Abstract
Gramicidin S (GS) is a nonribosomally synthesized decapeptide from Aneurinibacillus migulanus. Its pronounced antibiotic activity is attributed to amphiphilic structure and enables GS interaction with bacterial membranes. Despite its medical use for over 70 years, the peptide-lipid interactions of GS and its molecular mechanism of action are still not fully understood. Therefore, a comprehensive structural analysis of isotope-labeled GS needs to be performed in its biologically relevant membrane-bound state, using advanced solid-state nuclear magnetic resonance (NMR) spectroscopy. Here, we describe an efficient method for producing the uniformly 13C/15N-labeled peptide in a minimal medium supplemented by selected amino acids. As GS is an intracellular product of A. migulanus, we characterized the producer strain DSM 5759 (rough-convex phenotype) and examined its biosynthetic activity in terms of absolute and biomass-dependent peptide accumulation. We found that the addition of either arginine or ornithine increases the yield only at very high supplementing concentrations (1% and 0.4%, respectively) of these expensive 13C/15N-labeled amino acids. The most cost-effective production of 13C/15N-GS, giving up to 90 mg per gram of dry cell weight, was achieved in a minimal medium containing 1% 13C-glycerol and 0.5% 15N-ammonium sulfate, supplemented with only 0.025% of 13C/15N-phenylalanine. The 100% efficiency of labeling is corroborated by mass spectrometry and preliminary solid-state NMR structure analysis of the labeled peptide in the membrane-bound state.
INTRODUCTION
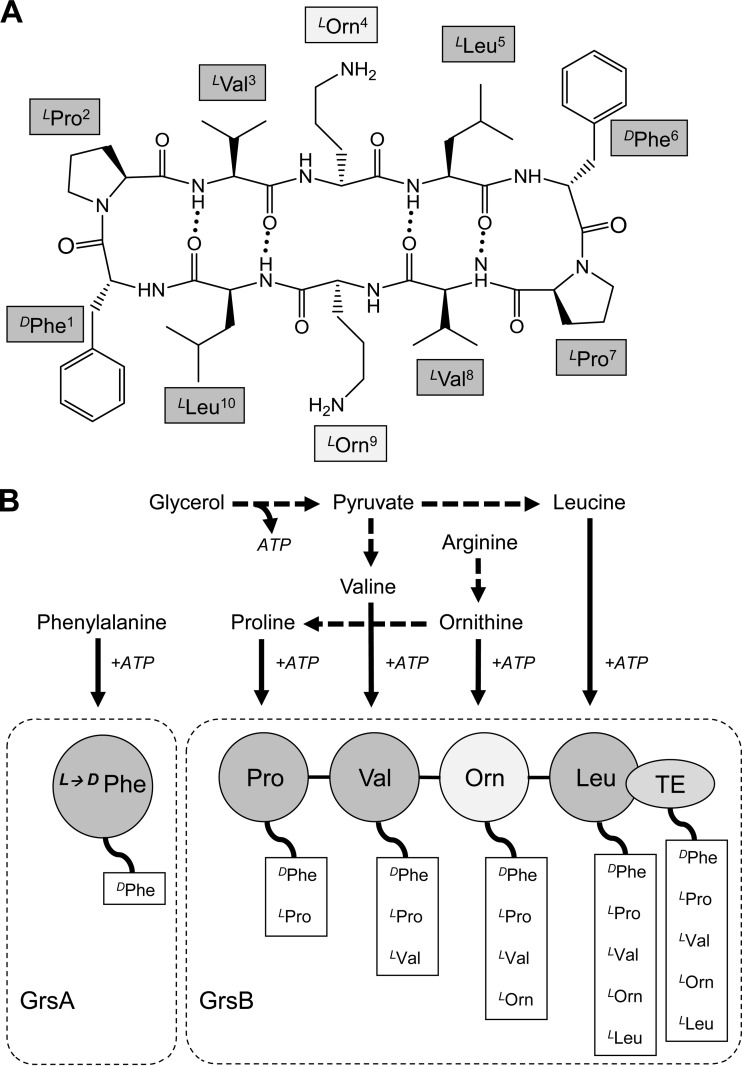
The cyclic decapeptide gramicidin S (GS) contains a repeat sequence of five amino acids (cyclo[DPhe-Pro-Val-Orn-Leu]2), including the unusual ornithine (Orn) and d-phenylalanine (DPhe) (Fig. 1A). Since its discovery about 70 years ago, GS has been used as a topical antibiotic in Russia and neighboring countries, exploiting its pronounced antimicrobial activity against Gram-positive bacteria (1, 2). For example, GS is the bioactive agent in GrammidinNeo, the lozenge against sore throat and mouth ulcer produced by the Russian JSC Valenta Pharmaceuticals. Notably, despite a long medication history, no clinical cases of bacterial resistance against GS have been reported, which suggest that this peptide may be an exceptionally promising resistance-free antibiotic (3).
FIG 1.
GS structure and nonribosomal GS biosynthesis. (A) Sequence, cyclic structure, and nomenclature of GS. (B) Involvement of amino acids in the nonribosomal GS biosynthesis and assembly of the protein complex consisting of GrsA and GrsB synthetases.
The biological activity of GS was first described as membranolytic, which is believed to determine its profound antimicrobial as well as hemolytic activities in vitro. Less acknowledged are GS activities not directly related to interaction with membrane bilayers: the peptide has been shown to inhibit membrane enzymes, such as bacterial NADH dehydrogenase and cytochrome bd terminal oxidase (4), and to delocalize the peripheral cell division regulator MinD, the lipid II biosynthesis protein MurG, and cytochrome c (5).
The main molecular target of GS is supposed to be the plasma membrane of susceptible bacterial species, where an accumulation of the amphiphilic peptide leads to a loss of the barrier function and an irreversible increase in permeability for ions and metabolites (6). This concentration-dependent effect leads to membrane depolarization (7) and an overall dysfunction of the cellular osmoregulation, which starts already at GS levels well below the respective MICs (8). However, the structure-function relationship of the peptide and its detailed molecular mechanism of action at the plasma membrane, i.e., its interaction with the lipid bilayer, are still lacking a consensus view. Over the last decades, the molecular structure of GS has been studied by a variety of biophysical methods, including nuclear magnetic resonance (NMR) in solution and X-ray crystallography. The peptide has an antiparallel β-sheet conformation, in which the two β-strands are fixed by two type-II′ β-turns and by up to four intramolecular hydrogen bonds (9, 10). The two reported crystal structures agree on the fold, but they differ significantly in the atomic details (11, 12). Surprisingly, GS does not readily produce X-ray quality single crystals; hence, highly artificial crystallization conditions were applied in both cases, in part explaining these discrepancies. The picture is somewhat more consistent for the NMR-derived structures that have been obtained in membrane-mimicking solutions, such as dimethyl sulfoxide (DMSO) (13) or a CHCl3-methanol mixture (14), but in most NMR studies GS derivatives have been used rather than real GS. Most significantly, GS is a membrane-active peptide; hence, functionally relevant structural information should be acquired when it is bound to a genuine lipid bilayer. To the best of our knowledge, except for a single computational study involving molecular dynamics simulation (MD) of GS in DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) (15), there is no atomic structure available so far for native GS embedded in a phospholipid membrane. Given the problems with crystallization, the only available method for solving the atomic structure of GS in native membranes is solid-state NMR spectroscopy, for which entirely unperturbed, uniformly 13C/15N-labeled molecules are required.
GS is synthesized nonribosomally by two complementary synthetases, GrsA and GrsB (Fig. 1B). The biosynthesis starts with the activation, thioesterification, and racemization of phenylalanine by GrsA synthetase. GrsB further catalyzes activation of the constituent amino acids that follow. Polymerization begins with the transfer of d-phenylalanine on GrsA to the 4-phosphopanthetheine residue of GrsB peptidyl carrier protein (pan-PCP) and proceeds through a sequential thiolation and transpeptidation reaction via pan-PCP on GrsB (16). Penultimately, two pentapeptides, one bound as an ester to the active-site serine of the terminal thioesterase domain (TE) and the second bound as a thioester to the adjacent pan-PCP, are ligated to a decapeptidyl-pan-PCP, which is finally cyclized on TE (17). The effectiveness of this unusual way of biosynthesis depends on multiple factors, such as growth temperature, medium composition, and phenotype variability; therefore, GS production is not a trivial task (18).
