA manipulations, and plasmids
The S. pombe strains used in this study were SP870 (h90 ade6-210 leu1-32 ura4-D18; from D. Beach), CHP428 (h+ ade6-210 his7-366 leu1-32 ura4-D18), 586 (h- leu1-32 ura4-D18 wee1∷ura4 plb1-1; from P. Nurse), SPPLB1-1 (h- ade6-210 leu1-32 ura4-D18 plb1-1; derived from a cross between 586 and CHP428), SPPLB1U (h90 ade6-210 leu1-32 ura4-D18 plb1∷ura4), DY114 (h90 ade6-210 leu132 ura4-D18 cyr1∷ura4; from M. Wigler), CHP459 (h+ ade6-M216 leu1-32 ura4∷fbp1-lacZ gpa2∷ura4), RWP29 (h+ leu1-32 ura4∷fbp1-lacZ git3∷GFP-kan; git3 is deleted with GFP-kan), CHP453 (h- leu1-32 ura4-D18 his7-366 pka1∷ura4), and RWP38 (h+ leu1-32 ura4∷fbp1-lacZ cgs1∷ura4). The plb1∷ura4 mutant, SPPLB1U, was constructed by transforming SP870 with an SphI-XmnI fragment of plb1∷ura4 isolated from the plasmid pBSplb1∷ura4. Standard yeast culture media and genetic methods were used (Rose et al. 1990; Alfa et al. 1993). S. pombe cultures were grown in either YEAU (0.5% yeast extract, 3% dextrose, 75 mg/l adenine, 75 mg/l uracil) or synthetic minimal medium (EMM) with appropriate auxotrophic supplements (Alfa et al. 1993). Yeast strains were transformed by the lithium acetate procedure (Rose et al. 1990).
pREP3xPlb1 contains the full-length plb1 cDNA under the control of the thiamine-repressible nmt1 promoter (Maundrell 1990) in the plasmid pREP3x (Forsburg 1993). This plasmid was isolated from an S. pombe cDNA library constructed in pREP3x (see below). pREP3xGit3 and pREP3xGpa2 likewise allow for expression of the git3 and gpa2 sequences, respectively, from the nmt1 promoter. pBSplb1∷ura4 was constructed as follows. A SalI-BamHI fragment of plb1 was isolated from pREP3xPlb1 and cloned into the corresponding sites of pBluescript II, producing pBSIIplb1. pBSIIplb1 was digested with HindIII and ligated to the HindIII fragment of the S. pombe ura4 gene, producing the plasmid pBSIIplb1∷ura4.
Cloning of the plb1 gene
The plb1-1 mutation was identified as a recessive background mutation responsible for a high osmolarity-sensitive growth defect in the wee1Δ strain 586. Briefly, 586 was crossed to CHP428 and diploids produced from this cross were sporulated and subjected to tetrad analysis. The high osmolarity sensitive growth defect segregated 2:2 and was independent of the wee1Δ mutation. An S. pombe cDNA library constructed in the plasmid pREP3x (a gift from P. Nurse) was screened for clones that suppress this osmosensitive mutation. More than 20 full-length clones of plb1 were isolated as a result of this screen. Disruption of plb1 with the ura4 gene resulted in an osmosensitive growth defect. Aplb1∷ura4 strain was crossed to the osmosensitive mutant isolated from 586. Diploids isolated from this cross, as well as all progeny produced from a tetrad dissection of several diploids, were unable to grow in high osmolarity media, indicating that the hyperosmolarity defective mutation originally found in 586 is allelic to plb1. Thus, this mutant allele of plb1 was designated plb1-1.
Analysis of osmotic stress-induced expression of gpd1
S. pombe cultures (25 ml) were grown in EMM to a density of about 6×106 cells/ml, then mixed with equal volumes of EMM or EMM with 3 M KCl (for a final concentration of 1.5 M KCl) and incubated for 30 or 90 min. prior to harvesting of cells by centrifugation. Cell pellets were washed once with yeast lysis buffer (YLB; 10 mM TRIS-HCl pH 7.4, 10 mM EDTA, 0.5% SDS), then resuspended in 400 μl YLB and frozen at −80°C. Then 300 μl of water-buffered acid phenol (AP) (Ambion) was pipetted on top of the frozen cells. The suspension was then incubated at 65 C for 20 min. with vortexing at 5-min intervals. Cells were then placed on ice for 10 min. The aqueous phase was removed and extracted once with 250 μl of AP, followed by extraction with 150 μl of chloroform. RNA was precipitated by adding 30 μl of 3 M sodium acetate (pH 5.2) and 1 ml of 95% ethanol, and incubating on ice for 5 min. RNA was pelleted by centrifugation, washed with 500 μl of 70% ethanol, dried, and resuspended in 100 μl of TE buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA) that had been treated with diethylpyrocarbonate. RNA was resolved by agarose gel electrophoresis and immobilized on nylon membranes for Northern blot analysis of gpd1 expression.
Results
A member of the fungal PLB family is required for viability of S. pombe cells under high osmolarity growth conditions
Our laboratory has been investigating the contributions of various cell cycle and morphological regulators to osmotic stress response in S. pombe. A wee1 deletion mutant that was being analyzed in our studies, 586, was found to carry a background mutation that caused a high osmolarity sensitive growth defect (see Materials and methods). A screen for S. pombe cDNAs capable of suppressing this defect resulted in the isolation of multiple full-length clones corresponding to a novel gene encoding a protein that exhibited a high degree of homology to members of the fungal PLB family of proteins (GenBank Accession No. AY235223). Since PLB encoding genes have not previously been described in S. pombe, we named this gene plb1, in accordance with the nomenclature that has been used for related genes in other fungal species. A BLAST search of the S. pombe genome database revealed that plb1 is one of five predicted genes encoding proteins highly homologous to fungal PLB enzymes (E values of 1×10−64 to 6×10−89; Fig. 1). An analysis of phylogenetic relationships revealed that PLB proteins in fungi in which more than one PLB enzyme has been identified (S. cerevisiae and C. albicans) or predicted from genome sequence data (S. pombe) exhibit a greater degree of structural similarity to other PLBs in the same species than to PLBs from other species (Fig. 1B). This finding suggests the existence of a single ancestral PLB and implies that PLB multiplicity within a given species evolved after the evolutionary split from a common fungal ancestor.
Fig. 1.
