ABSTRACT
Although Staphylococcus aureus is exposed to antimicrobial fatty acids on the skin, in nasal secretions, and in abscesses, a specific mechanism of inducible resistance to this important facet of innate immunity has not been identified. Here, we have sequenced the genome of S. aureus USA300 variants selected for their ability to grow at an elevated concentration of linoleic acid. The fatty acid-resistant clone FAR7 had a single nucleotide polymorphism resulting in an H121Y substitution in an uncharacterized transcriptional regulator belonging to the AcrR family, which was divergently transcribed from a gene encoding a member of the resistance-nodulation-division superfamily of multidrug efflux pumps. We named these genes farR and farE, for regulator and effector of fatty acid resistance, respectively. Several lines of evidence indicated that FarE promotes efflux of antimicrobial fatty acids and is regulated by FarR. First, expression of farE was strongly induced by arachidonic and linoleic acids in an farR-dependent manner. Second, an H121Y substitution in FarR resulted in increased expression of farE and was alone sufficient to promote increased resistance of S. aureus to linoleic acid. Third, inactivation of farE resulted in a significant reduction in the inducible resistance of S. aureus to the bactericidal activity of 100 μM linoleic acid, increased accumulation of [14C]linoleic acid by growing cells, and severely impaired growth in the presence of nonbactericidal concentrations of linoleic acid. Cumulatively, these findings represent the first description of a specific mechanism of inducible resistance to antimicrobial fatty acids in a Gram-positive pathogen.
IMPORTANCE Staphylococcus aureus colonizes approximately 25% of humans and is a leading cause of human infectious morbidity and mortality. To persist on human hosts, S. aureus must have intrinsic defense mechanisms to cope with antimicrobial fatty acids, which comprise an important component of human innate defense mechanisms. We have identified a novel pair of genes, farR and farE, that constitute a dedicated regulator and effector of S. aureus resistance to linoleic and arachidonic acids, which are major fatty acids in human membrane phospholipid. Expression of farE, which encodes an efflux pump, is induced in an farR-dependent mechanism, in response to these antimicrobial fatty acids that would be encountered in a tissue abscess.
INTRODUCTION
Staphylococcus aureus has a dichotomous relation with human hosts, being able to establish an asymptomatic commensal relationship, but is also historically known as a leading cause of human infectious morbidity and mortality. Significantly, death attributed to S. aureus in the United States is now comparable to mortality rates for AIDS, tuberculosis, and viral hepatitis (1–3). Not surprisingly, therefore, S. aureus has been the subject of intensive research on mechanisms of pathogenesis and acquisition and transfer of antibiotic resistance and of efforts to identify potential vaccine antigens (4–6). Until the late 1990s, much of this was directed toward hospital-associated strains of methicillin-resistant S. aureus (HA-MRSA) to address the anticipated emergence of superbugs that would be resistant to all clinically useful antibiotics (7, 8). However, a new threat emerged in the late 1990s with community-acquired MRSA (CA-MRSA). Although these strains evolved in the community setting, one notorious strain known as USA300 has achieved pandemic status across North America and is now the leading cause of S. aureus infections, irrespective of community or hospital origin (9, 10). This has engendered greater attention toward identifying mechanisms of S. aureus persistence on human hosts and of host-to-host transmission.
Approximately 25% of humans are persistently colonized by S. aureus, where the preferred site of colonization is the anterior nares, and among colonized individuals, the bacterium is also frequently recovered from other body sites, including the axillae, perineum, hands, chest, and limbs (11). Accordingly, the bacteria's ability to persist on skin is an important mediator of transmission, as underscored by the recent discovery that the hypertransmissible USA300 strain has overcome one of the innate defense barriers of the skin through horizontal gene transfer with Staphylococcus epidermidis to acquire resistance to toxic polyamines that restrict the growth of other S. aureus strains (12, 13). Other innate defense barriers of the skin include its acidic pH and antimicrobial fatty acids, foremost of which is sapienic acid that is released from triglycerides secreted by the sebaceous glands (14, 15). Nasal secretions also contain antimicrobial fatty acids, primarily linoleic, arachidonic, and palmitoleic acids or their corresponding cholesterol esters (16), and infected abscess tissue also contains abundant antimicrobial fatty acids (17, 18). Consequently, S. aureus is exposed to antimicrobial fatty acids not only during colonization but also during infection, and thus it is reasonable to hypothesize that S. aureus has evolved mechanisms of intrinsic resistance.
Among mechanisms that have been described are cell surface teichoic acids that can selectively restrict the access of palmitoleic acid to the cytoplasmic membrane (19) and a cell surface protein, IsdA, that is expressed in response to iron-limiting conditions and that also restricts the access of palmitoleic acid, or its isomer sapienic acid, to the cytoplasmic membrane (20). Other investigators have reported that tet38, encoding a major facilitator superfamily (MFS) efflux pump, promotes resistance to palmitoleic acid (21). Expression of tet38 was induced by palmitoleic acid but not by linoleic acid, which suggested that there could be distinct mechanisms for coping with different antimicrobial fatty acids. Importantly, linoleic acid is an essential fatty acid for humans, which must be obtained from dietary sources, and is an essential precursor for synthesis of arachidonic acid. These two unsaturated fatty acids comprise a major proportion of unsaturated fatty acids in membrane phospholipid (22, 23). Therefore, the ability to sense and respond to linoleic acid could represent a specific sensory mechanism to signal colonization or infection of a human host, and yet specific mechanisms for regulating gene expression and intrinsic resistance in response to linoleic acid have not been reported.
To address this, we drew from our previous observation that exposure of S. aureus USA300 to a subinhibitory (25 μM) concentration of linoleic acid caused a robust induction of secreted protease expression, which led to proteolytic processing of a secreted glycerol ester hydrolase, Geh (24). We subsequently noted that when S. aureus cultures were supplemented with a trilinolein triglyceride substrate, Geh activity quickly liberated growth-inhibitory concentrations of linoleic acid (25). Moreover, 50 μM free linoleic acid imposed a 10- to 12-h growth delay in cultures of S. aureus USA300, which was then followed by unimpeded exponential growth; similar results were obtained with 50 μM trilinolein in wild-type geh-proficient S. aureus USA300, whereas growth of a geh-deficient mutant was unaffected by 50 μM trilinolein (24, 25). From these observations, we hypothesized that, in addition to the induction of expression of secreted proteases, there should also be an inducible mechanism for resistance of S. aureus to linoleic acid.
In related studies, selection of S. aureus strains that were able to grow at elevated concentrations of glycopeptides led to the identification of point mutations in the vraS sensor of antimicrobial glycopeptides (26, 27). Therefore, we adopted a similar strategy by conducting comparative genome sequencing of USA300 clones that were selected for their ability to initiate growth without a lag phase when they were inoculated into medium containing 50 μM linoleic acid. We now provide the first description of a novel gene pair, farR-farE (fatty acid resistance), constituting divergently transcribed genes that, respectively, encode a regulator and effector of S. aureus resistance to linoleic and arachidonic acids.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A list of bacterial strains and plasmids that were used or constructed for this study is provided in Table 1. S. aureus cultures were maintained as frozen stocks (−80°C) in 20% glycerol and streaked on tryptic soy broth (TSB) agar when required. TSB was supplemented, when required, with 10 μg/ml of erythromycin or chloramphenicol for propagation of strains bearing resistance markers. Escherichia coli strains were grown on LB agar or in LB broth containing 100 μg/ml ampicillin when required. Unless otherwise stated, all cultures were grown at 37°C, and liquid cultures were incubated on an orbital shaking platform at 180 rpm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| USA300 LAC | Community-associated MRSA, wild-type strain cured of resistance plasmids | 24 |
| RN4220 | rK− mK+, capable of accepting foreign DNA | 32 |
| NE1393 | Transposon insertion in SAUSA300_2490, Ermr | 34 |
| NE2336 | Transposon insertion in SAUSA300_2489, Ermr | 34 |
| USA300 farR::ΦNE | USA300 LAC recipient of transposon from NE1393 | This study |
| USA300 farR::ΦNE(pLIfarR) | farR::ΦNE complemented with native farR, cloned in pLI50; Ermr Cmr | This study |
| USA300 farR::ΦNE(pCNfarR) | farR::ΦNE complemented with pCNfarR for cadmium-inducible expression, Ermr Cmr | This study |
| USA300 farE::ΦNE | USA300 LAC recipient of transposon from NE2336 | This study |
| USA300 farE::ΦNE(pLIfarE) | farE::ΦNE complemented with native farE, cloned in pLI50 | This study |
| USA300 farE::ΦNE(pLI50) | USA300 farE::ΦNE with empty pLI50 vector, Cmr | This study |
| USA300 Δtet38 | USA300 LAC with internal deletion of tet38 (SAUSA300_0139) | This study |
| USA300 Δtet38-farE::ΦNE | USA300 Δtet38 recipient of farE::ΦNE transposon insertion, Ermr | This study |
| E. coli DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| Plasmids | ||
| pLI50 | E. coli-S. aureus shuttle vector | 35 |
| pLIfarR | pLI50 with native farR gene | This study |
| pLIfarE | pLI50 with native farE gene | This study |
| pGYlux | E. coli-S. aureus shuttle vector harboring promoterless luxABCDE operon | 37 |
| pCN51 | E. coli-S. aureus shuttle vector with Pcad promoter for cadmium-inducible gene expression | 36 |
| pCN51c | pCN51 with ermC cassette replaced by cat194 cassette from pRN7146 | This study |
| pCN51farR | pCN51c with promoterless farR for cadmium-inducible expression of farR | This study |
| pKOR-1 | E. coli-S. aureus shuttle vector; contains Pxyl-tetO, expresses antisense secY RNA | 38 |
| pKORΔtet38 | pKOR-1 containing upstream and downstream flanking sequences for deletion of tet38 | This study |
For experimental purposes, inoculum cultures of S. aureus were prepared by transferring cells from a single colony into 13-ml polypropylene tubes containing 3 ml of TSB supplemented with antibiotic, as required, followed by overnight incubation. After determination of the optical density at 600 nm (OD600), aliquots of the overnight cultures were diluted into 25 ml of medium in 125-ml flasks to achieve an initial OD600 equivalent to 0.01. To supplement medium with different fatty acids, a 5 mM stock concentration was initially prepared in sterile TSB containing 1% dimethyl sulfoxide (DMSO) and then diluted into sterile TSB supplemented with 0.1% DMSO to achieve the desired concentration of fatty acids, ranging from 5 μM to 100 μM.
