Abstract
Neurons exploit local mRNA translation and retrograde transport of transcription factors to regulate gene expression in response to signaling events at distal neuronal ends. Whether epigenetic factors could also be involved in such regulation is not known. We report that the mRNA encoding the high-mobility group N5 (HMGN5) chromatin binding protein localizes to growth cones of both neuron-like cells and of hippocampal neurons, where it has the potential to be translated, and that HMGN5 can be retrogradely transported into the nucleus along neurites. Loss of HMGN5 function induces transcriptional changes and impairs neurite outgrowth, while HMGN5 overexpression induces neurite outgrowth and chromatin decompaction; these effects are dependent on growth cone localization of Hmgn5 mRNA. We suggest that the localization and local translation of transcripts coding for epigenetic factors couple the dynamic neuronal outgrowth process with chromatin regulation in the nucleus.
INTRODUCTION
The localization of mRNA coupled to local translation in axons and dendrites constitutes an efficient way for neuronal cells to control gene expression at high spatial and temporal resolution (1). High-throughput technologies have facilitated the identification of broad catalogues of mRNAs localized in axonal and dendritic compartments of neuronal cells (2).
The recent discovery of locally translated transcription factors that are retrogradely transported to the nucleus to elicit transcriptional programs controlling cell survival or death or specification of neuronal identity (3–7) has led to a new paradigm of neuronal gene regulation. Local synthesis coupled to retrograde transport of nuclear factors enables a constant cross talk between the cell periphery and the nucleus, instructing transcriptional programs in response to local cues (e.g., growth factors, neurotransmitters, extracellular matrix, injury, etc.). In addition to mRNAs encoding transcription factors, previous transcriptomic studies of purified neuronal processes have identified several axonal mRNAs encoding chromatin interacting and remodeling factors (8). However, the relevance of the axonal localization and, possibly, the local translation of such mRNAs have not been explored so far.
We previously identified ∼80 mRNAs localizing to the extending neurites of neuron-like N1E-115 cells (9), a mouse neuroblastoma cell line widely used as an in vitro system to study neuronal differentiation (10, 11). This model recapitulates the extension of neurites before axon-dendrite specification, which is the principal morphological characteristic of early neuronal differentiation (12). Using this model, we demonstrated that local mRNA translation not only is a feature of axons and dendrites but also occurs at early neuronal differentiation stages (9).
Among the neurite-enriched mRNAs in N1E-115 cells, we identified transcripts encoding nuclear proteins (9). One of these mRNAs encodes the high-mobility group N5 (HMGN5) chromatin binding protein. HMGN proteins bind the nucleosome core particle and compete with linker histone H1 for chromatin binding sites, therefore affecting chromatin structure and transcriptional activity (13). HMGN5 is the most recently characterized member of the HMGN family. Its structure comprises an N-terminal nuclear localization signal, a nucleosome binding domain (NBD), and a C-terminal acidic tail that is able to interact with the histone H1 C-terminal tail (14). In animals with impaired HMGN5 function, the transcriptional profiles of several organs, including brain, spleen, liver, and thymus, are affected (15). Although little is known about HMGN5 physiological functions, it has been suggested that HMGN5 might control cellular differentiation, glutathione metabolism, tumor progression, and cardiac function (14, 16, 17).
Here, we present evidence supporting a novel function of HMGN5 in controlling neurite outgrowth and chromatin structure in both neuroblastoma cells and mouse hippocampal neurons. We show that Hmgn5 mRNA growth cone localization is important for neurite outgrowth, and we suggest that the local synthesis coupled to retrograde transport of HMGN5 might serve as a mechanism to influence chromatin structure and function in response to signaling at distal neuronal ends.
MATERIALS AND METHODS
Cell culture and transfection.
Mouse N1E-115 cells (American Tissue Culture Collection; cell line established by cloning the C-1300 spontaneous mouse neuroblastoma tumor) were cultured and transfected as previously described (9). For Hmgn5 knockdown (KD), cells were transfected with 80 nM small interfering RNA (siRNA; Dharmacon siRNA SMARTpool Plus or a single Dharmacon siRNA [J-044143-05] for rescue experiments).
Neurite purification, RNA extraction, and RT-qPCR analysis.
Purification of total RNA from soma and neurite fractions of N1E-115 cells and reverse transcription (RT) were performed as previously described (9). Quantitative PCR (qPCR) was performed using the GoTaq qPCR master mix (Promega) with the primers indicated in Table S1 in the supplemental material. Rpl19 mRNA was used as a normalization control. Relative quantification was performed using the 2−ΔΔCT method (18).
Immunofluorescence and Western blotting.
N1E-115 cells and hippocampal neurons were fixed in 4% paraformaldehyde (Sigma-Aldrich) at 96 h posttransfection and at 3 days in vitro (DIV3) or DIV7, respectively, permeabilized, and stained as previously described (9). For Western blot analysis, protein lysates were run on NuPAGE 4 to 12% Bis-Tris gels (Life Technologies) and transferred to a polyvinylidene difluoride (PVDF) microporous membrane (Immobilon-FL), which was then incubated with primary antibodies, washed, and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. After the membrane was washed, the signal was revealed with the Amersham ECL Prime Western blotting detection reagent by autoradiography.
FISH.
Fluorescent in situ hybridization (FISH) was performed as previously described (9) by using digoxigenin RNA labeling mix (Roche), antidigoxigenin antibodies coupled to horseradish peroxidase (Roche), and tyramide-Alexa Fluor 546 (Invitrogen). The primers used to generate FISH probes are listed in Table S1 in the supplemental material.
Microscopy, image acquisition, and analysis.
Wide-field microscopy experiments were performed on an inverted Eclipse Ti microscope (Nikon). Phase-contrast live imaging and Dendra2 photobleaching experiments were previously described (9). For histone H1 and HMGN5 fluorescence recovery after photobleaching (FRAP) experiments, bleaching was performed with the 488-nm laser from a FRAP3D module (Roper Scientific). A spot of ∼3 μm in diameter was bleached with a 400-ms bleach pulse, and recovery epifluorescence images were collected in the green channel every 3 s for 200 s (for histone H1 FRAP) or in streaming mode for 26 s (for HMGN5 FRAP). Quantification of fluorescence recovery was done as previously described (19). For photoconversion experiments, distal neurites and growth cones of N1E-115 cells were photoconverted with a 1-s pulse of UV light (excitation filter, 377/50 nm; dichroic mirror, 409 nm [using Leica EL6000 fluorescence lamp]), and images were taken every 10 s for 2 min in a Leica DMI 6000 B inverted microscope in a temperature-controlled incubation chamber using the Leica Application Suite software. Where indicated, cells were treated with 1 mM EHNA [erythro-9-(2-hydroxy-3-nonyl)adenine] (Sigma-Aldrich) dissolved in water 1 h before imaging. To assess the effectiveness of the EHNA treatment, differentiated N1E-115 cells were treated with 20 nM LysoTracker green DND-26 (Life Technologies) 20 min before imaging, medium was changed, and images were taken every 5 s for 2.5 min with an inverted Eclipse Ti microscope (Nikon). To analyze neurite outgrowth, automated neurite segmentation was performed using MetaMorph software or the Simple Neurite Tracer plug-in of ImageJ. A neurite was defined as a cell process with a length of >1 cell body diameter. Quantification of staining or fluorescent protein intensities and Western blot band intensities was performed with either MetaMorph or ImageJ. Quantification of the number of heterochromatic foci in 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei was done using the Find Maxima process of ImageJ. Where fluorescence intensities are compared, images are all equally scaled. The outcome of the first repetition of many of the experiments (Fig. 4D, E, G, and H and 5A and B) presented was assessed in a blinded fashion.
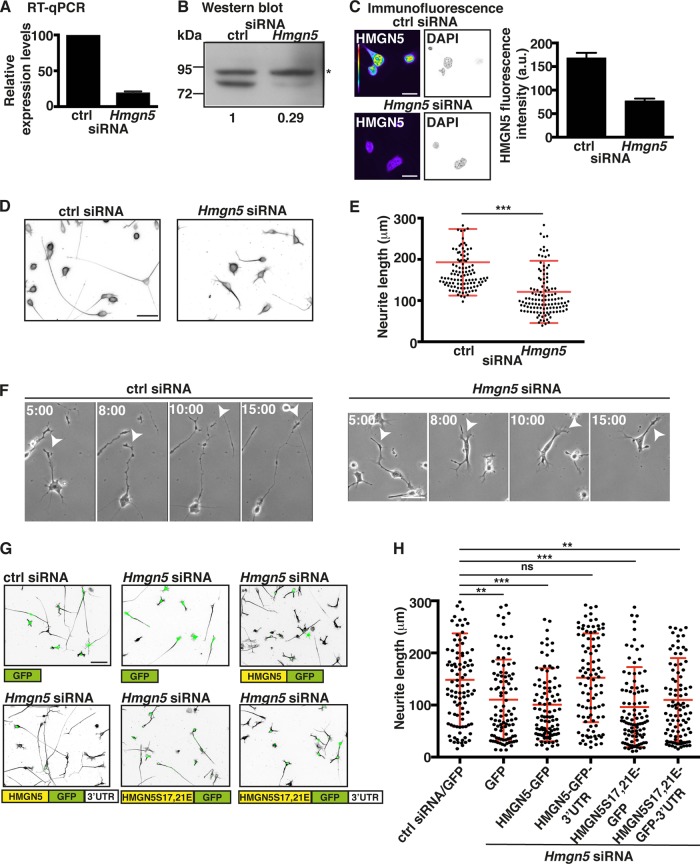
FIG 4.