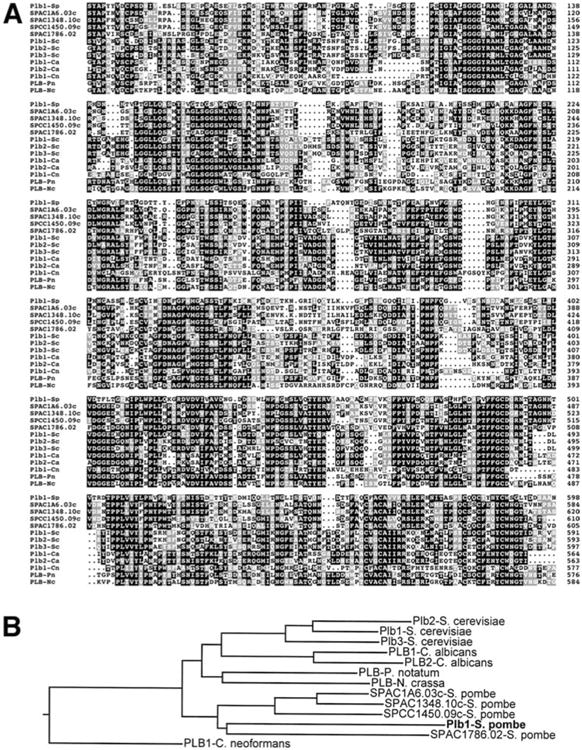
A, B Homology of S. pombe Plb1 to other fungal PLB proteins. A Amino acids 48–598 of the 613-amino acid S. pombe Plb1 protein (Plb1-Sp) were aligned with the corresponding sequences of other predicted S. pombe PLB proteins (S. pombe cosmid sequences SPAC1A6.03c, SPAC1348.10c, SPCC1450.09c, and SPAC1786.02) and characterized or predicted PLB homologs from S. cerevisiae (Plb1-Sc, Plb2-Sc, and Plb3-Sc; Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999), C. albicans (Plb1-Ca and Plb2-Ca; Leidich et al. 1998; Sugiyama et al. 1999), C. neoformans (Plb1-Cn; Cox et al. 2001), P. notatum (PLB-Pn; Masuda et al. 1991), and Neurospora crassa (PLB-Nc, GenBank Accession No. AAC03052) using the MegAlign program in Lasergene (DNAStar). Identical amino acids are indicated by black boxes, while similar amino acids are denoted by gray boxes. B Phylogenetic relationships among fungal PLB proteins. The dendrogram is based on the alignment shown in Fig. 2 and was constructed using the MegAlign program (http://link.springer.de/link/sevice/journals/00438/index.htm)
The plb1 gene was disrupted with an S. pombe ura4 cassette in the wild-type S. pombe strain SP870. plb1∷ura4 (plb1Δ) mutants exhibited no obvious growth defects on either complex rich medium (YEAU) or synthetic minimal medium (EMM) under normal growth conditions (Fig. 2, left panels). However, they were inviable on YEAU or EMM containing 1.2 M KCl (Fig. 2, right panels). These results demonstrate that the plb1 gene is required for cell viability in a high osmolarity growth environment. Microscopic analyses were performed to determine whether plb1Δ cells exhibit morphological defects under normal or hyperosmotic culture conditions. Under normal growth conditions, plb1Δ cells exhibited no obvious morphological defects, except that they divided, on average, at a length about 15% shorter than that of wild-type S. pombe cells (Fig. 3A). In hyperosmotic media, wild-type S. pombe cells divide at a shorter length than they do under normal growth conditions (Kishimoto and Yamashita 2000) (Fig. 3B, left panel). In contrast, we observed that septated cells in plb1Δ cultures incubated for 18 h in high osmolarity media were typically longer than wild-type cells grown under the same conditions (Fig. 3B, right panel). We also observed that in a large proportion of dividing plb1Δ cells, the septum was positioned substantially off-center (difference in length between the two daughter cells >15%), a defect that was rarely observed in wild-type S. pombe cultures (Fig. 3B, right panel). Incubation in hyperosmotic media also resulted in a higher incidence of cells with multiple septa (about 4% of septated cells) in plb1Δ cultures than in cultures of wild-type S. pombe cells (<0.5% of septated cells). plb1Δ cultures arrested growth in YEAU+1.2 M KCl with a similar frequency of septated cells as that observed in non-arrested cultures of wild-type S. pombe cells (about 9% of total cells). About 10% of plb1Δ cells were clearly necrotic (shrunken and/or lysed) after 18 h in YEAU+KCl, and the frequency of necrotic cells increased with prolonged incubation of the cultures (data not shown).
Fig. 2.
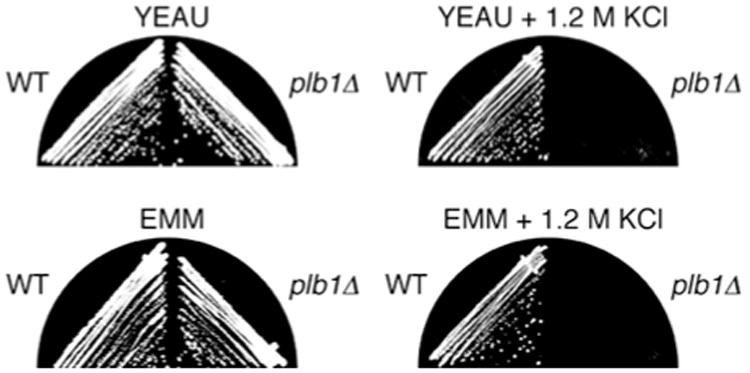
The plb1 gene is required for viability of S. pombe cells on high osmolarity media. Wild type and plb1Δ S. pombe strains were streaked onto YEAU and EMM or on the same media supplemented with 1.2 M KCl, as indicated, and incubated at 30°C for 2-6 days (YEAU, 2 days; EMM, 3 days; YEAU+KCl, 4 days, EMM+KCl, 6 days). Wild-type cells grew on both YEAU and EMM plates containing 1.2 M KCl, whereas plb1Δ cells did not
Fig. 3.