Selection and comparative genome sequencing of FAR clones.
As reported previously, when an overnight culture of S. aureus USA300 was inoculated into fresh TSB containing 50 μM linoleic acid, there was a 10- to 12-h lag phase, followed by unimpeded exponential growth (24). Therefore, to promote the selection of fatty acid-resistant (FAR) clones, seven separate flasks of S. aureus USA300 were subjected to two consecutive cycles of growth to stationary phase in TSB–50 μM linoleic acid, after which samples of each culture were plated for isolation of single colonies. Colonies from each plate were screened to identify fatty acid-resistant clones that could initiate growth without a lag phase when inoculated into TSB–50 μM linoleic acid. A single FAR clone was then selected from each of the seven separate biologic replicates for comparative genome sequencing. For controls, two single colonies of USA300 were selected after two consecutive cycles of growth in TSB alone.
For comparative genome sequencing, genomic DNA was extracted from S. aureus using previously described protocols (28, 29). All samples for comparative genome sequencing were processed at the London Regional Genomics Centre (Robarts Research Institute, London, Ontario, Canada [http://www.lrgc.ca]) using an Ion Torrent Personal Genome Machine (PGM) (Life Technologies, Carlsbad, CA) and 316 chips. Briefly, genomic DNA was quantified using a Qubit and Qubit double-stranded DNA (dsDNA) high-sensitivity assay (Life Technologies, Carlsbad, CA). Samples then underwent fragmentation and adapter and bar code ligation as per an Ion Xpress Fragment Library kit (catalog number 4469142, revision B) and size selection using a Pippin Prep system (Sage Science, Beverly, MA). The size of the final libraries was verified using an Agilent 2100 Bioanalyzer and a High Sensitivity DNA kit (Agilent Technologies Inc., Palo Alto, CA). Bar-coded libraries were pooled at equimolar concentrations, based on Qubit values, and the template dilution factor (TDF) for the final pooled library was calculated using molarity determined via quantitative PCR (qPCR) with an Ion Library Quantification kit (catalog number 4468802). Diluted libraries were processed as per the Ion OneTouch template kit (catalog number 4468007, revision B) for automated clonal amplification and sequenced using an Ion Express Template 200 kit (catalog number 4474280), Enrichment Station, and an Ion Sequencing 200 kit (catalog number 4471999, revision B). Sequence reads were mapped to the genome of S. aureus USA300 (FPR3757) (30) using CLC Genomics Workbench, version 7.0 (Boston, MA), and automated detection of single nucleotide polymorphisms (SNPs) was conducted using the neighborhood quality standard algorithm (31).
Strain and plasmid construction.
Techniques for genetic manipulation of S. aureus were conducted according to established guidelines (32) and as described in our previous work (24, 25, 33). The University of Nebraska transposon mutant library (34) was used as a source of transposon insertions that inactivated SAUSA300_2490 (NE1393) and SAUS300_2489 (NE2336). These were transferred into plasmid-cured USA300 strain LAC, creating USA300 farR::ΦNE and USA300 farE::ΦNE, respectively (Table 1). All recombinant plasmids were first constructed as shuttle vectors in E. coli DH5α. The integrity of plasmids isolated from E. coli was confirmed by restriction enzyme digestion and nucleotide sequencing of the cloned DNA fragments prior to electroporation into S. aureus RN4220 as an intermediate host. From S. aureus RN4220, the individual plasmids were then introduced, via electroporation, into S. aureus USA300 or isogenic derivatives as required. Primers used for PCR amplification of gene segments that were required for plasmid construction are listed in Table S1 in the supplemental material.
Plasmid pLI50 (35) was used to complement mutations in SAUSA300_2490 (farR) and SAUSA300_2489 (farE). To complement farE, a 2.8-kb fragment was amplified by PCR of genomic DNA from S. aureus USA300 with forward and reverse primers farE_F1 and farE_R1. Similarly, a 1.2-kb product containing the native farR gene was amplified with primers farR_F1 and farR-R1. The PCR products were digested with KpnI and SacI and ligated into pLI50, which had been digested with the same enzymes. To construct pCN51farR in which expression of farR is dependent on the cadmium-inducible Pcad promoter, we first excised the ermC cassette from pCN51 by digestion with AvrII and XhoI and replaced it with a 1.0-kb AvrII-XhoI fragment containing the cat194 cassette from pRN7146 (36). The resulting pCN51c plasmid was then digested with BamHI and AscI and ligated to a 605-nucleotide (nt) BamHI-AscI fragment containing the promoterless farR gene, which was generated by PCR with primers CNfarR_F and CNfarR_R. To construct pGYfarE::lux, in which expression of the luciferase operon is driven from the farE promoter, a 396-bp fragment containing the intergenic segment between SAUSA300_2490 and SAUSA300_2489 (farE) was amplified with primers GYfarE_F and GYfarE_R and cloned into the BamHI and SalI sites of pGYlux (37).
A markerless in-frame deletion of tet38 (SAUSA300_0139), encoding a major facilitator efflux pump, was constructed using pKOR-1 according to established protocols (25, 38). Briefly, sequences flanking the tet38 locus were amplified by PCR using primers tet38-5′F and tet38-5′R to generate the upstream arm and primers tet38-3′F and tet38-3′R to generate the downstream arm. The upstream and downstream flanking arms were digested with SacII, ligated to one another, and then recombined into the temperature-sensitive pKOR-1 vector using attB1 and attB2 sites incorporated into the flanking sequences by the respective tet38-5′F and tet38-3′R primers. The resulting pKOR-1Δtet38 vector was first passaged through S. aureus RN4220 before being introduced into USA300 by electroporation. The correct deletion of codons 42 through 439 of the tet38 gene was confirmed by PCR and DNA sequence analysis. The resulting USA300 Δtet38 strain was then used as a recipient for phage transduction, using USA300 farE::ΦNE as a donor (Table 1), to create USA300 Δtet38-farE::ΦNE.
Assays of growth and bactericidal activity.
For growth and bactericidal assays, inoculum cultures were supplemented with antibiotic where required, and these cultures were then inoculated into medium that lacked antibiotics to assess growth or bactericidal activity in the presence of antimicrobial fatty acids. For growth assays, flasks containing medium at a 1:5 ratio of medium volume to flask size and supplemented with the concentrations of fatty acid indicated in the figures or figure legends were inoculated to an initial OD600 of 0.01, and samples were withdrawn at hourly intervals for determination of the OD600. All cultures were grown in triplicate or quadruplicate as specified in individual figure legends. For bactericidal assays, the overnight inoculum cultures were first subcultured into 25 ml of fresh TSB alone to prepare noninduced cells or in TSB containing 20 μM subinhibitory fatty acid to allow induction of intrinsic resistance mechanisms. After growth to mid-exponential phase (OD600 of 0.5), these inoculum cultures were then inoculated into triplicate or quadruplicate flasks of fresh TSB (OD600 of 0.01; approximately 2 × 106 CFU/ml) containing a 100 μM bactericidal concentration of fatty acid. The cultures were then incubated with shaking at 37°C, and aliquots were withdrawn at hourly intervals for preparation of serial dilutions in sterile TSB. Subsequently, 10-μl aliquots from each dilution were spotted in quadruplicate onto TSB agar plates, and colonies were counted after 24 h of incubation. The mean of each quadruplicate technical replicate was entered as a single data point for each flask, from which the mean and standard deviation of the biologic replicate flasks were determined.
Assay for uptake of [14C]linoleic acid.
Assays for growth and uptake of [14C]linoleic acid were conducted according to an established protocol (39), with modifications, to evaluate the influence of farE on accumulation of [14C]linoleic acid in S. aureus cells. Briefly, quadruplicate cultures of S. aureus USA300 or the isogenic USA300 farE::ΦNE complemented with empty pLI50 vector or pLIfarE were grown in TSB–20 μM linoleic acid to an OD600 of approximately 0.3 to allow induction of farE. The cultures were then supplemented with an additional 50 μM dose of linoleic acid and returned to the shaker. After 30 min of exposure to 50 μM linoleic acid, aliquots were withdrawn and supplemented with 0.2 μCi/ml of [14C]linoleic acid. Aliquots of 200 μl were then removed at intervals of 1, 2, 5, and 10 min, and samples from each replicate were simultaneously filtered onto 0.45-μm-pore-size membrane filter discs using a vacuum manifold. The filters were then washed twice with 4 ml of 0.1 M phosphate buffer, pH 7.0, containing 1% Triton X-100 and, after a drying step, placed in scintillation vials containing 4 ml of Cytoscint scintillation cocktail (Fisher Scientific). Accumulated [14C]linoleic acid was then quantified using a Beckman LS 6500 scintillation system. Data are expressed as picomoles of [14C]linoleic acid accumulated per microgram of total cell lysate protein in each sample.
farE::lux reporter gene assays.
Inoculum cultures harboring pGYfarE::lux or pGYlux control plasmid were subcultured into triplicate or quadruplicate flasks of TSB or TSB supplemented with different fatty acids to achieve an initial OD600 of 0.01. The cultures were incubated at 37°C with orbital shaking, and samples were withdrawn at hourly intervals for OD600 determinations. For quantification of luminescence, four 200-μl aliquots of each sample were added to 96-well white, opaque flat-bottom plates (Greiner Bio-one). After each well was supplemented with 20 μl of 0.1% (vol/vol) decanal in 40% ethanol, luminescence measurements were immediately taken on a BioTek Synergy H4 Hybrid Reader (BioTek, Winooski, VT) with 1 s of integration and a gain of 200. Data values were recorded as relative light units (RLU), corrected for background by subtraction of values recorded from cultures harboring the empty pGYlux vector. The data points were standardized for differences in growth by dividing RLU values by the recorded OD600 values of the cultures when samples were withdrawn.
Data analyses.
Data points for growth, viability, and luciferase reporter gene assays were plotted and analyzed using Graph Pad Prism, version 6.0f. Significant differences at specific time points were determined by unpaired one-tailed Student's t tests.