Hmgn5 mRNA localization modulates neurite outgrowth in N1E-115 cells. (A to C) N1E-115 cells were transfected with control (ctrl) or Hmgn5 siRNA, and knockdown (KD) efficiency was monitored by RT-qPCR (A), Western blotting (B), and immunofluorescence staining (C). Rpl19 mRNA serves as an internal control for RT-qPCR (A), while the upper unspecific band of ∼85 kDa in panel B marked with an asterisk serves as a loading control for Western blotting. (A) n = 3 RNA preparations; (C) n = 8 to 12 cells, mean ± SEM. In panel C, HMGN5 staining is shown in pseudocolor, with warm and cold colors representing, respectively, high and low fluorescence intensity, while DAPI staining is shown in IBW contrast. Scale bar, 20 μm. (D) Representative micrographs in IBW contrast of α-tubulin-stained N1E-115 cells transfected with ctrl or Hmgn5 siRNA. Scale bar, 50 μm. (E) Neurite length measurement of N1E-115 cells transfected with ctrl or Hmgn5 siRNA (n = 120 cells from three independent experiments, mean ± SD). Statistical significance was evaluated by a Kolmogorov-Smirnov test (***, P < 0.001). (F) Neurite outgrowth dynamics of ctrl and Hmgn5 siRNA-transfected N1E-115 cells analyzed by phase-contrast time-lapse microscopy. Arrowheads point to neurite protrusion/retraction events. Scale bars, 50 μm. Time scale is in hours:minutes. (G) Representative micrographs of α-tubulin-stained N1E-115 cells transfected with siRNA and GFP rescue constructs. α-Tubulin staining is shown in IBW contrast, while GFP signal is shown in green. Scale bar, 50 μm. (H) Neurite length measurement of N1E-115 cells transfected with ctrl or Hmgn5 siRNA and with rescue constructs (n = 100 cells from three independent experiments, mean ± SD). Statistical significance was evaluated by a Kolmogorov-Smirnov test (**, P < 0.01; ***, P < 0.001; ns, not significant).
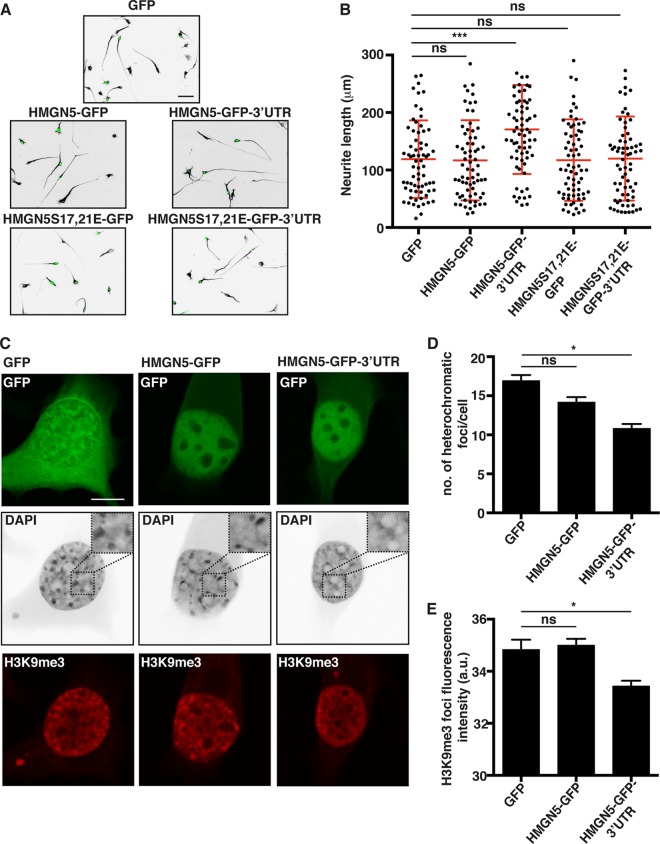
FIG 5.
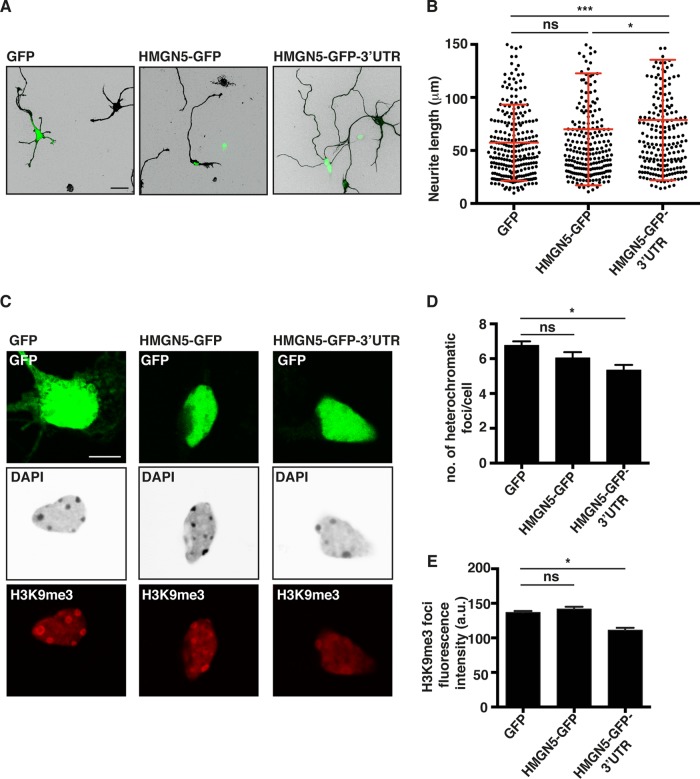
HMGN5 promotes neurite outgrowth and chromatin decompaction in a 3′-UTR-dependent manner. (A) Representative micrographs of α-tubulin-stained N1E-115 cells transfected with GFP, HMGN5-GFP, HMGN5-GFP-3′ UTR, HMGN5S17,21E-GFP, or HMGN5S17,21E-GFP-3′ UTR. α-Tubulin staining is shown in IBW contrast, while GFP signal is shown in green. Scale bar, 50 μm. (B) Neurite length measurement of N1E-115 cells transfected with the different GFP constructs (n = 70 cells from three independent experiments, mean ± SD). Statistical significance was evaluated by a Kolmogorov-Smirnov test (***, P < 0.001; ns, not significant). (C) Representative confocal micrographs of DAPI- and H3K9me3-stained N1E-115 cells transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR. DAPI staining is shown in IBW contrast, H3K9me3 is shown in red, and GFP signal is shown in green. A confocal plane in the middle of the soma was chosen as the most appropriate focus on nuclei. Images were collected on the same day with identical exposure and scaling settings. Scale bar, 10 μm. (D) Measurement of the number of heterochromatic foci in the DAPI staining of cells transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR (n = 60 cells over three independent experiments, mean ± SEM). Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05; ns, not significant). (E) Quantification of mean fluorescence intensity of H3K9me3 foci in N1E-115 cells transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR (n = 160 to 230 foci, mean ± SEM). Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05; ns, not significant).
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 6 software. A Kolmogorov-Smirnov test was used to compare neurite length distributions, while a paired or unpaired (according to the type of experiment) two-tailed t test was used for parametric distributions. Normality of the distributions was assessed with SPSS Statistics software (IBM).
Primary neuron isolation, transfection, and culture.
Mice were maintained on a 12-h day-night cycle with adequate food and water under specific pathogen-free conditions and according to Swiss federal regulations under license number AF-ZH. The day of vaginal plug was considered embryonic day 0 (E0). Primary neurons were isolated from mouse embryos at E18.5, transfected using the Amaxa Nucleofector II system (6 μg of plasmid DNA and/or 50 pmol of siRNA for 5 × 106 to 10 × 106 cells) and analyzed at DIV3 or DIV7. Only green fluorescent protein (GFP)-positive cells were used for analysis. Neurons were cultured as previously described (9).
Antibodies and plasmids.
The following antibodies were used for Western blotting and immunofluorescence: anti-α tubulin (Sigma), anti-H3K9me3 (Abcam), anti-GFP (Roche), antidoublecortin (Santa Cruz Biotechnology), anti-βIII tubulin (Abcam), anti-MAP2 (Millipore), anti-SMI312 (Covance). Secondary HRP-conjugated antibodies were from GE Healthcare, while secondary Alexa Fluor fluorophore-conjugated antibodies were from Invitrogen. Anti-HMGN5 antibody was described previously (20). The PalX2-Dendra2 construct (21) was flanked at the 3′ end with the Hmgn5 3′ untranslated region (UTR). The HMGN5-Dendra2-3′ UTR construct was generated by removing the palmitoylation sequence and inserting the Hmgn5 coding sequence 5′ to Dendra2 and the Hmgn5 3′ UTR 3′ to Dendra2. HMGN5 overexpression constructs were generated by cloning the Hmgn5 coding sequence into pEGFP-N1 (BD Biosciences Clontech) at the 5′ end of enhanced GFP (EGFP), while Hmgn5 3′ UTR was inserted at the 3′ end of EGFP. Dendra2, GFP, and HMGN5-GFP constructs bear the ∼200-nucleotide 3′ UTR derived from the pEGFP-N1 vector. All constructs bear SV40 polyadenylation signal. The Hmgn5 coding sequence was made siRNA resistant by site-directed mutagenesis. Overexpression and rescue constructs for primary neuron experiments were obtained by subcloning sequences of HMGN5-EGFP-3′ UTR into the pCAG vector. For generating FISH probes, the 3′-UTR sequence of Hmgn5 and the coding sequence of EGFP were cloned into pBluescript II KS(+/−) (Agilent Technologies). The EGFP-H1 construct was described previously (22), while the HMGN5-mRuby2 expression constructs were obtained by cloning the mRuby2 coding sequence in the place of EGFP in the HMGN5-GFP expression constructs. All primers used for cloning are listed in Table S1 in the supplemental material. Plasmid maps are available upon request.