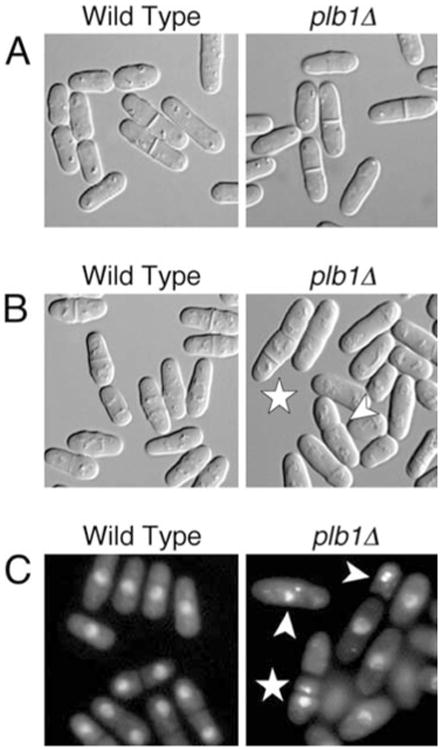
A–C Microscopic analysis of the plb1Δ mutant. A Photomicrographs of wild-type (left) and plb1Δ (right) cells grown in YEAU to mid-log phase. Wild-type cells divided at an average cell length of 15.6 μm, while plb1Δ cells divided at an average length of 13.3 μm. B Wild-type (left) and plb1Δ (right) cells were grown in YEAU to mid-log phase, then subcultured into YEAU containing 1.2 M KCl and incubated for 18 h prior to microscopy. Arrowheads point to cells with misplaced and/or multiple septa. C Wild-type and plb1Δ cultures treated as in B were stained with DAPI to visualize nuclei by indirect fluorescence microscopy. Arrowheads point to cells with hypercondensed (intense, compact nuclear signal) and/or fragmented nuclei. The star indicates a cell in which the nucleus has been cut by the septum. This same cell also has two septa, and one of the three cell compartments lacks a nucleus
Nuclear staining experiments revealed that osmotic stress caused a substantial incidence of nuclear defects in plb1Δ cultures (Fig. 3C). More than 10% of plb1Δ cells exhibited hypercondensed nuclei, whereas less than 2% of cells had hypercondensed nuclei in wild-type S. pombe cultures. We also observed a low incidence of nuclear cutting by the septum (“cut” phenotype) in plb1Δ cultures, as well as instances in which septation had occurred without nuclear segregation, resulting in daughter cells (or cell compartments in the case of multi-septated cells) without a nucleus. These nuclear segregation defects were not observed in cultures of wild-type S. pombe cells grown in high osmolarity media. Perhaps as a consequence of nuclear cutting, fragmented nuclei were also commonly observed in plb1Δ cultures, but not in cultures of wild-type cells. Taken together, these observations indicate that loss of Plb1 function results in marked defects in the execution of mitosis and cytokinesis under conditions of osmotic stress.
Plb1 is required for normal osmotic stress-induced gene expression
Experiments were performed to determine whether Plb1 is required for osmotic stress-induced gene expression. The glycerol-3-phosphate dehydrogenase gene, gpd1, has been shown to be strongly induced by osmotic stress in S. pombe (Ohmiya et al. 1995). To determine whether Plb1 is required for osmotic stress-induced expression of gpd1, we grew wild-type and plb1Δ S. pombe cultures in EMM to mid-log phase, then subcultured them into either EMM or EMM containing 1.5 M KCl and incubated for a further 30 or 90 min. The cultures were then harvested, and RNA was isolated and subjected to Northern analysis of gpd1 expression. As shown in Fig. 4, osmotic stress-induced expression of gpd1 was greatly reduced in plb1Δ cells in comparison to wild-type S. pombe cells. The fact that Plb1 is required for osmotic stress-induced gene expression suggests a role for this protein in mediating osmotic stress-induced signal transduction in S. pombe cells. The modest increase in gpd1 expression observed in the plb1Δ mutant in response to osmotic stress may be an indication that one or more of the other PLB enzymes in S. pombe shares overlapping functions with Plb1.
Fig. 4.
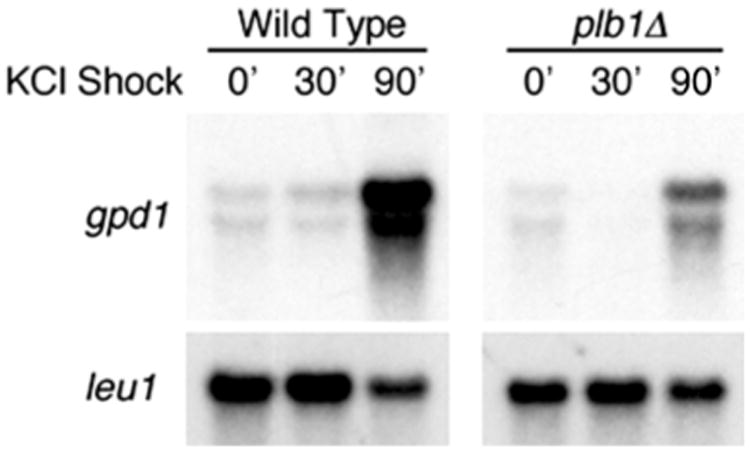
The plb1Δ mutant is defective in osmotic stress-induced expression of the gpd1 gene. Wild-type and plb1Δ S. pombe cultures were grown in EMM to mid-log phase, then mixed with equal volumes of either EMM or EMM containing 3 M KCl (for final concenrtation of 1.5 M KCl) and incubated for 30 or 90 min prior to the isolation of RNA and subsequent Northern blot analysis for expression of gpd1. Membranes were stripped and reprobed with the leu1 gene to check for equivalence of RNA loading
Plb1 is not required for survival upon exposure to oxidative or heat stress
The major stress-responsive mitogen-activated protein kinase (MAPK) homolog in S. pombe, Spc1 (also known as Sty1 and Phh1), is required for cellular responses to multiple environmental stresses, including not only osmotic stress but also oxidative, heat, and UV stresses (Millar et al. 1995; Shiozaki and Russell 1995; Kato et al. 1996; Millar 1999). Experiments were performed to determine if Plb1, like Spc1, is required for survival of S. pombe following exposure to oxidative or heat stresses. To induce oxidative stress, we incubated wild-type, plb1Δ, and spc1Δ cultures in YEAU containing 40 mM H2O2 for 1 h, while heat stress was imposed by incubating cultures at 48 C for 10 min. After the respective stress treatments, cells were diluted into fresh medium and plated on YEAU. As shown in Fig. 5, plb1Δ cells survived exposure to both oxidative and heat stresses that were toxic to spc1Δ cells. Thus, unlike Spc1, Plb1 function is not required to survive exposure to oxidative or heat stress in S. pombe.
Fig. 5.
