Abstract
The cyclic nucleotide gated channel (CNGC) is suggested to be one of the important calcium conducting channels. Nevertheless, genome-wide identification and systemic functional analysis of CNGC gene family in crop plant species have not yet been conducted. In this study, we performed genome-wide identification of CNGC gene family in the economically important crop tomato (Solanum lycopersicum L.) and analyzed function of the group IVb SlCNGC genes in disease resistance. Eighteen CNGC genes were identified in tomato genome, and four CNGC loci that were misannotated at database were corrected by cloning and sequencing. Detailed bioinformatics analyses on gene structure, domain composition and phylogenetic relationship of the SlCNGC gene family were conducted and the group-specific feature was revealed. Comprehensive expression analyses demonstrated that SlCNGC genes were highly, widely but differently responsive to diverse stimuli. Pharmacological assays showed that the putative CNGC activators cGMP and cAMP enhanced resistance against Sclerotinia sclerotiorum. Silencing of group IVb SlCNGC genes significantly enhanced resistance to fungal pathogens Pythium aphanidermatum and S. sclerotiorum, strongly reduced resistance to viral pathogen Tobacco rattle virus, while attenuated PAMP- and DAMP-triggered immunity as shown by obvious decrease of the flg22- and AtPep1-elicited hydrogen peroxide accumulation in SlCNGC-silenced plants. Additionally, silencing of these SlCNGC genes significantly altered expression of a set of Ca2+ signaling genes including SlCaMs, SlCDPKs, and SlCAMTA3. Collectively, our results reveal that group IV SlCNGC genes regulate a wide range of resistance in tomato probably by affecting Ca2+ signaling.
Keywords: cyclic nucleotide gated channel (CNGC), tomato, genome-wide identification, resistance, PAMP-triggered immunity, Sclerotinia sclerotiorum
Introduction
The cyclic nucleotide gated channels (CNGCs) are ligand-gated cation channels localized typically in plasma membrane (Chin et al., 2009; Ma et al., 2009). Plant CNGCs are composed of cytoplasmic C-terminal calmodulin binding (CaMB) domain and cyclic nucleotide-binding (CNB) domain as well as N-terminal hexa-transmembrane (TM) domains. The TM domains form a pore to facilitate cation transport (Zelman et al., 2012). Individual plant CNGC genes were identified in several plant species including barley (Schuurink et al., 1998), Arabidopsis (Kohler et al., 1999; Kohler and Neuhaus, 2000) and tobacco (Arazi et al., 1999). Genome-wide identification of CNGC gene family was also conducted in Arabidopsis (Maser et al., 2001), rice (Bridges et al., 2005), Populus trichocarpa (Ward et al., 2009), pear (Chen et al., 2015), a moss (Physcomitrella patens) and some algae (Verret et al., 2010; Zelman et al., 2013). The Arabidopsis CNGC gene family is comprised of 20 members that are divided into groups I, II, III, IVa, and IVb according to their phylogenetic relationship (Maser et al., 2001). Recently, a plant CNGC-specific motif spanning the phosphate binding cassette (PBC) and hinge region within CNB domain of CNGC proteins is predicted. This motif identifies CNGCs but no other protein and thus represents an efficient tool to identify plant CNGCs, which has been validated in several species such as Arabidopsis, rice, P. patens and Selaginella moellendorffii (Zelman et al., 2012, 2013). Based on animal research, it has been suggested that plant CNGCs might conduct calcium. They might be activated by direct binding of cyclic nucleotides such as cAMP and cGMP to the CNB domain and inhibited by binding of CaM to the CaMB domain (Chin et al., 2009; Wang et al., 2013). It has been also suggested that upon activation, they promote influx of the calcium into the cytosol (Kaplan et al., 2007; Ma and Berkowitz, 2011). As a matter of fact, recent electrophysiological studies provide evidences that AtCNGC18 functions as a Ca2+-permeable divalent cation-selective channel and is activated by cGMP and cAMP in HEK293T cells (Gao et al., 2014). Additionally, AtCNGC18 is activated by a calcium-dependent protein kinase AtCPK32 in Xenopus laevis oocytes (Zhou et al., 2014).
The plant CNGCs are involved in numerous biological functions varying from plant development and stress tolerance (Kaplan et al., 2007) to disease resistance (Abdel-Hamid et al., 2011; Ma, 2011). As regulators of plant development and stress tolerance, AtCNGC1 is involved in Ca2+ uptake (Ma et al., 2006); a P. patens CNGC (PpCNGCb) and its Arabidopsis ortholog (AtCNGC2) are essential to thermotolerance (Finka et al., 2012); AtCNGC3 contributes to seed germination (Gobert et al., 2006); AtCNGC6 mediates heat induced Ca2+ influx and the acquisition of thermotolerance (Gao et al., 2012); AtCNGC10 is associated with plant growth (Borsics et al., 2007); AtCNGC16 and AtCNGC18 play a role in pollen fertility under stress and pollen tip growth respectively (Frietsch et al., 2007; Tunc-Ozdemir et al., 2013); while AtCNGC19 and AtCNGC20 are related to salt tolerance (Kugler et al., 2009). Additionally, genetics studies have unveiled the important role of four Arabidopsis CNGC genes, AtCNGC2, AtCNGC4, AtCNGC11, and AtCNGC12, in plant disease resistance. The AtCNGC2 mutant, defense, no death 1 (dnd1) exhibits diminished hypersensitive response (HR) development but shows enhanced basal resistance to Pectobacterium carotovorum (Ahn, 2007) as well as R gene-conferred resistance to avirulent pathogens with accumulation of salicylic acid (SA; Yu et al., 1998; Clough et al., 2000). AtCNGC2 promotes Ca2+ current to produce nitric oxide (NO) which leads to HR generation in response to pathogen (Ali et al., 2007; Ma and Berkowitz, 2011). The AtCNGC4 mutant, defense, no death 2 (DND2)/hypersensitive response-like lesion mimic 1 (HLM1) depicts a lesion mimic phenotype, shows constitutive pathogenesis-related (PR) gene expression and accumulates high level of SA (Balague et al., 2003; Jurkowski et al., 2004). This mutant displays resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 but fails to induce HR after infection with avirulent strains of Pst and Xanthomonas campestris pv. campestris (Balague et al., 2003; Jurkowski et al., 2004). Mutation of NEC1, an AtCNGC4 homolog in barley, leads to similar phenotypes to dnd2/hlm1 (Rostoks et al., 2006; Keisa et al., 2011). AtCNGC2 and AtCNGC4 seem to work in the same signaling pathway in regulation of pathogen defense and floral transition (Chin et al., 2013). Additionally, the Arabidopsis mutant constitutive expresser of pathogenesis related genes 22 (cpr22) in which a chimeric gene of AtCNGC11 and AtCNGC12 is generated, exhibits enhanced resistance to Hyaloperonospora arabidopsidis (formerly Peronospora parasitica; Yoshioka et al., 2001, 2006). The described evidence undoubtedly supports an essential role of CNGCs in plant defense. However, the evidence is mainly obtained from Arabidopsis. Studies in other plant species should promote understanding of CNGC functions.
In this study, we conducted genome-wide identification of CNGCs in the economically important crop tomato (Solanum lycopersicum L.) and explored function of the group IVb SlCNGC genes in various types of disease resistance. We demonstrated that tomato CNGC family is comprised of 18 members and group IVb SlCNGCs are required for a wide range of disease resistance in tomato.
Materials and Methods
Identification of SlCNGC Genes
The 20 Arabidopsis CNGC protein sequences were obtained from TAIR1 and were then used to TBLASTN search the tomato genome database2. All retrieved non-redundant sequences were collected and subjected to domain analysis using the programs Pfam3, SMART4 and CDD5. Those containing both a cyclic nucleotide-binding domain [CNBD, cNMP_binding family (PF00027)] and a TM or ion transport protein domain [ITP, Ion_trans family (PF00520)] were recognized as CNGC proteins. The PBC and hinge region within the CNB domain of all SlCNGCs were analyzed to check whether they carried a similar motif as suggested for plant CNGCs (Zelman et al., 2013).
Cloning of SlCNGC5, SlCNGC6, and SlCNGC15
Leaves of 2-months-old tomato plants were collected from cultivars Heinz, Money maker and Suhong 2003. Total RNA extraction and reverse transcription were performed as before (Li et al., 2012). The obtained cDNA was used to amplify the SlCNGCs. As we expect that SlCNGC6 is the combination of Solyc03g007260.2.1 and Solyc03g007250.2.1, which are annotated to be truncated N-part and C-part of a complete CNGC, we designed F and R primers corresponding to the beginning of Solyc03g007260.2.1 and the end of Solyc03g007250.2.1 respectively. For amplification of genes SlCNGC5 and SlCNGC15, we designed F and R primers corresponding to the beginning and the end of Solyc06g051920.2.1 and Solyc03g098210.2.1 respectively. The primers were listed in Supplementary Table S1. PCR products were ligated into T-vector and sequenced. Sequences of the cloned SlCNGCs were compared with the corresponding loci in tomato genome database using GeneDoc program (Nicholas et al., 1997). The cloned SlCNGC sequences have been deposited at GenBank with accession numbers KJ499456, KJ499457, and KJ499458 for SlCNGC5, SlCNGC6, and SlCNGC15 respectively.
Gene Structure and Phylogenetic Analyses of CNGCs
Tomato and Arabidopsis CNGC protein sequences were aligned using clustalX 2.01 program (Larkin et al., 2007). The phylogenetic tree between these CNGCs was constructed using MEGA 5.0 (Tamura et al., 2011) with maximum likelihood (ML) method and bootstrap of 1000. The exon/intron structure of CNGC genes was analyzed online using the gene structure display server (GSDS)6.
Cis-Acting Element Prediction of the SlCNGC Genes
Cis-acting element of 1 kb upstream sequence of the SlCNGC genes was predicted with Signal Scan search program in the PLACE database7.
Gene Expression Analyses by qRT-PCR
Quantitative real time RT-PCR (qRT-PCR) analyses and consequent statistical data analyses were conducted as described (Zhao et al., 2013). The primers used in qRT-PCR analyses were listed (Supplementary Table S1). The primers for amplification of the SlCNGC genes were designed from their 5′ and 3′ UTR regions to ensure the gene targeting specificity.
Plant Treatments and Inoculations
Tomato (cv. Money maker) plants were grown in growth room at 28°C with 16 h light/8 h dark photoperiod. For SlCNGC gene expression analyses, leaves of 2-months-old plants were sprayed with diverse chemicals including 380 μM benzothiadiazole (BTH), 10 mM ethephon (ETH), 200 μM jasmonic acid (JA), 375 μM oxalic acid (OA) or sterilized water as control.