Under optimal growth conditions, the nonribosomal biosynthesis of GS proceeds error free and is more cost-effective than chemical peptide synthesis. High-yield microbiological production of GS by fermentation of the natural producer Aneurinibacillus migulanus requires a growth medium that contains a large amount of amino nitrogen. This condition makes uniformly 13C/15N-labeled media very costly. Furthermore, the overall yields from nonribosomal peptide biosynthesis depend strongly on the individual amino acids supplemented, as they deviate in this respect from the “normal” biosynthesis involving ribosomes. Indeed, standard approaches using media that are fully supplemented with stable isotopes—as developed for the production of recombinant ribosomally produced proteins in Escherichia coli—are not suitable for GS fermentation. These peculiarities of the growth conditions for the GS producer strain prompted us to develop a new, cost-effective medium for preparing uniformly 13C/15N isotope-labeled GS. In this paper, we describe the production of suitably labeled GS by fermentation of the natural producer strain A. migulanus DSM 5759 in media supplemented with stable 13C/15N isotopes. Application of 13C/15N-labeled GS could be also useful for investigations of GS interactions with membrane proteins and surrounding phospholipids as well as for structural studies of the GS-based nanofibers (19).
MATERIALS AND METHODS
Materials.
Yeast extract with 5% amino nitrogen, agar for microbiology, d-pantothenic acid as a calcium salt, commercial GS, unlabeled glycerol, l-amino acids (phenylalanine, arginine, histidine, ornithine, glutamic acid, methionine), pyridoxine hydrochloride, and matrices for matrix-assisted laser desorption–ionization (MALDI) mass spectrometry were obtained from Sigma-Aldrich (Munich, Germany). Bacto tryptone with 4 to 6% amino nitrogen and Noble agar were purchased from Becton, Dickinson & Co. (Heidelberg, Germany). 13C-labeled glycerol, 15N-labeled ammonium sulfate (both with >99% isotope enrichment), and the uniformly 13C/15N-labeled amino acid l-phenylalanine (>98% for both isotopes) were obtained from Euriso-Top GmbH (Saarbrücken, Germany). Inorganic salts, solvents, and other chemicals were of the highest quality available. Water was purified with a MilliQ Biocell system (Merck Millipore, Darmstadt, Germany) and used for all solutions, including media.
Phenotype control of producer strain.
Aneurinibacillus migulanus DSM 5759 was received from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). It had been characterized earlier to consist of an entirely rough phenotype with a convex center, which is capable of GS production (18). Spores were obtained in NBYS medium, containing the following (in g/liter): Bacto tryptone (5.0), meat extract (3.0), yeast extract (5.0), MgCl2·6H2O (0.2), CaCl2·2H2O (0.1), MnCl2·4H2O (0.01), and FeCl3·6H2O (0.0002). The first three salts (salt solution 1) and a solution of FeCl3·6H2O in 0.01 M HCl (salt solution 2) were prepared separately as 1,000-fold concentrated stocks. NBYS cultures were grown 48 to 50 h and washed with sterile water (8,000 × g at 4°C for 10 min). Suspensions of spores and vegetative cells were heated for 20 min at 80°C to destroy vegetative cells, followed by washing with sterile ultrapure water. Concentrated spore suspensions were stored in sterile water with 30% glycerol at −20°C. Due to phenotype instability, the spore suspension was always used to first inoculate yeast peptone (YP) medium (18, 20) with Bacto tryptone and yeast extract (each 50 g/liter), i.e., under conditions that should promote the development of rough phenotypes (18). Subsequent plating of this culture onto the surface of LBY agar (10 g/liter Bacto tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, and 30 g/liter microbiological agar) was used as a control to check the colony morphology.
Fermentation media and inoculation material.
The compositions of the chemically defined media with glycerol, which were supplemented with different compounds as nitrogen sources, are summarized in Table 1. Ultrapure water and 10-fold-concentrated stock solutions of glycerol were autoclaved, and 10-fold-concentrated amino acids solutions, phosphate buffer, Tris-HCl (pH 7.4 at 25°C), and 40-fold-concentrated solutions of d-pantothenate and pyridoxine hydrochloride, as well as 1,000-fold-concentrated salt solutions 1 and 2 (see above), were sterilized by membrane filtration (cellulose membrane filter with pore diameter of 0.22 μm; Sarstedt, Nümbrecht, Germany). Chemically defined media were prepared by combining aliquots of these stocks. Each 40 ml of medium (in 350-ml Erlenmeyer flasks) was inoculated either with cells from the overnight preculture, grown in LBY liquid medium, or with cells from 3- to 5-day-old colonies of A. migulanus DSM 5759. Rough individual colonies from LBY agar were resuspended in 150 mM NaCl. Inoculation volumes were determined by measurement of the optical density at 660 nm (OD660) and adjusted to 105 CFU/ml. All experiments were done at least in duplicate or with more repetitions at a temperature of 40°C, which is optimal for A. migulanus (1).
TABLE 1.
Composition and GS yield in different chemically defined mediaa used in this work
| Component or GS yield | Content (g/liter) in: |
|||
|---|---|---|---|---|
| GATb | G4/4c | GATF1 | GATF2 | |
| Glycerol | 10.0 | 10.0 | 10.0 | 10.0 |
| (NH4)2SO4 | 5.0 | 5.0 | 5.0 | |
| l-Arginine | 10.0 | |||
| l-Phenylalanine | 1.0 | 1.0 | 0.25 | |
| l-Methionine | 0.5 | |||
| l-Histidine | 1.3 | |||
| K2HPO4 | 6.5 | 6.5 | 6.5 | 6.5 |
| KH2PO4 | 1.7 | 1.7 | 1.7 | 1.7 |
| Tris-HCl | 6.0 | 6.0 | 6.0 | |
| MgCl2·6H2O | 0.2 | 0.2 | 0.2 | 0.2 |
| CaCl2·2H2O | 0.1 | 0.1 | 0.1 | 0.1 |
| MnCl2·4H2O | 0.01 | 0.01 | 0.01 | 0.01 |
| FeCl3·6H2O | 0.0002 | 0.0002 | 0.0002 | 0.0002 |
| GS yieldd (mg/liter) | 109 ± 24 | 1356 ± 156 | 173 ± 24 | 263 ± 35 |
GAT, basal minimal medium; G4/4, medium found to be optimal for GS production; GATF1 and GATF2, phenylalanine-containing GAT media. GATF2 was found to be the optimal minimal medium and the most cost-effective in terms of 13C/15N-GS production.
Analogous to 1% G2T6 medium used in reference 22, but reduced here in ammonium sulfate and Tris.
Analogous to fructose-containing medium from reference 24.
GS average yield ± standard deviation over several (n) independent fermentation experiments (for GAT, n = 6; for G4/4, n = 10; for GATF1, n = 6; for GATF2, n = 4).
Extraction of GS.
Aliquots of 1 ml were removed from the fermentation flasks every 2 to 4 h after the OD660 reached a value of 2 to 3. The cells were immediately centrifuged (5,000 × g at ambient temperature for 10 min) and frozen at −20°C. Later, the obtained pellets were resuspended in the preextraction solution (150 mM NaCl and 20 mM HCl) and incubated at 80°C for 15 min to facilitate the extraction of GS. The suspensions were diluted 1:1 with absolute ethanol, and GS was extracted by agitation for 1 h at 30°C. The cell debris was removed by centrifugation, and the amount of GS in the 50% ethanolic extract was determined either using a bioassay or directly by chromatography.
Quantification of GS using a bioassay.