Microarray analysis.
Microarray analysis of N1E-115 cells was performed in triplicate. The cRNA target was synthesized and amplified using the WT expression kit (Ambion) and then fragmented and biotin labeled using the WT terminal labeling and control kit (Affymetrix) starting from 270 ng total RNA. For each sample, 12 μg of cRNA was used to generate cDNA. For each sample, 3.75 μg of cDNA was fragmented. All synthesis reactions were carried out in 0.2-ml tubes using a PCR machine (TProfessional Trio; Biometra, Gottingen, Germany) to ensure the highest possible degree of temperature control. The hybridization cocktail containing fragmented biotin-labeled target DNA at a final concentration of 25 ng/μl was transferred into the Affymetrix GeneChip mouse gene 2.0 ST array (Affymetrix) and incubated at 45°C on a rotator in a hybridization oven 640 (Affymetrix) for 17 h at 60 rpm. The arrays were washed and stained on a fluidics station 450 (Affymetrix) by using the hybridization wash and stain kit (Affymetrix) using fluidics procedure FS450_0002. The GeneChip products were processed with an Affymetrix GeneChip scanner 3000 7G (Affymetrix). DAT image files of the microarrays were generated using the Affymetrix GeneChip command console (Affymetrix). Data were imported into R (version 3.0.2; R Core Team) and normalized with robust multiarray averaging (RMA) (23). For each Entrez gene, only one probe set with the highest variance across the data set was selected (using the genefilter and annotation packages from Bioconductor repository version 2.13 [24]). A moderated version of the t test (limma package) was used to identify the differentially expressed genes. Obtained P values were corrected for multiple testing using the Benjamini and Hochberg method.
Immunoprecipitation of GFP-tagged proteins and mass spectrometry analysis.
A total of 1.5 × 106 N1E-115 cells were transfected with the GFP fusion constructs and differentiated on laminin-coated petri dishes. Immunoprecipitation of GFP-tagged proteins was performed in triplicate using the GFP-Trap_A kit (Chromotek) by following the manufacturer's protocol. Mass spectrometry analysis was conducted as previously described (25) by using Lysyl endopeptidase (Wako). The mass spectrometry raw data were analyzed by the MaxQuant 1.3 software package. The tandem mass spectrometry (MS/MS) spectra were searched against the mouse UniProt database. Carbamylation (C) was selected as the fixed modification. Variable modifications were phosphorylation (STY) and oxidation (M).
Accession numbers.
Proteomic data have been deposited in ProteomeXchange (26) with accession number PXD001070. Microarray data have been deposited in the Gene Expression Omnibus (GEO) database with accession number GSE57185.
RESULTS
Hmgn5 mRNA localizes to the growth cones of N1E-115 cells by virtue of a 3′-UTR localization signal.
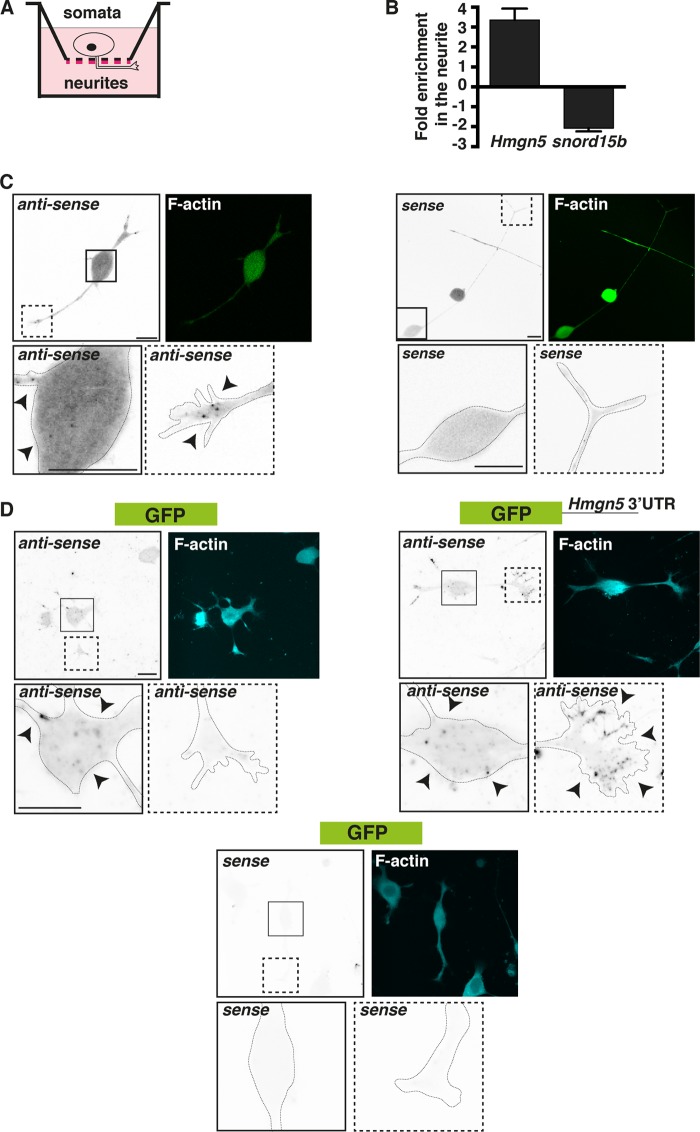
In our previous microarray analysis, we found mouse Hmgn5 mRNA to be enriched in the neurites of differentiated N1E-115 cells (9). These cells exhibit neurite outgrowth in response to serum deprivation and laminin/integrin signaling (27). We confirmed the neurite enrichment of Hmgn5 mRNA by performing quantitative real-time PCR (qRT-PCR) on total RNA extracted from neurite and soma fractions of differentiated N1E-115 cells (Fig. 1A and B). As a quality control for the neurite/soma purification strategy, we evaluated the levels of a noncoding RNA (snord15b) that in our previous analysis showed a strong somatic enrichment (9). Fluorescent in situ hybridization (FISH) with riboprobes antisense to Hmgn5 mRNA showed that Hmgn5 mRNA localizes to bright punctate structures in the growth cones of N1E-115 cells, similarly to other known localized mRNAs (4, 9). This punctate pattern suggests the association of mRNAs in ribonucleoprotein (RNP) particles (Fig. 1C, black arrowheads), which have been proposed to be responsible for regulating mRNA transport and/or translation (28). Such punctate structures are also visible for Hmgn5 mRNA in the soma (Fig. 1C), where bulk protein synthesis usually occurs. The sense control riboprobe displayed a lower and more diffuse signal than the antisense probe and the absence of any punctate structure (Fig. 1C). Since the localization and translation of mRNAs are often controlled by sequences residing in mRNA 3′ untranslated regions (UTRs) (29), we assessed whether the mouse Hmgn5 3′ UTR contains a localization signal by fusing it to a GFP reporter and performing FISH with a riboprobe antisense to GFP mRNA. Appending the Hmgn5 3′ UTR to the GFP mRNA recapitulated the punctate growth cone localization pattern observed with endogenous Hmgn5 (Fig. 1D, black arrowheads; observed in 21/30 growth cones), while GFP mRNA without the Hmgn5 3′ UTR shows diffuse staining throughout the growth cone (Fig. 1D; observed in 23/30 growth cones), suggesting no RNP association. As observed with Hmgn5 mRNA, punctate structures are visible for GFP mRNA both with and without the Hmgn5 3′ UTR in the soma (Fig. 1D). We concluded that Hmgn5 mRNA localizes to growth cones of N1E-115 cells and that the Hmgn5 3′ UTR contains a growth cone mRNA localization signal.
FIG 1.
Hmgn5 mRNA localizes to N1E-115 cell growth cones by virtue of a 3′-UTR localization signal. (A) Schematics of the neurite purification technique. N1E-115 cells were allowed to extend neurites on a 3-μm microporous filter coated with laminin on the bottom part (red lines). Neurites growing in the bottom filter surface were then biochemically separated from the cell bodies. (B) Total RNA was purified from neurite and soma equivalents (n = 3; mean ± standard error of the mean [SEM]), reverse transcribed, and used in RT-qPCR analysis with primers specific for Hmgn5 and snord15b RNA (positive control for a somatic noncoding RNA). (C) Confocal fluorescence micrographs of FISH with riboprobes antisense and sense (negative control) to Hmgn5 mRNA. FISH signal is represented in inverted black and white (IBW) contrast, while F-actin staining is represented in green. Arrowheads indicate punctate structures. The insets correspond to magnifications of either cell bodies (continuous line) or growth cones (dashed line). Scale bars, 20 μm. (D) Confocal fluorescence micrographs of FISH with riboprobes antisense and sense (negative control) to GFP mRNA and GFP mRNA fused to Hmgn5 3′ UTR. FISH signal is represented in IBW contrast, while F-actin staining is represented in cyan. Arrowheads indicate punctate structures. The insets correspond to magnifications of either cell bodies (continuous line) or growth cones (dashed line). Scale bars, 20 μm.
Hmgn5 3′ UTR supports growth cone translation in N1E-115 cells.