RESULTS
Identification of single nucleotide polymorphisms in linoleic acid-resistant variants of S. aureus.
The preferred site of S. aureus colonization of humans is the anterior nares, where concentrations of linoleic acid in nasal secretions can reach 40 to 50 μM (16). These values correlate with our previous work, where 50 μM linoleic acid caused a 10- to 12-h lag phase in growth of USA300, followed by unimpeded exponential growth (24). Following up on this, we observed that when stationary-phase cells from a primary culture grown in TSB–50 μM linoleic acid were reinoculated into the same medium, growth resumed without a lag phase (see Fig. S1 in the supplemental material). To determine if this was due to the selection of genetic variants with increased resistance to linoleic acid, stationary-phase cells from this second culture were plated on TSB agar for selection of single colonies. From these, we identified several that could initiate growth without a lag phase when they were inoculated into TSB–50 μM linoleic acid. Seven such fatty acid-resistant (FAR) clones were subjected to comparative genome sequencing, and two of these, designated FAR6 and FAR7, had an identical single nucleotide polymorphism (SNP): a C → T transition that alters the H121 codon (CAT) to Y (TAT) in a putative transcriptional regulator encoded by SAUSA300_2490 (30). FAR6 had a second SNP in a pyruvate oxidase encoded by cidC. Therefore, we focused on FAR7, which had just one SNP in SAUSA300_2490, and resequencing of this gene in USA300 and FAR7 confirmed the unique SNP in FAR7.
Description of the farE-farR locus.
We hypothesize that SAUSA300_2490 and a divergently transcribed gene, SAUSA300_2489, respectively, comprise a regulator and effector gene pair that we have designated farR and farE, to denote predicted functions as a regulator and effector of fatty acid resistance. These assignments are supported by bioinformatics analyses. farR encodes a 182-amino-acid protein, with an N-terminal TetR family DNA binding domain (2.33e−4) and overall similarity to the AcrR cluster of orthologous groups of proteins (6.52e−9). In Gram-negative bacteria, AcrR regulators control expression of efflux pumps belonging to the AcrB family, which are often encoded by divergently transcribed genes, as with acrR-acrABC in E. coli (40) and the orthologous mtrR-mtrCDE arrangement in Neisseria gonorrhoeae (41). Similarly, farE is divergently transcribed from farR and encodes an 822-amino-acid protein that is annotated as a drug exporter of the resistance-nodulation-division (RND) superfamily (30), to which AcrB and orthologous efflux pumps are also assigned (42). Genome annotation also assigns FarE to the MMPL (mycobacterial membrane proteins, large) family of proteins, on the basis of homology to large membrane proteins of Mycobacterium tuberculosis that transport mycolic acids to the cell surface (43). Using protein structural modeling programs HHPRED and PHYRE2 (44, 45), FarR was predicted with greater than 99% confidence to resemble known AcrR family regulators, including PfmR and FadR of Thermus thermophilus, which control expression of genes involved in fatty acid synthesis and metabolism (46, 47), and MtrR, an efflux pump regulator of Neisseria gonorrhoeae (41, 48). Likewise, 80% of the FarE amino acid sequence was modeled with 100% confidence on the structure of AcrB from E. coli (49).
farR is required for inducible resistance to linoleic acid.
We hypothesized that farR should regulate expression of farE in response to antimicrobial fatty acids, which was addressed by constructing a farE::lux reporter, where expression of the lux operon is under transcriptional control of the farE promoter. When USA300(pGYfarE::lux) was cultured in TSB, there was a modest peak of luciferase activity in early exponential growth, which quickly dissipated (Fig. 1). However, in TSB supplemented with 20 μM linoleic acid, luciferase activity was strongly induced in early exponential-phase cells and again dissipated as the cells progressed toward stationary phase. Importantly, no induction was observed in USA300 farR::ΦNE cells. Although USA300 farR appeared to exhibit superior growth to wild-type USA300 in TSB–20 μM linoleic acid (Fig. 1), our further analysis of this phenomenon uncovered that it reflects a growth penalty that is imposed on USA300 by forced expression of the luxABCDE genes. This was evident from a growth comparison of USA300 harboring either pGYfarE::lux or empty pGYlux in TSB–20 μM linoleic acid, where cells carrying pGYfarE::lux exhibited significantly slower growth than USA300 carrying the empty vector (see Fig. S2 in the supplemental material).
FIG 1.
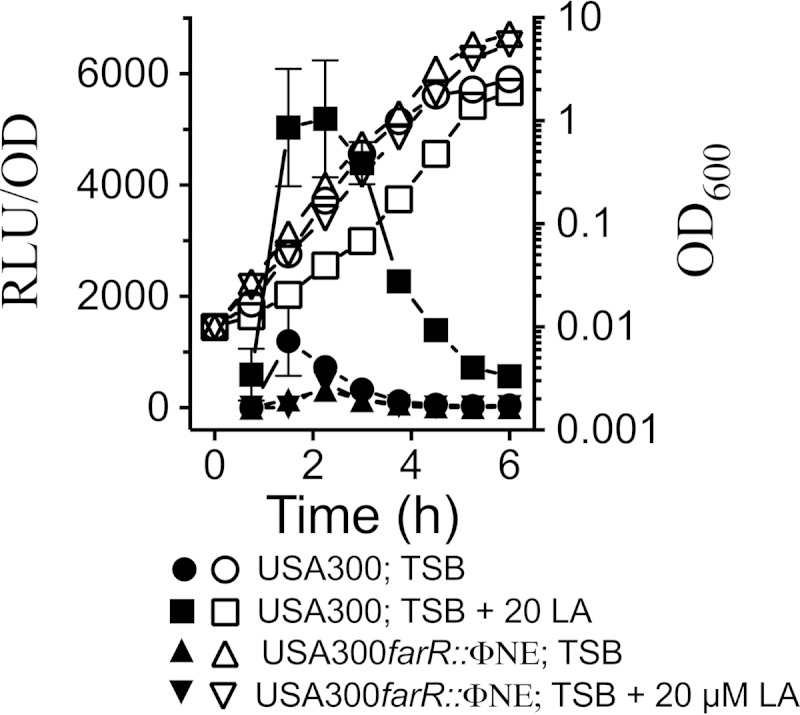
Linoleic acid induces expression of farE. Growth (OD600; open symbols) and relative luminescence units (RLU/OD; closed symbols) of USA300 and USA300 farR::ΦNE, harboring the pGYfarE::lux reporter vector, are charted. USA300 was grown in TSB or in TSB–20 μM linoleic acid (LA); USA300 farR::ΦNE was grown in TSB or in TSB–20 μM linoleic acid. Each value represents the mean and standard deviation of results of three separate cultures, and each culture was subjected to quadruplicate luminescence readings at each time point.
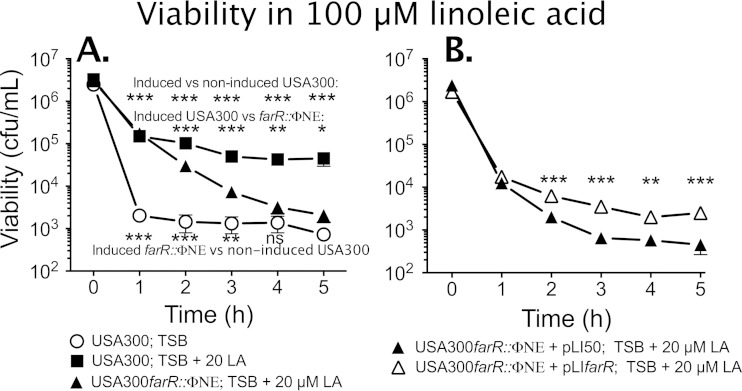
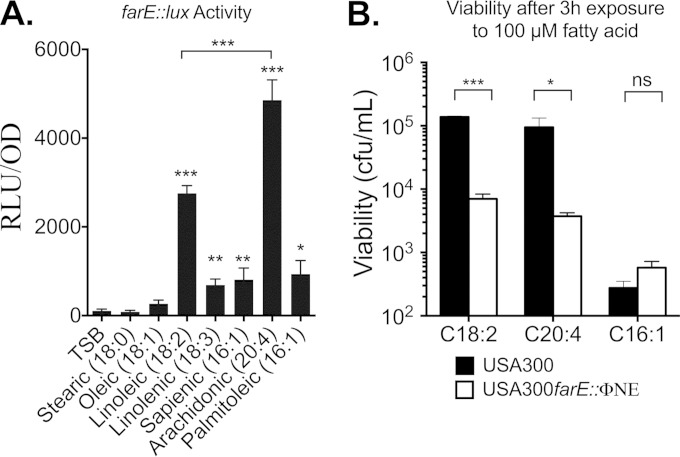
These assays suggested that USA300 should exhibit inducible resistance to the antimicrobial activity of linoleic acid. It was previously reported that exponential-phase cells of S. aureus were significantly more sensitive to the bactericidal activity of antimicrobial fatty acids than stationary-phase cells (17), which we confirmed in a preliminary experiment (see Fig. S3 in the supplemental material). Therefore, to assess inducible resistance, USA300 and USA300 farR::ΦNE were grown to mid-exponential phase in TSB (noninduced) or TSB–20 μM linoleic acid (induced) and then diluted to 106 CFU/ml in fresh TSB containing 100 μM linoleic acid. Noninduced USA300 suffered a >3-log loss of viability after 1 h of exposure to 100 μM linoleic acid (Fig. 2A), while the induced cells retained significantly greater viability at all time points such that there was only an approximate 40-fold loss of viability after 5 h. Furthermore, the induced USA300 farR::ΦNE cells exhibited a significantly greater loss of viability than induced USA300 after 2 h and onwards. Although the induced USA300 farR::ΦNE cells initially retained significantly greater viability than noninduced USA300 cells, they exhibited a progressive loss of viability such that after 4 h of exposure, the remaining viable cells did not significantly differ from noninduced USA300 cells.
FIG 2.