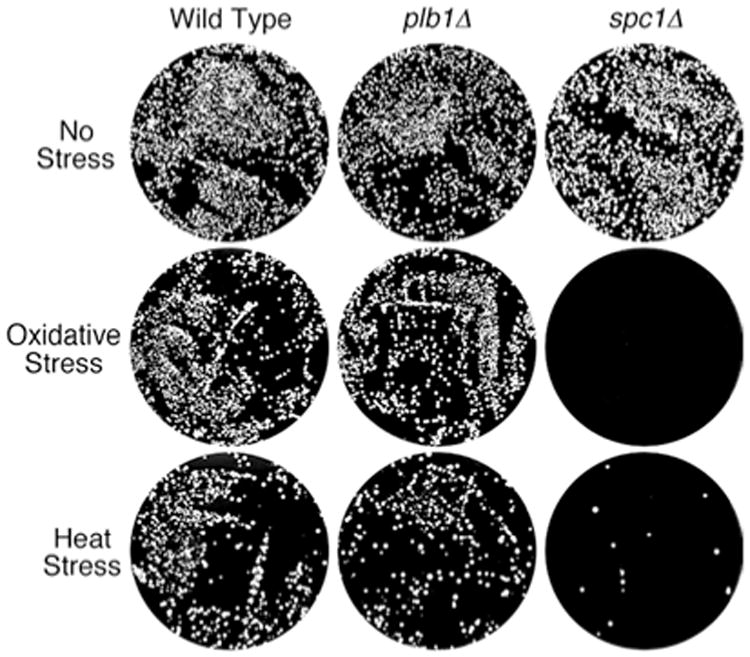
Plb1 is not required for survival upon exposure to oxidative or heat stress. Wild-type, plb1Δ, and spc1Δ cells were grown in YEAU to mid-log phase. Part of each culture was subjected to oxidative stress by adding hydrogen peroxide to a concentration of 40 mM, incubating for 1 h, then diluting into fresh YEAU and plating onto YEAU agar (about 5000 cells/plate). Another portion of each culture was subjected to heat shock by incubating at 48 C for 10 min, then diluting into cold YEAU before plating on YEAU agar (about 5000 cells/plate). Wild-type and plb1Δ cultures each survived oxidative and heat stresses, whereas spc1Δ cultures exhibited a much lower frequency of post-stress survival. The top row shows plates of unstressed control cultures
Plb1 contributes to nutrient-dependent mating repression in S. pombe
To determine if plb1Δ cells exhibit cold- or temperature-dependent phenotypes, we grew wild-type and plb1Δ cultures on YEAU at 20°C and 37°C, respectively. The plb1Δ mutant exhibited no obvious phenotypic defects at 37°C (data not shown). However, at 20°C, plb1Δ cultures grew more slowly than wild-type S. pombe cultures (Fig. 6A). Microscopic analysis revealed that plb1Δ cells mated at a very high frequency on the nutrient-rich YEAU media, in contrast to wild type cells, in which mating response was substantially inhibited on YEAU (Fig. 6B). Thus, the slow growth defect observed for plb1Δ cells at 20°C is likely to be caused by enhanced mating activity. Mating response in S. pombe is normally dependent on nutrient deprivation, and, in particular, starvation for glucose and nitrogen (Nielsen and Davey 1995; Stettler et al. 1996). Mating response is inhibited under nutrient-rich conditions by a cAMP-dependent pathway in S. pombe, and cells lacking adenylate cyclase activity mate promiscuously on rich media (Kawamukai et al. 1991; Nielsen and Davey 1995). Our finding that plb1Δ cells bypass nutritional repression of mating response suggests a potential functional link between Plb1 and the cAMP pathway. This speculation is supported by the results described in the following sections.
Fig. 6.
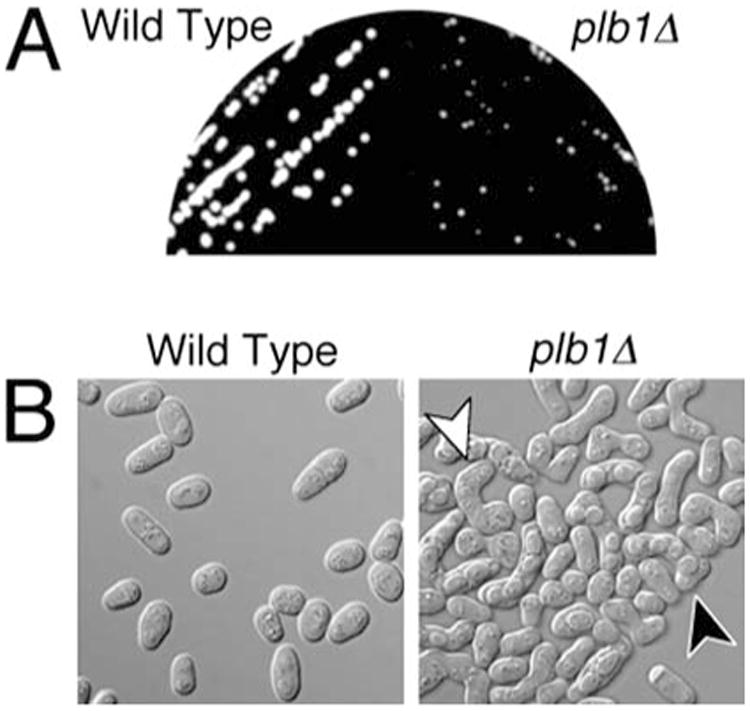
A, B The plb1Δ mutant exhibits a cold-sensitive defect in nutrient-dependent mating repression. A Wild type (left) and plb1Δ (right) cells were streaked onto YEAU, and incubated at 20°C for 5 days prior to photographing a group of isolated colonies of each culture. B Photomicrographs of cells from the cultures shown in A. The plb1Δ culture (right panel) exhibited very high mating activity, as indicated by the high frequency of zygotes (e.g., note the cell marked by the white arrowhead) and asci (e.g., note the cell indicated by the black arrowhead). Zygotes and asci were relatively rare in the wild-type culture (left panel)
The cAMP pathway is required for survival of osmotic stress in S. pombe
Given our observation that the plb1Δ mutant exhibits a severe defect in nutrient-mediated mating repression, experiments were performed to determine whether the cAMP pathway, like Plb1, contributes to osmotic stress response in S. pombe. S. pombe possesses a single adenylate cyclase-encoding gene, cyr1 (Young et al. 1989), as well as a single gene, pka1, for a cAMP-dependent protein kinase (Maeda et al. 1994). Adenylate cyclase activity is positively regulated by several proteins in S. pombe, including the G protein alpha subunit encoded by the gpa2 gene, and a predicted seven-transmembrane-helix protein encoded by the git3 gene (Isshiki et al. 1992; Nocero et al. 1994; Welton and Hoffman 2000). The single negative regulatory subunit of Pka1 in S. pombe is encoded by the cgs1 gene (DeVoti et al. 1991). To determine whether the cAMP pathway is required for survival of S. pombe cells in high osmolarity medium, we compared the growth of wild type, cyr1Δ, git3Δ, gpa2Δ, and pka1Δ mutants in YEAU and YEAU containing 1.2 M KCl. As shown in Fig. 7A, cyr1Δ, gpa2Δ, and pka1Δ mutants were all inviable on YEAU+KCl, while growth of the git3Δ mutant was strongly impaired. As might be expected, the cgs1Δ mutant grew about as well as wild-type S. pombe cells on YEAU+KCl. These results demonstrate that the cAMP pathway is indeed required for survival of S. pombe cells in a high osmolarity growth environment.