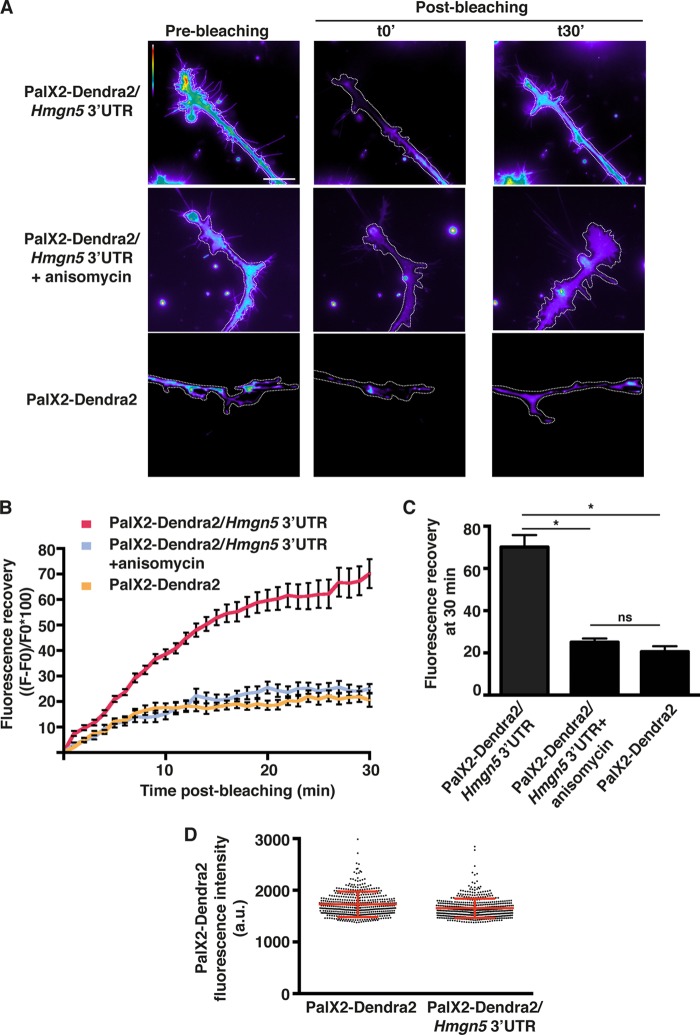
The 3′ UTRs of localized mRNAs usually drive local mRNA translation by virtue of their association with RNA binding proteins (RBPs) (29–31). To assess whether Hmgn5 3′ UTR can support local translation of a reporter mRNA, we fused it to a Dendra2 reporter (PalX2-Dendra2–Hmgn5 3′ UTR) that contains two palmitoylation signals. This targets Dendra2 to the plasma membrane and therefore limits its diffusion to 50 μm/h (32). Thus, fluorescent PalX2-Dendra2 signals arising in the growth cone shortly after photobleaching reflect newly synthesized proteins rather than protein transport from the soma (>50 μm away from the bleached growth cone). Distal neurite segments of transfected N1E-115 cells were bleached with intense green light, and fluorescence recovery was measured for 30 min. For each condition, only protruding growth cones were selected. A significantly higher fluorescence recovery was observed for the PalX2-Dendra2–Hmgn5 3′ UTR construct than for PalX2-Dendra2 (Fig. 2A to C; see also Movie S1 in the supplemental material), while both constructs showed similar expression levels before being bleached (Fig. 2D). The fluorescence recovery was dependent on new translation since it was abrogated by incubation with the translation inhibitor anisomycin (Fig. 2A to C; see Movie S1 in the supplemental material). Thus, Hmgn5 3′ UTR supports local mRNA translation in distal neurite segments and growth cones of N1E-115 cells.
FIG 2.
Hmgn5 3′ UTR supports growth cone translation. (A) Representative micrographs of live N1E-115 cells transfected with PalX2-Dendra2 reporters bearing Hmgn5 or no 3′ UTR pre- and postbleaching. The PalX2-Dendra2 signal was bleached in distal neurites, and growth cones and fluorescence recovery kinetics were acquired using time-lapse microscopy over 30 min. The bleached region corresponds to the whole neurite segment present in the field of view. Images are color-coded so that warm and cold colors represent, respectively, high and low fluorescence intensity. Scale bar, 20 μm. (B) The increase in average fluorescence intensity with respect to the average fluorescence intensity immediately after bleaching (F − F0) is represented as a percentage of the initial postbleaching intensity (F0) for each time point. Where indicated, cells were treated with 40 μM anisomycin 30 min before imaging. n = 8 cells per condition over three independent experiments, mean ± SEM. (C) Fluorescence recovery after 30 min is plotted as a bar graph. Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05; ns, not significant). (D) Quantification of PalX2-Dendra2 expression levels in cells transfected with PalX2-Dendra2 reporters bearing Hmgn5 or no 3′ UTR prebleaching. Data are presented as a dot plot where every dot corresponds to a cell measurement (n = 500 cells). The red bars correspond to means ± standard deviation (SD).
HMGN5 can be retrogradely transported along neurites of N1E-115 cells.
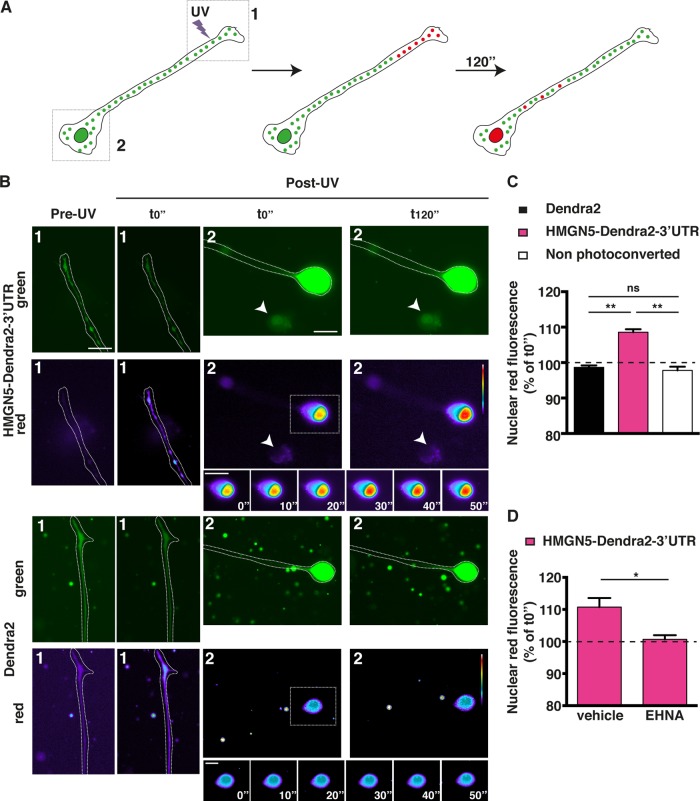
As previously shown for locally synthesized CREB, SMAD, STAT3, and ATF4 transcription factors (4–7), we hypothesized that locally translated HMGN5 might be trafficked back to the cell nucleus. To test this hypothesis, we transfected N1E-115 cells with an HMGN5-Dendra2 fusion followed by Hmgn5 3′ UTR. We then locally photoconverted Dendra2 (from green to red) in distal neurite segments and growth cones (>50 μm away from the soma) with UV light and monitored the nuclear accumulation of the red signal over 2 min after photoconversion (Fig. 3A). We assumed that if HMGN5 is actively retrogradely transported along the neurites of N1E-115 cells, it should accumulate in the nucleus faster than the Dendra2 protein alone by virtue of passive diffusion, as shown for CREB and SMAD (4, 5). Consistently, we observed an accumulation of nuclear red fluorescence when cells were transfected with HMGN5-Dendra2-3′ UTR in comparison to Dendra2 alone (Fig. 3B and C). Nuclei belonging to cells that did not undergo photoconversion but were located in the same field of view as the photoconverted nuclei (white arrowheads in Fig. 3B) were used as controls for nonspecific nuclear photoconversion during imaging of Dendra2 or HMGN5-Dendra2-3′ UTR (Fig. 3B and C). Treatment with the dynein inhibitor EHNA abolished the accumulation of nuclear red fluorescence, demonstrating that HMGN5 retrograde transport occurs via dynein motor proteins (Fig. 3D), as previously reported for the transcription factors SMAD and STAT3 (5, 6). The effectiveness of the EHNA treatment in abolishing retrograde transport was confirmed by assessing the movement of acidic organelles using the LysoTracker dye (see Movies S2 and S3 and Fig. S1 in the supplemental material). These data indicate that HMGN5 can be retrogradely transported from the neurite to the nucleus of N1E-115 cells.
FIG 3.
HMGN5 can be retrogradely transported from the neurite to the nucleus in a dynein-dependent fashion. (A) Schematics of the photoconversion experiment. The Dendra2 green signal was photoconverted to red with UV illumination in the distal part of the neurite (>50 μm away from the soma), and the accumulation of red signal in the nucleus was measured over 2 min. The boxed regions correspond to the field of views shown in panel B, with region 1 corresponding to the distal neurites and region 2 corresponding to proximal neurites and cell bodies. (B) Representative micrographs pre- and post-UV conversion of cells expressing Dendra2 or HMGN5-Dendra2-3′ UTR. Dendra2 is shown in green, while photoconverted red Dendra2 is shown in pseudocolor, with warm and cold colors representing, respectively, high and low fluorescence intensity. Note that the neurites in the post-UV micrographs are slightly out of focus to appropriately focus on nuclei and that neurite images of the Dendra2 construct have been rotated 90° to fit the figure layout. A time series of an inset zooming in on the nucleus is also shown. Scale bars, 20 μm. (C) Measurement of accumulation of red nuclear signal 2 min post-UV photoconversion. Nonphotoconverted cells in the same field as the photoconverted ones were used as controls for a nonspecific increase in red fluorescence (arrowheads in panel B). Percentage increase in red fluorescence for every cell was normalized against the average percent increase of nonphotoconverted cells. n = 17 to 24 cells over three independent experiments, mean ± SEM. Statistical significance was evaluated by a two-tailed unpaired t test (**, P < 0.01; ns, not significant). (D) Measurement of accumulation of red nuclear signal 2 min post-UV photoconversion in cells transfected with the HMGN5-Dendra2-3′ UTR construct and treated either with vehicle or with 1 mM EHNA 1 h before imaging. n = 19 cells over four independent experiments, mean ± SEM. Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05).