Sensitivity of USA300 and USA300 farR::ΦNE to the bactericidal activity of 100 μM linoleic acid (LA). (A) USA300 or USA300 farR::ΦNE challenge cells were grown to mid-exponential phase in TSB or in TSB–20 μM linoleic acid and then diluted to 106 CFU/ml in TSB containing 100 μM linoleic acid. Viability was monitored at hourly intervals. (B) USA300 farR::ΦNE was complemented with empty pLI50 vector or pLIfarR and assayed for viability in 100 μM linoleic acid after initial growth in TSB–20 μM linoleic acid. All data points represent the means ± standard deviations of viability determinations from quadruplicate cultures. Significant differences in viability at each time point were determined by an unpaired one-tailed Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant).
To validate a role for farR, USA300 farR::ΦNE was complemented with empty pLI50 or pLIfarR harboring farR and its native promoter to determine whether pLIfarR could restore inducible resistance. Accordingly, when preinduced by growth in 20 μM linoleic acid, USA300 farR::ΦNE(pLIfarR) retained significantly greater viability after 2 h of exposure to 100 μM linoleic acid than USA300 farR::ΦNE(pLI50) (Fig. 2B). Nevertheless, pLIfarR did not appear to restore the level of inducible resistance to that of wild-type USA300, which retained approximately 105 CFU/ml viable cells after 5 h of exposure (Fig. 2A). We reasoned that this could be due to two variables: first, farR might be expressed at a high level from its native promoter on a multicopy plasmid; second, the FarR protein could engage nucleotide sequences on pLIfarR, which contained the entire farE-farR intergenic segment, and these in trans interactions could limit the ability of FarR to regulate farE on the chromosome. To overcome these limitations, we expressed farR using the cadmium-inducible Pcad promoter and observed an approximate 100-fold difference in viability when USA300 farR::ΦNE(pCNfarR) cells were exposed to 100 μM linoleic acid in the presence or absence of 10 μM cadmium (see Fig. S4 in the supplemental material). Cumulatively, these data support the contention that farR is required to manifest an inducible resistance phenotype in S. aureus USA300.
farE contributes to persistence and growth of S. aureus in the presence of linoleic acid.
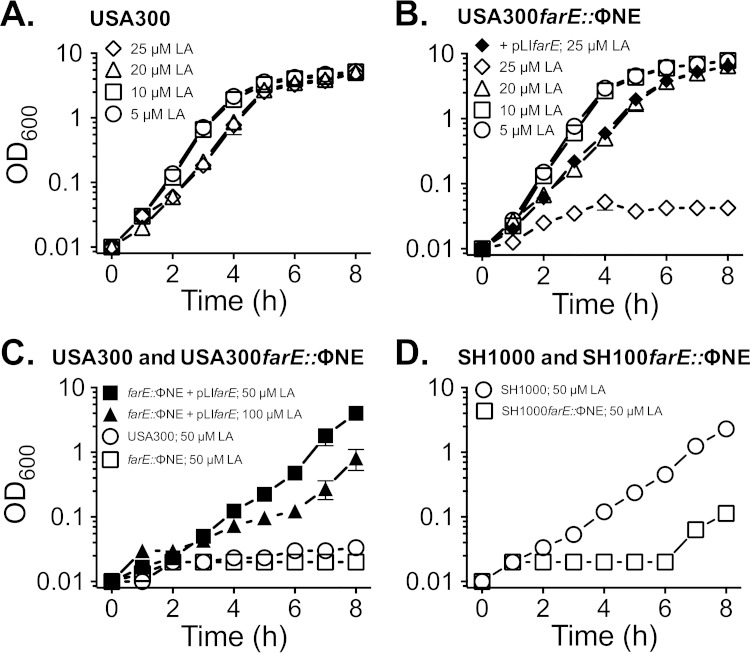
We previously established that USA300 could grow in TSB containing 25 μM linoleic acid, whereas 50 μM linoleic acid imposed a 10- to 12-h lag phase. Our current reporter gene assays also established that farE was induced by growth in TSB containing 20 μM linoleic acid. Therefore, we expected that farE would be required to support growth of S. aureus USA300 at an upper threshold of 25 μM linoleic acid and that induction of farE would confer protection against challenge of S. aureus with a 100 μM bactericidal concentration. To address the growth requirement, USA300 or USA300 farE::ΦNE was cultured in TSB containing 5, 10, 20, or 25 μM linoleic acid. USA300 was not adversely affected by 5 or 10 μM linoleic acid but exhibited slower growth in 20 or 25 μM linoleic acid (Fig. 3A). Comparatively, USA300 farE::ΦNE exhibited similar behavior at 5, 10, and 20 μM linoleic acid but was unable to initiate growth over 8 h of incubation in 25 μM linoleic acid (Fig. 3B). Furthermore, when USA300 farE::ΦNE was complemented with pLIfarE, we observed growth restoration not only in 25 μM linoleic acid (Fig. 3B) but also in up to 100 μM linoleic acid (Fig. 3C). In contrast wild-type USA300 was unable to grow in 50 μM linoleic acid (Fig. 3C).
FIG 3.
Mutation of farE::ΦNE enhances sensitivity of S. aureus to toxicity of linoleic acid. Growth of USA300 (A) or USA300 farE::ΦNE (B) in TSB supplemented with 5 μM, 10 μM, 20 μM, or 25 μM linoleic acid and that of USA300 farE::ΦNE(pLIfarE) in TSB–25 μM linoleic acid were measured. (C) Growth of USA300 or USA300 farE::ΦNE in TSB–50 μM linoleic acid and growth of USA300 farE::ΦNE(pLIfarE) in 50 μM or 100 μM linoleic acid. (D) Growth of S. aureus SH1000 or SH1000 farE::ΦNE in TSB–50 μM linoleic acid. Each data point represents the mean value of triplicate (A, C, and D) or quadruplicate (B) cultures.
To ensure that the role of farE was not dependent on factors that are uniquely associated with the CA-MRSA strain USA300 genetic background, we transduced farE::ΦNE into S. aureus SH1000, which is a methicillin-susceptible laboratory strain that has the same multilocus sequence type (MLST) as USA300 (50). Although SH1000 exhibited somewhat greater intrinsic resistance to linoleic acid, as evident from its ability to grow in TSB–50 μM linoleic acid, SH1000 farE::ΦNE exhibited an extended lag phase, with no obvious growth over 6 h (Fig. 3D). Therefore, farE promotes growth of both MRSA and methicillin-susceptible S. aureus (MSSA) strains at elevated concentrations of linoleic acid.
To evaluate the role of farE in promoting inducible resistance, USA300 and USA300 farE::ΦNE were grown in TSB alone or in TSB containing 20 μM linoleic acid prior to subculture into 100 μM linoleic acid (Fig. 4). Consistent with farE not being appreciably expressed in noninduced cells, the noninduced USA300 and USA300 farE::ΦNE cultures both suffered a rapid loss of viability on exposure to 100 μM linoleic acid. However, when the cells were grown under inducing conditions prior to challenge with 100 μM linoleic acid, USA300 exhibited only a 10- to 40-fold loss of viability over 5 h and retained significantly greater viability at all time points than USA300 farE::ΦNE. Interestingly, the induced USA300 farE::ΦNE challenge cells still retained significantly greater viability than noninduced USA300, which suggests that factors in addition to farE may also promote inducible resistance. Cumulatively, these data confirm that farE contributes to the inducible resistance of S. aureus to the bactericidal activity of 100 μM linoleic acid and is also required to support growth in as low as 25 μM linoleic acid. It further appears that resistance is proportional to farE expression, as suggested by the ability of pLIfarE to support growth of USA300 farE::ΦNE at concentrations of linoleic acid that could not be tolerated by USA300 (Fig. 3C).
FIG 4.
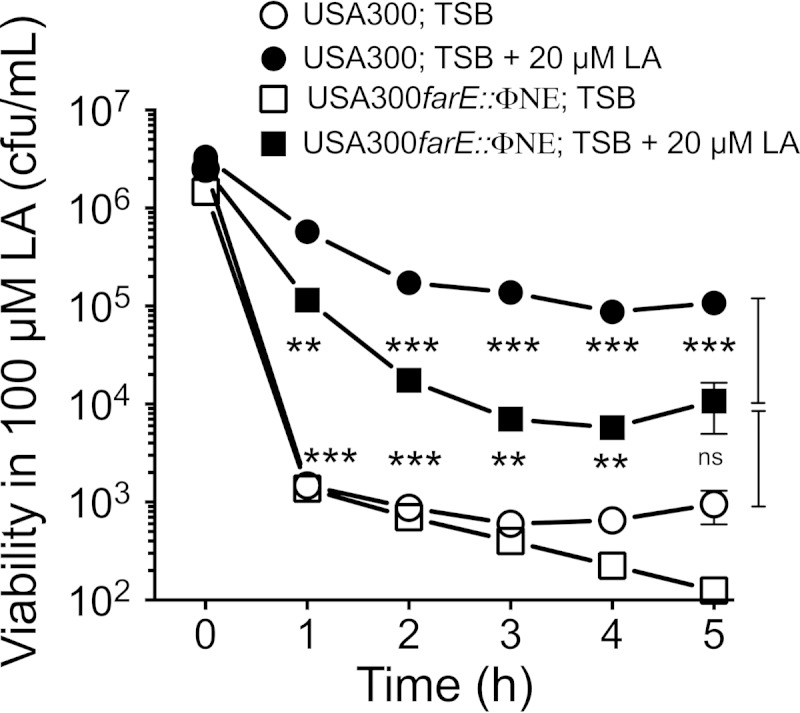
Sensitivity of USA300 and USA300 farE::ΦNE cells to the bactericidal activity of 100 μM linoleic acid. Cells of USA300 or USA300 farE::ΦNE were exposed to 100 μM linoleic acid after growth to mid-exponential phase in TSB or in TSB–20 μM linoleic acid. Each data point represents the mean value of quadruplicate cultures. P values are indicated by asterisks (**, P < 0.01; ***, P < 0.001; ns, nonsignificant).
The FAR7 clone exhibits increased expression of farE.
FAR7 is distinguished from USA300 by an SNP in farR that changes H121 to Y in the gene product. This clone was selected for its ability to grow without a lag phase in TSB–50 μM linoleic acid, and our data suggest that this should be due to increased expression of farE as a consequence of the SNP in farR. This was confirmed by conducting farE::lux reporter gene assays in both USA300 and FAR7 (Fig. 5). When grown in TSB, FAR7 exhibited significantly greater luciferase activity than USA300, and during growth in TSB–20 μM linoleic acid, the luciferase activity in FAR7 significantly exceeded that of USA300. Therefore, the SNP that causes an H121Y substitution in FarR results in a constitutive level of farE expression during growth in TSB and permits a significantly greater induced level of expression than could otherwise be achieved in USA300.