Fig. 7.
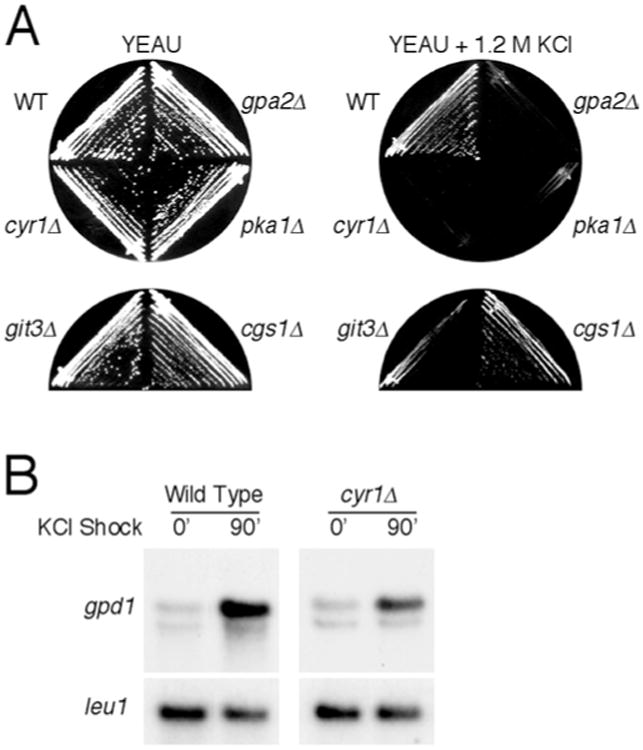
A, B Involvement of the cAMP pathway in osmotic stress response. A The cAMP pathway is required for viability of S. pombe cells in high osmolarity medium. Wild-type, cyr1Δ, gpa2Δ, git3Δ, pka1Δ, and cgs1Δ cells were streaked onto YEAU (left) or YEAU containing 1.2 M KCl (right) and incubated at 30°C for 2 and 4 days, respectively, prior to photographing the plates. Wild-type and cgs1Δ cells grew on YEAU+KCl, whereas the other strains were either inviable (cyr1Δ, gpa2Δ, and pka1Δ) or strongly growth impaired (git3Δ) on the same medium. B Adenylate cyclase is required for full induction of gpd1 expression in response to osmotic stress. Wild-type and cyr1Δ S. pombe cultures were grown in EMM to mid-log phase, then mixed with equal volumes of either EMM or EMM containing 3 M KCl (for a final concentration of 1.5 M KCl) and incubated for 90 min, prior to isolation of RNA and subsequent Northern blot analyses for expression of gpd1. Membranes were stripped and reprobed with the leu1 gene as a standard for RNA loading
We next determined whether adenylate cyclase function is required for osmotic stress-induced gene expression. Wild-type and cyr1Δ cells were incubated in EMM to mid-log phase, then subcultured into EMM+1.5 M KCl and incubated for 90 min to induce hyperosmotic shock, prior to harvesting of the cultures and isolation of RNA for Northern analysis of gpd1 expression. As shown in Fig. 7B, cyr1Δ cells exhibited a modestly reduced level of osmotic stress-induced expression of gpd1 relative to wild-type S. pombe cells. However, the difference was not as great as that observed between wild-type cells and the plb1Δ mutant (see Fig. 4).
Loss of Plb1 function is suppressed by gain of function in the cAMP pathway
We next sought to determine whether gain of function in the cAMP pathway can suppress loss of Plb1 function or vice versa. Genetic studies indicate that adenylate cyclase can be activated in S. pombe by overexpression of the gpa2 gene (Nocero et al. 1994; Welton and Hoffman 2000). As shown in Fig. 8, plb1Δ cells transformed with a plasmid in which gpa2 is overexpressed from the strong nmt1 promoter (Maundrell 1990) were capable of growing on EMM containing 1.2 M KCl, whereas cells transformed with a control plasmid were not. We determined further that overexpression of the git3 gene is likewise capable of suppressing the plb1Δ mutation (Fig. 8). These results demonstrate that loss of Plb1 function can be rescued by gain of function in the cAMP pathway. In reciprocal experiments, we determined that overexpression of plb1 could not rescue the osmosensitive growth defect of the adenylate cyclase null mutant (data not shown). Taken together, these results are consistent with a model in which Plb1 functions upstream of adenylate cyclase in mediating osmotic stress response in S. pombe.
Fig. 8.