The concentration of GS was determined using an agar plate diffusion bioassay with Bacillus subtilis ATCC 6633 as a test organism. The initial GS extract was diluted 2-, 4-, 5-, or 10-fold, and 50 μl of each dilution was pipetted into 9-mm wells that had been punched into an agar plate containing spores of the test organism (inoculation dose, 104 CFU/ml). The nutrient agar for the plates (5 g/liter Bacto tryptone, 3 g/liter beef extract powder, 20 g/liter KCl, 15 g/liter agar [Difco Agar Noble; Becton, Dickinson and Co.], pH 7.0, set at 25°C) was prepared according to the method described in reference 20. Calibration curves for the radius of growth inhibition were obtained from high-performance liquid chromatography (HPLC) traces of pure GS. The range of GS concentrations from the 50% ethanolic extract showed a linear concentration dependence over the range of 25 to 250 mg/liter. The pure solvent (50% ethanol) was used as a negative control, as it did not inhibit bacterial growth. To facilitate GS diffusion through the agar, the plates were refrigerated at 4°C for 22 h before incubation at 37°C for 20 h. Absolute yields (in milligrams per liter) were used for calculation of specific biomass-dependent GS production (in milligrams per gram, dry cell weight), according to the method described in reference 20.
Quantification of GS and phenylalanine consumption using chromatography.
To determine low concentrations of GS by reverse-phase (RP)-HPLC, a 0.5-ml aliquot of each extract was first dried in a Univapo 100H vacuum concentrator (Froebel, Lindau, Germany) and then redissolved in 50 μl of 50% ethanol, to obtain a concentration 10-fold higher than that of the initial extract. Analytical RP-HPLC was performed on a chromatographic system from Jasco Germany GmbH (Gross-Umstadt, Germany) equipped with a diode array detector. A reverse-phase analytical Grace Vydac C18 column (4.6 by 250 mm) from Grace Davison Discovery Sciences (Deerfield, IL, USA) was used, with a linear water/acetonitrile gradient (relevant part of the gradient: 45 to 90% B in 15 min) at 35°C. The mixture of 10% acetonitrile and 90% water was used as “solvent A,” while “solvent B” was composed of 90% acetonitrile and 10% water; both solvents were supplemented with 5 mM HCl as the ion-pairing agent. For all samples, a constant injection volume of 20 μl was applied. The total area of the GS peak as determined by the Jasco ChromPass software at 257 nm was used as a measure of concentration. Two individual calibration curves were constructed for the ranges of 10 to 100 mg/liter and 0.1 to 1 g/liter.
To determine the residual concentration of phenylalanine in the culture medium, the biomass was centrifuged and 20 μl of the supernatant was directly loaded onto the analytical column. A linear gradient of 0.1% CF3COOH versus 10% methanol was used on the same analytical column as described above at 40°C. The calibration curve was constructed with concentrations of l-phenylalanine ranging from 0.1 to 1.2 g/liter.
RESULTS
Phenotype control and optimization of the inoculation procedure.
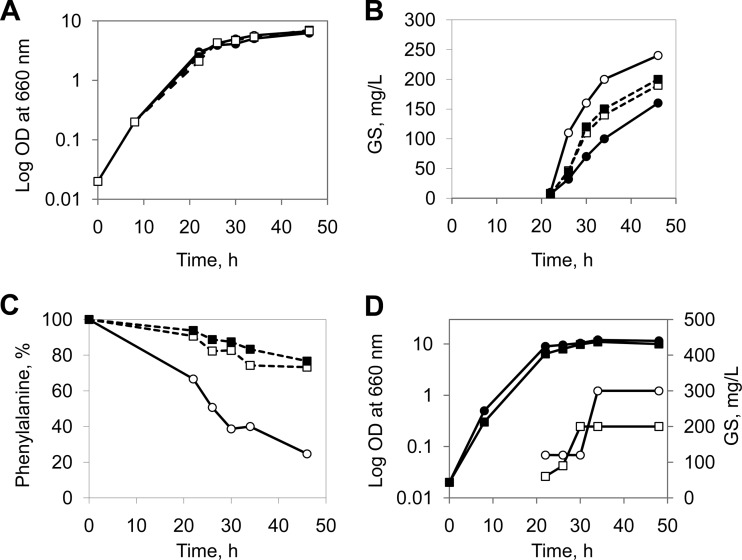
Since the phenotype variations of GS producer strains determine the GS production (18), the phenotype control of inoculation material for each experiment is required. The GS fermentation medium can be inoculated by using either spores (21), an overnight preculture (20), or a cell suspension from bacterial colonies (Fig. 2A). Our multiple attempts to inoculate directly with spores (data not shown) showed this approach to be rather irreproducible. Inoculation with an overnight preculture led to faster exponential growth but slower GS biosynthesis than inoculation with cells obtained from colonies (Fig. 2B). A maximum GS yield of about 200 mg/liter was reached in 48 h by the former inoculation strategy, whereas the latter approach needed only 28 to 30 h for the same yield in the GATF1 medium (glycerol-ammonium sulfate-Tris-phenylalanine) supplemented with 0.1% arginine. Based on these results, in all further experiments colonies were used for inoculation.
FIG 2.
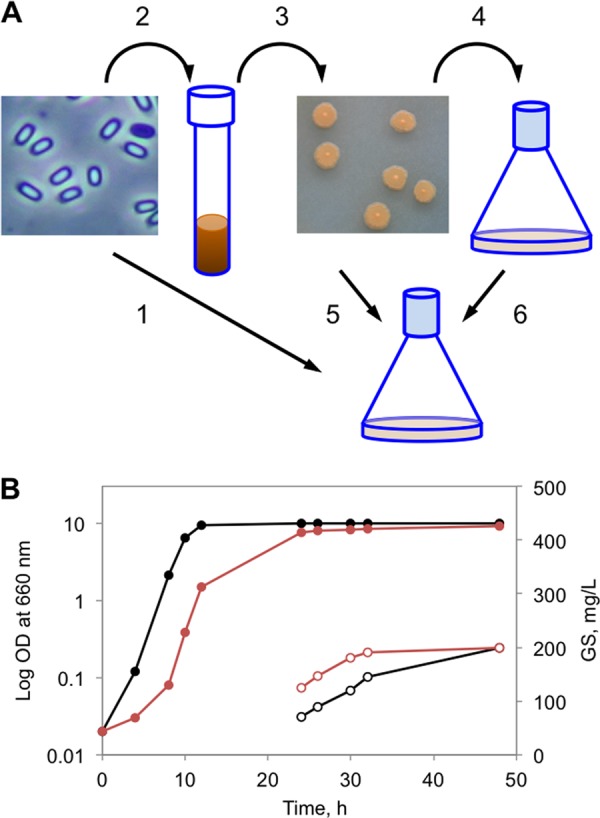
Impact of inoculation on GS fermentation. (A) General scheme for inoculation: 1, direct application of spores; 2, spore germination in YP medium; 3, control of the colony morphology; 4, preparation of the preculture; 5, inoculation of the fermentation medium with colony cells; 6, inoculation of the fermentation medium with the preculture. (B) Influence of inoculation on the growth profile (solid circles, left axis) and GS production (empty circles, right axis). Black, inoculation with the preculture (steps 2, 3, 4, and 6); red, inoculation with colony cells (steps 2, 3, and 5).
Effects of amino acids and vitamins.
Fermentation of GS in the basal minimal GAT medium (glycerol-ammonium sulfate-Tris, without any amino acids) even at the optimal growth temperature of 40°C resulted in exceptionally poor GS yields (Table 1). On the other hand, amino nitrogen-rich G4/4 medium, which contains phenylalanine, arginine, histidine, and methionine, was found to facilitate peptide biosynthesis very well.