For both SlCNGC gene expression and disease resistance evaluation analyses, tomato plants were inoculated with a variety of pathogens. Sclerotinia sclerotiorum was grown at 22°C on potato dextrose agar (PDA) medium for 2 days. The PDA plugs of 3 mm at diameter were taken from the colony outside circle that contained most active young mycelia, and then were stuck mycelial side down onto the tomato leaves. The inoculated plants were grown under high relative humidity for 24 and 4 h for disease resistance and expression analyses respectively. Inoculation with another fungal pathogen Pythium aphanidermatum and the bacterial pathogens Pseudomonas syringae pv. tomato (Pst) DC3000 and Xanthomonas oryzae pv. oryzae (Xoo) was performed as described (Li et al., 2012; Zhao et al., 2013).
For SlCNGC gene expression analyses, all the chemical-treated and pathogen-inoculated leaves were sampled at 4 h post treatment/inoculation and stored at -80°C.
Pharmacological Studies
The chemicals (Sigma, USA) dibromo-cGMP (db-cGMP), dibutyryl-cAMP (db-cAMP), sodium metavanadate anhydrous (NaVO3), alloxan monohydrate and lanthanum (III) chloride hydrate (LaCl3) were dissolved into sterilized ddH2O while ethyleneglycol-bis-(2-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) into 1 M NaOH as stock solution. They were diluted with sterilized ddH2O in turn to 1 mM, 100 μM, 50 μM, 1 mM, 1 mM and 1 mM respectively, and infiltrated into tomato leaves with sterilized needless syringe. The infiltrated areas were immediately inoculated with Ss as mentioned above. The disease symptoms of the inoculated plants were investigated. Diameter of the disease lesions was measured by a ruler. In case of irregular lesions, the shortest and longest diameter of the same lesions was measured respectively and the average of them serves as the diameter of the lesions for statistical analysis.
Virus-Induced Gene Silencing (VIGS) Analyses
The SlCNGC gene members are highly conserved among each other. Therefore care was taken to ensure the specificity to target an individual gene member. To achieve this, the VIGS target fragment of SlCNGC16 and SlCNGC17 were designed from 299 and 312 bp of 5′ UTR region while that of SlCNGC18 corresponded to 144 bp of coding sequence. The cloning sites EcoRI and Bam HI were added to the end of the F and R primers for target fragment amplification respectively (Supplementary Table S1). These fragments were cloned and ligated into the TRV VIGS vector pYL156, which were subsequently electroporated into Agrobacterium tumefaciens strain GV3101 for VIGS analyses. VIGS analyses were conducted with vacuum-infiltration delivery approach as described (Wang et al., 2006; Cai et al., 2007) except that recombinant pYL156 with insertion of an eGFP fragment instead of an empty pYL156 was used as control to alleviate viral symptom (Cheng et al., 2012). At about 3 weeks post agro-infiltration, plants were inoculated with different pathogens and disease was investigated as described above. For each pathogen at least six silenced plants were examined. The experiments were conducted three times independently.
Detection of PAMP/DAMP-Triggered H2O2 in Tomato Leaves
The peptides AtPep1 and flg22 with purity of over 95% were synthesized by China Peptides Company Limited. Tomato leaf disks of 3 mm at diameter were dipped in 200 μl of distilled water in a 96-well plate in dark over night. Water was replaced with 200 μl solution containing 100 μM luminal (Sigma-Aldrich) and 1 μg of horseradish peroxidase. AtPep1 (10 nM) and flg22 (100 nM) were added and immediately H2O2 were measured for 30 min as luminescence using a luminometer (Glomax 96 microplate luminometer, Promega).
Results
Treatment of cNMPs and Ca2+ Channel Related Chemicals Altered Resistance Against Sclerotinia sclerotiorum in Tomato
Pharmacological approach was employed to investigate the role of calcium channels in plant disease resistance. For this purpose a set of Ca2+ concentration regulators and putative Ca2+ channel activators and inhibitors including Ca2+ chelator EGTA; Ca2+ channel blocker LaCl3; Ca2+ ATPase blocker NaVO3; putative CNGC activators lipophilic dibromo-cGMP and dibutyryl-cAMP and adenylyl cyclase inhibitor alloxan (Ali et al., 2007; Ma et al., 2009; Qi et al., 2010), were infiltrated into tomato leaves, and subsequently effect of these chemicals on resistance to the important necrotrophic pathogen S. sclerotiorum was evaluated. Compared with controls, the necrosis disease symptom of tomato leaves infiltrated with 1 mM dibromo-cGMP, 100 μM dibutyryl-cAMP (Ma et al., 2009; Qi et al., 2010), and 50 μM NaVO3 was obviously less severe (Supplementary Figure S1A), indicated as significantly smaller diameter of lesions, 0.9 and 1.0 cm in cGMP- and cAMP-treated leaves while near 1.3 cm in non-treated control leaves respectively at 44 hpi (Supplementary Figure S1B). These results suggest that Ca2+ channels, which are regulated by cNMPs such as CNGCs, may play a role in tomato resistance to S. sclerotiorum.
This finding and the current status that function of CNGC has not been systemically studied in crop plant species raise our interest to identify at genome-wide level the CNGC family in economically important crop tomato and to dissect their roles in disease resistance.
Identification of CNGC Genes in Tomato Genome
The 20 Arabidopsis CNGC protein sequences collected from TAIR8 were used to search against tomato genome with TBLASTN at Sol Genomics Network9. Consequently total 22 SlCNGC candidate genes were retrieved in tomato genome, which were subsequently analyzed for domain and motif composition. Those bear both a cyclic nucleotide-binding domain (CNBD) and a TM domain or an ion transporter (ITP) domain as well as a recently suggested plant CNGC specific motif (Zelman et al., 2013) are recognized as CNGC proteins. Domain composition analyses using three different databases including10, SMART11 and CDD12 revealed that out of the 22 sequences, five lacked one of the required domains. Among them, Solyc03g007250.1.1 and Solyc03g007260.2.1 contained a CNB domain and an ITP domain respectively. Moreover, they are adjacent on chromosome 3 with a small insertion of 383 bp. We thus suspect that mistake exists in these sequences and/or their annotations and they should be actually a full-length CNGC protein, which was later confirmed by our PCR cloning and sequencing results that will be described below. Similarly, another two sequences Solyc06g010180.1.1 and Solyc06g010190.1.1 carried a CNB domain and an ITP domain respectively, and located on the same chromosome (No. 6) but with a much larger insertion of 4506 bp. CDD analysis demonstrated that this fragment contained a GAG-pre-integrase domain (gag_pre-integrs [pfam13976]), an integrase core domain (rve [pfam00665]) and a gag-polypeptide of LTR copia-type retrotransposon (UBN2_2 [pfam14227]) (Supplementary Figure S2), indicating that a retrotransposon-mediated sequence insertion might have resulted in loss of a functional CNGC protein. Additionally, searching of a recently suggested plant CNGC specific motif in the PBC and hinge region (Zelman et al., 2013) revealed that the whole PBC and hinge region were deleted in Solyc06g051920.2.1 while the PBC only was deleted in Solyc03g098210.2.1 (Supplementary Figure S3). However, cloning of these two genes from tomato cv. Heinz 1706 and subsequent sequencing analyses demonstrated that the actual genes contained the whole PBC and hinge region, and thus the sequences deposited in SGN database were incorrect (see below for detail). Therefore, these two sequences are indeed full length tomato CNGCs. Finally, 18 full-length and three truncated CNGCs were identified in tomato genome (Table 1, Supplementary Table S2), which is less than in Arabidopsis (20; Supplementary Table S3; Maser et al., 2001). We named the SlCNGCs in ascending order in accordance with group numbers on the basis of phylogenetic relationship (see below for detail) for easier and better understanding of their functions.
Table 1.
The SlCNGC gene family identified in this study.
| Group | Gene symbol | Gene locus | Chromosomal position | Protein size (aa) | Intron | Domain organization | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Pfam |
SMART | CDD | ||||||||
| Significant | Insignificant | |||||||||
| I | SlCNGC1 | Solyc01g095770.2.1 | ch01 78724016-78720407 | 706 | 8 | ITP, CNBD | – | cNMP, 5TMD, | CAP_ED | |
| SlCNGC2 | Solyc05g050380.2.1 | ch05 59634104-59638697 | 607 | 7 | ITP, CNBD | – | cNMP, 3TMD, | ITP, CAP_ED | ||
| SlCNGC3 | Solyc05g050350.1.1 | ch05 59612050-59608116 | 715 | 7 | ITP, CNBD | – | cNMP, 5TMD, | ITP, CAP_ED | ||
| SlCNGC4 | Solyc05g050360.2.1 | ch05 59616314-59621717 | 719 | 7 | ITP, CNBD | – | cNMP, 5TMD, | ITP, CAP_ED | ||
| SlCNGC5 | Corrected Solyc06g051920.2.1 |
Corrected ch06 32082202-32086782 |
708 | 8 | ITP, CNBD | — | cNMP, 3TMD, | ITP, CAP_ED | ||
| SlCNGC6 | Corrected (Solyc03g007260.2.1 + Solyc03g007250.1.1) | Corrected ch03 1817349-1813353 + ch03 1812970-1812278 |
708 | 6 | ITP, CNBD | – | cNMP, 5 TMD | ITP, CAP_ED | ||
| II | SlCNGC7 | Solyc07g005590.2.1 | ch07 481459-487527 | 735 | 6 | ITP, CNBD | – | cNMP, 5TMD | ITP, CAP_ED | |
| SlCNGC8 | Solyc12g010010.1.1 | ch12 3144264-3150904 | 692 | 5 | ITP, CNBD | IQ∗ | cNMP, 5TMD | ITP, CAP_ED | ||
| SlCNGC9 | Solyc03g116850.2.1 | ch03 60167174-60162079 | 690 | 6 | ITP, CNBD | IQ∗ | cNMP, 5TMD | ITP, CAP_ED | ||
| III | SlCNGC10 | Solyc11g069580.1.1 | ch11 51297223-51293074 | 707 | 6 | ITP, CNBD | IQ∗ | cNMP, 6 TMD | ITP, CAP_ED | |
| SlCNGC11 | Solyc09g007840.2.1 | ch09 1376961-1373797 | 712 | 6 | ITP, CNBD | – | cNMP, 5TMD | ITP, CAP_ED | ||
| SlCNGC12 | Solyc07g006510.2.1 | ch07 1308919-1317697 | 720 | 6 | ITP, CNBD | IQ∗ | cNMP, 6TMD | ITP, CAP_ED | ||
| SlCNGC13 | Solyc08g069140.2.1 | ch08 55418170-55422079 | 726 | 6 | ITP, CNBD | – | cNMP, 5TMD | ITP, CAP_ED | ||
| SlCNGC14 | Solyc03g114110.2.1 | ch03 58195300-58189455 | 678 | 5 | ITP, CNBD | – | cNMP, 5TMD | CAP_ED | ||
| IVa | SlCNGC15 | Corrected Solyc03g098210.2.1 |
Corrected ch03 54020551-54006686 |
763 | 12 | CNBD | – | cNMP, 6TMD | ITP, CAP_ED | |
| IVb | SlCNGC16 | Solyc02g088560.2.1 | ch02 45190069-45186246 | 708 | 7 | CNBD | IC∗, IQ∗ | cNMP, 6TMD, | CAP_ED | |
| SlCNGC17 | Solyc10g006800.2.1 | ch10 1246964-1240517 | 628 | 8 | CNBD | GP41∗ | cNMP, 6TMD | CAP_ED | ||
| SlCNGC18 | Solyc12g005400.1.1 | ch12 242060-236128 | 686 | 7 | CNBD | DUF4056∗ | cNMP, 6TMD | CAP_ED, Neocle∗ | ||
ITP, ion transport protein; CNBD, cyclic nucleotide-binding domain; IQ, IQ calmodulin-biding motif; IC, ion channel; GP41, glycoprotein 41; DUF, domain of unknown function; cNMP, cyclic nucleotide-monophosphate binding domain; TMD, transmembrane domain; CAP_ED, effector domain of CAP family. The asterisks showed in the table indicate the insignificant or incomplete domains.