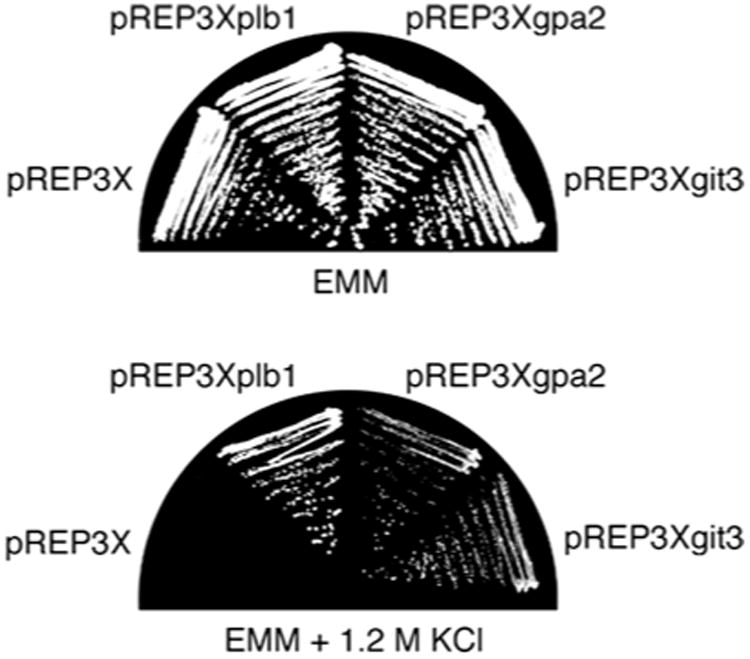
The osmosensitive growth defect of the plb1Δ mutant is rescued by overproduction of the adenylate cyclase activators Gpa2 and Git3. The plb1Δ mutant was transformed with plasmids for overexpression of plb1 (pREP3xplb1), gpa2 (pREP3xgpa2), or git3 (pREP3xgit3), as well as the empty vector pREP3x. Transformants were grown on EMM plates, then subcultured onto either EMM (top) or EMM containing 1.2 M KCl (bottom) and incubated at 30 C for 3 and 6 days, respectively
Discussion
We have described the characterization of a new member of the fungal PLB protein family, Plb1, in the fission yeast S. pombe. Our results demonstrate that Plb1 is required for survival of S. pombe cells under high osmolarity growth conditions, for normal osmotic stress-induced gene expression, and for nutrient-mediated repression of sexual differentiation. This last phenotype is shared by mutants in the cAMP pathway in S. pombe, and this prompted us to investigate whether the cAMP pathway, like Plb1, is required for osmotic stress response. We determined that the cAMP pathway is indeed essential for survival of osmotic stress in S. pombe, thus demonstrating a previously unrecognized role for cAMP in this organism. Furthermore, we found that loss of Plb1 function could be suppressed by gain of function in the cAMP pathway but not vice versa. Taken together, our findings suggest that the effects of Plb1 in S. pombe may be mediated, at least in part, by the cAMP pathway.
The molecular mechanisms by which Plb1 function contributes to osmotic stress response in S. pombe remain to be elucidated. One possibility is that Plb1 is involved in regulating the production of lipid or lipid-derived second messengers, such as arachidonic acid and inositol triphosphate, perhaps in concert with other phospholipid/lipid-modifying enzymes. This is an attractive hypothesis, given our genetic data suggesting a functional link between Plb1 and the G protein-dependent adenylate cyclase in S. pombe. An alternative model is that the actions of Plb1 lead to changes in membrane lipid composition that induce stimulatory conformational changes in membrane-associated mediators of osmotic stress response. In considering these possible roles for Plb1, it is noteworthy that the putative adenylate cyclase activator, Git3, is a predicted seven-transmembrane-helix protein and that adenylate cyclase itself is a peripheral membrane protein (Kawamukai et al. 1991; Welton and Hoffman 2000). As we have shown that loss of Plb1 is rescued by overproduction of Git3, this protein may be positively regulated by Plb1-mediated phospholipid metabolism. It remains to be determined whether the cAMP pathway is the sole mediator of Plb1 activity in S. pombe cells. Our observation that plb1Δ cells exhibit a substantially greater defect in osmotic stress-induced expression of the gpd1 gene than that observed in cells lacking adenylate cyclase raises the possibility that Plb1 may have additional targets.
Unlike mutants defective in the function for the Spc1 stress-responsive MAPK pathway, plb1Δ cells do not undergo cell cycle-specific growth arrest in response to osmotic stress, nor are they hypersensitive to heat or oxidative stress. In addition, under normal growth conditions, mutants in the Spc1 pathway are delayed in G2/M progression (Shiozaki and Russell 1995), whereas plb1Δ cells undergo this transition in a modestly accelerated fashion, as judged by the average length at which cells undergo division. The marked phenotypic differences between mutants in the Spc1 pathway and plb1Δ mutants suggest that Plb1 and Spc1 may function in separate pathways. This idea is supported by our results suggesting a functional link between Plb1 and cAMP pathway, as the cAMP pathway has been shown to act in opposition to the Spc1 pathway in the regulation of glucose responsive gene expression in S. pombe (Neely and Hoffman 2000). While osmotic stress does not cause a cell cycle-specific block in plb1 mutant cultures, a substantial frequency of defects in mitosis and/or cytokinesis can be observed, namely a relatively high incidence of cells with hypercondensed nuclei, as well as lower frequencies of cells with mis-segregated, cut, and/ or fragmented nuclei and cells with multiple septa. These observations suggest that Plb1 function may be required for proper regulation of some aspects of mitosis and cytokinesis under high osmolarity growth conditions.
The biological functions of PLB proteins in eukaryotic cells are poorly understood. Although PLB activities have been isolated from mammalian cells, their biological functions are unknown, as published studies to date have been mostly limited to analyses of enzymatic activities in vitro and the tissue distribution of a single molecularly cloned PLB isozyme (Gassama-Diagne et al. 1989; Delagebeaudeuf et al. 1998). PLB enzyme activities have also been isolated from plants (Matsuda and Hirayama 1979; Kim et al. 1994) and a variety of insect and reptile venoms (Doery and Pearson 1964; Shiloah et al. 1973; Bernheimer et al. 1987; Matuszek et al. 1994; Abe et al. 2000). S. pombe Plb1 belongs to a structurally unique class of fungal PLB enzymes, which, in addition to PLB activity, possess lysophosp-holipase and lysophospholipid transacylase activities. Several fungal species, including S. pombe (this study), S. cerevisiae (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999), and C. albicans (Leidich et al. 1998; Sugiyama et al. 1999), possess multiple PLB isozymes. The substantial conservation and reiteration of these proteins in fungi suggests that they are likely to have important biological functions. However, prior studies have failed to provide significant insights into the cellular functions of fungal PLB proteins, except that these enzymes have been implicated as virulence factors in the pathogenic fungi C. albicans and C. neoformans (Ibrahim et al. 1995; Cox et al. 2001; Mukherjee et al. 2001). The role(s) of PLB enzymes in fungal virulence have yet to be established, although it has been speculated that they may digest host cell membranes (some PLB enzyme activity is secreted) or catalyze the release of lipid second messengers necessary for establishing and/or maintaining the invasive growth phase (Ghannoum 2000; Noverr et al. 2001). In considering a possible signaling role for PLB enzymes in fungal pathogenicity, it is worth noting that, in C. albicans, adenylate cyclase activity is required for fungal virulence and, in particular, for the conversion from a non-invasive yeast phase of growth to a highly invasive filamentous growth phase (Rocha et al. 2001). It will be of great interest to determine whether adenylate cyclase is a target of PLB activity in pathogenic fungi, as appears to be the case in S. pombe.