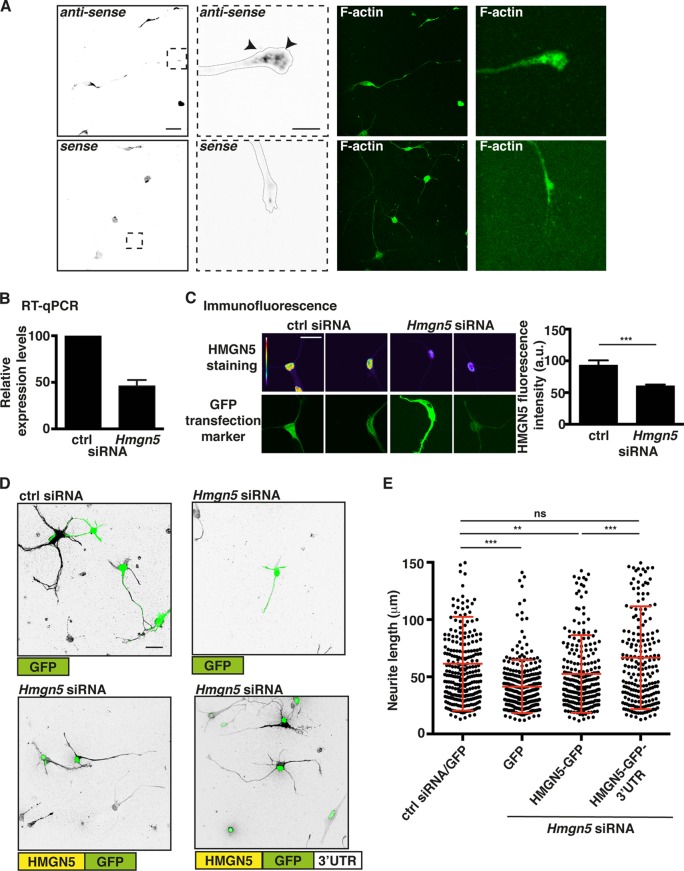
Hmgn5 KD causes transcriptional changes and impairs neurite outgrowth in N1E-115 cells.
Since HMGN5 modulates the transcriptome of different cell types and organs (15, 33), we performed transcriptome analysis of N1E-115 cells transfected with Hmgn5 small interfering RNAs (siRNAs) and compared them to N1E-115 cells transfected with control (ctrl) siRNAs. To identify transcripts specifically linked to neurite outgrowth, we first differentiated our cells by serum starvation and detached and allowed them to extend neurites for 24 h on a laminin substrate. Assessment of knockdown (KD) efficiency by qRT-PCR, Western blotting, and immunofluorescence indicated an ∼80% reduction of Hmgn5 mRNA level and an ∼70% reduction of HMGN5 protein level (Fig. 4A to C). Additionally, we evaluated the control siRNA-transfected cells that were allowed to extend neurites on the laminin substrate only for 4 h and therefore exhibit short neurites, similar in size to those of Hmgn5 knockdown cells allowed to extend neurites for 24 h (see Movie S4 in the supplemental material). We found that 31 genes were significantly affected by Hmgn5 KD (see Fig. S2A, column “kd vs ctrl,” in the supplemental material), and we confirmed the microarray results by qRT-PCR (see Fig. S2B). Among these 31 genes, 18 were also affected by the extent of neurite outgrowth (see Fig. S2A, column “4 h vs 24 h”). Interestingly, all of these genes except one showed matching changes in Hmgn5 KD and the extent of neurite outgrowth (i.e., genes downregulated upon Hmgn5 KD were also less expressed in cells exhibiting short compared to long neurites). We also noticed that Hmgn5 KD influences the expression of genes (e.g., Pltp, Adam12, Prkg1, Plxna4, Capn6, Cnn2, septin3) that are known to be involved in the control of cytoskeletal dynamics (34–40).
These findings led us to test whether HMGN5 controls the neurite outgrowth process in N1E-115 cells. We found that Hmgn5 KD causes an ∼40% reduction of neurite length in N1E-115 cells (Fig. 4D and E). In these cells, neurite outgrowth is first characterized by an initial stage where multiple neurites stochastically protrude and retract, followed by the stabilization of one or two neurites that grow continuously (41). Phase-contrast time-lapse analysis revealed that Hmgn5 KD cells displayed repeated collapse events and had difficulties in establishing steady neurite outgrowth (Fig. 4F; see also Movie S4 in the supplemental material). To evaluate whether the growth cone mRNA localization and translation of Hmgn5 mRNA are important for the regulation of neurite outgrowth, we rescued the KD phenotype by reexpressing different siRNA-resistant versions of HMGN5. The HMGN5-GFP constructs contain only Hmgn5 coding sequence (CDS) fused to GFP and will solely lead to somatic translation. In contrast, the HMGN5-GFP-3′ UTR constructs contain also Hmgn5 3′ UTR and will additionally localize and be translated in the growth cone (see Fig. S3A in the supplemental material). We found that only HMGN5-GFP-3′ UTR was able to fully rescue the neurite outgrowth defect caused by Hmgn5 KD (Fig. 4G to H), indicating that proper Hmgn5 mRNA localization is required for the regulation of neurite outgrowth in N1E-115 cells. To explore the existence of potential HMGN5 functions other than its chromatin binding function, we mutated two residues in the HMGN5 nucleosome binding domain (NBD) that are known to mediate chromatin binding (serines 17 and 21 mutated to glutamic acids, hence named the S17,21E mutant). HMGN5S17,21E mutants have been previously shown to no longer be able to bind the nucleosome core particle (42). HMGN5S17,21E-GFP-3′ UTR failed to rescue the KD phenotype, indicating that HMGN5 function in N1E-115 cells is exerted through its binding to chromatin (Fig. 4G to H). The expression levels of the different HMGN5-GFP constructs were approximately equal (see Fig. S3B in the supplemental material), suggesting that the observed effects were exclusively due to the molecular properties of the exogenously expressed proteins. These results show that both HMGN5 chromatin binding function and Hmgn5 mRNA growth cone localization are important for the modulation of neurite outgrowth in N1E-115 cells.
HMGN5 stimulates neurite outgrowth and modulates chromatin dynamics in N1E-115 cells in a 3′-UTR-dependent manner.
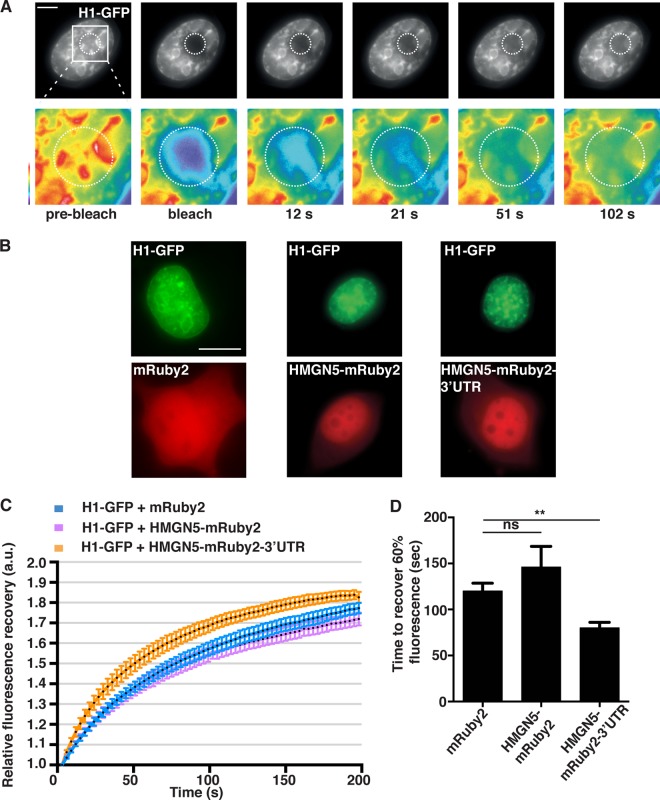
We next overexpressed the various HMGN5 fusion constructs in N1E-115 cells. Opposite to what we observed upon KD, the overexpression of HMGN5 increased neurite outgrowth by ∼30%, and this effect was dependent both on the proper growth cone localization and on the chromatin binding function of the exogenously expressed mRNA and protein (Fig. 5A and B). Since the chromatin binding activity of HMGN5 is required for neurite outgrowth, we analyzed the effect of HMGN5 overexpression on chromatin structure and dynamics. HMGN5 overexpression in fibroblasts, U2OS cells, 293T cells, and a mouse pituitary cell line triggers global chromatin decompaction, as revealed by DAPI and heterochromatin marker staining (42). To evaluate chromatin structure, we used DAPI staining and quantitated the number of heterochromatic foci. We found that HMGN5-mediated chromatin decompaction (i.e., reduction or disappearance of dense heterochromatic foci) was strictly dependent on growth cone localization of Hmgn5 mRNA (Fig. 5C and D; see also Fig. S4A and B in the supplemental material). Similarly, overexpression of HMGN5-GFP-3′ UTR but not HMGN5-GFP caused a reduction of H3K9me3 heterochromatic foci and a decrease in H3K9me3 focal staining intensity (Fig. 5C and E), indicating a rearrangement of heterochromatin. Fluorescence recovery after photobleaching (FRAP) and cross-linking experiments have previously revealed that HMGN5 interacts with linker histone H1 and reduces its binding to chromatin (22, 42). To test whether this was also the case in N1E-115 cells, we performed FRAP experiments on cells expressing both histone H1-GFP and HMGN5 (Fig. 6; see Movie S5 in the supplemental material). We observed that only the HMGN5-mRuby2-3′ UTR construct was able to influence the mobility of histone H1 (by decreasing its chromatin residency time, thus accelerating its fluorescence recovery [Fig. 6C and D]), while the HMGN5-mRuby2 construct did not have any effect (Fig. 6C and D). This implies that HMGN5-mRuby2-3′ UTR is more efficient in counteracting histone H1-mediated chromatin compaction than HMGN5-mRuby2. Taken together, our DAPI/H3K9me3 staining and FRAP experiments indicate that growth cone-translated and retrogradely transported HMGN5 influences chromatin structure and dynamics in N1E-115 cells.