FIG 5.
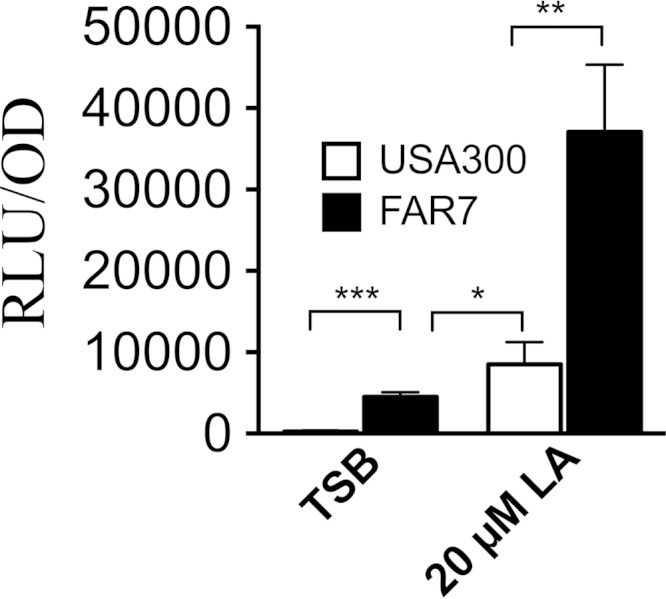
The FAR7 SNP causes enhanced induction of farE expression. The cultures were grown in TSB or TSB–20 μM linoleic acid (LA) as indicated. Data are expressed as relative luminosity units (RLU), standardized to one OD600 unit. Values represent the means of four replicates from each of four independent cultures. Measurements were taken from triplicate cultures when OD600 values reached approximately 0.5, and P values are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
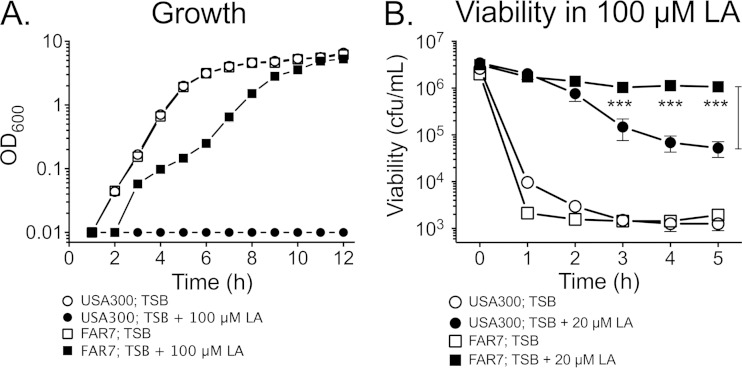
An H121Y substitution in FarR is sufficient for increased resistance to linoleic acid.
Since USA300 and FAR7 are differentiated on the basis of an SNP that causes an H121Y substitution in FarR, we expected that this alone would be sufficient to promote increased resistance to linoleic acid. Accordingly, although FAR7 and USA300 exhibited no difference in growth when cultured in TSB, FAR7 was uniquely able to grow in TSB–100 μM linoleic acid (Fig. 6A). In bactericidal assays, noninduced USA300 and FAR7 suffered similar rapid losses of viability when exposed to 100 μM linoleic acid (Fig. 6B). Therefore, although there is some constitutive expression of farE during growth of FAR7 in TSB, this is not sufficient to promote resistance to 100 μM linoleic acid. However, when the assay was conducted with cells grown under inducing conditions, FAR7 did not exhibit any significant loss of viability over 5 h of exposure to 100 μM linoleic acid and exhibited significantly greater retention of viability from 3 to 5 h than USA300 (Fig. 6B). These observations are consistent with our farE::lux assays, where FAR7 exhibited a significantly higher induced level of farE expression than USA300, and support the contention that increased expression of farE correlates with increased resistance.
FIG 6.
FAR7 is more resistant than USA300 to linoleic acid. (A) Growth analysis of USA300 and FAR7 cultured in TSB or in TSB–100 μM linoleic acid. (B) Bactericidal activity of 100 μM linoleic acid measured with USA300 or FAR7 challenge cells, prepared by growth to mid-exponential phase in TSB or in TSB–100 μM linoleic acid. Each data point represents the mean value of triplicate cultures. P values for comparison of induced USA300 and induced FAR7 cells are indicated by asterisks (***, P < 0.001).
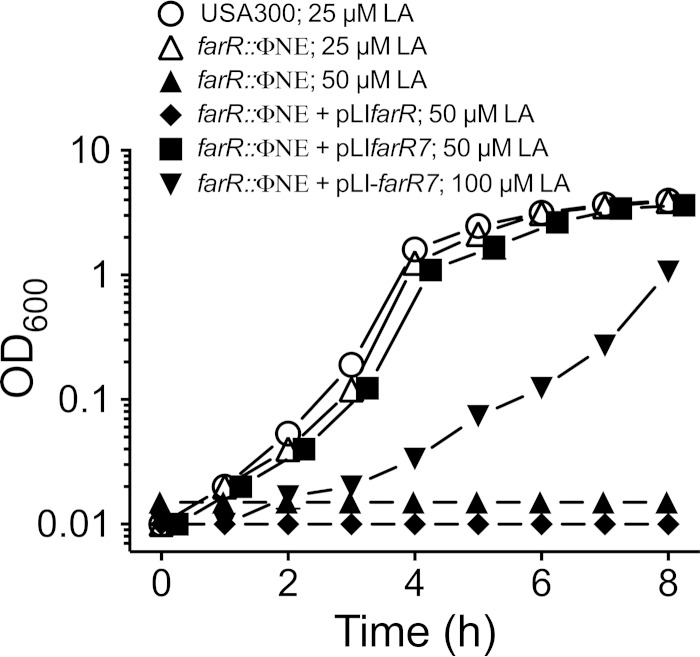
To further define the impact of the H121Y substitution, USA300 farR::ΦNE was transformed with pLI50 harboring wild-type farR or the variant farR7 allele derived from FAR7. With no complementation, USA300 farR::ΦNE exhibited no growth over 8 h of incubation in TSB–50 μM linoleic acid, and cells complemented with wild-type pLIfarR were also unable to grow (Fig. 7). However, cells complemented with the variant farR7 allele acquired the ability to grow in 50 μM linoleic acid and also, at a reduced rate, in 100 μM linoleic acid. Therefore, an SNP that introduces an H121Y substitution in FarR is alone sufficient to confer increased resistance of S. aureus toward linoleic acid, presumably due to increased expression of farE.
FIG 7.
The variant farR7 allele, but not wild-type farR, enables USA300 farR::ΦNE to grow at inhibitory concentrations of linoleic acid. USA300 was grown in TSB–25 μM linoleic acid, USA300 farR::ΦNE was grown in 25 μM or 50 μM LA, USA300 farR::ΦNE(pLIfarR) was grown in 50 μM linoleic acid, and USA300 farR::ΦNE(pLIfarR7) was grown in 50 μM or 100 μM linoleic acid. All data points represent the mean values of triplicate cultures.
Role of farE in resistance to other UFFA.
Although farE is induced by and promotes resistance to linoleic acid, S. aureus would be exposed to a changing diversity and abundance of free fatty acids, dependent on the context within the human body. In a tissue abscess, pus contains high concentrations of unsaturated free fatty acids (UFFA), which could be derived from triglyceride (18, 51) or human cell membrane phospholipid, where each of the major unsaturated fatty acids, oleic (C18:1), linoleic (C18:2), and arachidonic (C20:4) acid, comprises approximately 13 to 15% of the total fatty acid content (22, 23). Conversely, although sapienic acid or its isomer palmitoleic acid (C16:1) do not comprise a major proportion of the fatty acid profile of phospholipid, sapienic acid is the major unsaturated fatty acid in human sebum, both as free fatty acid and in sebum triglyceride (14, 52). Therefore, to better understand the biological role of farE, we evaluated the specificity of farE induction by these different fatty acids and the extent to which farE confers resistance to other fatty acids.
To evaluate the specificity of induction, USA300(pGYfarE::lux) was grown to an OD600 of ∼0.5 in TSB or in TSB supplemented with 20 μM fatty acid, followed by an assay of luciferase activity (Fig. 8A). There were significant differences in the abilities of different 18-carbon-chain-length fatty acids to induce farE::lux such that no induction was observed with saturated stearic acid (C18:0) or oleic acid (C18:1), while linoleic acid (C18:2) was a strong inducer. Strikingly, arachidonic acid (C20:4) promoted a significantly higher level of expression than linoleic acid, while linolenic acid (C18:3) together with palmitoleic acid (C16:1) and its isomer sapienic acid each facilitated an intermediate level of expression which was significantly greater than that of TSB alone but significantly less than the levels of linoleic and arachidonic acids.
FIG 8.
Influence of different antimicrobial fatty acids on induction of farE or viability of S. aureus USA300 and USA 300 farE::ΦNE. (A) Quantification of pGYfarE::lux-dependent luciferase activity in S. aureus USA300 grown to an OD600 of 0.5 in TSB alone or in TSB supplemented with 20 μM fatty acid, as indicated. Each value represents the mean of quadruplicate measurements from each of four replicate cultures. P values indicate significant differences compared to growth in TSB alone or a significant difference between growth with linoleic and arachidonic acids. (B) Bactericidal activity of 100 μM linoleic acid (C18:2), arachidonic acid (C20:4), or palmitoleic acid (C16:1) toward USA300 or USA300 farE::ΦNE cells. The inoculum cultures were grown to an OD600 of 0.5 in TSB supplemented with 20 μM concentrations of the respective fatty acids prior to challenge with a 100 μM bactericidal concentration. Asterisks indicate P values of significant differences between values for USA300 and USA300 farE::ΦNE. Each value represents the mean viability determination from quadruplicate cultures. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
Consistent with the modest induction by 20 μM palmitoleic acid, when a bactericidal assay was conducted with USA300 and USA300 farE::ΦNE cells that were preinduced by growth in 20 μM palmitoleic acid, there were no significant differences in retention of viability after exposure to 100 μM palmitoleic acid (Fig. 8B). However, when this assay was performed with arachidonic acid, USA300 retained significantly greater viability after 2 h of exposure than USA300 farE::ΦNE (Fig. 8B). Therefore, farE appears to have a primary role in mediating resistance to linoleic and arachidonic acids, which are the most effective inducers of farE expression.