The SlCNGCs were basic proteins with a size of 607 amino acids (aa; SlCNGC2) ∼763 aa (SlCNGC15) that is dominated by about 700 aa (Table 1), which is similar to AtCNGCs (Supplementary Table S3; Maser et al., 2001).
Domain and Motif Composition of SlCNGC Family
Domain composition analyses using databases Pfam, SMART and CDD revealed that in addition to the two main domains CNBD and TM or ITP, some tomato and Arabidopsis CNGC proteins contained several other insignificantly matched or incomplete domains and motifs (Table 1, Supplementary Table S3). These included IQ calmodulin-binding motif (PF00612) widely distributed in five SlCNGCs and seven AtCNGCs, a domain for GP41 family of envelop proteins from a variety of retroviruses (PF00517) in SlCNGC17 and AtCNGC2, TMEMspv1-c74-12 (PF11044) in a TM protein expressed by Plectrovirus spv1-c74 in AtCNGC2, and neocle (nucleoplasmin) domain (pfam03066) at N termini of SlCNGC18 and AtCNGC4, DUF4056 (PF13265) in SlCNGC18 and DUF4414 (pfam14377) in AtCNGC18. Whether these domains are actually functional awaits further experimental proof.
We further analyzed the PBC and hinge region within the CNB domain to check whether SlCNGCs carried similar motif as suggested for plant CNGCs (Zelman et al., 2013). The alignment of the PBC and hinge region of 18 SlCNGCs revealed a conserved motif in SlCNGCs as [LI]-X(2)-[GSNC]-X-[FYA]-X-G-X-E-LL-X-W-X-[LI]-X(7,13)-[LI]-P-X(1,5)-S-X(9)-[VIT]-E-[AST]-F-X-[LV] (Figure 1). Comparison of this motif with the one suggested by Zelman et al. (2013) for plant CNGCs, [LI]-X(2)-[GS]-X-[VFIYS]-X-G-X(0,1)-[DE]-L-[LI]-X-[WN]-X(6,32)-[SA]-X(9)-[VTI]-[EN]-[AG]-F-X-[LI], demonstrated that except for minor variation in some positions such as the 4th, 49th and the last positions, the motif for SlCNGCs generally fitted and was more conserved than that for plant CNGCs. All five essential amino acids G, L, S, E, and F in earlier version (Zelman et al., 2012) or three G, L, and F in their revised version (Zelman et al., 2013) of motif for plant CNGCs, presented in SlCNGCs as well. Furthermore, another four amino acids E, L, W, and P, which were not conserved in plant CNGCs, identically existed in all SlCNGCs (Figure 1). Alignment of the PBC and hinge region of 20 AtCNGCs gave rise to a motif as [LI]-X(2)-[GS]-X-[FVYSI]-X-G-X(0,1)-[ED]-LL-X-[WN]-X(0,1)-[L0]-X(4,14)-P-X(1,5)-S-X-[RAS]-X(7)-[VIT]-E-[AG]-F-X-[LI] with identical existence of seven amino acids G, L, L, P, S, E, and F (Supplementary Figure S4). Comparison of the PBC and hinge region of tomato and Arabidopsis CNGCs demonstrated that tomato CNGCs contained E and W two more identically existed amino acids in PBC (Figure 1, Supplementary Figures S4 and S5). These data reveal that the 18 SlCNGCs are most probably true CNGCs, and the PBC of CNB domain is more conserved within SlCNGCs than within AtCNGCs. Additionally, based on alignment of 38 tomato and Arabidopsis CNGCs, we predicted a more specific plant CNGC motif for the PBC and hinge region than the one suggested previously: [LI]-X(2)-[GSNC]-X-[FYVSIA]-X-G-X(0,1)-[ED]-LL-X-[WN]-X(0,1)-[LI0]-X(4,14)-P-X(1,5)-S-X(9)-[VIT]-E-[AGST]-F-X-[LIV] (Supplementary Figure S5). Whether this motif fits CNGCs of other plant species remains further analyses.
FIGURE 1.
The cyclic nucleotide gated channel (CNGC)-specific motif spanning the phosphate binding cassette (PBC) and hinge region within CNB domain of 18 SlCNGCs. The CNGC-specific motifs for all plant species (Zelman et al., 2013) or Arabidopsis and tomato only are shown at top of alignment respectively. The square brackets “[ ]” indicate the amino acids allowed in this position of motif, “X” represents any amino acid, while round brackets “( )” denote the number of amino acids. The names and residue positions from the N-termini of SlCNGCs are indicated to the left of the motif. The PBC and hinge region are indicated by black lines above sequences. Residues in white highlighted in black indicate >90% conservation among SlCNGCs. The red and black asterisks above the alignment indicate 100% conservation among all tomato and Arabidopsis CNGCs and among tomato CNGCs only respectively. The SlCNGC-specific motifs were aligned by ClustalW and generated by MEGA5 program.
Correction and Re-Annotation of Four Loci Deposited in SGN Database
As mentioned above Solyc03g007260.2.1 and Solyc03g007250.1.1 are localized on the same chromosome (No. 3) with a small insertion of 383 bp, which makes us hypothesize that they are actually two parts of the same CNGC gene. To confirm it, we designed forward and reverse primers separately starting from the start codon of Solyc03g007260.2.1 and the stop codon of Solyc03g007250.1.1 respectively, and finally the full-length sequence containing both parts was amplified by RT-PCR from tomato cv. Heinz 1706. Sequencing result revealed that the cloned gene carried an ORF of 2127 bp, rather not the total length of the insertion and Solyc03g007260.2.1 and Solyc03g007250.1.1 (2294 bp). It encoded for a protein of 708 aa (GenBank accession no. KJ499457, Supplementary Figure S6). Domain composition analysis showed that this protein possessed both CNB and IPT domains, and thus is a CNGC protein (Table 1). In accordance with the phylogenetic relationship with Arabidopsis CNGCs, we name this gene member as SlCNGC6. To check the possible variation of this gene in different tomato cultivars, we used the same primers to amplify this gene from the other two tomato cultivars Money maker and Suhong 2003, and obtained identical products to that from Heinz 1706 except one bp non-synonymous mutation (G1642 to C1642 resulting in C548 to S548) in Suhong 2003 (data not shown). Our data clearly show that SlCNGC6 exists generally in tomato cultivars, and sequences and annotation of Solyc03g007260.2.1 and Solyc03g007250.1.1 deposited in SGN are incorrect.
Additionally, sequence analysis demonstrated that the whole PBC and hinge region or PBC only was deleted in Solyc06g051920.2.1 and Solyc03g098210.2.1 respectively (Supplementary Figure S3). To ensure the sequences, we cloned both full-length genes from Heinz 1706 with forward and reverse primers separately designed to start from the start codon and stop at the stop codon of Solyc06g051920.2.1 and Solyc03g098210.2.1 respectively. Consequently, we cloned ORFs of 2127 and 2292 bp. Sequencing results demonstrated that these cloned sequences were identical to Solyc06g051920.2.1 and Solyc03g098210.2.1 except that they were 207 and 111 bp longer respectively (GenBank accession no. KJ499456 and KJ499458, Supplementary Figures S7 and S8). Sequence comparison results revealed that our cloned sequences included the whole PBC and hinge regions (Figure 1, Supplementary Figure S5). We name these two genes as SlCNGC5 and SlCNGC15 respectively according to the phylogenetic relationship with Arabidopsis CNGCs. Our experimental data unanimously show that sequences of Solyc06g051920.2.1 and Solyc03g098210.2.1 deposited in SGN are not correct.
Gene Structure and Chromosomal Location of SlCNGC Gene Family
To investigate further relationship between SlCNGC and AtCNGC genes, we comparatively analyzed their exon/intron structure (Supplementary Figure S9, Supplementary Table S4). In both SlCNGC and AtCNGC genes, the exon/intron structure predicted by GSDS exhibited group-specific feature in aspects of intron number and phase. Groups I, II, III, and IVb showed similar features, carrying 4∼8 phase 2 or 0 introns except SlCNGC5, while group IVa, carrying over nine introns, some of them belonging to phase 1 type (Supplementary Figure S9). These data suggested that expression and/or function of group IVa CNGCs might be different from the other CNGCs. When the exon/intron structure of the same group CNGC genes from tomato and Arabidopsis was compared, they displayed significant difference in intron size, which of tomato CNGC genes was dramatically larger than that of Arabidopsis counterparts. In addition, slight difference in the number and/or phase of introns of CNGC genes from the two species also existed (Supplementary Figure S9).
The 18 SlCNGC genes were distributed on 11 out of totally 12 chromosomes except chromosome 4 of tomato genome (Table 1). Four chromosomes bore two or more SlCNGC genes. Chromosomes 3 and 5 carried four and three SlCNGCs respectively, while chromosomes 7 and 12 carried two SlCNGC genes respectively. The other seven SlCNGC genes were solely located on seven different chromosomes respectively. Therefore, generally speaking, SlCNGC genes are evenly distributed in the whole tomato genome.