However, the addition of such uniformly 13C/15N-labeled amino acids for producing an NMR sample would be extremely expensive. Since the two media differ in their nitrogen sources, besides the total amount of nitrogen, it appears that the GS yield is determined by the way in which nitrogen is supplied. As the next step, we therefore compared urea, ammonium chloride, and ammonium sulfate as a nitrogen source in a glycerol-containing minimal medium for A. migulanus strain DSM 5759. Only ammonium sulfate was able to support GS production, in agreement with previous observations (22) for the type strain ATCC 9999. Notably, the authors reported a yield of 100 mg/liter GS with 12 g/liter ammonium sulfate, but we also checked results with 10 and 5 g/liter and obtained the same 100-mg/liter yield independent of concentration. Obviously, using less ammonium sulfate is more economical for labeling experiments, so we kept this lower concentration. Moreover, being also a sulfur source, the addition of ammonium sulfate justifies the omission of 13C/15N-methionine from the fermentation medium. Next, we proceeded to supplement the minimal GAT medium separately with each of the remaining three amino acids (Phe, Arg, His) used in the G4/4 medium, with the aim to reduce their amounts. Due to its known function of being the direct inducer of the nonribosomal synthesis of GS (23, 24), phenylalanine was initially maintained in all tested media at the recommended concentration of 0.1% (but see below) (23). A positive effect of arginine and histidine can be clearly seen in Fig. 3, as the GS yield almost tripled in the case of 1% arginine. Such increase in production is apparently most pronounced for the amino-nitrogen-rich amino acids, since an experiment with supplemented glutamate, on the other hand, facilitated the initial growth but dramatically reduced the peptide production in GATF1 (Fig. 3B and C), although glutamate in bacteria is a source for biosynthesis of arginine, ornithine, and proline.
FIG 3.
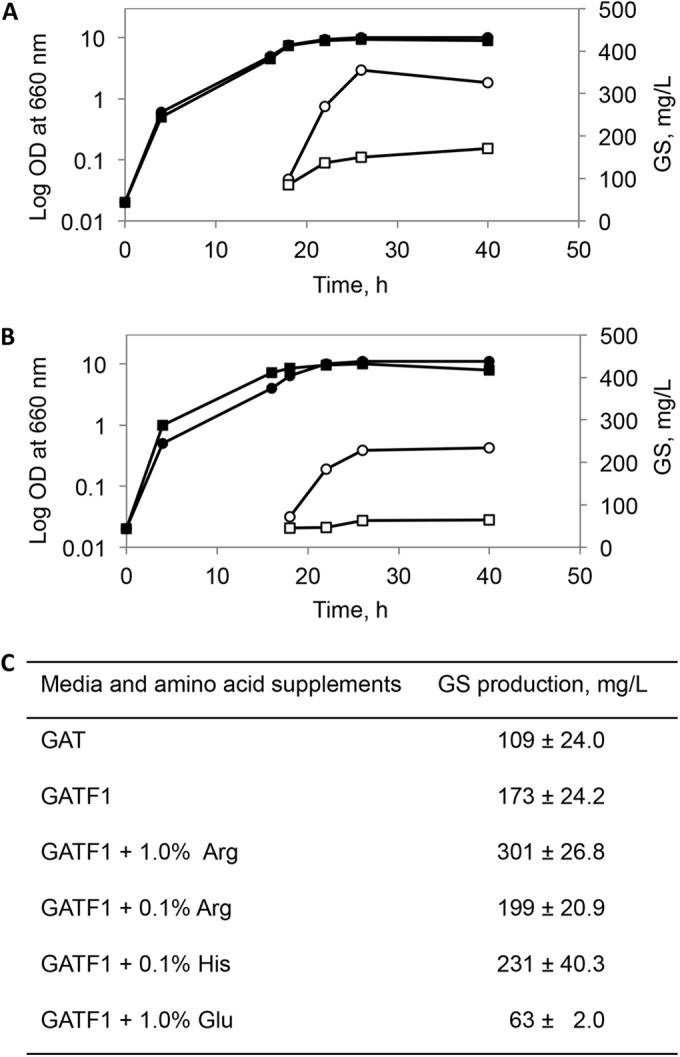
Growth and GS production in GATF1 medium supplemented with selected amino acids. Monitoring of growth (filled signs, left axis) and GS production (empty signs, right axis) in the presence of 1% arginine (circles) and 0.1% arginine (squares) (A) and in the presence of 0.1% histidine (circles) and 1% glutamate (squares) (B). The curves presented are mutually comparable, since they belong to one set of experiments in which the cultures were inoculated with the same cell suspension. (C) Mean values of the maximal GS production as an average over several independent experiments (for GAT and GATF1, n = 6; for GATF1 with 1% glutamate and GATF1 with 1% arginine, n = 2; for GATF1 with 0.1% histidine and GATF1 with 0.1% arginine, n = 5).
Vitamin B5 (calcium d-pantothenate) is a specific precursor of 4′-phosphopantetheine and a constituent of coenzyme A, which acts as a prosthetic group in many acyl carrier proteins. As an ubiquitous cofactor, coenzyme A plays a central role in the energy metabolism and in general in the anabolic processes. In bacteria that produce peptides by nonribosomal peptide synthesis, coenzyme A also participates in the transferase reactions, e.g., switches the GS synthetase of A. migulanus from its inactive apo- to the active holo-form (25). Our results in GATF1 media revealed that calcium pantothenate on the one hand exhibited the expected concentration-dependent stimulatory effect on the initiation of GS production, but on the other hand it also promoted the overall growth of A. migulanus and did not increase significantly the final GS yield (Fig. 4A). Biosynthesis of GS requires 10 ATP molecules to synthesize a single peptide (Fig. 1B), which leads to significant competition with the other energy-consuming processes in the cell (growth, division, etc.). Over the tested concentration range below 70 μM, we observed that the two opposing stimuli balanced each other; hence, the actual biomass-proportional yield of GS remained unchanged at about 60 mg/g dry cell weight. However, when supplemented with 70 μM calcium d-pantothenate, the absolute yield of GS in milligrams per liter increased to about 30% already at 30 h of fermentation (Fig. 4A). These observations suggest that GS yields are influenced not just by the presence of amino acids but also by the metabolic status of the producing cells. This supposition can be further supported, at least with respect to the anabolic/catabolic balance of amino acids, by testing the action of pyridoxine hydrochloride. In bacterial cells, pyridoxine hydrochloride is transformed into pyridoxal phosphate (vitamin B6) in an ATP-consuming enzymatic reaction. B6 is a key cofactor in catabolic reactions of amino acids like transamination and decarboxylation. Thus, in a way similar to that of the B5 action above, competition for ATP may affect GS biosynthesis.
FIG 4.
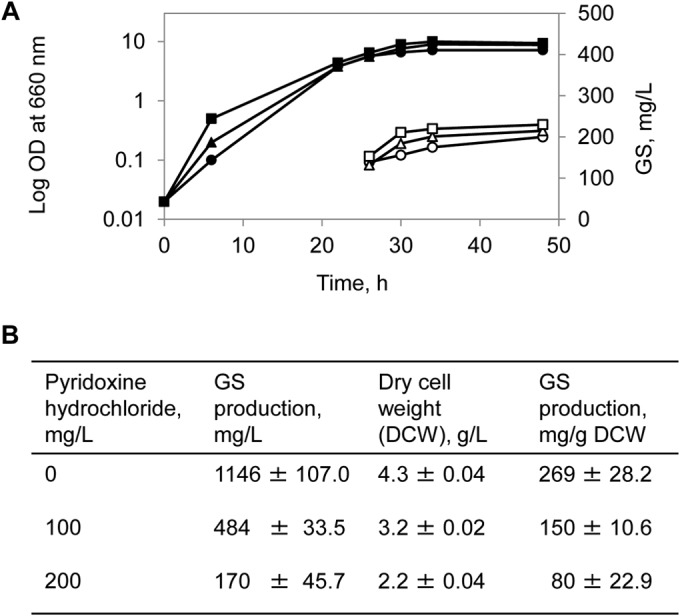
Impact of vitamins on growth and GS production. (A) Monitoring of growth (filled signs, left axis) and GS production (empty signs, right axis) in GATF1 medium with calcium pantothenate (vitamin B5) added in a final concentration of either 0 μM (circles), 20 μM (triangles), or 70 μM (squares). (B) Growth and GS production in G4/4 medium supplemented with pyridoxine hydrochloride (vitamin B6) calculated from two independent experiments.
As summarized in Fig. 4B, bacterial growth and GS biosynthesis dropped to the levels typical for GATF1 medium when pyridoxine hydrochloride was added at a high concentration (200 mg/liter) to the G4/4 medium, which had been otherwise optimal for biosynthesis (Table 1).
Optimization of phenylalanine concentration.