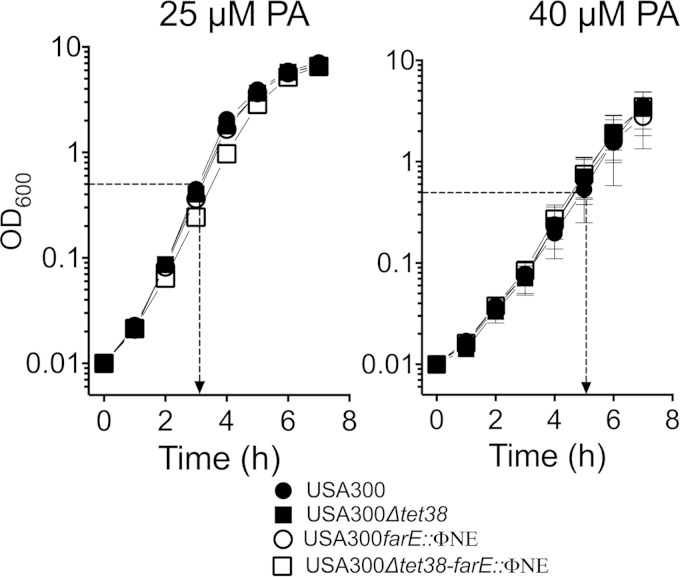
Although farE did not promote resistance to palmitoleic acid, we nevertheless observed a significant induction of expression by 20 μM palmitoleic acid (Fig. 8A); moreover, FAR7 was able to grow in TSB containing 50 μM palmitoleic acid, whereas USA300 could not (see Fig. S5 in the supplemental material). This suggested that farE could still promote resistance to palmitoleic acid if it was expressed at a sufficiently high level. Furthermore, it was recently reported that tet38, which encodes a major facilitator superfamily efflux pump, was induced by palmitoleic acid and contributed to resistance (21). Therefore, we considered that one efflux pump might compensate for the loss of another, which could obfuscate the phenotype of USA300 farE::ΦNE when it was tested with palmitoleic acid. To address this, we constructed a markerless Δtet38 mutation in USA300, which was assayed for growth in TSB supplemented with 25 μM or 40 μM palmitoleic acid. The higher concentration imposed a lower growth rate, as evident from a time of 5 h being required for USA300 to achieve an OD600 of 0.5 compared to approximately 3 h in 25 μM palmitoleic acid (Fig. 9). Nevertheless, there were no discernible differences in growth between USA300 and the individual USA300 Δtet38 or USA300 farE::ΦNE mutants or the combined USA300 Δtet38-farE::ΦNE double mutant. Therefore, neither farE nor tet38 exerted a significant impact on resistance to palmitoleic acid under the conditions that we tested.
FIG 9.
Effect of farE::ΦNE and Δtet38 mutations on growth of S. aureus in the presence of 25 μM or 40 μM palmitoleic acid (PA). USA300, USA300 farE::ΦNE, USA300 Δtet38, and USA300 Δtet38-farE::ΦNE were grown in TSB supplemented with 25 μM or 40 μM palmitoleic acid (PA), as indicated. The dotted line with the arrow depicts the time of growth at which the OD600 reached 0.5.
Inactivation of farE promotes increased uptake of [14C]linoleic acid.
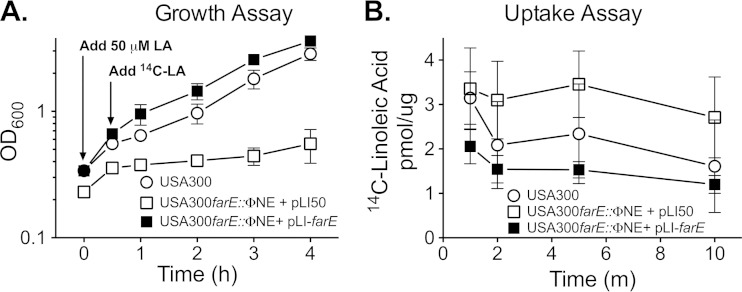
Although many bacteria can derive energy from exogenous fatty acids through an inducible β-oxidation pathway (53), S. aureus lacks this ability, and its primary means of coping with exogenous fatty acids is through incorporation into phospholipid (19, 39, 54, 55). Since our data suggest that FarE promotes efflux of fatty acids, we expected that inactivation of farE would promote increased uptake of exogenous fatty acid. Prior to quantifying uptake of [14C]linoleic acid, we first conducted a mock assay to evaluate the ability USA300 and USA300 farE::ΦNE to recover from exposure to an abrupt increase in the concentration of linoleic acid. Cultures were grown to an OD600 of 0.3 in TSB supplemented with subinhibitory 20 μM linoleic acid to allow induction of farE in USA300, and the cells were then challenged with a 50 μM dose of linoleic acid, followed by monitoring of the OD600 value. After 30 min, USA300, USA300 farE::ΦNE(pLI50), and USA300 farE::ΦNE(pLIfarE) exhibited evidence of continued growth (Fig. 10A). However, beyond 30 min, growth of USA300 farE::ΦNE(pLI50) was severely impaired, whereas USA300 continued to grow, and USA300 farE::ΦNE(pLIfarE) exhibited superior recovery. These data confirm that farE contributes to the ability of S. aureus USA300 to recover from an abrupt increase in the concentration of exogenous linoleic acid and that conditions of the assay were not bactericidal.
FIG 10.
Growth (A) and uptake of [14C]linoleic acid (B) following exposure of S. aureus USA300 and USA300 farE::ΦNE to an increase in concentration of linoleic acid. In panel A, quadruplicate cultures of USA300, USA300 farE::ΦNE(pLI50), or USA300 farE::ΦNE(pLIfarE) were grown in TSB–20 μM linoleic acid to an OD600 of approximately 0.2 to 0.3. The cultures were then supplemented with an additional 50 μM dose of linoleic acid, and growth (OD600) was measured after 30 min and then at hourly intervals. When this experiment was conducted for the purpose of quantifying uptake of [14C]linoleic acid, the cultures were supplemented with 0.20 μCi/ml of [14C]linoleic acid ([14C]-LA) at the 30-min time point, and aliquots of culture were processed for quantification of [14C]linoleic acid uptake at intervals of 1, 2, 5, and 10 min. Each data point represents the mean and standard deviation of values of quadruplicate samples.
We next wished to address the question of whether FarE was responsible for actively extruding linoleic acid from the S. aureus cell. To do this, we performed uptake assays using [14C]linoleic acid. We performed these assays on cells that were treated the same as for the growth experiments described in the legend of Fig. 10A, and cultures were supplemented with [14C]linoleic acid 30 min after challenge with 50 μM linoleic acid. Strikingly, USA300 farE::ΦNE complemented with pLIfarE exhibited the least accumulation of [14C]linoleic acid, while USA300 farE::ΦNE harboring the empty pLI50 vector exhibited the greatest accumulation; wild-type USA300 exhibited intermediate accumulation (Fig. 10B). Importantly, this reflected an inverse correlation between recovery of growth after exposure to 50 μM linoleic acid and accumulation of [14C]linoleic acid. Specifically, USA300 farE::ΦNE(pLIfarE) exhibited the least accumulation of [14C]linoleic acid, and its growth was not adversely affected; in contrast, USA300 farE::ΦNE(pLI50) exhibited the greatest accumulation, and its growth was severely impaired while wild-type USA300 exhibited intermediate growth and accumulation kinetics. These data support the contention that FarE-mediated efflux of unsaturated free fatty acids is required to support growth of S. aureus at elevated concentrations of antimicrobial fatty acid.
DISCUSSION
Through comparative genome sequencing of S. aureus USA300 variants that were selected for enhanced resistance to linoleic acid, we identified a regulator of fatty acid resistance, farR, and an effector of fatty acid resistance, farE, and this is, to our knowledge, the first description of a dedicated and inducible mechanism of S. aureus resistance to antimicrobial fatty acids. These genes bear similarity to the acrR and acrB paradigm in E. coli, where acrR and acrB were discovered through in vitro selection of acriflavine-resistant mutants, which mapped to the acr locus (40, 56, 57). The emergence of antibiotic resistance in Gram-negative bacteria has also been attributed to the in vivo selection of mutations in the transcriptional repressor acrR which promote increased expression of the efflux pump encoded by acrB (58–60). Similarly, we discovered farR through in vitro selection of USA300 variants with increased resistance to linoleic acid. As with many proteins that possess an N-terminal TetR DNA binding domain, protein structural modeling and homology searches indicate that FarR belongs to the TetR/AcrR family of regulators, while FarE belongs to the RND family of multidrug efflux pumps, which include AcrB.
In addition to our own work, which supports a role for FarE as an efflux pump, other researchers using a different approach with S. aureus COL demonstrated that an amino acid substitution in FarE (SACOL2566) promotes resistance to a newly described oxadiazole family of antibiotics (61). In E. coli, polymorphisms that cause amino acid substitutions in AcrB can also accrue during in vitro selection of strains that are resistant to fluoroquinolone antibiotics (62), and in these examples, it is likely that resistance is due to amino acid substitutions that expand the substrate specificity of the efflux pump (61, 62). However, although AcrB family efflux pumps have been most extensively characterized as mediators of multidrug resistance, we contend that the primary function of FarE is to promote efflux of antimicrobial fatty acids that would be encountered during colonization or within a tissue abscess. This is consistent with the belief that members of the AcrB family, which are encoded by the core genome, evolved to promote efflux of host-derived toxic compounds, including bile salts and fatty acids (63–68).