Phylogenetic Relationship between Tomato and Arabidopsis CNGCs
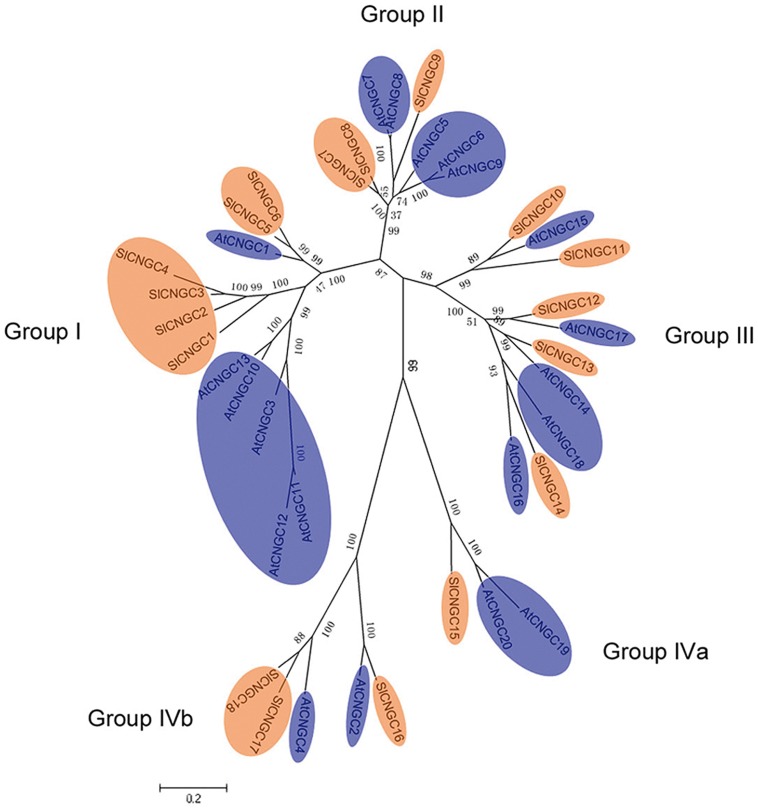
The relationship among SlCNGC proteins was observed by generating the unrooted ML phylogenetic tree. The 18 SlCNGCs were clustered in the tree into five groups (Supplementary Figure S10) as reported for AtCNGCs (Maser et al., 2001). To highlight the relationship between AtCNGCs and SlCNGCs, the ML phylogenetic tree was generated among 20 AtCNGCs (Maser et al., 2001) and 18 SlCNGCs. The SlCNGCs gathered with AtCNGCs into five groups (Figure 2). For each group of AtCNGCs tomato homologs existed. The size of each SlCNGC groups was unequal. The largest groups, groups I and III contained six and five members respectively; while the smallest one group IVa had just one member (Figure 2, Supplementary Figure S10). Compared with the group architecture of Arabidopsis CNGCs, group IVb of SlCNGCs gained one member; while groups II and IVa of SlCNGCs lost two and one member(s) respectively, which resulted in two member reduction in total in tomato CNGC family. In groups I and II, the tomato and Arabidopsis members clustered separately, implying that CNGC genes might have evolved separately in these two species.
FIGURE 2.
Phylogentic tree of the SlCNGC and AtCNGC proteins. The tree was created using Clustalx program by maximum liklihood (ML) method with bootstrap of 1000 in MEGA5. The SlCNGCs and AtCNGCs were shown in orange and blue respectively.
Cis-acting Element Composition of the SlCNGC Genes
In order to obtain preliminary clues for SlCNGC gene function, cis-acting elements in 1 kb upstream region of the SlCNGC genes were predicted in PLACE database. This divulged that SlCNGC promoter sequences generally carried a variety of predicted cis-acting elements that are responsive to various hormones such as ABA, auxin, GA, ethylene and cytokinin, biotic and abiotic factors and numerous WRKY transcription factors. However, the pattern of cis-acting elements in promoters of the different SlCNGC genes was distinct. Some elements widely distributed in promoters of almost all SlCNGC genes. Among them were nine elements responsive to ABA, auxin, GA, ethylene, cytokinin, biotic and abiotic factors respectively and four WRKY binding sites (Table 2). On the contrary, some elements were uniquely present in one SlCNGC gene. These included seven ABA responsive, two auxin responsive, one GA responsive and two biotic factor responsive elements. In addition, a Ca2+/CaMB element CGCGBOXAT existed in the promoters of SlCNGC2 and SlCNGC11, suggesting that these two SlCNGC genes might be feedback regulated by Ca2+/calmodulin. Interestingly, promoters of SlCNGC17 and SlCNGC18, two group IVb SlCNGC genes, contained biotic factor-regulated elements, indicating that these SlCNGC genes might play important roles in regulating plant disease resistance. Collectively, the element-profiling analysis revealed that SlCNGC genes might be regulated widely by a variety of factors including phytohormones, biotic and abiotic factors, and the underlying regulation mechanisms vary for different SlCNGC members.
Table 2.
Predicted cis-acting elements in 1 kb upstream region of the SlCNGC gene family.
| Regulator | Cis-acting element | Signal sequence | Code | Number of elements in 18 SlCNGCs |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||||
| ABA | ABREATCONSENSUS | YACGTGGC | S000406 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ACGTABREMOTIFA2OSEM | ACGTGKC | S000394 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| ABREATRD22 | RYACGTGGYR | S000013 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| ATHB6COREAT | CAATTATTA | S000399 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| DRE2COREZMRAB17 | ACCGAC | S000402 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| DPBFCOREDCDC3 | ACACNNG | S000292 | 1 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| MYCATRD22 | CACATG | S000174 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | |
| MYB2CONSENSUSAT | YAACKG | S000409 | 2 | 0 | 1 | 0 | 1 | 2 | 3 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | |
| MYBATRD22 | CTAACCA | S000175 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| PYRIMIDINEBOXHVEPB1 | TTTTTTCC | S000298 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | |
| PROXBBNNAPA | CAAACACC | S000263 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| RYREPEATBNNAPA | CATGCA | S000264 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| RYREPEATVFLEB4 | CATGCATG | S000102 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Auxin | ARFAT | TGTCTC | S000270 | 0 | 0 | 1 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ASF1MOTIFCAMV | TGACG | S000024 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | |
| AUXREPSIAA4 | KGTCCCAT | S000026 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| GGTCCCATGMSAUR | GGTCCCAT | S000360 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| NTBBF1ARROLB | ACTTTA | S000273 | 1 | 0 | 4 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 3 | 0 | 4 | 1 | 1 | 2 | |
| GA | GARE1OSREP1 | TAACAGA | S000419 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GARE2OSREP1 | TAACGTA | S000420 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| GAREAT | TAACAAR | S000439 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 4 | 0 | 0 | 0 | 4 | 2 | 1 | 1 | 1 | |
| MYBGAHV | TAACAAA | S000181 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 2 | 1 | 1 | 0 | |
| PYRIMIDINEBOXOSRAMY1A | CCTTTT | S000259 | 3 | 1 | 0 | 3 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 1 | |
| TATCCAOSAMY | TATCCA | S000403 | 1 | 0 | 3 | 2 | 0 | 2 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | |
| TATCCACHVAL21 | TATCCAC | S000416 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| ETH | ERELEE4 | AWTTCAAA | S000037 | 1 | 1 | 0 | 0 | 2 | 1 | 2 | 0 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| LECPLEACS2 | TAAAATAT | S000465 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 2 | 0 | 0 | |
| ARR1AT | NGATT | S000454 | 9 | 11 | 11 | 10 | 14 | 15 | 21 | 10 | 21 | 12 | 13 | 8 | 9 | 13 | 5 | 11 | 7 | 12 | |
| Cytokinin | CPBCSPOR | TATTAG | S000491 | 0 | 0 | 1 | 0 | 1 | 3 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 0 | 2 | 2 |
| Ca2+/Calmodulin binding | CGCGBOXAT | VCGCGB | S000501 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Biotic | BOXLCOREDCPAL | ACCWWCC | S000492 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| CACGTGMOTIF | CACGTG | S000042 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| ELRECOREPCRP1 | TTGACC | S000142 | 1 | 0 | 2 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| GT1GMSCAM4 | GAAAAA | S000453 | 2 | 2 | 1 | 7 | 2 | 2 | 1 | 0 | 1 | 3 | 3 | 5 | 7 | 1 | 6 | 3 | 7 | 3 | |
| MYB1LEPR | GTTAGTT | S000443 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| QARBNEXTA | AACGTGT | S000244 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SEBFCONSSTPR10A | YTGTCWC | S000391 | 1 | 0 | 0 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Abiotic | MYCATERD1 | CATGTG | S000413 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 |
| MYCCONSENSUSAT | CANNTG | S000407 | 10 | 6 | 41 | 8 | 4 | 10 | 6 | 8 | 6 | 4 | 10 | 6 | 6 | 10 | 2 | 10 | 0 | 0 | |
| MYB2AT | TAACTG | S000177 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WRKY transcription factor | WBBOXPCWRKY1 | TTTGACY | S000310 | 1 | 1 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| WBOXATNPR1 | TTGAC | S000390 | 4 | 2 | 5 | 4 | 2 | 3 | 4 | 4 | 4 | 3 | 1 | 2 | 2 | 6 | 0 | 0 | 0 | 1 | |