FIG 6.
HMGN5 decreases histone H1 binding to chromatin in a 3′-UTR-dependent manner. (A) Representative micrographs of the FRAP experiment. The black-and-white panels represent histone H1-GFP signal, and the pseudocolor images represent magnifications of the bleached area indicated by a dotted circle, with warm and cold colors representing, respectively, high and low fluorescence intensity. Scale bar, 3 μm. (B) Representative micrographs of cells expressing histone H1-GFP and either mRuby2, HMGN5-mRuby2, or HMGN5-mRuby2-3′ UTR. Scale bar, 10 μm. (C) Fluorescence recovery curves of histone H1-GFP-expressing cells transfected with the different Ruby constructs. The curves represent averages ± SEM from n = 11 cells over three independent experiments. After quantification of absolute fluorescence recovery (see Materials and Methods), the postbleach fluorescence intensity was normalized to 1 for every cell analyzed and relative fluorescence recovery was calculated. (D) Bar graph showing the time required to recover 60% of histone H1-GFP fluorescence intensity. Statistical significance was evaluated by a two-tailed paired t test (**, P < 0.01; ns, not significant).
Hmgn5 mRNA localization affects HMGN5 NBD phosphorylation and chromatin residency time.
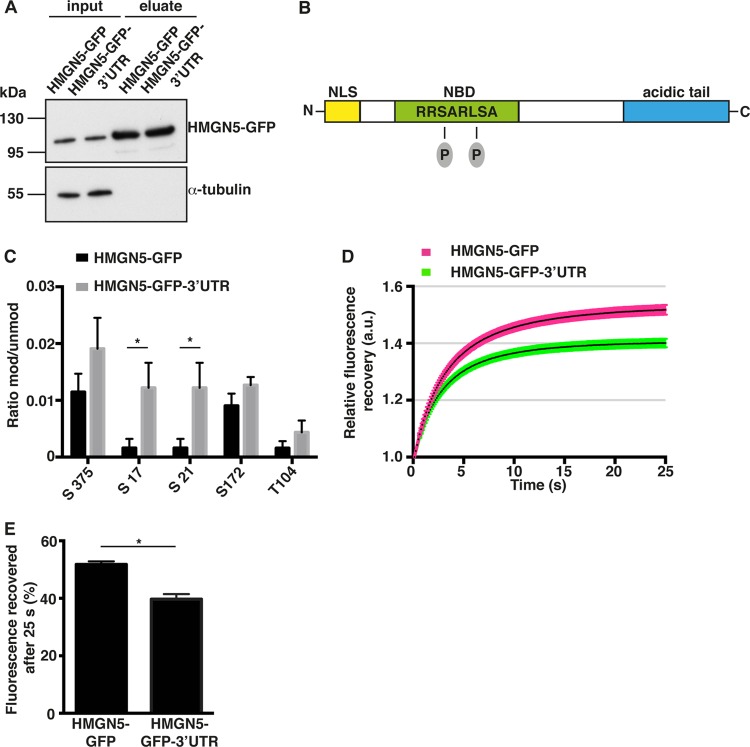
Locally translated proteins might be endowed with different biochemical properties than their cell body-translated counterparts (1). Since the function of HMGN proteins is regulated by posttranslational modifications such as phosphorylation and acetylation (43), we hypothesized that locally synthesized HMGN5 might bear different posttranslational modifications compared to the protein synthesized in the soma. To test this hypothesis, we performed GFP immunoprecipitation followed by mass spectrometry analysis of cell lysates expressing either HMGN5-GFP or HMGN5-GFP-3′ UTR constructs (Fig. 7A). The HMGN5 protein synthesized from the construct bearing Hmgn5 3′ UTR showed 7-fold-higher levels of phosphorylation of serines 17 and 21 in the NBD with respect to HMGN5 synthesized from a somatic mRNA (Fig. 7B and C). Other phosphorylation sites outside the NBD were not significantly affected. Since phosphorylation of HMGN NBD has been shown to affect HMGN affinity for the nucleosome core particle and for other interactors (13, 44), we performed a FRAP analysis to measure intranuclear mobility of HMGN5-GFP expressed from somatically or growth cone-translated mRNAs (HMGN5-GFP or HMGN5-GFP-3′ UTR) (42). HMGN5-GFP showed a faster recovery of fluorescence after photobleaching than HMGN5-GFP-3′ UTR, indicating that HMGN5 synthesized from a growth cone localizing mRNA has a higher chromatin residency time (Fig. 7D and E). A comparable difference in the extent of fluorescence recovery (∼10%) was previously observed for mouse HMGN5 mutants showing different chromatin binding affinities, supporting the physiological relevance of the magnitude of the effect we observe in N1E-115 cells (22). These results suggest that growth cone mRNA localization and translation influence HMGN5 phosphorylation and chromatin binding properties to regulate neurite outgrowth in N1E-115 cells.
FIG 7.
Hmgn5 mRNA localization affects HMGN5 NBD phosphorylation and chromatin residency time. (A) Western blot analysis of N1E-115 cell lysates expressing HMGN5-GFP or HMGN5-GFP-3′ UTR before (lanes “input”) and after (lanes “eluate”) immunoprecipitation with anti-GFP antibodies. A total of 5 μl of input and 20 μl of eluate samples were loaded on the gel. (B) Schematic representation of HMGN5 domain structure. NLS, nuclear localization signal; NBD, nucleosome binding domain. The two detected phosphorylated serines in the nucleosome binding domain are depicted. (C) Results of the mass spectrometry analysis of the GFP immunoprecipitates. Only HMGN5 residues for which a posttranslational modification was identified are shown. The y axis reports the ratio between the intensity of the modified peptide versus the unmodified peptide (n = 3 independent immunoprecipitation experiments, mean ± SEM). Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05). (D) Fluorescence recovery curves after photobleaching of cells expressing HMGN5-GFP and HMGN5-GFP-3′ UTR. The curves represent averages ± SEM from n = 35 to 38 cells over four independent experiments. After quantification of absolute fluorescence recovery (see Materials and Methods), the postbleach fluorescence intensity was normalized to 1 for every cell analyzed, and relative fluorescence recovery was calculated. (E) Bar graph showing the percentage of fluorescence recovered 25 s after bleaching. Statistical significance was evaluated by a two-tailed paired t test (*, P < 0.05).
Hmgn5 mRNA localization controls neurite outgrowth and chromatin structure in hippocampal neurons.
Next we investigated Hmgn5 mRNA localization and protein function in mouse hippocampal neurons, a well-known system to study neuronal outgrowth and the establishment of neuronal polarity (45). In culture, the round shape of dissected neurons is rapidly broken and several neurites are sequentially established. These neurites extend as the cells differentiate and, after 7 days, a single axon and several dendrites can be observed (12, 45). FISH analysis revealed localization of Hmgn5 mRNA in growth cones of hippocampal neurons at 3 days in vitro (DIV3) and showed that Hmgn5 3′ UTR functions as a growth cone localization element in this cell system (Fig. 8A; see also Fig. S5, arrowheads, in the supplemental material). Hmgn5 KD in hippocampal neurons (Fig. 8B and C) led to reduced neurite length at DIV3 (Fig. 8D and E). Since only a small proportion of neurons (∼10%) showed clear axon/dendrite specification at DIV3 in our hands, we referred to all cell processes with a length over 1 cell body diameter as “neurites.” The Hmgn5 KD phenotype was rescued by exogenous HMGN5 expression only when the Hmgn5 3′ UTR was present in the expression construct (Fig. 8D and E). Using antineurofilament SMI312 staining, no defect in axonal specification at either DIV3 (for the small percentage of neurons that already present an axon at this stage) or DIV7 was observed, indicating that HMGN5 might function exclusively as a regulator of neurite length and not of axonal specification (see Fig. S6 in the supplemental material). To explore whether HMGN5 is able to modify chromatin structure in hippocampal neurons, we overexpressed the different HMGN5 constructs. Only HMGN5-GFP-3′ UTR was able to induce a statistically significant increase in neurite length in hippocampal neurons (Fig. 9A and B) and chromatin decompaction and reorganization, as quantified by counting the number of heterochromatic foci and the average staining intensity of H3K9me3 foci (Fig. 9C to E). We conclude that, as observed in N1E-115 cells, Hmgn5 mRNA growth cone localization and HMGN5 function are involved in modulating neurite outgrowth and chromatin compaction in hippocampal neurons.
FIG 8.