It is especially significant that expression of farE was most strongly induced by linoleic and arachidonic acids. Since S. aureus cannot synthesize unsaturated fatty acids (69), our data suggest that farE is induced as part of a signaling pathway that is activated by host-specific unsaturated free fatty acids. In other work, bactericidal assays conducted with human nasal secretions established that cholesterol esters of linoleic and arachidonic acids were the principal components with bactericidal activity toward Pseudomonas aeruginosa, which does not colonize the nose, but that they did not affect viability of S. aureus (16); linoleic acid is also the principal antimicrobial fatty acid in homogenates of murine tissue abscesses (18, 51). Although arachidonic acid was not identified as a major fatty acid in abscess homogenates, it is a major unsaturated fatty acid in erythrocyte and leukocyte membrane phospholipid (23, 70), from which it is released by phospholipases at sites of infection and rapidly converted to inflammatory mediators (71). Therefore, the induction of farE in response to linoleic and arachidonic acids may represent an evolutionary feature that contributes to the success of S. aureus as a human pathogen.
Our observations are consistent with a requirement for FarE in maintaining membrane homeostasis when S. aureus is exposed to host-derived antimicrobial unsaturated free fatty acids. Since S. aureus cannot degrade exogenous fatty acids through β-oxidation, its primary means of coping with exogenous fatty acids is through incorporation into membrane phospholipid, which involves a novel fatty acid kinase pathway whereby phosphorylated fatty acid is directly incorporated into glycerol-3-phosphate (54, 55). This in itself may represent a primary means of detoxifying long-chain unsaturated free fatty acids, which promote loss of membrane integrity and cell death if allowed to accumulate in the cytoplasmic membrane (19). Importantly, S. aureus cannot synthesize unsaturated fatty acids and maintains membrane fluidity through synthesis of branched-chain fatty acids, primarily anteiso-C15 (69). From these considerations, we can envision two scenarios whereby FarE would be required under such conditions.
First, although some bacteria cease the de novo synthesis of fatty acids when provided with an exogenous supply of unsaturated fatty acids, this does not occur in S. aureus, which continues to synthesize fatty acids (72). However, under such conditions, there is reduced incorporation of endogenously synthesized anteiso-C15 into phospholipid, likely due to displacement or competition from the exogenous unsaturated fatty acid (72, 73). Consequently, it is likely that unutilized metabolites will accumulate, which could be dealt with through an efflux mechanism, and at least one study has proposed that the primary function of an RND family efflux pump is to promote efflux of fatty acids that are replaced as a result of membrane damage or phospholipid turnover (74). Second, although incorporation of unsaturated fatty acids into phospholipid may comprise an effective means of detoxification, it would also promote an increase in membrane fluidity which, if too severe, would compromise membrane function. In this context, we note from our analysis of uptake of [14C]linoleic acid that USA300 farE::ΦNE cells exhibited significantly greater uptake of [14C]linoleic acid than wild-type USA300 cells (Fig. 10B). Therefore, although growth of USA300 farE::ΦNE cells was impaired under these conditions (Fig. 10A), the cells continued to accumulate [14C]linoleic acid, which suggests that there is sufficient metabolic capacity to incorporate unsaturated fatty acid into phospholipid at a level that is beyond the tolerance for proper membrane function. Consequently, FarE function could also be required under such conditions to ensure that incorporation of unsaturated fatty acid into phospholipid does not exceed a level of tolerance for membrane fluidity.
Although our data supported a role for farE in mediating resistance to linoleic and arachidonic acids, it did not confer resistance to palmitoleic acid, which is consistent with there being distinct mechanisms for resistance to unsaturated fatty acids of 16- and 18-carbon chain lengths. First, S. aureus exhibits a differential capacity to incorporate exogenous unsaturated 16- or 18-carbon fatty acids into membrane phospholipid. Oleic acid (C18:1) is directly incorporated into phospholipid (72), but palmitoleic acid must first be extended by the S. aureus fatty acid biosynthesis machinery, in a rate-limiting step, to produce C18:1, which is then incorporated into phospholipid (19). Perhaps due to the less efficient incorporation of C16:1 fatty acids into phospholipid, S. aureus has evolved some capacity to exclude entry of palmitoleic and sapienic acids into the cytoplasm due to cell surface teichoic acids and the low iron-induced cell surface protein IsdA, which functions as a filtering mechanism to restrict penetration through the cell wall (19, 20). Other investigators also reported that a major facilitator superfamily efflux pump encoded by tet38 promoted resistance to palmitoleic acid (21), and although we were not able to confirm this through construction of a USA300 Δtet38 deletion mutant, it may be that tet38 functions in a strain-specific context.
It is further relevant to these considerations that expression of tet38 was induced primarily by palmitoleic acid and much less effectively by linoleic acid, whereas we observed the opposite response for induction of farE. Importantly, with our identification of an SNP in farR that promotes increased expression of farE, we have provided the first mechanistic description of an efflux pump that is specifically induced in response to antimicrobial fatty acids in S. aureus and, at a broader level, in Gram-positive bacteria. FarR belongs to the TetR/AcrR family of transcriptional regulators, which usually repress transcription of divergent genes by means of an N-terminal DNA binding domain that recognizes a specific operator site in the promoter segment of a target gene, and the affinity of this interaction is modulated by a C-terminal domain that binds a small inducing ligand (75, 76). In a relevant example, FadR of Thermus thermophilus represses expression of genes required to degrade fatty acids, which are derepressed upon binding of an acyl-coenzyme A (CoA) ligand to FadR (46). However, although farE is induced by antimicrobial fatty acids, we cannot yet conclude that farR is alone sufficient to regulate farE. If FarR functioned strictly as a repressor, then inactivation of farR should have caused derepression of farE. However, this was not observed, and farR was in fact needed for induction of farE (Fig. 1). Conversely, FAR7 exhibited a constitutive measure of farE expression, attributed to the H121Y substitution in FarR, which also conferred a significantly higher induced level of farE expression than could be achieved in wild-type USA300 (Fig. 8B).
As this substitution is not within the N-terminal DNA binding domain, which spans amino acids 28 to 61 of FarR, it should not directly affect the DNA binding function. However, in a potentially related example, FadR represses expression of genes required for β-oxidation of fatty acids, and the conformation of amino acids 106 to 119 in the C-terminal domain underwent a significant shift on binding of fatty acid, including R109, which had an important role in maintaining the DNA-binding affinity even though it is not within the N-terminal DNA binding domain (77). Therefore, the H121Y substitution in FarR could still affect the function of the N-terminal DNA binding domain; alternatively, it may affect the ability of FarR to form functional oligomers, typically dimers or tetramers, which is another characteristic trait of the TetR family of regulators (75, 78).
Although most TetR regulators repress expression of divergently transcribed genes (75, 78), our observation that FarR is required for induction of farE is not unprecedented, and FarR may resemble a limited number of TetR regulators that trigger a broader cellular response to environmental insults (78–83). In one such example, the SczA metal ion-dependent transcriptional regulator of Streptococcus pneumoniae (82) binds to a specific operator site to repress transcription of a target gene in the absence of zinc, but when zinc is present, it binds to a different DNA segment upstream of the regulated gene to activate transcription. Alternatively, FarR may still function as repressor of farE in the absence of inducer, and then in the presence of exogenous fatty acid it may serve to promote expression of a positive-acting transcription factor that is needed to activate farE. This would partially conform to the AcrR-AcrB paradigm, where AcrR ensures that acrB is not expressed in the absence of an inducing stimulus, but other positive-acting factors are required to activate acrB (84–86). With these considerations in mind, work is in progress to determine the mechanism of FarR-dependent regulation of gene expression through analysis of its interaction with different fatty acids and target promoters and the scope of genes that are affected by this interaction.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NSERC Discovery grants to M.J.M. and D.E.H. and by funding from the Schulich School of Medicine and Dentistry to M.J.M. J.E.T.S. is the recipient of an Ontario Graduate Scholarship award, and J.C.K. is the recipient of a Frederick Banting and Charles Best Canada Graduate Scholarship from the Canadian Institutes of Health Research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02607-14.