| WBOXHVISO1 | TGACT | S000442 | 5 | 2 | 3 | 3 | 3 | 1 | 4 | 3 | 4 | 4 | 0 | 3 | 1 | 4 | 1 | 2 | 1 | 1 | |
| WBOXNTERF3 | TGACY | S000457 | 5 | 2 | 5 | 4 | 3 | 2 | 7 | 3 | 7 | 6 | 0 | 5 | 2 | 4 | 2 | 2 | 3 | 1 | |
| WRKY71OS | TGAC | S000447 | 9 | 4 | 9 | 5 | 4 | 7 | 7 | 9 | 7 | 7 | 4 | 7 | 3 | 9 | 4 | 5 | 4 | 5 | |
| WBOXNTCHN48 | CTGACY | S000508 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | |
Expression Pattern of the SlCNGC Genes in Response to Diverse Stimuli
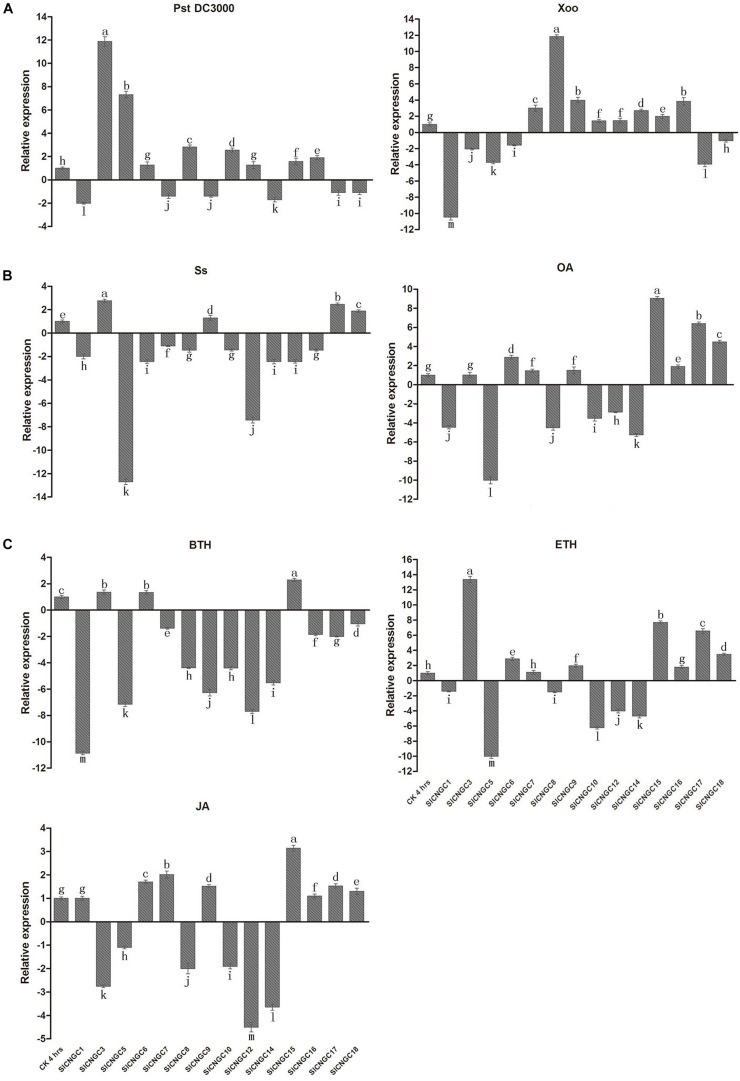
By quantitative real-time PCR (qRT-PCR) analysis, the expression patterns of SlCNGC genes were examined in early response (4 hpi) to various defense signaling molecules or their analogs or compounds from which they are released, including ethylene-releasing compound ETH, JA and a biologically active analog of SA BTH, the pathogen S. sclerotiorum (Ss)-secreted phytotoxin OA and the pathogens Ss, Pst DC3000 and Xoo in tomato (Figure 3). The specific primers used for qRT-PCR were listed in Supplementary Table S1. The gene expression patterns were distinct among different SlCNGC genes in response to diverse stimuli. When viewed from the stimuli, Ss inoculation and BTH treatment down-regulated expression of most of the SlCNGC genes, while the other stimuli up-regulated expression of eight SlCNGC genes but down-regulated expression of the other six SlCNGC genes. When analyzed from the SlCNGC genes, expression of group III SlCNGC genes was suppressed by BTH, ETH, JA, and OA treatments and Ss inoculation while was not significantly altered or induced by Pst DC3000 and Xoo inoculation except SlCNGC14 by Pst DC3000 inoculation. For group IVb genes, SlCNGC17 and SlCNGC18 exhibited same expression patterns, both up-regulated by ETH, JA, and OA treatment and Ss inoculation while down-regulated by BTH treatment and Pst DC3000 and Xoo inoculation. The remaining member SlCNGC16 displayed similar expression pattern except responsiveness to three pathogens, which was identical to SlCNGC15, the sole member of group IVa. For groups I and II, different members showed distinct expression patterns in response to the stimuli. SlCNGC5 was generally highly responsive to almost all stimuli tested in this study. SlCNGC3 specially and highly responded to Pst DC3000 inoculation and ETH treatment, while SlCNGC1 and SlCNGC8 highly responded to Xoo inoculation and OA and BTH treatments. The expression data indicated that some SlCNGC genes may play a role in regulating plant disease resistance.
FIGURE 3.
Expression profiles of 14 SlCNGC genes. Expression profiles of the SlCNGC genes in response to Pst DC3000 and Xoo inoculation (A), oxalic acid (OA) treatment and Ss inoculation (B) and hormone treatments (C) were detected by qRT-PCR with the gene-specific primers listed in Supplementary Table S1. The small letters indicated the significant difference among expression level of the SlCNGC genes in treated samples and non-treated CK and data represent the mean ± SE of three independent experiments (p < 0.05, DMRT).
Silencing of Group IVb SlCNGC Genes Significantly Enhanced Resistance to Necrotrophic Pathogens but Reduced Resistance to Viral Pathogen TRV
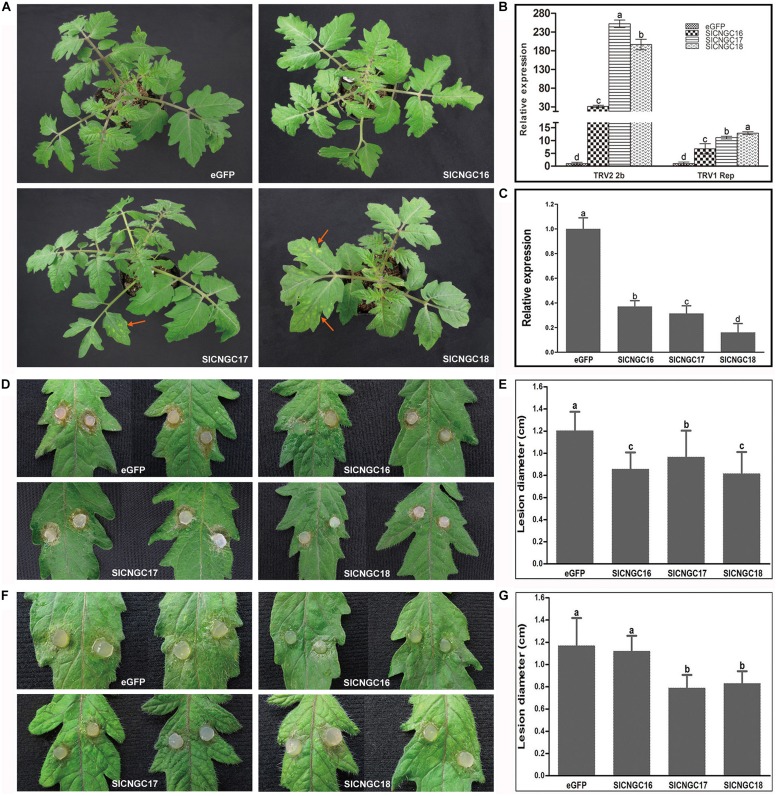
Further detailed functional analyses of group IVb SlCNGC genes in plant disease resistance were performed employing virus-induced gene silencing (VIGS) technique. To ensure the target specificity of silencing, the gene-specific UTR sequences of the SlCNGC genes were used for VIGS analyses. They were ligated into the TRV silencing vector pYL156 for silencing analyses, while non-silenced eGFP fragment-inserted recombinant pYL156 vector was used as a negative control (Cheng et al., 2012). Three weeks after agro-infiltration for silencing, the tomato plants agro-infiltrated for pYL156::SlCNGC17 and pYL156::SlCNGC18 showed clear mosaic symptoms in leaves; those for pYL156::SlCNGC16 showed very mild mosaic symptoms, while those for control vector pYL156::eGFP grew normally Figure 4A). Gene expression analysis revealed that transcripts of TRV1 replicase gene accumulated 6, 11, and 12 folds higher in SlCNGC16-, SlCNGC17- and SlCNGC18-silenced plants respectively than control plants. More strikingly, transcripts of TRV2 2b gene accumulated 31, 252, and 196 folds higher in SlCNGC16-, SlCNGC17-, and SlCNGC18-silenced plants respectively than control plants (Figure 4B).
FIGURE 4.
Silencing of group IVb SlCNGC genes in tomato plants reduced resistance to Tobacco rattle virus (TRV) and enhanced resistance to Sclerotinia sclerotiorum and Pythium aphanidermatum. (A) Phenotypes of the SlCNGC-silenced plants. Gene silencing analyses were performed using TRV-based vectors. TRV viral symptoms (indicated with red arrows) were more severe in the SlCNGC17- and SlCNGC18-silenced plants compared with the eGFP-control plants. Photographs were taken 3 weeks post agro-inoculation. (B) Detection of transcripts of the TRV1 replicase gene and TRV2 2b gene in silenced plants by qRT-PCR. (C) Gene expression anaysis by qRT-PCR to detect SlCNGC gene silencing efficiency. (D) Disease symptoms of the SlCNGC-silenced plants after inoculated with S. sclerotiorum (Ss). Photograhps were taken at 24 hpi. (E) Statistical anaysis of disease severity in Ss inoculated plants. (F) Disease symptoms of the SlCNGC-silenced plants after inoculated with P. aphanidermatum. Photograhps were taken at 14 hpi. (G) Statistical anaysis of disease severity in P. aphanidermatum inoculated plants. For each pathogen at least six silenced plants were examined. The experiments were conducted three times independently. The data in all statistical analyses represent the mean ± SD of three experiments. Significant difference is indicated as small letters (p < 0.05, DMRT).
To clarify whether the SlCNGC genes had been efficiently silenced in the agro-infiltrated plants, transcripts of these genes in the agro-infiltrated plants were quantified with qRT-PCR. Result showed that transcripts of SlCNGC16, SlCNGC17, and SlCNGC18 in the agro-infiltrated plants dropped to 37, 31, and 17%, respectively, of that in control plants (Figure 4C). These results demonstrated that silencing of these SlCNGC genes resulted in the TRV viral symptoms and higher level of virus accumulation. Together, these data indicated that the group IVb SlCNGC genes, especially SlCNGC17 and SlCNGC18 positively regulate tomato resistance to TRV.
To further probe functions of group IVb SlCNGCs in disease resistance, the silenced tomato plants were inoculated with a set of different types of pathogens. The non-host bacterial semi-biotrophic pathogen, Xoo, the host bacterial semi-biotrophic pathogen Pst DC3000 and the host fungal necrotrophic pathogens Ss and P. aphanidermatum were tested. Non-host resistance to Xoo in silenced tomato plants for all three SlCNGC genes was similar to that in control plants, as both plants developed clear HR necrosis in infiltrated areas at 14 hpi (Supplementary Figure S11A). Resistance to Pst DC3000 was also not altered significantly in silenced plants when compared with the control plants. Both plants showed severe necrosis at 48 hpi (Supplementary Figure S11B). However, when inoculated with Ss, necrotic symptoms of the leaves of the SlCNGC-silenced plants were obviously less severe than that of the eGFP-control plants (Figure 4D). The lesions in the SlCNGC-silenced plants, lower than 1.0 cm in diameter, were statistically significantly smaller than those in control plants, 1.2 cm in diameter at 24 hpi (Figure 4E).