The optimal phenylalanine concentration for GS production has been previously reported as 0.1% (23), based on the range of tested concentrations of 1, 0.5, 0.1, and 0.01%. We challenged this value by examining in more detail the concentration range from 0.1 to 0.025%, and we monitored full-growth curves to assess the biomass and process of GS accumulation. The initial growth during the first 26 h slowed down in proportion to the given concentration of phenylalanine (Fig. 5A). However, further cultivation led to an inversion of the biomass accumulation, as the higher OD values were reached in the presence of the lower phenylalanine concentration. Surprisingly, the lowest tested concentration of phenylalanine stimulated also the highest GS production (Fig. 5B). This effect can be explained by the efficiency of phenylalanine consumption. In the concentration range from 0.1 to 0.05% (6 to 3 mM), only about 20% of the amino acid was consumed. In the fermentation culture with 0.025% (1.5 mM), on the other hand, the phenylalanine uptake by the cells reached almost 80% (Fig. 5C). Given that the highest yields were obtained under these conditions, this low concentration was found to be optimal for GS biosynthesis in minimal medium. This particular medium (GAT + 0.025% of phenylalanine) was named GATF2 and was used for the final optimization step.
FIG 5.
Optimization of phenylalanine concentration and impact of ornithine. Growth profile (A) and GS production (B) in GAT medium, supplemented with different concentrations of phenylalanine: 0% (filled circles, solid line), 0.025% (empty circles, solid line), 0.05% (filled squares, dotted line), 0.1% (empty squares, dotted line). (C) Phenylalanine consumption in the fermentation medium as monitored by HPLC. (D) Growth profile (solid signs, left axis) and GS production (empty signs, right axis) in GATF2 medium only (squares) and in GATF2 medium supplemented with 0.4% ornithine (circles).
Stimulation of GS production by ornithine.
Since arginine is a precursor of ornithine and proline, which are immediate components of GS (Fig. 1B), as a next step we studied the effects obtained by direct addition of ornithine. The new GATF2 medium was supplemented in a concentration range from 0.025 to 0.4%. As expected, a general promotion of both biomass accumulation and GS production was observed (Fig. 5D). In the presence of ornithine, the production of GS almost doubled already in the exponential growth phase. This effect was maintained during the idiophase (the main stage of secondary metabolite production according to reference 26), reaching almost 160% of the GS yields in basal minimal GAT medium. Notably, the idiophase itself was much more pronounced in the presence of ornithine.
Analytical procedures and characterization of the 13C/15N-labeling.
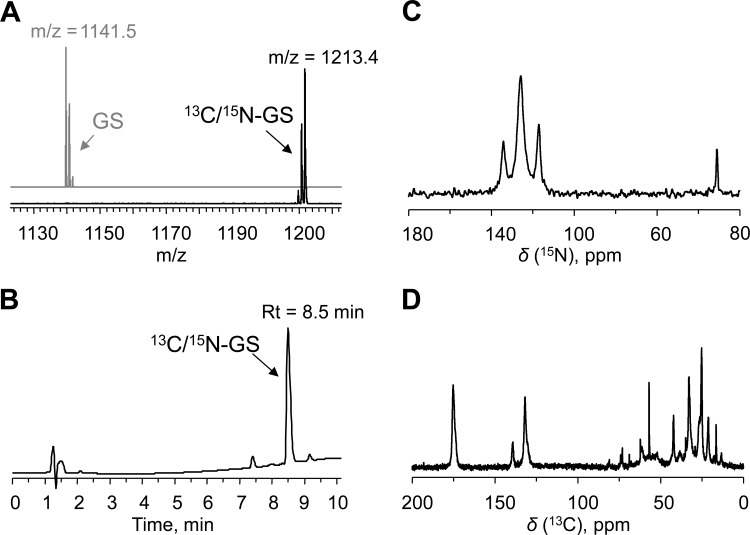
We applied standard physicochemical methods for characterizing the yield and labeling efficiency of the peptide. In particular, quantitative determination of the absolute amounts of peptide was done by HPLC, while matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry was used to quantify the extent of 13C/15N incorporation (Fig. 6A and B). A bioassay, based on the antimicrobial activity in an agar diffusion assay, was used to additionally determine the functional activity of the GS-containing solutions.
FIG 6.
Physicochemical characterization of the 13C/15N-GS produced here. (A) MALDI-TOF spectra of commercial unlabeled GS (upper [gray], theoretical m/z = 1141.7) in comparison with the produced 13C/15N-GS (lower [black], theoretical m/z = 1213.7), showing 100% labeling efficiency. Experimental m/z values are shown. (B) Typical RP-HPLC trace recorded at 220 nm (GS peak is indicated). (C, D) Solid-state MAS-NMR spectra of 13C/15N-GS reconstituted into bilayers of 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine at a peptide/lipid ratio of 1/20, measured at 25°C: 15N-NMR spectrum (C); 13C-NMR spectrum (D).
Gratifyingly, quantification results were found to agree between the two methods used. The comparison of our product as obtained under ultimately optimized conditions against commercial unlabeled GS indeed confirms 100% labeling efficiency (Fig. 6A). This very high degree of labeling for a biological producing system in the case of GS may be due to the nonribosomal nature of the biosynthesis. Nonetheless, as all 60 of the carbons and all 12 of the nitrogens of GS were isotope substituted and very few partially labeled species were detected, we conclude that our production protocol was successful.
The ultimate aim of producing 13C/15N-GS is to perform a comprehensive multidimensional solid-state NMR structure analysis of the membrane-bound peptide. To demonstrate that such a sample has been successfully obtained, we have collected NMR spectra of the fully labeled peptide that was reconstituted in lipid bilayers. The 1-dimensional solid-state magic angle spinning (MAS) 13C- and 15N-NMR data (Fig. 6C and D) show that all expected signals are present, indicating again the complete labeling, with impurity levels below 1%.
Solid-state NMR spectroscopy.
Solid-state NMR was used for a preliminary investigation of the structure of 13C/15N-labeled peptide in multilamellar lipid vesicles. A set of phase-alternated recoupling irradiation scheme (PARIS) and dipolar assisted rotational resonance (DARR) spectra were collected with mixing times ranging from 20 to 60 ms, and a representative example is presented in Fig. S1 in the supplemental material. Since the NMR spectra contained only one set of resonances for each residue, we conclude that the symmetry of the cyclic structure was maintained in the membrane-bound state under the solid-state NMR conditions.
The assignment of the observed 13C resonances was based on the expected intra-residue cross-peak patterns from the individual amino acids of GS and on the characteristic chemical shift regions of the corresponding nuclei. The assigned chemical shifts of GS are listed in Table S1 in the supplemental material, and the results of the dihedral angle prediction by TALOS+ are shown in Table S2 in the supplemental material.
DISCUSSION
The prime objective of this study was to optimize the laboratory-scale fermentation of GS in chemically well-defined minimal media with the strain Aneurinibacillus migulanus DSM 5759. We consider this a practical example of producing a nonribosomally synthesized peptide and achieving a high yield of 13C/15N-labeled GS in the most economical way. To reach this microbiological aim, we have evaluated both the inoculation and fermentation strategies and the composition of the growth medium. The production of GS was characterized by quantifying not only the absolute yield but also the labeling efficiency, such that the best production method could be selected by ultimately judging the price of the isotope-labeled media used to obtain the product. The selection of the producer strain was based on the following criteria. There are several commonly available strains known as GS producers: the type strain Aneurinibacillus migulanus (formerly Bacillus brevis) ATCC 9999 (DSM 2895), two other strains of A. migulanus maintained in DSMZ, i.e., DSM 5658 and DSM 5759 (27), and the Bacillus brevis strain Nagano (28). We have previously described the correlation between colony morphology, the extent of growth, spore formation, cultivation temperature, and GS production (18). We have explained the strong phenotype dissociation into nonproducing phenotypes of the strains available in culture collections by their cultivation at inappropriate temperatures (<40°C) and unsuitable media. Among the above-mentioned strains in our preliminary screening studies, the entirely rough (producing) phenotype of A. migulanus DSM 5759 showed the highest GS yields in the chemically defined G4/4 medium and was therefore used for the present investigation.