REFERENCES
- 1.Boucher H, Miller LG, Razonable RR. 2010. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis 51(Suppl 2):S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- 2.Kochanek KD, Xu J, Murphy SL, Minino AM, Kung HC. 2012. Deaths: final data for 2009. Natl Vital Stat Rep 60:1–117. [PubMed] [Google Scholar]
- 3.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. 2012. Predictors of mortality in Staphylococcus aureus bacteremia. Clin Microbiol Rev 25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLeo FR, Diep BA, Otto M. 2009. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin North Am 23:17–34. doi: 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archer GL. 1998. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 6.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 7.de Lencastre H, Oliveira D, Tomasz A. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol 10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson H. 2002. “Superbug” hurdles key drug barrier. Nature 418:469. doi: 10.1038/418469b. [DOI] [PubMed] [Google Scholar]
- 9.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, McDanel JS, Doern GV. 2014. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 35:285–292. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 11.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 12.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planet PJ, Larussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 Coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4(6):e00889-13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takigawa H, Nakagawa H, Kuzukawa M, Mori H, Imokawa G. 2005. Deficient production of hexadecenoic acid in the skin is associated in part with the vulnerability of atopic dermatitis patients to colonization by Staphylococcus aureus. Dermatology 211:240–248. doi: 10.1159/000087018. [DOI] [PubMed] [Google Scholar]
- 15.Cork MJ, Robinson DA, Vasilopoulos Y, Ferguson A, Moustafa M, MacGowan A, Duff GW, Ward SJ, Tazi-Ahnini R. 2006. New perspectives on epidermal barrier dysfunction in atopic dermatitis: gene-environment interactions. J Allergy Clin Immunol 118:3–23. doi: 10.1016/j.jaci.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 16.Do TQ, Moshkani S, Castillo P, Anunta S, Pogosyan A, Cheung A, Marbois B, Faull KF, Ernst W, Chiang SM, Fujii G, Clarke CF, Foster K, Porter E. 2008. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J Immunol 181:4177–4187. doi: 10.4049/jimmunol.181.6.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Z, Ge S, Chamberlain NR, Kapral FA. 1993. Growth cycle-induced changes in sensitivity of Staphylococcus aureus to bactericidal lipids from abscesses. J Med Microbiol 39:58–63. doi: 10.1099/00222615-39-1-58. [DOI] [PubMed] [Google Scholar]
- 18.Shryock TR, Kapral FA. 1992. The production of bactericidal fatty acids from glycerides in staphylococcal abscesses. J Med Microbiol 36:288–292. doi: 10.1099/00222615-36-4-288. [DOI] [PubMed] [Google Scholar]
- 19.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. 2012. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol 194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke SR, Mohamed R, Bian L, Routh AF, Kokai-Kun JF, Mond JJ, Tarkowski A, Foster SJ. 2007. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Truong-Bolduc QC, Villet RA, Estabrooks ZA, Hooper DC. 2014. Native efflux pumps contribute resistance to antimicrobials of skin and the ability of Staphylococcus aureus to colonize skin. J Infect Dis 209:1485–1493. doi: 10.1093/infdis/jit660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min Y, Blois A, Geppert J, Khalil F, Ghebremeskel K, Holmsen H. 2014. Dietary fat intake, circulating and membrane fatty acid composition of healthy Norwegian men and women. J Hum Nutr Diet 27:69–75. doi: 10.1111/jhn.12105. [DOI] [PubMed] [Google Scholar]
- 23.Koehrer P, Saab S, Berdeaux O, Isaico R, Gregoire S, Cabaret S, Bron AM, Creuzot-Garcher CP, Bretillon L, Acar N. 2014. Erythrocyte phospholipid and polyunsaturated fatty acid composition in diabetic retinopathy. PLoS One 9:e106912. doi: 10.1371/journal.pone.0106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arsic B, Zhu Y, Heinrichs DE, McGavin MJ. 2012. Induction of the staphylococcal proteolytic cascade by antimicrobial fatty acids in community acquired methicillin resistant Staphylococcus aureus. PLoS One 7:e45952. doi: 10.1371/journal.pone.0045952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadieux B, Vijayakumaran V, Bernards MA, McGavin MJ, Heinrichs DE. 2014. Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J Bacteriol 196:4044–4056. doi: 10.1128/JB.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato Y, Suzuki T, Ida T, Maebashi K. 2010. Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J Antimicrob Chemother 65:37–45. doi: 10.1093/jac/dkp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renzoni A, Andrey DO, Jousselin A, Barras C, Monod A, Vaudaux P, Lew D, Kelley WL. 2011. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One 6:e21577. doi: 10.1371/journal.pone.0021577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickerson NN, Joag V, McGavin MJ. 2008. Rapid autocatalytic activation of the M4 metalloprotease aureolysin is controlled by a conserved N-terminal fungalysin-thermolysin-propeptide domain. Mol Microbiol 69:1530–1543. doi: 10.1111/j.1365-2958.2008.06384.x. [DOI] [PubMed] [Google Scholar]
- 29.Yeung M, Balma-Mena A, Shear N, Simor A, Pope E, Walsh S, McGavin MJ. 2011. Identification of major clonal complexes and toxin producing strains among Staphylococcus aureus associated with atopic dermatitis. Microbes Infect 13:189–197. doi: 10.1016/j.micinf.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 31.Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 32.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. doi: 10.1016/0076-6879(91)04029-N. [DOI] [PubMed] [Google Scholar]
- 33.Nickerson N, Ip J, Passos DT, McGavin MJ. 2010. Comparison of staphopain A (ScpA) and B (SspB) precursor activation mechanisms reveals unique secretion kinetics of proSspB (staphopain B), and a different interaction with its cognate staphostatin, SspC. Mol Microbiol 75:161–177. doi: 10.1111/j.1365-2958.2009.06974.x. [DOI] [PubMed] [Google Scholar]
- 34.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CY, Iandolo JJ. 1986. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J Bacteriol 166:385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mesak LR, Yim G, Davies J. 2009. Improved lux reporters for use in Staphylococcus aureus. Plasmid 61:182–187. doi: 10.1016/j.plasmid.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Greenway DL, Dyke KG. 1979. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J Gen Microbiol 115:233–245. doi: 10.1099/00221287-115-1-233. [DOI] [PubMed] [Google Scholar]
- 40.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 41.Hagman KE, Shafer WM. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 177:4162–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blair JM, Smith HE, Ricci V, Lawler AJ, Thompson LJ, Piddock LJ. 2015. Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J Antimicrob Chemother 70:424–431. doi: 10.1093/jac/dku380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacheco SA, Hsu FF, Powers KM, Purdy GE. 2013. MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. J Biol Chem 288:24213–24222. doi: 10.1074/jbc.M113.473371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program PHYRE. Proteins 70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 46.Agari Y, Agari K, Sakamoto K, Kuramitsu S, Shinkai A. 2011. TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation. Microbiology 157:1589–1601. doi: 10.1099/mic.0.048017-0. [DOI] [PubMed] [Google Scholar]
- 47.Agari Y, Sakamoto K, Kuramitsu S, Shinkai A. 2012. Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8. J Bacteriol 194:4630–4641. doi: 10.1128/JB.00668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann KM, Williams D, Shafer WM, Brennan RG. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J Bacteriol 187:5008–5012. doi: 10.1128/JB.187.14.5008-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 50.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shryock TR, Dye ES, Kapral FA. 1992. The accumulation of bactericidal lipids in staphylococcal abscesses. J Med Microbiol 36:332–336. doi: 10.1099/00222615-36-5-332. [DOI] [PubMed] [Google Scholar]
- 52.Wille JJ, Kydonieus A. 2003. Palmitoleic acid isomer (C16:1Δ6) in human skin sebum is effective against gram-positive bacteria. Skin Pharmacol Appl Skin Physiol 16:176–187. doi: 10.1159/000069757. [DOI] [PubMed] [Google Scholar]
- 53.Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. 2009. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol 191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. 2014. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol Microbiol 92:234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. 2014. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci U S A 111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamura H. 1968. Genetic determination of resistance to acriflavine, phenethyl alcohol, and sodium dodecyl sulfate in Escherichia coli. J Bacteriol 96:987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura H. 1965. Gene-controlled resistance to acriflavine and other basic dyes in Escherichia coli. J Bacteriol 90:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneiders T, Amyes SG, Levy SB. 2003. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother 47:2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olliver A, Valle M, Chaslus-Dancla E, Cloeckaert A. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett 238:267–272. doi: 10.1111/j.1574-6968.2004.tb09766.x. [DOI] [PubMed] [Google Scholar]
- 60.Webber MA, Talukder A, Piddock LJ. 2005. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob Agents Chemother 49:4390–4392. doi: 10.1128/AAC.49.10.4390-4392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao Q, Vakulenko S, Chang M, Mobashery S. 2014. Mutations in mmpL and in cell wall stress stimulon contribute to resistance to oxadiazole antibiotics in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 58:5841–5847. doi: 10.1128/AAC.03501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu JH, Pan YS, Yuan L, Wu H, Hu GZ, Chen YX. 2013. Genetic variations in the active efflux pump genes acrA/B and tolC in different drug-induced strains of Escherichia coli CVCC 1547. Genet Mol Res 12:2829–2836. doi: 10.4238/2013.August.8.3. [DOI] [PubMed] [Google Scholar]
- 63.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindemann A, Koch M, Pessi G, Muller AJ, Balsiger S, Hennecke H, Fischer HM. 2010. Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum. FEMS Microbiol Lett 312:184–191. doi: 10.1111/j.1574-6968.2010.02115.x. [DOI] [PubMed] [Google Scholar]
- 65.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 66.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 67.Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun 71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh VK, Hattangady DS, Giotis ES, Singh AK, Chamberlain NR, Stuart MK, Wilkinson BJ. 2008. Insertional inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl Environ Microbiol 74:5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anel A, Naval J, Gonzalez B, Torres JM, Mishal Z, Uriel J, Pineiro A. 1990. Fatty acid metabolism in human lymphocytes. I. Time-course changes in fatty acid composition and membrane fluidity during blastic transformation of peripheral blood lymphocytes. Biochim Biophys Acta 1044:323–331. [DOI] [PubMed] [Google Scholar]
- 71.Yoshikai Y. 2001. Roles of prostaglandins and leukotrienes in acute inflammation caused by bacterial infection. Curr Opin Infect Dis 14:257–263. doi: 10.1097/00001432-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 72.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. 2011. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci U S A 108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller DJ, Zhang YM, Subramanian C, Rock CO, White SW. 2010. Structural basis for the transcriptional regulation of membrane lipid homeostasis. Nat Struct Mol Biol 17:971–975. doi: 10.1038/nsmb.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adebusuyi AA, Foght JM. 2011. An alternative physiological role for the EmhABC efflux pump in Pseudomonas fluorescens cLP6a. BMC Microbiol 11:252. doi: 10.1186/1471-2180-11-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol 7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 77.Fujihashi M, Nakatani T, Hirooka K, Matsuoka H, Fujita Y, Miki K. 2014. Structural characterization of a ligand-bound form of Bacillus subtilis FadR involved in the regulation of fatty acid degradation. Proteins 82:1301–1310. doi: 10.1002/prot.24496. [DOI] [PubMed] [Google Scholar]
- 78.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Croxatto A, Chalker VJ, Lauritz J, Jass J, Hardman A, Williams P, Camara M, Milton DL. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J Bacteriol 184:1617–1629. doi: 10.1128/JB.184.6.1617-1629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Novakova R, Homerova D, Feckova L, Kormanec J. 2005. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 151:2693–2706. doi: 10.1099/mic.0.28019-0. [DOI] [PubMed] [Google Scholar]
- 81.Lin YH, Miyamoto C, Meighen EA. 2000. Purification and characterization of a luxO promoter binding protein LuxT from Vibrio harveyi. Protein Expr Purif 20:87–94. doi: 10.1006/prep.2000.1285. [DOI] [PubMed] [Google Scholar]
- 82.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. 2007. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol 65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 83.Christen S, Srinivas A, Bahler P, Zeller A, Pridmore D, Bieniossek C, Baumann U, Erni B. 2006. Regulation of the Dha operon of Lactococcus lactis: a deviation from the rule followed by the TetR family of transcription regulators. J Biol Chem 281:23129–23137. doi: 10.1074/jbc.M603486200. [DOI] [PubMed] [Google Scholar]
- 84.Bratu S, Landman D, George A, Salvani J, Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother 64:278–283. doi: 10.1093/jac/dkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ruiz C, Levy SB. 2014. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J Antimicrob Chemother 69:390–399. doi: 10.1093/jac/dkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bailey AM, Paulsen IT, Piddock LJ. 2008. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother 52:3604–3611. doi: 10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.