When investigated with another important necrotrophic pathogen P. aphanidermatum, results similar to Ss were observed except SlCNGC16. After inoculation with P. aphanidermatum, pathogen-driven necrosis in the leaves of SlCNGC17- and SlCNGC18-silenced plants were obviously less severe than that of the eGFP-control plants (Figure 4F). The necrotic lesions in the silenced plants, about 0.8 cm in diameter, were strikingly smaller than those in control plants, near 1.2 cm in diameter at 14 hpi (Figure 4G). However, leaves of the SlCNGC16-silenced plants displayed necrotic lesions approximately with similar size to those of the eGFP-control plants (Figures 4F,G). These data revealed that the group IVb SlCNGCs negatively regulates resistance to necrotrophic pathogens Ss and P. aphanidermatum in tomato plants.
Collectively, our results reveal that the group IVb SlCNGC genes play important roles in regulation of plant disease resistance to a variety of pathogens in a gene- and target pathogen-dependent manner. SlCNGC17 and SlCNGC18 negatively regulate resistance to necrotrophic pathogens such as Ss and P. aphanidermatum while positively regulate resistance to viral pathogens such as TRV in tomato plants. However, SlCNGC16 mainly plays a role in regulating resistance to the necrotrophic pathogen Ss.
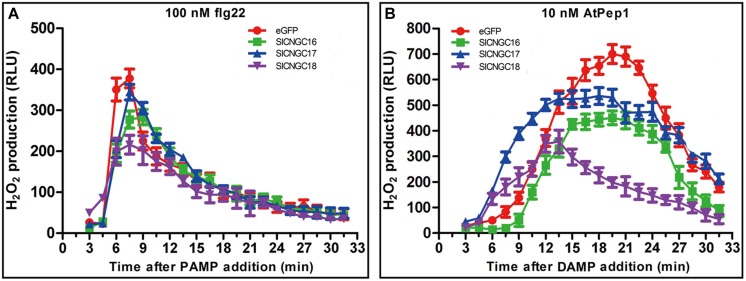
Silencing of Group IVb SlCNGC Genes Reduced PAMP- and DAMP-Triggered Hydrogen Peroxide Accumulation
To further explore functions of SlCNGCs in disease resistance, role of group IVb SlCNGC genes in pathogen-associated molecular pattern (PAMP)- and damage-associated molecular pattern (DAMP)-triggered immunity was examined. AtPep1 (10 nM) and flg22 (100 nM) were applied to leaf disks (3 mm at diameter) of the SlCNGC-silenced and non-silenced (control) plants. Subsequently the dynamics of hydrogen peroxide (H2O2) accumulation, which was indicated as relative luminescence (RLU), was measured. In response to flg22, H2O2 increased rapidly and peaked at 7 min post application (mpa) to 377 RLU and subsequently decreased quickly to reach the basal level within about 30 mpa in control plants (Figure 5A). In the SlCNGC-silenced plants, H2O2 accumulation displayed similar dynamics but the peak value was lower, which differed depending on the SlCNGC gene that was silenced. In the SlCNGC16- and SlCNGC18-silenced plants, H2O2 peaked to 277 and 216 RLU respectively, which was significantly lower than control. However, in the SlCNGC17-silenced plants, H2O2 peaked to 344 RLU, which was only slightly lower than control (Figure 5A). These data supported a positive role of group IVb SlCNGC genes, especially SlCNGC16 and SlCNGC18 in PAMP-triggered immunity (PTI). After application of AtPep1, H2O2 accumulation peaked to 700 RLU at 20 mpa in control plants, 451 RLU at 20 mpa in the SlCNGC16-silenced plants, 530 RLU at 20 mpa in the SlCNGC17-silenced plants while 349 RLU at 15 mpa in the SlCNGC18-silenced plants (Figure 5B), demonstrating that silencing of group IVb SlCNGC genes, especially SlCNGC16 and SlCNGC18, strongly reduced the AtPep1-triggered H2O2 accumulation, and therefore suggesting that the group IVb SlCNGC genes are required for PTI amplification triggered by plant endogenous DAMPs.
FIGURE 5.
Silencing of group IVb SlCNGC genes in tomato plants reduced PAMP/DAMP-induced accumulation of H2O2. Production of H2O2 induced by 100 nM flg22 (A) and 10 nM AtPep1 (B) was detected using a luminal-based assay in leaf disks of the SlCNGC16-, SlCNGC17-, and SlCNGC18-silenced plants and the eGFP non-silenced control plants. Data are shown as relative luminal units (RLU) and represent the mean ± SE of three independent experiments.
Silencing of Group IVb SlCNGC Genes Reduced Expression of the Defense-Related Ca2+ Signaling Genes
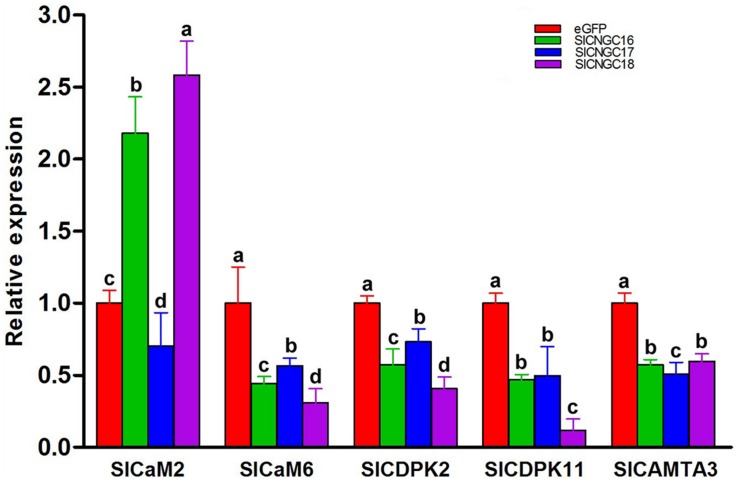
To find out molecular mechanism of group IVb SlCNGC genes in regulating plant disease resistance, we examined expression of a series of resistance-related Ca2+ signaling genes to clarify whether they are involved in group IVb SlCNGCs-mediated resistance regulation. The genes under this expression analysis included two calmodulin genes SlCaM2 and SlCaM6, two calcium-dependent protein kinase genes SlCDPK2 and SlCDPK11, which are the tomato homologs of Arabidopsis AtCPK2 and AtCPK11, and one CaM-binding transcription activator gene SlCAMTA3. All these genes play an important role in regulating disease resistance (Du et al., 2009; Boudsocq et al., 2010; Zhao et al., 2013). The expression result depicted that silencing of all three group IVb SlCNGC genes SlCNGC16, SlCNGC17, and SlCNGC18 unanimously reduced the expression of SlCaM6, SlCDPK2, SlCDPK11, and SlCAMTA3, but differently affected the expression of SlCaM2. Silencing of SlCNGC16 and SlCNGC18 increased its expression by 2.2 and 2.6 folds respectively, while silencing of SlCNGC17 reduced its expression by 70% (Figure 6). These results indicated that CaMs, CDPKs, and CAMTAs may play a role in the group IVb SlCNGCs-mediated resistance.
FIGURE 6.
Expression of a set of Ca2+ signaling genes in group IVb SlCNGC-silenced tomato plants. Expression of SlCaM2, SlaM6, SlCDPK2, SlCDPK11, and SlCAMTA3 genes in the SlCNGC16-, SlCNGC17-, and SlCNGC18-silenced plants and the eGFP non-silenced control plants was analyzed by qRT-PCR with gene-specific primers listed in Supplementary Table S1. Small letters denote the significant difference between expression values of the silenced plants and those of the eGFP-control plants (p < 0.05, DMRT). Data represent the mean ± SE of three independent experiments.
Discussion
Tomato and Arabidopsis CNGC Families
Cyclic nucleotide gated channel is suggested to be Ca2+ conducting channel in plants, yet, surprisingly, genome-wide identification and functional study so far are mainly conducted in the model plants Arabidopsis. In this study, we have identified CNGC family in the economically important crop plant tomato. Through bioinformatics and experimental analyses, we finally identified 18 tomato CNGCs. It is unexpected that members of CNGCs in tomato are less than in Arabidopsis, which contains 20 members (Maser et al., 2001), considering that the genome size of tomato is much larger than Arabidopsis. It seems unlikely that our identification is incomplete, since only 22 sequences were retrieved when we performed BLAST searching with all 20 AtCNGCs. All 22 retrieved sequences had been analyzed in detail. More importantly, for those annotated to be truncated, confirmation experiments were conducted such as RT-PCR cloning and sequencing. Consequently, we have corrected four loci in SGN tomato database, including completion of Solyc06g051920.2.1 and Solyc03g098210.2.1 sequences and naming them as SlCNGC5 and SlCNGC15 respectively, and reannotating two truncated sequences Solyc03g007260.2.1 and Solyc03g007250.1.1 as a full-length CNGC (SlCNGC6; Table 1, Supplementary Figures S6–S8). Then what are the reasons to cause smaller CNGC family in tomato than in Arabidopsis? We find that a gag-polypeptide of LTR copia-type retrotransposon presents between two truncated sequences Solyc06g010180.1.1 and Solyc06g010190.1.1 (Supplementary Figure S2). Moreover, SlCNGC17 contains a domain homologous to GP41 family of retroviral envelop protein (Table 1). Therefore, it is possible that retrotransposons and retroviruses disrupt CNGC genes and lead to loss of full-length CNGC gene members in tomato genome. Additionally, smaller CNGC family in tomato than in Arabidopsis could be ascribed to relatively more conserved genomes among the members of Solanaceae family, including tomato (Mueller et al., 2005). Moreover, genome size of rice and pear is significantly larger than that of Arabidopsis but they are comprised of only 16 and 21 CNGC genes respectively (Nawaz et al., 2014; Chen et al., 2015). This indicates that the phenomenon that a species with a larger genome contains similar or even less CNGC genes in comparison with Arabidopsis also occurs in plant species other than tomato. The cause and possible effect of this phenomenon remains further dissection.
The phylogenetic and gene structure analyses manifest that generally tomato CNGC genes exhibit group-wise similar structure and clusters to Arabidopsis counterparts (Supplementary Table S4; Figure 2, Supplementary Figures S9 and S10). However, CNGC genes of the two species discriminate in the following aspects. Firstly, the group I and II AtCNGCs and SlCNGCs cluster separately, demonstrating that the CNGC genes of these two species might evolve independently during evolution. Furthermore, intron numbers of groups II, IVa, and IVb differs between the two species, and group IVa SlCNGC (SlCNGC15) bore significantly more phase 1 type of introns than its Arabidopsis counterparts AtCNGC19 and AtCNGC20. Additionally, size of introns of SlCNGCs is much larger than that of AtCNGCs (Supplementary Table S4, Supplementary Figure S9), suggesting tomato CNGC genes might be more highly expressed than and functionally discriminate from Arabidopsis CNGCs as suggested for other genes in Arabidopsis and rice (Ren et al., 2006).