The elucidation of an essential growth function for a PLB enzyme in S. pombe makes this genetically tractable yeast an ideal model organism for gaining new insights into cellular and molecular functions of PLB enzymes in fungi and perhaps other eukaryotes as well. Our results also point to the use of S. pombe as a powerful tool for the screening of potential inhibitors of fungal PLBs, which, by virtue of their unique structure and important roles in fungal pathogenesis, have significant promise as targets for antifungal drug therapy.
Acknowledgments
This work was supported by research grants GM46226 from the National Institutes of Health (C.S.H.) and SR99-263 from the Pharmacia Corporation (S.M.).
Contributor Information
P. Yang, Department of Molecular Genetics and Program in Genes and Development, M.D. Anderson Cancer Center, University of Texas, 1515 Holcombe Blvd., Houston, TX 77030, USA, Tel.: +1-713-7452032, Fax: +1-713-7944394
H. Du, Department of Molecular Genetics and Program in Genes and Development, M.D. Anderson Cancer Center, University of Texas, 1515 Holcombe Blvd., Houston, TX 77030, USA, Tel.: +1-713-7452032, Fax: +1-713-7944394
C.S. Hoffman, Biology Department, Boston College, 140 Commonwealth Ave., Chestnut Hill, MA 02467, USA
S. Marcus, Email: smarcus@mdacc.tmc.edu, Department of Molecular Genetics and Program in Genes and Development, M.D. Anderson Cancer Center, University of Texas, 1515 Holcombe Blvd., Houston, TX 77030, USA. Tel.: +1-713-7452032, Fax: +1-713-7944394.
References
- Abe T, Sugita M, Fujikura T, Hiyoshi J, Akasu M. Giant hornet (Vespa mandarinia) venomous phospholipases. The purification, characterization and inhibitory properties by biscoclaurine alkaloids. Toxicon. 2000;38:1803–1816. doi: 10.1016/s0041-0101(00)00109-4. [DOI] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1993. [Google Scholar]
- Ansell GB, Hawthorne JN. Catabolism. In: Ansell GB, Hawthorne JN, editors. Phospholipids. Elsevier; Amsterdam: 1964. pp. 152–174. [Google Scholar]
- Bernheimer AW, Linder R, Weinstein SA, Kim KS. Isolation and characterization of a phospholipase B from venom of Collett's snake, Pseudechis colletti. Toxicon. 1987;25:547–554. doi: 10.1016/0041-0101(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Capper EA, Marshall LA. Mammalian phospholipases A(2): mediators of inflammation, proliferation and apoptosis. Prog Lipid Res. 2001;40:167–197. doi: 10.1016/s0163-7827(01)00002-9. [DOI] [PubMed] [Google Scholar]
- Cockcroft S. Signalling roles of mammalian phospholipase D1 and D2. Cell Mol Life Sci. 2001;58:1674–1687. doi: 10.1007/PL00000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Delagebeaudeuf C, Gassama-Diagne A, Nauze M, Ragab A, Li RY, Capdevielle J, Ferrara P, Fauvel J, Chap H. Ectopic epididymal expression of guinea pig intestinal phospholipase B. Possible role in sperm maturation and activation by limited proteolytic digestion. J Biol Chem. 1998;273:13407–13414. doi: 10.1074/jbc.273.22.13407. [DOI] [PubMed] [Google Scholar]
- DeVoti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 1991;10:3759–3768. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doery HM, Pearson JE. Phospholipase B in snake venoms and bee venom. Biochem J. 1964;92:599–602. doi: 10.1042/bj0920599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrst H, Oskouian B, Kuypers FA, Saba JD. The PLB2 gene of Saccharomyces cerevisiae confers resistance to lysophosphatidylcholine and encodes a phospholipase B/lysophospholipase. Biochemistry. 1999;38:5864–5871. doi: 10.1021/bi9824590. [DOI] [PubMed] [Google Scholar]
- Gassama-Diagne A, Fauvel J, Chap H. Purification of a new, calcium-independent, high molecular weight phospholipase A2/ lysophospholipase (phospholipase B) from guinea pig intestinal brush-border membrane. J Biol Chem. 1989;264:9470–9475. [PubMed] [Google Scholar]
- Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cambronero J, Keire P. Phospholipase D: a novel major player in signal transduction. Cell Signal. 1998;10:387–397. doi: 10.1016/s0898-6568(97)00197-6. [DOI] [PubMed] [Google Scholar]
- Hanel A, Gelb M. Multiple enzymatic activities of the human cytosolic 85-kDa phospholipase A2: hydrolytic reactions and acyl transfer to glycerol. Biochemistry. 1995;34:7807–7818. doi: 10.1021/bi00024a004. [DOI] [PubMed] [Google Scholar]
- Ibrahim AS, Mirbod F, Filler SG, Banno Y, Cole GT, Kitajima Y, Edwards JE, Jr, Nozawa Y, Ghannoum MA. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- James SR, Downes CP. Structural and mechanistic features of phospholipases C: effectors of inositol phospholipid-mediated signal transduction. Cell Signal. 1997;9:329–336. doi: 10.1016/s0898-6568(96)00175-1. [DOI] [PubMed] [Google Scholar]
- Kato T, Jr, Okazaki K, Murakami H, Stettler S, Fantes PA, Okayama H. Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 1996;378:207–212. doi: 10.1016/0014-5793(95)01442-x. [DOI] [PubMed] [Google Scholar]
- Kawamukai M, Ferguson K, Wigler M, Young D. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell Regul. 1991;2:155–164. doi: 10.1091/mbc.2.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Lee HJ, Lee Y. Detection of two phospholipase A2(PLA2) activities in leaves of higher plant Vicia faba and comparison with mammalian PLA2's. FEBS Lett. 1994;343:213–218. doi: 10.1016/0014-5793(94)80558-x. [DOI] [PubMed] [Google Scholar]
- Kishimoto N, Yamashita I. Multiple pathways regulating fission yeast mitosis upon environmental stresses. Yeast. 2000;16:597–609. doi: 10.1002/(SICI)1097-0061(200005)16:7<597::AID-YEA556>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Lee KS, Patton JL, Fido M, Hines LK, Kohlwein SD, Paltauf F, Henry SA, Levin DE. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem. 1994;269:19725–19730. [PubMed] [Google Scholar]
- Leidich SD, Ibrahim AS, Fu Y, Koul A, Jessup C, Vitullo J, Fonzi W, Mirbod F, Nakashima S, Nozawa Y, Ghannoum MA. Cloning and disruption of CaPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- Leslie CC. Kinetic properties of a high molecular mass arachidonoyl-hydrolyzing phospholipase A2 that exhibits lysophospholipase activity. J Biol Chem. 1991;266:11366–11371. [PubMed] [Google Scholar]