Hmgn5 mRNA localization modulates neurite outgrowth in hippocampal neurons. (A) Confocal fluorescence micrographs of FISH with riboprobes antisense and sense (negative control) to Hmgn5 mRNA on DIV3 hippocampal neurons. FISH signal is represented in IBW contrast, while F-actin staining is represented in green. Arrowheads indicate punctate structures. Scale bar, 20 μm for whole-cell micrographs, 5 μm for growth cone micrographs. (B and C) Hippocampal neurons were transfected with ctrl or Hmgn5 siRNA, and KD efficiency was monitored at DIV3 by RT-qPCR (B) and immunofluorescence staining (C). (B) n = 2 RNA preparations; (C) n = 22 to 25 cells, mean ± SEM. In panel C, HMGN5 staining is shown in pseudocolor, with warm and cold colors representing, respectively, high and low fluorescence intensity, while GFP signal (identifying transfected neurons) is shown in green. Statistical significance was evaluated by a Kolmogorov-Smirnov test (***, P < 0.001). Scale bar, 20 μm. (D) Representative confocal micrographs of doublecortin-stained hippocampal neurons transfected with ctrl or Hmgn5 siRNA and with rescue constructs. Doublecortin staining is shown in IBW contrast, while GFP signal is shown in green. Scale bar, 20 μm. (E) Neurite length measurement of DIV3 hippocampal neurons transfected with ctrl or Hmgn5 siRNA and with rescue constructs (n = 200 to 300 neurites from 92 to 105 cells over three independent experiments, mean ± SD). Statistical significance was evaluated by a Kolmogorov-Smirnov test (**, P < 0.01; ***, P < 0.001; ns, not significant).
FIG 9.
HMGN5 promotes neurite outgrowth and controls chromatin structure in hippocampal neurons. (A) Representative confocal micrographs of βIII-tubulin-stained hippocampal neurons transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR. βIII-tubulin staining is shown in IBW contrast, while GFP signal is shown in green. Scale bar, 20 μm. (B) Neurite length measurement of DIV3 hippocampal neurons transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR (n = 220 to 260 neurites from 85 to 90 cells from three independent experiments, mean ± SD). Statistical significance was evaluated by a Kolmogorov-Smirnov test (***, P < 0.001; ns, not significant). (C) Representative confocal micrographs of DAPI- and H3K9me3-stained hippocampal neurons transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR. GFP signal is shown in green, DAPI staining is shown in IBW contrast, and H3K9me3 staining is shown in red. The confocal plane was chosen as the best focus on nuclei. Images were collected on the same day with identical exposure and scaling settings. Scale bar, 5 μm. (D) Measurement of the number of heterochromatic foci in the DAPI staining of hippocampal neurons transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR (n = 65 cells over three independent experiments, mean ± SEM). Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05; ns, not significant). (E) Quantification of mean fluorescence intensity of H3K9me3 foci in hippocampal neurons transfected with GFP, HMGN5-GFP, or HMGN5-GFP-3′ UTR (n = 20 to 40 foci, mean ± SEM). Statistical significance was evaluated by a two-tailed unpaired t test (*, P < 0.05; ns, not significant).
DISCUSSION
Local translation and retrograde transport of transcription factors in neuronal cells have recently been proposed to couple distal signaling events, such as growth factor application to growth cones, to transcriptional changes in the nucleus (8). Transcripts encoding chromatin regulators have also been identified in axonal terminals, but whether their proper localization and function are involved in such a growth cone-to-nucleus signaling program in neuronal development has not been explored. Importantly, regulation of higher-order chromatin architecture has been shown to play a key role in controlling gene expression during neuronal development (46). Here, we show that the growth cone localization of the mRNA encoding the chromatin binding protein HMGN5 modulates neurite outgrowth. Hmgn5 KD impairs neurite outgrowth, while HMGN5 overexpression stimulates it in both neuroblastoma cells and primary hippocampal neurons (Fig. 4, 5, 8, and 9). Both the rescue of the KD and the overexpression phenotypes are strictly dependent on (i) the presence of Hmgn5 3′ UTR in the expression construct (Fig. 4, 5, 8, and 9), which is able to support growth cone localization (Fig. 1D; see also Fig. S3A and S4 in the supplemental material) and translation (Fig. 2), and (ii) the chromatin binding ability of HMGN5 (Fig. 4 and 5). These results strengthen previous observations showing the importance of proper subcellular mRNA localization for both initial neurite (9) and later axonal (47, 48) outgrowth.
While indications of possible neurological functions have been ascribed to HMGN1 and HMGN3 (49–51), HMGN5 has not been implicated in brain physiology so far. Loss of HMGN5 function in the brain induces transcriptional changes in genes implicated in nervous system development and cell morphogenesis (15). As observed for HMGN1 (49), we also detect high HMGN5 expression levels in neurogenic areas of the mouse brain (F. Moretti and C. Rolando, unpublished observations). Furthermore, Hmgn5 mRNA has been detected in preparations from the synaptic neuropil (52). These observations suggest an in vivo role of HMGN5 in neuronal differentiation, which awaits further characterization. In neuron-like N1E-115 cells, transcriptional profiling reveals that Hmgn5 KD affects the expression of 31 genes, many of which are involved in the control of cytoskeletal dynamics (see Fig. S2A in the supplemental material). Furthermore, many of these genes are also temporally modulated during the neurite outgrowth process (see Fig. S2A). These findings suggest a role for HMGN5 in transcriptional regulation of cytoskeleton-regulating genes during the neuronal outgrowth process, a possibility that is in agreement with the inability of Hmgn5 KD cells to extend stable neurites (see Movie S4 in the supplemental material).
The transcriptional changes observed upon Hmgn5 KD might be a direct consequence of HMGN5-mediated chromatin structure rearrangements. Indeed, in line with previous observations in different cell types (22, 42), we show that HMGN5 overexpression affects chromatin structure in both neuroblastoma cells as well as hippocampal neurons (Fig. 5 and 9). Specifically, HMGN5 overexpression leads to chromatin decompaction (Fig. 5 and 9) and reduced linker histone H1 binding to chromatin, as evidenced by the decrease in its chromatin residence time (Fig. 6). These phenotypes strictly depend on growth cone localization of Hmgn5 mRNA (Fig. 5, 6, and 9). We find that HMGN5 synthesized from an mRNA that is locally translated in the growth cone is extensively phosphorylated at serines 17 and 21 in its NBD in comparison to somatically synthesized HMGN5 (Fig. 7B and C). Consistently with the NBD being the major determinant of the interaction of HMGN proteins with the nucleosome core particle, and with this interaction being specifically regulated by Ser17/21 phosphorylation (13, 44), our FRAP analysis indicates that growth cone-synthesized, phosphorylated HMGN5 has a higher chromatin residency time than its somatically synthesized counterpart (Fig. 7D and E). We therefore propose that phosphorylation-induced, tighter chromatin binding of growth cone-synthesized HMGN5 enables the regulation of chromatin dynamics necessary to control transcriptional programs impinging on neurite outgrowth. To further investigate the significance of HMGN5 NBD phosphorylation, which is strictly conserved among HMGN family members, the use of phosphomimetic point mutants would be of great help. However, in the context of HMGN proteins, there are several limitations inherent to this approach. In HMGN1, Ser17/21 phosphorylation also modulates its interaction with 14-3-3 proteins involved in multiple intracellular targeting processes (44). Thus, HMGN5 NBD phosphorylation might regulate not only HMGN5 chromatin binding affinity but also additional regulatory steps. Furthermore, at least for HMGN1, phosphomimetic mutants have been shown not to fully behave as their phosphorylated counterparts (44), and both phosphomimetic and conservative amino acid substitutions abolish chromatin binding (53). Overall, the inability of the HMGN5S17,21E mutant to modulate neurite outgrowth (Fig. 4G and H and 5A and B) suggests that HMGN5 NBD phosphorylation might play an important role in neuronal differentiation, which awaits further characterization.
What is the significance of the requirement of such a complex spatiotemporal signaling mechanism that involves (i) transport of Hmgn5 mRNA to the growth cone, (ii) local translation, and (iii) retrograde HMGN5 transport to the nucleus? In vitro, the neuronal differentiation process is highly stochastic and involves successive cycles of neurite outgrowth and collapse that occur on timescales of hours (see Movie S4 in the supplemental material) (12). As proposed before (54), cell-intrinsic mechanisms must exist to coordinate such a dynamic neurite outgrowth process with protein synthesis through regulation of transcription. We therefore hypothesize that the spatiotemporal regulation of HMGN5 function might be part of a mechanism that links cell morphodynamics with the control of transcription through chromatin regulation at relevant time scales. Interestingly, several other mRNAs encoding chromatin regulators have been identified in axons of different neuronal subtypes. These include, for example, those for additional members of the high-mobility group protein family (e.g., Hmgb1, Hmgb2, Hmgn1, Hmgn2, and Hmgn3), members of the SWI/SNF chromatin remodeling complex (e.g., Smarca2, Smarca5, and Arid1a), and histone-modifying enzymes (e.g., Jmjdc1a, Jmjdc1c, Ash1l, and Fbxl10) (8). Furthermore, a recent study identified mRNAs encoding different histone H4 variants localized in dendrites of CA1 pyramidal neurons (55). This suggests that modulation of chromatin structure and function via local translation and retrograde transport of epigenetic regulators might constitute a general growth cone-to-nucleus signaling mechanism during neuronal development. Our results thus pave the way for the characterization of additional chromatin binding proteins that might be locally synthesized in neuronal processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Philippe Demougin for technical help with the microarray analysis, Katrin Martin for help with photobleaching experiments, Bernhard Bettler for the gift of anti-MAP2 antibody, the members of the Pertz lab for discussion, and Valeria Cavalli and Primo Schär for useful comments on the manuscript.
This work was supported by an EMBO Long-Term Postdoctoral Fellowship to F.M. and by a grant of the Swiss National Science Foundation to O.P.
F.M. and O.P. conceived the experiments. F.M., C.R., and M.W. performed the experiments. F.M., M.W., and O.P. analyzed the results. R.I. performed analysis of microarray experiments. J.R. and A.V.K. performed and analyzed mass spectrometry experiments. M.B. and V.T. provided vital reagents and intellectual contribution to the project. F.M. and O.P. wrote the paper with input from the other coauthors.
We declare that no conflicts of interest exist.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00133-15.