Taken together, identification of tomato CNGC family establishes a basis to dissect the functions of CNGCs in crop plant species.
Function of SlCNGCs
Cyclic nucleotide gated channels are involved in regulation of various biological processes such as growth and development, environmental stresses and plant defense responses (Chin et al., 2009; Ma, 2011). In the present study, we focused on dissection of the role of tomato CNGCs in disease resistance by conducting four levels of analyses, including prediction of cis-elements in promoter of SlCNGC genes, expression profiling of SlCNGC genes in response to diverse defense molecules and pathogens, pharmacological analyses on effect of cNMPs and Ca2+ channel activators/blockers on disease resistance and reverse genetics functional analyses of SlCNGC genes. All these analyses reveal that SlCNGCs are required for a wide range of disease resistances, and the effect is pathosystem-dependent. Regarding the expression profile, we found that treatment with BTH, a biological analog of SA, repressed the expression of SlCNGC16, the ortholog of AtCNGC2, and SlCNGC17 and SlCNGC18, the orthologs of AtCNGC4. In contrast, treatments with both ETH and JA induced the expression of these group IVb CNGC genes in tomato (Figure 3). Our finding is similar to what have been observed in Arabidopsis, i.e., expression of both AtCNGC2 and AtCNGC4 are repressed by SA treatment while induced by MeJA treatment (Moeder et al., 2011). Moreover, we observed that expression of the three group IVb SlCNGC genes is distinct in response to the same pathogen inoculation, and all these genes respond distinctly to different types of pathogens. SlCNGC16 is upregulated by inoculation with the biotrophic bacterial pathogens Pst DC3000 and Xoo, while is downregulated by inoculation with the necrotrophic fungal pathogen Ss, whereas the expression of SlCNGC17 and SlCNGC18 is just the opposite (Figure 3). Furthermore, VIGS analyses demonstrated that SlCNGC17 and SlCNGC18 are required for resistance to both Ss and P. aphanidermatum; while SlCNGC16 is involved in resistance only to Ss but not to P. aphanidermatum (Figure 4). Our results suggest that group IVb SlCNGC genes play a role in both SA and JA/ETH signaling, however, which signaling pathway is activated and play a key role in defense is dependent on the pathosystem and therefrom the CNGC gene activated, as reported for group IVb AtCNGCs (Yoshioka et al., 2001; Balague et al., 2003; Genger et al., 2008; Moeder et al., 2011). Additionally, AtCNGC11 and AtCNGC12 are not essential to PTI. However, expression of AtCNGC2 and AtCNGC4 is suppressed after flg22 treatment (Moeder et al., 2011). Nevertheless, the role of AtCNGC2 and AtCNGC4 in PTI remains unclear. Besides, the respond of these genes to DAMPs is not yet analyzed. In this study, we found that silencing of group IVb SlCNGC genes reduced flg22- and AtPEP1-elicited hydrogen peroxide accumulation (Figures 5A,B), indicating that group IVb SlCNGC genes play a role in both PTI and DTI. Together with demonstration of the function of group IVb SlCNGC genes in tomato resistance to Xoo, Ss and P. aphanidermatum, our results extend the role of group IVb CNGCs in plant disease resistance.
As mentioned above, we found that tomato is responsive to AtPep1, which leads to accumulation of H2O2 (Figure 5). This suggests that a Pep1 receptor, most probably the receptor of the tomato ortholog of AtPep1, exists in tomato genome and is functional to recognize AtPep1 as well as its tomato homologs. This is conceivable considering the following findings. Firstly, it has been found that a Pep receptor could recognize several Peps sharing only moderate sequence identity. For example, the Arabidopsis Pep1 receptor PEPR1 can recognize other four Peps as well (Yamaguchi et al., 2006), which show only 35∼65% sequence identity to AtPep1 (Huffaker et al., 2006). Additionally, the signaling pathway downstream the recognition of Peps by PEPRs are conserved in plant species of different families. It has been reported that the downstream signaling initiated by recognition of AtPep1 by AtPEPR1 in Arabidopsis is also observed in the AtPEPR1 transgenic tobacco when supplied with AtPep1 (Yamaguchi et al., 2006). Finally, an ortholog of AtPEP1 has been identified in tomato. This ortholog is 39% identical to sequence of AtPep1. Moreover, it is involved in resistance to Pythium dissotocum through signaling pathways mediated by jasmonic acid/ethylene (JA/ET; Trivilin et al., 2014), as reported for AtPep1 (Huffaker et al., 2006).
It is notable that in the pharmacological study, supply with putative CNGC activators cAMP and cGMP, thus expected to activate CNGCs, enhance tomato resistance to Ss (Supplementary Figure S1), which seems to be contradictory to the result from the CNGC-VIGS analyses that silencing of group IVb SlCNGC genes, thus expected to decrease the activity of these CNGCs, enhance the same resistance (Figures 4D,E). One explanation for this contradiction is that cAMP and cGMP generally activate the whole CNGC family in tomato, and some of them play a role opposite to group IVb CNGCs in regulating this resistance. The CNGC gene-dependent function has been observed in this study. Members of SlCNGC gene family respond in expression variously to the same pathogen inoculation (Figure 3). Furthermore, VIGS analysis demonstrates that SlCNGC17 and SlCNGC18, but not SlCNGC16 play a role in resistance to P. aphanidermatum (Figures 4F,G); while SlCNGC16 and SlCNGC18, but not SlCNGC17 function in flg22-elicited hydrogen peroxide accumulation (Figure 5A). Another possibility is the pleiotropism of cAMP and cGMP. They activate not only CNGCs but also other targets, which function opposite to the CNGCs in the resistance. For instance, beside CNGCs, cAMP activates cAMP-dependent protein kinases (PKAs) and guanine nucleotide exchange factors (GEFs) for small GTPase as well (Bos, 2003). It is possible that some PKAs and/or GEFs may regulate resistance in contrast to CNGCs. Similar observation has been reported for P. patens CNGCb gene. When an abrupt, continuous heat treatment at 34 or 38°C was applied, the 24°C-grown P. patens CNGCb mutant does not show reduced Ca2+ influx and decreased cytosolic Ca2+, in contrast, they display a stronger Ca2+ influx and elevated cytoplasmic Ca2+ concentration. This might be attributed to the promotive effect of PpCNGCb mutation on opening of other related Ca2+ channels (Finka et al., 2012). Interestingly, tomato CNGC16, CNGC17, and CNGC18 are the orthologs of Arabidopsis CNGC2 and CNGC4 (Figure 2), which are the orthologs of PpCNGCb (Finka et al., 2012). Therefore, whether similar mechanism occurs for tomato group IVb CNGCs deserves to be further analyzed.
Functional Mechanisms of SlCNGCs
The electrophysiological studies have suggested that some CNGCs such as AtCNGC18 are Ca2+-permeable channels (Gao et al., 2014; Zhou et al., 2014), thus we examined the effect of SlCNGCs on expression of the Ca2+ signaling related genes. The results demonstrated that silencing of group IVb SlCNGC genes reduced the expression of SlCaM6, SlCDPK2, SlCDPK11, and SlCAMTA3 but differentially altered the expression pattern of SlCaM2, i.e., silencing of SlCNGC16 and SlCNGC18 up-regulated while silencing of SlCNGC17 down-regulated the expression of SlCaM2 (Figure 6). Our previous results reveal that silencing of SlCaM2 and SlCaM6 enhances expression of SlCNGC17 but reduces expression of the SlCNGC18 and both reduce resistance to P. aphanidermatum (Zhao et al., 2013). Collectively, these results indicate the complexity of interactions between CNGC and CaM during disease resistance. In addition, CNGCs regulate disease resistance through promoting SlCDPK2, SlCDPK11, and SlCAMTA3. Besides, silencing of group IVb SlCNGC genes obviously reduces PAMP- and DAMP-triggered hydrogen peroxide accumulation (Figure 5), demonstrating that these CNGCs affect PTI and its amplification through regulating PAMP- and DAMP-triggered hydrogen peroxide accumulation. More detailed functional mechanisms of SlCNGCs remain further dissection.
Conclusion
Cyclic nucleotide gated channels are multifunctional and have been supposed to be calcium conducting channels. Nevertheless, genome-wide identification of plant CNGC gene family has been conducted only in four flowering plant species Arabidopsis, rice, pear and Populus trichocarpa, a moss P. patens and some alga species. Furthermore, systemic functional analysis of plant CNGC genes has not been well performed except in Arabidopsis. In this study, we identified CNGC gene family in the economically important crop tomato (Solanum lycopersicum L.) and analyzed function of the group IV SlCNGC genes in disease resistance, representing the first genome-wide identification and functional analysis of CNGC gene family in dicotyldons crop species. Importantly, the tomato CNGC gene family is not only identified through complex bioinformatics analyses but also confirmed by PCR cloning and sequencing. We correct four CNGC loci that were misannotated at database. We demonstrate that gene structure, domain composition and phylogenetic relationship of the SlCNGC gene family are group-specific. The tomato CNGC gene family contains less members, carries different exon/intron gene structures but own similar groups when compared with Arabidopsis CNGCs. We conduct comprehensive expression analyses and reveal that SlCNGC genes are highly and widely responsive to diverse stimuli in a gene-dependent manner. Employing pharmacological assays and VIGS technique, we further unravel the function of the group IVb SlCNGC genes in disease resistance. Silencing of these SlCNGC genes significantly enhances resistance to fungal pathogens P. aphanidermatum and S. sclerotiorum, strongly reduces resistance to viral pathogen Tobacco rattle virus, while attenuates PAMP- and DAMP-triggered immunity as shown by obvious decrease of flg22- and AtPEP1-elicited hydrogen peroxide accumulation in SlCNGC-silenced plants. Finally, this work indicates that the group IVb SlCNGC genes regulate a wide range of resistance to diverse pathogens in tomato probably via affecting Ca2+ signaling including SlCaMs, SlCDPKs, and SlCAMTA3. Collectively, this work identifies CNGC gene family in tomato genome, provides some insights into functions of tomato CNGCs and thus makes a platform to further elucidate the functions of plant CNGCs.
Author Contributions
MS and Y-P X conducted the bioinformatics, phylogenetic and pharmacological analyses. MS, WL and J-PW carried out the gene expression, ROS detection and VIGS analysis. MS and Y-P X designed and analyzed all statistical data. X-Z C conceived of the study, and participated in its design and coordination. X-Z C and MS prepared the manuscript.