Here, we were able to demonstrate that the reproducibility of GS production depends strongly on the nature of the inoculation material. Some essential differences in GS production had previously been noted when the fermentation media were inoculated with spores (21) or with vegetative cells from a liquid preculture (24, 29). We suggest that this phenomenon can be attributed to the pronounced phenotype instability of A. migulanus (18). Only by careful phenotype control or by inoculation directly with cells from rough colonies (Fig. 2) is it possible to obtain reproducible fermentation results.
A comprehensive understanding of the GS biosynthesis pathway in the producer cells provides important clues for optimizing the isotope-labeling protocols. A major peculiarity of the GS biosynthesis is its nonribosomal nature. This means that the optimal conditions may differ considerably from the standard isotope-enrichment approaches commonly used for the production of recombinant proteins, e.g., in E. coli. Therefore, it was not surprising that none of the standard isotope-supplemented media developed for the production of recombinant proteins in E. coli (Celtone, Martek-9, Spectra-9) could promote growth and GS biosynthesis in A. migulanus (data not shown).
A. migulanus utilizes two nonribosomal peptide synthetases for GS biosynthesis, both having a modular architecture (17). GrsA synthetase functions as a phenylalanine-racemase and catalyzes the ATP-dependent transformation of l-phenylalanine into d-phenylalanine (30). Epimerization initiates the biosynthesis and naturally requires phenylalanine (23, 24), which explains the direct role of phenylalanine as an inducer. Indeed, we found that the addition of even low phenylalanine concentrations (1.5 mM) to the minimal fermentation medium could significantly (>150%) enhance the peptide biosynthesis in our experiments (Fig. 4B). GrsB synthetase is responsible for the sequential peptide bond formation of all five constituent amino acids, in each of the two equivalent pentapeptide chains. The five peptidyl-carrier proteins (thiolation domains) of both GS synthetases play a key role in peptide biosynthesis. It is known that the posttranslational modification of these domains through the attachment of 4′-phosphopantetheine converts the inactive apo-proteins into their active holo-forms. The activated amino acids are then attached covalently as thioesters to the SH groups of 4′-phosphopantetheine, and only in this form are they able to form the peptide bond in the subsequent condensation reaction (31). Indeed, we observed that supplementation of GATF1 minimal medium with the 4′-phosphopantetheine precursor (vitamin B5) (Fig. 3A) facilitates the bacterial growth and the initial biosynthesis of GS (absolute yield after 30 h). However, this did not lead to an increase in biomass-dependent GS production. This is because GS is produced only by those cells that have reached a certain age (idiophase), i.e., immediately after the specific growth rate has started to decrease (32). Since 4′-phosphopantetheine not only is necessary for GS biosynthesis but also stimulates many reactions of the primary metabolism, it appears that a significant fraction of the cell population remained in the exponential growth phase, i.e., in the nonproducing tropophase (26). Therefore, despite the large biomass values obtained with the high concentration of vitamin B5, the final yield of GS did not increase significantly, mainly as a result of the lowered specific biosynthetic activity of the whole bacterial population. The transition of A. migulanus cells into the GS-producing idiophase has been shown to be induced by depletion of one or more essential nutrients (33). In the chemically defined minimal medium, phosphorus and sulfur limitation, for instance, led to the highest specific activities of the GS synthetases and enhanced relative GS production (34). All starvation-induced processes in Bacillus species, such as sporulation, competence development, or production of secondary metabolites, are mutually competitive and are regulated at the level of gene expression (35).
Being a secondary or “special” metabolite, which is not essential for bacterial growth but rather needed for environmental survival (33), GS is produced only under specific nutritional conditions and depends strongly on a high level of amino nitrogen in the fermentation medium (36). A record overproduction of GS up to 2.5 g/liter has been previously achieved in the complex amino nitrogen-rich YP medium (20), but this medium is not available in a uniformly 13C/15N-supplemented form. To develop a chemically defined minimal medium, the nutrition components should therefore be carefully evaluated within the limited set of available and affordable isotope-labeled chemicals. For example, A. migulanus cannot grow with glucose as a sole carbon source (37). Therefore, glucose-containing minimal media, which are generally used for uniform labeling, cannot be employed to produce GS. A. migulanus can utilize only fructose, glycerol, or inositol as a single carbon source (22); hence, only these substrates in the isotope-enriched form can be used. In the chemically defined medium with glycerol, the best nitrogen sources are ammonium phosphate, ammonium sulfate (22), or ammonium succinate (21). However, these salts, especially ammonium sulfate, increase the acidity of the medium, which could be a reason for the low GS yields noted earlier (36). Acidification can be compensated for by adjusting the buffer: addition of 12 g/liter Tris-HCl at pH 7.4 into the medium with glycerol and ammonium sulfate supports the growth and increases the yield of GS compared to the Tris-free medium (20). For this reason, we used the GAT (glycerol-ammonium sulfate-Tris) medium as the basal medium for optimizing the cost-effective minimal conditions, lowered here in ammonium sulfate and Tris contents. Since Tris is not consumed in the metabolic reactions, it can be used without isotope enrichment.
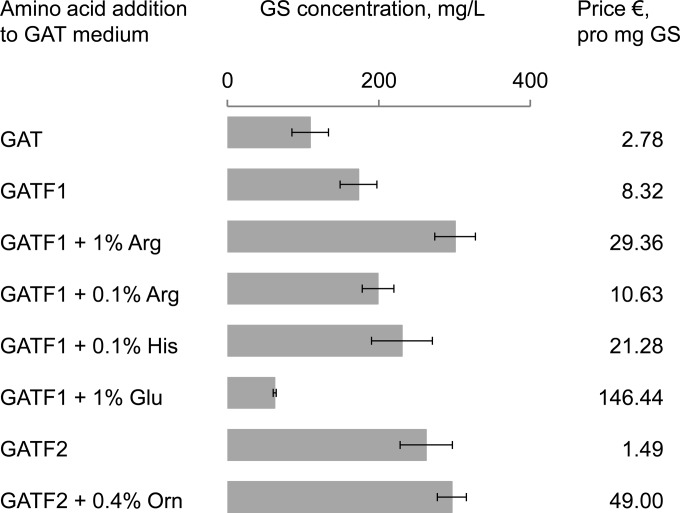
It has been previously demonstrated that GS biosynthesis can be stimulated by the addition of phenylalanine, arginine, histidine, and methionine to the fermentation media, using a chemically defined F4/4 medium (23, 38). In this F4/4 medium, fructose and the four amino acids serve as the sole sources of carbon, nitrogen, and sulfur (23, 24). Since uniformly 13C-labeled fructose was not available at the time when we started our work, we optimized the production of GS within the framework of the minimal glycerol-based GAT medium (see above) and a G4/4 medium, which is identical to F4/4 except that glycerol replaces the fructose (29). Strategically, we supplemented GAT with the same four amino acids as in F4/4, with the exception of methionine, which is needed only as a sulfur source and is therefore redundant in the presence of ammonium sulfate. We were able to show (Fig. 5A and B) that the phenylalanine concentration can be reduced down to 0.025% (1.5 mM) for the highest GS production in minimal GAT basal medium. The addition of arginine to the medium stimulated GS production as expected, presumably due to two effects: a stabilization of the GS synthetase (38) and an induction of the arginase that produces ornithine and proline in A. migulanus (39). A similar degree of stimulation was observed when ornithine was supplemented (Fig. 5D). However, a significant increase in the specific biosynthetic activity occurred only at rather high concentrations (0.4 and 1%, respectively, for ornithine and arginine), which negatively affects the production costs in view of the price for these 13C/15N-labeled amino acids (Fig. 7).
FIG 7.
Typical GS yields and respective mean costs of GS production in various GAT-derived media, supplemented with uniformly 13C/15N-enriched amino acids. The price estimation is based on the catalogue prices for the isotope-labeled amino acids by CortecNet (Voisins-Le-Bretonneux, France).