- Loo RW, Conde-Frieboes K, Reynolds LJ, Dennis EA. Activation, inhibition, and regiospecificity of the lysophospholipase activity of the 85-kDa group IV cytosolic phospholipase A2. J Biol Chem. 1997;272:19214–19219. doi: 10.1074/jbc.272.31.19214. [DOI] [PubMed] [Google Scholar]
- Maeda T, Watanabe Y, Kunitomo H, Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J Biol Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- Masuda N, Kitamura N, Saito K. Primary structure of protein moiety of Penicillium notatum phospholipase B deduced from the cDNA. Eur J Biochem. 1991;202:783–787. doi: 10.1111/j.1432-1033.1991.tb16433.x. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Hirayama O. Purification and properties of a lipolytic acyl-hydrolase from potato leaves. Biochim Biophys Acta. 1979;573:155–165. doi: 10.1016/0005-2760(79)90182-6. [DOI] [PubMed] [Google Scholar]
- Matuszek MA, Hodgson WC, King RG, Sutherland SK. Some enzymic activities of two Australian ant venoms: a jumper ant Myrmecia pilosula and a bulldog ant Myrmecia pyriformis. Toxicon. 1994;32:1543–1549. doi: 10.1016/0041-0101(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Merkel O, Fido M, Mayr JA, Pruger H, Raab F, Zandonella G, Kohlwein SD, Paltauf F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J Biol Chem. 1999;274:28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- Millar JB. Stress-activated MAP kinase (mitogen-activated protein kinase) pathways of budding and fission yeasts. Bio-chem Soc Symp. 1999;64:49–62. [PubMed] [Google Scholar]
- Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Seshan KR, Leidich SD, Chandra J, Cole GT, Ghannoum MA. Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology. 2001;147:2585–2597. doi: 10.1099/00221287-147-9-2585. [DOI] [PubMed] [Google Scholar]
- Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol Cell Biol. 2000;20:6426–6434. doi: 10.1128/mcb.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O, Davey J. Pheromone communication in the fission yeast Schizosaccharomyces pombe. Semin Cell Biol. 1995;6:95–104. doi: 10.1016/1043-4682(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Nocero M, Isshiki T, Yamamoto M, Hoffman CS. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein alpha subunit encoded by gpa2 (git8) Genetics. 1994;138:39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nishizaki T, Enomoto T, Itoh H. A long-lasting facilitation of hippocampal neurotransmission via a phospholipase A2 signaling pathway. Life Sci. 2001;68:2885–2891. doi: 10.1016/s0024-3205(01)01072-4. [DOI] [PubMed] [Google Scholar]
- Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001;69:2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya R, Yamada H, Nakashima K, Aiba H, Mizuno T. Osmoregulation of fission yeast: cloning of two distinct genes encoding glycerol-3-phosphate dehydrogenase, one of which is responsible for osmotolerance for growth. Mol Microbiol. 1995;18:963–973. doi: 10.1111/j.1365-2958.1995.18050963.x. [DOI] [PubMed] [Google Scholar]
- Pei ZD, Williamson JR. Mutations at residues Tyr771 and Tyr783 of phospholipase C-gamma1 have different effects on cell actin-cytoskeleton organization and cell proliferation in CCL-39 cells. FEBS Lett. 1998;423:53–56. doi: 10.1016/s0014-5793(98)00063-5. [DOI] [PubMed] [Google Scholar]
- Reynolds L, Hughes L, Louis A, Kramer R, Dennis E. Metal ion and salt effects on the phospholipase A2, lysophospholipase, and transacylase activities of human cytosolic phospholipase A2. Biochim Biophys Acta. 1993;1167:272–280. doi: 10.1016/0005-2760(93)90229-3. [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1990. [Google Scholar]
- Saito K, Sugatani J, Okumura T. Phospholipase B from Penicillium notatum. Methods Enzymol. 1991;197:446–456. doi: 10.1016/0076-6879(91)97170-4. [DOI] [PubMed] [Google Scholar]
- Sharp JD, et al. Serine 228 is essential for catalytic activities of 85-kDa cytosolic phospholipase A2. J Biol Chem. 1994;269:23250–23254. [PubMed] [Google Scholar]
- Shiloah J, Klibansky C, de Vries A, Berger A. Phospholipase B activity of a purified phospholipase A from Vipera palestinae venom. J Lipid Res. 1973;14:267–278. [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- Stettler S, Warbrick E, Prochnik S, Mackie S, Fantes P. The wis1 signal transduction pathway is required for expression of cAMP- repressed genes in fission yeast. J Cell Sci. 1996;109:1927–1935. doi: 10.1242/jcs.109.7.1927. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Nakashima S, Mirbod F, Kanoh H, Kitajima Y, Ghannoum MA, Nozawa Y. Molecular cloning of a second phospholipase B gene, CaPLB2 from Candida albicans. Med Mycol. 1999;37:61–67. [PubMed] [Google Scholar]
- Takemori H, Zolotaryov FN, Ting L, Urbain T, Komatsubara T, Hatano O, Okamoto M, Tojo H. Identification of functional domains of rat intestinal phospholipase B/lipase. Its cDNA cloning, expression, and tissue distribution. J Biol Chem. 1998;273:2222–2231. doi: 10.1074/jbc.273.4.2222. [DOI] [PubMed] [Google Scholar]
- Tevzadze GG, Mushegian AR, Esposito RE. The SPO1 gene product required for meiosis in yeast has a high similarity to phospholipase B enzymes. Gene. 1996;177:253–255. doi: 10.1016/0378-1119(96)00261-2. [DOI] [PubMed] [Google Scholar]
- Wang A, Dennis EA. Mammalian lysophospholipases. Biochim Biophys Acta. 1999;1439:1–16. doi: 10.1016/s1388-1981(99)00063-3. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang C, Sang Y, Zheng L, Qin C. Determining functions of multiple phospholipase Ds in stress response of Arabidopsis. Biochem Soc Trans. 2000;28:813–816. [PubMed] [Google Scholar]
- Welton RM, Hoffman CS. Glucose monitoring in fission yeast via the Gpa2 galpha, the git5 Gbeta and the git3 putative glucose receptor. Genetics. 2000;156:513–5121. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D, Riggs M, Field J, Vojtek A, Broek D, Wigler M. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1989;86:7989–7993. doi: 10.1073/pnas.86.20.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]