Acknowledgments
This work was financially supported by grants from the Genetically Modified Organisms Breeding Major Projects (no. 2014ZX0800905B), the Special Fund for Agro-scientific Research in the Public Interest (no. 201103016), the Program for Changjiang Scholars and Innovative Research Team in University (no. IRT0943), the SRFDP (no. 20110101110092) and the Program for New Century 151 Talents of Zhejiang Province.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00303/abstract
References
- Abdel-Hamid H., Chin K., Moeder W., Yoshioka K. (2011). High throughput chemical screening supports the involvement of Ca2+ in cyclic nucleotide-gated ion channel-mediated programmed cell death in Arabidopsis. Plant Signal. Behav. 6 1817–1819 10.4161/psb.6.11.17502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn I. P. (2007). Disturbance of the Ca2+/calmodulin-dependent signalling pathway is responsible for the resistance of Arabidopsis dnd1 against Pectobacterium carotovorum infection. Mol. Plant Pathol. 8 747–759 10.1111/j.1364-3703.2007.00428.x [DOI] [PubMed] [Google Scholar]
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., Von Bodman S., et al. (2007). Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19 1081–1095 10.1105/tpc.106.045096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Sunkar R., Kaplan B., Fromm H. (1999). A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 20 171–182 10.1046/j.1365-313x.1999.00588.x [DOI] [PubMed] [Google Scholar]
- Balague C., Lin B., Alcon C., Flottes G., Malmstrom S., Kohler C., et al. (2003). HLM1 an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15 365–379 10.1105/tpc.006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsics T., Webb D., Andeme-Ondzighi C., Staehelin L. A., Christopher D. A. (2007). The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana. Planta 225 563–573 10.1007/s00425-006-0372-3 [DOI] [PubMed] [Google Scholar]
- Bos J. L. (2003). Epac: a new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 4 733–738 10.1038/nrm1197 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M. R., Mccormack M., Lee H., Shan L., He P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464 418–422 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D., Fraser M. E., Moorhead G. B. (2005). Cyclic nucleotide binding proteins in the Arabidopsis thaliana and Oryza sativa genomes. BMC Bioinformatics 6:6 10.1186/1471-2105-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Wang C., Xu Y., Xu Q., Zheng Z., Zhou X. (2007). Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Res. 125 169–175 10.1016/j.virusres.2006.12.016 [DOI] [PubMed] [Google Scholar]
- Chen J., Yin H., Gu J., Li L., Liu Z., Jiang X., et al. (2015). Genomic characterization, phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear (Pyrus bretchneideri Rehd.). Genomics 105 39–52 10.1016/j.ygeno.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Cheng W. S., Xu Q. F., Li F., Xu Y. P., Cai X. Z. (2012). Establishment of a suitable control vector for Tobacco rattle virus-induced gene silencing analysis in Nicotiana benthamiana. J. Zhejiang Univ. (Agric. Life Sci.) 38 10–20 10.3785/j.issn.1008-9209 [DOI] [Google Scholar]
- Chin K., Defalco T. A., Moeder W., Yoshioka K. (2013). The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol. 163 611–624 10.1104/pp.113.225680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., Moeder W., Yoshioka K. (2009). Biological roles of cyclic-nucleotide-gated ion channels in plants: what we know and don’t know about this 20 member ion channel family. Botany 87 668–677 10.1139/B08-147 [DOI] [Google Scholar]
- Clough S. J., Fengler K. A., Yu I. C., Lippok B., Smith R. K., Jr., Bent A. F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 97 9323–9328 10.1073/pnas.150005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457 1154–1158 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Finka A., Cuendet A. F., Maathuis F. J., Saidi Y., Goloubinoff P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24 3333–3348 10.1105/tpc.112.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietsch S., Wang Y. F., Sladek C., Poulsen L. R., Romanowsky S. M., Schroeder J. I., et al. (2007). A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. U.S.A. 104 14531–14536 10.1073/pnas.0701781104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Han X., Wu J., Zheng S., Shang Z., Sun D., et al. (2012). A heat-activated calcium-permeable channel – Arabidopsis cyclic nucleotide-gated ion channel 6 – is involved in heat shock responses. Plant J. 70 1056–1069 10.1111/j.1365-313X.2012.04969.x [DOI] [PubMed] [Google Scholar]
- Gao Q. F., Fei C. F., Dong J. Y., Gu L. L., Wang Y. F. (2014). Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 7 739–743 10.1093/mp/sst174 [DOI] [PubMed] [Google Scholar]
- Genger R. K., Jurkowski G. I., Mcdowell J. M., Lu H., Jung H. W., Greenberg J. T., et al. (2008). Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol. Plant Microbe Interact. 21 1285–1296 10.1094/mpmi-21-10-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A., Park G., Amtmann A., Sanders D., Maathuis F. J. (2006). Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 57 791–800 10.1093/jxb/erj064 [DOI] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C. A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103 10098–10103 10.1073/pnas.0603727103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski G. I., Smith R. K., Jr., Yu I. C., Ham J. H., Sharma S. B., Klessig D. F., et al. (2004). Arabidopsis DND2 a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520 10.1094/mpmi.2004.17.5.511 [DOI] [PubMed] [Google Scholar]
- Kaplan B., Sherman T., Fromm H. (2007). Cyclic nucleotide-gated channels in plants. FEBS Lett. 581 2237–2246 10.1016/j.febslet.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Keisa A., Kanberga-Silina K., Nakurte I., Kunga L., Rostoks N. (2011). Differential disease resistance response in the barley necrotic mutant nec1. BMC Plant Biol. 11:66 10.1186/1471-2229-11-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C., Merkle T., Neuhaus G. (1999). Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18 97–104 10.1046/j.1365-313X.1999.00422.x [DOI] [PubMed] [Google Scholar]
- Kohler C., Neuhaus G. (2000). Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471 133–136 10.1016/S0014-5793(00)01383-1 [DOI] [PubMed] [Google Scholar]
- Kugler A., Kohler B., Palme K., Wolff P., Dietrich P. (2009). Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 9:140 10.1186/1471-2229-9-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li W., Xu Y. P., Zhang Z. X., Cao W. Y., Li F., Zhou X., et al. (2012). Identification of genes required for nonhost resistance to Xanthomonas oryzae pv. oryzae reveals novel signaling components. PLoS ONE 7:e42796 10.1371/journal.pone.0042796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. (2011). Roles of Ca2+ and cyclic nucleotide gated channel in plant innate immunity. Plant Sci. 181 342–346 10.1016/j.plantsci.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Ma W., Ali R., Berkowitz G. A. (2006). Characterization of plant phenotypes associated with loss-of-function of AtCNGC1 a plant cyclic nucleotide gated cation channel. Plant Physiol. Biochem. 44 494–505 10.1016/j.plaphy.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Ma W., Berkowitz G. A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190 566–572 10.1111/j.1469-8137.2010.03577.x [DOI] [PubMed] [Google Scholar]
- Ma W., Qi Z., Smigel A., Walker R. K., Verma R., Berkowitz G. A. (2009). Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 106 20995–21000 10.1073/pnas.0905831106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., Sze H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126 1646–1667 10.1104/pp.126.4.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W., Urquhart W., Ung H., Yoshioka K. (2011). The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 4 442–452 10.1093/mp/ssr018 [DOI] [PubMed] [Google Scholar]
- Mueller L. A., Solow T. H., Taylor N., Skwarecki B., Buels R., Binns J., et al. (2005). The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 138 1310–1317 10.1104/pp.105.060707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z., Kakar K. U., Saand M. A., Shu Q. Y. (2014). Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics 15:853 10.1186/1471-2164-15-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K. B., Nicholas H., Deerfield D. (1997). GeneDoc: analysis and visualization of genetic variation. Embnew. News 4:14. [Google Scholar]
- Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C. A., et al. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1 a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. U.S.A. 107 21193–21198 10.1073/pnas.1000191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. Y., Vorst O., Fiers M. W., Stiekema W. J., Nap J. P. (2006). In plants, highly expressed genes are the least compact. Trends Genet. 22 528–532 10.1016/j.tig.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Rostoks N., Schmierer D., Mudie S., Drader T., Brueggeman R., Caldwell D. G., et al. (2006). Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Mol. Genet. Genomics 275 159–168 10.1007/s00438-005-0073-9 [DOI] [PubMed] [Google Scholar]
- Schuurink R. C., Shartzer S. F., Fath A., Jones R. L. (1998). Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. U.S.A. 95 1944–1949 10.1073/pnas.95.4.1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivilin A. P., Hartke S., Moraes M. G. (2014). Components of different signalling pathways regulated by a new orthologue of AtPROPEP1 in tomato following infection by pathogens. Plant Pathol. 63 1110–1118 10.1111/ppa.12190 [DOI] [Google Scholar]
- Tunc-Ozdemir M., Tang C., Ishka M. R., Brown E., Groves N. R., Myers C. T., et al. (2013). A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol. 161 1010–1020 10.1104/pp.112.206888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F., Wheeler G., Taylor A. R., Farnham G., Brownlee C. (2010). Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 187 23–43 10.1111/j.1469-8137.2010.03271.x [DOI] [PubMed] [Google Scholar]
- Wang C., Cai X., Wang X., Zheng Z. (2006). Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Funct. Plant Biol. 33 347–355 10.1071/FP05096 [DOI] [PubMed] [Google Scholar]
- Wang Y. F., Munemasa S., Nishimura N., Ren H. M., Robert N., Han M., et al. (2013). Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 163 578–590 10.1104/pp.113.225045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Maser P., Schroeder J. I. (2009). Plant ion channels: gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 71 59–82 10.1146/annurev.physiol.010908.163204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1 an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 103 10104–10109 10.1073/pnas.0603729103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Kachroo P., Tsui F., Sharma S. B., Shah J., Klessig D. F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26 447–459 10.1046/j.1365-313X.2001.2641039.x [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H. G., Kachroo P., Masmoudi K., Berkowitz G., et al. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18 747–763 10.1105/tpc.105.038786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I. C., Parker J., Bent A. F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. U.S.A. 95 7819–7824 10.1073/pnas.95.13.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelman A. K., Dawe A., Berkowitz G. A. (2013). “Identification of cyclic nucleotide gated channels using regular expressions,” in Cyclic Nucleotide Signaling in Plants: Methods and Protocols, Methods in Molecular Biology, ed. Gehring C. (New York, NY: Springer; ), 207–224. [DOI] [PubMed] [Google Scholar]
- Zelman A. K., Dawe A., Gehring C., Berkowitz G. A. (2012). Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 3:95 10.3389/fpls.2012.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu W., Xu Y. P., Cao J. Y., Braam J., Cai X. Z. (2013). Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 13:70 10.1186/1471-2229-13-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Lan W., Jiang Y., Fang W., Luan S. (2014). A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant 7 369–376 10.1093/mp/sst125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.