REFERENCES
- 1.Jung H, Gkogkas CG, Sonenberg N, Holt CE. 2014. Remote control of gene function by local translation. Cell 157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung H, O'Hare CM, Holt CE. 2011. Translational regulation in growth cones. Curr Opin Genet Dev 21:458–464. doi: 10.1016/j.gde.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett LE, Sul JY, Takano H, Van Bockstaele EJ, Haydon PG, Eberwine JH. 2006. Region-directed phototransfection reveals the functional significance of a dendritically synthesized transcription factor. Nat Methods 3:455–460. doi: 10.1038/nmeth885. [DOI] [PubMed] [Google Scholar]
- 4.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. 2008. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol 10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji SJ, Jaffrey SR. 2012. Intra-axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron 74:95–107. doi: 10.1016/j.neuron.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL, Fainzilber M. 2012. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J 31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. 2014. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell 158:1159–1172. doi: 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji SJ, Jaffrey SR. 2013. Axonal transcription factors: novel regulators of growth cone-to-nucleus signaling. Dev Neurobiol 74:245–258. doi: 10.1002/dneu.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feltrin D, Fusco L, Witte H, Moretti F, Martin K, Letzelter M, Fluri E, Scheiffele P, Pertz O. 2012. Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol 10:e1001439. doi: 10.1371/journal.pbio.1001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marler KJ, Kozma R, Ahmed S, Dong JM, Hall C, Lim L. 2005. Outgrowth of neurites from NIE-115 neuroblastoma cells is prevented on repulsive substrates through the action of PAK. Mol Cell Biol 25:5226–5241. doi: 10.1128/MCB.25.12.5226-5241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yakubchyk Y, Abramovici H, Maillet JC, Daher E, Obagi C, Parks RJ, Topham MK, Gee SH. 2005. Regulation of neurite outgrowth in N1E-115 cells through PDZ-mediated recruitment of diacylglycerol kinase zeta. Mol Cell Biol 25:7289–7302. doi: 10.1128/MCB.25.16.7289-7302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva JS, Dotti CG. 2002. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci 3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 13.Kugler JE, Deng T, Bustin M. 2012. The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochim Biophys Acta 1819:652–656. doi: 10.1016/j.bbagrm.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rochman M, Malicet C, Bustin M. 2010. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta 1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kugler JE, Horsch M, Huang D, Furusawa T, Rochman M, Garrett L, Becker L, Bohla A, Holter SM, Prehn C, Rathkolb B, Racz I, Aguilar-Pimentel JA, Adler T, Adamski J, Beckers J, Busch DH, Eickelberg O, Klopstock T, Ollert M, Stoger T, Wolf E, Wurst W, Yildirim AO, Zimmer A, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Garfinkel B, Orly J, Ovcharenko I, Bustin M. 2013. High mobility group N proteins modulate the fidelity of the cellular transcriptional profile in a tissue- and variant-specific manner. J Biol Chem 288:16690–16703. doi: 10.1074/jbc.M113.463315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciappio ED, Krausz KW, Rochman M, Furusawa T, Bonzo JA, Tessarollo L, Gonzalez FJ, Bustin M. 2014. Metabolomics reveals a role for the chromatin-binding protein HMGN5 in glutathione metabolism. PLoS One 9:e84583. doi: 10.1371/journal.pone.0084583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. 2015. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun 6:6138. doi: 10.1038/ncomms7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Phair RD, Misteli T. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 20.Shirakawa H, Landsman D, Postnikov YV, Bustin M. 2000. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem 275:6368–6374. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- 21.Welshhans K, Bassell GJ. 2011. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci 31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malicet C, Rochman M, Postnikov Y, Bustin M. 2011. Distinct properties of human HMGN5 reveal a rapidly evolving but functionally conserved nucleosome binding protein. Mol Cell Biol 31:2742–2755. doi: 10.1128/MCB.05216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho BS, Irizarry RA. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turriziani B, Garcia-Munoz A, Pilkington R, Raso C, Kolch W, von Kriegsheim A. 2014. On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology 3:320–332. doi: 10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarner S, Kozma R, Ahmed S, Lim L. 2000. Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells. Mol Cell Biol 20:158–172. doi: 10.1128/MCB.20.1.158-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiebler MA, Bassell GJ. 2006. Neuronal RNA granules: movers and makers. Neuron 51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Andreassi C, Riccio A. 2009. To localize or not to localize: mRNA fate is in 3′ UTR ends. Trends Cell Biol 19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Iacoangeli A, Tiedge H. 2013. Translational control at the synapse: role of RNA regulators. Trends Biochem Sci 38:47–55. doi: 10.1016/j.tibs.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornberg H, Holt C. 2013. RNA-binding proteins and translational regulation in axons and growth cones. FrontNeurosci 7:81. doi: 10.3389/fnins.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fivaz M, Meyer T. 2003. Specific localization and timing in neuronal signal transduction mediated by protein-lipid interactions. Neuron 40:319–330. doi: 10.1016/S0896-6273(03)00634-2. [DOI] [PubMed] [Google Scholar]
- 33.Rochman M, Taher L, Kurahashi T, Cherukuri S, Uversky VN, Landsman D, Ovcharenko I, Bustin M. 2011. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res 39:4076–4087. doi: 10.1093/nar/gkq1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonami K, Kurihara Y, Arima S, Nishiyama K, Uchijima Y, Asano T, Sorimachi H, Kurihara H. 2011. Calpain-6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J Cell Sci 124:1214–1223. doi: 10.1242/jcs.072561. [DOI] [PubMed] [Google Scholar]
- 35.Rozenblum GT, Gimona M. 2008. Calponins: adaptable modular regulators of the actin cytoskeleton. Int J Biochem Cell Biol 40:1990–1995. doi: 10.1016/j.biocel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Weirich CS, Erzberger JP, Barral Y. 2008. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol 9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 37.Xie Y, Ding YQ, Hong Y, Feng Z, Navarre S, Xi CX, Zhu XJ, Wang CL, Ackerman SL, Kozlowski D, Mei L, Xiong WC. 2005. Phosphatidylinositol transfer protein-alpha in netrin-1-induced PLC signalling and neurite outgrowth. Nat Cell Biol 7:1124–1132. doi: 10.1038/ncb1321. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi N, Sundberg C, Kveiborg M, Moghadaszadeh B, Asmar M, Dietrich N, Thodeti CK, Nielsen FC, Moller P, Mercurio AM, Albrechtsen R, Wewer UM. 2003. ADAM12 induces actin cytoskeleton and extracellular matrix reorganization during early adipocyte differentiation by regulating beta1 integrin function. J Cell Sci 116:3893–3904. doi: 10.1242/jcs.00699. [DOI] [PubMed] [Google Scholar]
- 39.Yuasa K, Nagame T, Dohi M, Yanagita Y, Yamagami S, Nagahama M, Tsuji A. 2012. cGMP-dependent protein kinase I is involved in neurite outgrowth via a Rho effector, rhotekin, in Neuro2A neuroblastoma cells. Biochem Biophys Res Commun 421:239–244. doi: 10.1016/j.bbrc.2012.03.143. [DOI] [PubMed] [Google Scholar]
- 40.Yaron A, Huang PH, Cheng HJ, Tessier-Lavigne M. 2005. Differential requirement for plexin-A3 and -A4 in mediating responses of sensory and sympathetic neurons to distinct class 3 semaphorins. Neuron 45:513–523. doi: 10.1016/j.neuron.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Pertz OC, Wang Y, Yang F, Wang W, Gay LJ, Gristenko MA, Clauss TR, Anderson DJ, Liu T, Auberry KJ, Camp DG II, Smith RD, Klemke RL. 2008. Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc Natl Acad Sci U S A 105:1931–1936. doi: 10.1073/pnas.0706545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. 2009. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell 35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pogna EA, Clayton AL, Mahadevan LC. 2010. Signalling to chromatin through post-translational modifications of HMGN. Biochim Biophys Acta 1799:93–100. doi: 10.1016/j.bbagrm.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Prymakowska-Bosak M, Hock R, Catez F, Lim JH, Birger Y, Shirakawa H, Lee K, Bustin M. 2002. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol Cell Biol 22:6809–6819. doi: 10.1128/MCB.22.19.6809-6819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dotti CG, Sullivan CA, Banker GA. 1988. The establishment of polarity by hippocampal neurons in culture. J Neurosci 8:1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh J, Gage FH. 2005. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol 17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. 2013. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci 126:90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo S, Kim HH, Kim P, Donnelly CJ, Kalinski AL, Vuppalanchi D, Park M, Lee SJ, Merianda TT, Perrone-Bizzozero NI, Twiss JL. 2013. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3′ untranslated region AU-rich regulatory element. J Neurochem 126:792–804. doi: 10.1111/jnc.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, Marino-Ramirez L, Meshorer E, Landsman D, Bustin M. 2013. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol Cell Biol 33:3377–3389. doi: 10.1128/MCB.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West KL, Castellini MA, Duncan MK, Bustin M. 2004. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol Cell Biol 24:3747–3756. doi: 10.1128/MCB.24.9.3747-3756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abuhatzira L, Shamir A, Schones DE, Schaffer AA, Bustin M. 2011. The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice. J Biol Chem 286:42051–42062. doi: 10.1074/jbc.M111.300541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. 2012. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda T, Catez F, Gerlitz G, Bustin M. 2008. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol 28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albus CA, Rishal I, Fainzilber M. 2013. Cell length sensing for neuronal growth control. Trends Cell Biol 23:305–310. doi: 10.1016/j.tcb.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Ainsley JA, Drane L, Jacobs J, Kittelberger KA, Reijmers LG. 2014. Functionally diverse dendritic mRNAs rapidly associate with ribosomes following a novel experience. Nat Commun 5:4510. doi: 10.1038/ncomms5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.