As mentioned above, the pronounced phenotype instability of the GS producer strains causes major reproducibility problems in fermentation studies. Not only the inoculation material but also some nutrition factors (medium composition) contribute to the dissociation of the GS-producing rough phenotypes into the undesirable smooth ones. For example, supplementation of histidine and arginine stabilizes the smooth variant and should thus lead to a rough-to-smooth dissociation over prolonged cultivation (40). This in turn means that the addition of arginine (or histidine) can have a dual effect on GS biosynthesis: immediately, it should have a positive effect by stimulating the cell-specific biosynthesis, but in the long term it will have a negative effect due to dissociation of the bacterial population. Even without the latter process, an arginine-induced stimulation is not worthwhile when judging the high price of 13C/15N-labeled Arg (Fig. 7). We therefore recommend a simple GATF2 medium (glycerol–ammonium sulfate–Tris–0.025% phenylalanine) as the most cost-effective choice for the biosynthesis of 13C/15N-labeled GS. For instance, compared to the use of standard GAT medium (no amino acids), the savings with GATF2 (using 13C/15N-labeled phenylalanine) amount to about €500 per gram of labeled peptide.
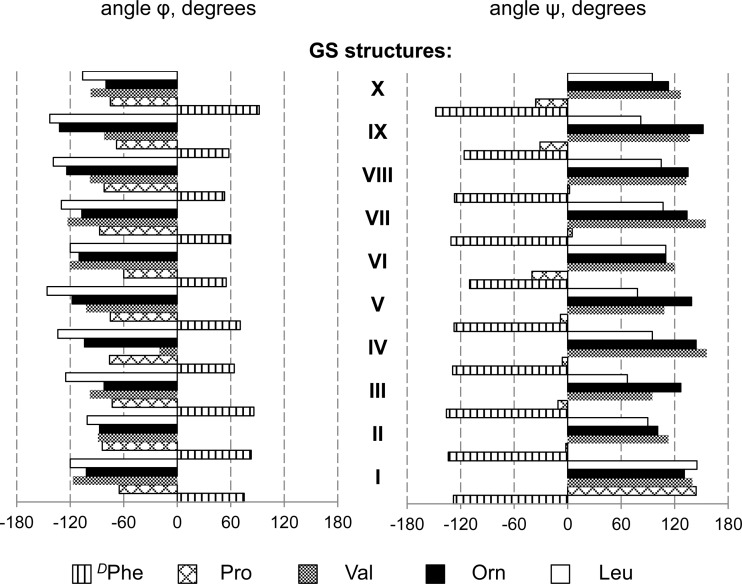
Finally, the solid-state NMR experiments of uniformly 13C/15N-labeled GS in lipid membranes have not only served to prove the quality of the desired product but also delivered a set of preliminary structural data to calculate the functionally relevant membrane-bound conformation of GS. We were able to demonstrate that MAS-NMR investigations are possible in a solid-state mode and to draw several structural conclusions. We achieved almost complete assignment of the GS backbone and of the 13Cα/13Cβ chemical shifts. Since the 13Cα and 13Cβ chemical shifts of each residue depend largely on the local backbone and side chain conformations (41), we were able to cross-compare the existing GS structures (derived from X-ray crystallography, from NMR in solution, and from theoretical studies) with the new data on GS determined here in proper lipid bilayers. Even though the number of structural constraints is limited and additional experiments are needed to obtain a complete atomic-resolution model, it is clear from Fig. 8 that the overall picture of the short double-stranded β-sheet that is flanked by β-turns remains unchanged, as previously obtained in isotropic solutions and in crystals. This similarity suggests that the GS backbone has a considerable conformational rigidity, which implies further that it was justified in three studies (42–44) to assume that the conformation of monomeric GS remains essentially the same upon realignment into an oligomeric β-barrel pore.
FIG 8.
Backbone torsion angles for each of the amino acids in the GS peptide, as determined by various structural methods. GS structures are denoted with Roman numerals: I, GS in 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine (DM-O-PC), as determined in this work by solid-state NMR; II, GS in DMPC, calculated by all-atom MD (15); III, GS in DMSO, calculated by all-atom MD (45); IV, GS in methanol, calculated by all-atom MD in reference 46; V, GS in DMSO, determined by liquid-state NMR (13); VI, GS in CHCl3-methanol, determined by solution state NMR (14); VII, GS in crystal, grown in the presence of urea and HCl (11); VIII, GS in crystal, grown in the presence of TFA and HCl (12); IX, free GS structure (in vacuo) as predicted in reference 9; X, free GS structure (in vacuo) as predicted in reference 10.
Further structural refinement will be possible by evaluating the obtained NMR data in combination with molecular dynamics simulations and 13C chemical shift calculations at the quantum-chemical level of theory and by additional 15N-NMR measurements, but this is within the scope of a separate publication. The current paper is focused on the critical process of producing 13C/15N-labeled GS, whereupon a fermentation procedure was developed. The method allows 100% labeling efficiency and is cost-effective, as was desired for being practically useful. We suggest that similar approaches could be successful for production of isotope-edited material of other nonribosomally synthesized peptides.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00229-15.
REFERENCES
- 1.Gause GF, Brazhnikova MG. 1944. Gramicidin S origin and mode of action. Lancet 244:715–716. doi: 10.1016/S0140-6736(00)88377-4. [DOI] [Google Scholar]
- 2.Salgado J, Grage SL, Kondejewski LH, Hodges RS, McElhaney RN, Ulrich AS. 2001. Membrane-bound structure and alignment of the antimicrobial β-sheet peptide gramicidin S derived from angular and distance constraints by solid state 19F-NMR. J Biomol NMR 21:191–208. doi: 10.1023/A:1012946026231. [DOI] [PubMed] [Google Scholar]
- 3.Polin AN, Egorov NS. 2003. Structural and functional characteristics of gramicidin S in connection with its antibiotic activity. Antibiot Khimioterap 48:29–32. (In Russian.) [PubMed] [Google Scholar]
- 4.Mogi T, Kita K. 2009. Gramicidin S and polymyxins: the revival of cationic cyclic peptide antibiotics. Cell Mol Life Sci 66:3821–3826. doi: 10.1007/s00018-009-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, Gust R, Albada HB, Penkova M, Kramer U, Erdmann R, Metzler-Nolte N, Straus SK, Bremer E, Becher D, Brotz-Oesterhelt H, Sahl HG, Bandow JE. 2014. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A 111:E1409–E1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korolev PN, Bulgakova VG, Polin AN, Korolev NP, Mil'gram VD. 1988. The action of gramicidin S on the ionic permeability of bilayer lipid membranes. Nauchnye Doki Vyss Shkoly Biol Nauki 7:31–35. (In Russian.) [PubMed] [Google Scholar]
- 7.Wu M, Maier E, Benz R, Hancock REW. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Santis P, Liquori AM. 1971. Conformation of gramicidin S. Biopolymers 10:699–710. doi: 10.1002/bip.360100408. [DOI] [PubMed] [Google Scholar]
- 10.Dygert M, Go N, Scheraga HA. 1975. Use of a symmetry condition to compute the conformation of gramicidin S. Macromolecules 8:750–761. doi: 10.1021/ma60048a016. [DOI] [PubMed] [Google Scholar]
- 11.Tishchenko GN, Andrianov VI, Vainstein BK, Woolfson MM, Dodson E. 1997. Channels in the gramicidin S-with-urea structure and their possible relation to transmembrane ion transport. Acta Crystallogr D 53:151–159. doi: 10.1107/S0907444995000916. [DOI] [PubMed] [Google Scholar]
- 12.Llamas-Saiz AL, Grotenbreg GM, Overhand M, van Raaij MJ. 2007. Double-stranded helical twisted β-sheet channels in crystals of gramicidin S grown in the presence of trifluoroacetic and hydrochloric acids. Acta Crystallogr D 63:401–407. doi: 10.1107/S0907444906056435. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Sugar IP, Krishna NR. 1995. A variable target intensity-restrained global optimization (VARTIGO) procedure for determining three-dimensional structures of polypeptides from NOESY data: application to gramicidin S. J Biomol NMR 5:37–48. doi: 10.1007/BF00227468. [DOI] [PubMed] [Google Scholar]
- 14.Ovchinnikov YA, Ivanov VT, Bystrov VF, Miroshnikov AI, Shepel EN, Abdullaev ND, Efremov ES, Senyavina LB. 1970. The conformation of gramicidin S and its N,N′-diacetyl derivative in solutions. Biochem Biophys Res Commun 39:217–225. doi: 10.1016/0006-291X(70)90781-3. [DOI] [PubMed] [Google Scholar]
- 15.Mihailescu D, Smith JC. 2000. Atomic detail peptide-membrane interactions: molecular dynamics simulation of gramicidin S in a DMPC bilayer. Biophys J 79:1718–1730. doi: 10.1016/S0006-3495(00)76424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finking R, Marahiel MA. 2004. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol 58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 17.Hoyer KM, Mahlert C, Marahiel MA. 2007. The iterative gramicidin S thioesterase catalyzes peptide ligation and cyclization. Chem Biol 14:13–22. doi: 10.1016/j.chembiol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berditsch M, Afonin S, Ulrich AS. 2007. The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl Environ Microbiol 73:6620–6628. doi: 10.1128/AEM.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal SM, Roy A, Mahata D, Migliolo L, Nolasco DO, Franco OL. 2014. Functional and structural insights on self-assembled nanofiber-based novel antibacterial ointment from antimicrobial peptides, bacitracin and gramicidin S. J Antibiot 67:771–775. doi: 10.1038/ja.2014.70. [DOI] [PubMed] [Google Scholar]
- 20.Matteo CC, Glade M, Tanaka A, Piret J, Demain AL. 1975. Microbiological studies on the formation of gramicidin S synthetases. Biotechnol Bioeng 42:129–142. [Google Scholar]
- 21.Korshunov VV, Egorov NS. 1962. A synthetic medium for the development of Bacillus brevis var. GB and production of gramicidin C. Mikrobiologiia 31:515–519. [PubMed] [Google Scholar]
- 22.Vandamme EJ, Demain AL. 1976. Nutrition of Bacillus brevis ATCC 9999, producer of gramicidin S. Antimicrob Agents Chemother 10:265–273. doi: 10.1128/AAC.10.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demain AL, Matteo CC. 1976. Phenylalanine stimulation of gramicidin S formation. Antimicrob Agents Chemother 9:1000–1003. doi: 10.1128/AAC.9.6.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JHD, Yang L, Demain AL. 1984. Further studies on the role of phenylalanine in gramicidin S biosynthesis by Bacillus brevis. J Biotechnol 1:81–94. doi: 10.1016/0168-1656(84)90011-7. [DOI] [Google Scholar]
- 25.Mootz HD, Finking R, Marahiel MA. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem 276:37289–37298. doi: 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- 26.Martin JF, Demain AL. 1980. Control of antibiotic biosynthesis. Microbiol Rev 44:230–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto K, Fujita R, Kato Y, Asahara M, Yokota A. 2004. Reclassification of Brevibacillus brevis strains NCIMB 13288 and DSM 6472 (=NRRL NRS-887) as Aneurinibacillus danicus sp. nov. and Brevibacillus limnophilus sp. nov. Int J Syst Evol Microbiol 54:419–427. doi: 10.1099/ijs.0.02906-0. [DOI] [PubMed] [Google Scholar]
- 28.Saito F, Hori K, Kanda M, Kurotsu T, Saito Y. 1994. Entire nucleotide sequence for Bacillus brevis Nagano Grs2 gene encoding gramicidin S synthetase 2: a multifunctional peptide synthetase. J Biochem 116:357–367. [DOI] [PubMed] [Google Scholar]
- 29.Fang A, Pierson DL, Mishra SK, Koenig DW, Demain AL. 1997. Gramicidin S production by Bacillus brevis in simulated microgravity. Curr Microbiol 34:199–204. doi: 10.1007/s002849900168. [DOI] [PubMed] [Google Scholar]
- 30.Kratzschmar J, Krause M, Marahiel MA. 1989. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol 171:5422–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mootz HD, Marahiel MA. 1997. Biosynthetic systems for nonribosomal peptide antibiotic assembly. Curr Opin Chem Biol 1:543–551. doi: 10.1016/S1367-5931(97)80051-8. [DOI] [PubMed] [Google Scholar]
- 32.Lykov VP, Bodnar IV, Khovrychev MP, Polin AN. 1986. Biosynthesis of the antibiotic gramicidin S by a Bacillus brevis culture associated with a change in growth kinetics. Mikrobiologiia 55:792–795. [PubMed] [Google Scholar]
- 33.Katz E, Demain AL. 1977. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol Rev 41:449–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matteo CC, Cooney CL, Demain AL. 1976. Production of gramicidin S synthetases by Bacillus brevis in continuous culture. J Gen Microbiol 96:415–422. doi: 10.1099/00221287-96-2-415. [DOI] [PubMed] [Google Scholar]
- 35.Marahiel MA, Nakano MM, Zuber P. 1993. Regulation of peptide antibiotic production in Bacillus. Mol Microbiol 7:631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 36.Udalova TP, Fedorova RI. 1965. The effect of various nutrient compounds upon gramicidin formation in Bacillus brevis var. GB. Mikrobiologiia 34:631–635. [PubMed] [Google Scholar]
- 37.Takagi H, Shida O, Kadowaki K, Komagata K, Udaka S. 1993. Characterization of Bacillus brevis with descriptions of Bacillus migulanus sp. nov., Bacillus choshinensis sp. nov., Bacillus parabrevis sp. nov., and Bacillus galactophilus sp. nov. Int J Syst Bacteriol 43:221–231. doi: 10.1099/00207713-43-2-221. [DOI] [PubMed] [Google Scholar]
- 38.Poirier A, Demain AL. 1981. Arginine regulation of gramicidin S biosynthesis. Antimicrob Agents Chemother 20:508–514. doi: 10.1128/AAC.20.4.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanda M, Ohgishi K, Hanawa T, Saito Y. 1997. Arginase of Bacillus brevis Nagano: purification, properties, and implication in gramicidin S biosynthesis. Arch Biochem Biophys 344:37–42. doi: 10.1006/abbi.1997.0174. [DOI] [PubMed] [Google Scholar]
- 40.Zharikova GG, Lukin AA. 1966. Effect of some factors on dissociation in Bacillus brevis var. GB. Mikrobiologiia 35:100–106. (In Russian.) [PubMed] [Google Scholar]
- 41.Vila JA, Baldoni HA, Scheraga HA. 2009. Performance of density functional models to reproduce observed 13Cα chemical shifts of proteins in solution. J Comput Chem 30:884–892. doi: 10.1002/jcc.21105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonin S, Dürr UHN, Wadhwani P, Salgado J, Ulrich AS. 2008. Solid state NMR structure analysis of the antimicrobial peptide gramicidin S in lipid membranes: concentration-dependent re-alignment and self-assembly as a β-barrel. Top Curr Chem 273:139–154. doi: 10.1007/128_2007_20. [DOI] [PubMed] [Google Scholar]
- 43.Afonin S, Glaser RW, Sachse C, Salgado J, Wadhwani P, Ulrich AS. 2014. 19F NMR screening of unrelated antimicrobial peptides shows that membrane interactions are largely governed by lipids. Biochim Biophys Acta 1838:2260–2268. doi: 10.1016/j.bbamem.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Koch K, Afonin S, Ieronimo M, Berditsch M, Ulrich AS. 2012. Solid-state 19F-NMR of peptides in native membranes. Top Curr Chem 306:89–118. doi: 10.1007/128_2011_162. [DOI] [PubMed] [Google Scholar]
- 45.Mihailescu D, Smith JC. 1999. Molecular dynamics simulation of the cyclic decapeptide antibiotic, gramicidin S, in dimethyl sulfoxide solution. J Phys Chem B 103:1586–1594. doi: 10.1021/jp983674t. [DOI] [Google Scholar]
- 46.Babii O, Afonin S, Berditsch M, Reibetaer S, Mykhailiuk PK, Kubyshkin VS, Steinbrecher T, Ulrich AS, Komarov IV. 2014. Controlling biological activity with light: diarylethene-containing cyclic peptidomimetics. Angew Chem Int Ed 53:3392–3395. doi: 10.1002/anie.201310019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.