Abstract
Flies may act as potential vectors for the spread of resistant bacteria to different environments. This study was intended to evaluate the presence of Escherichia coli strains resistant to cephalosporins in flies captured in the areas surrounding five broiler farms. Phenotypic and molecular characterization of the resistant population was performed by different methods: MIC determination, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and phylotyping. The presence of extended-spectrum beta-lactamase (ESBL) genes, their plasmid location, and the mobile genetic elements involved in their mobilization were studied. Additionally, the presence of 35 genes associated with virulence was evaluated. Out of 682 flies captured, 42 yielded ESBL-producing E. coli. Of these isolates, 23 contained blaCTX-M-1, 18 contained blaCTX-M-14, and 1 contained blaCTX-M-9. ESBL genes were associated mainly with the presence of the IncI1 and IncFIB replicons. Additionally, all the strains were multiresistant, and five of them also harbored qnrS. Identical PFGE profiles were found for E. coli isolates obtained from flies at different sampling times, indicating a persistence of the same clones in the farm environment over months. According to their virulence genes, 81% of the isolates were considered avian-pathogenic E. coli (APEC) and 29% were considered extraintestinal pathogenic E. coli (ExPEC). The entrance of flies into broiler houses constitutes a considerable risk for colonization of broilers with multidrug-resistant E. coli. ESBLs in flies reflect the contamination status of the farm environment. Additionally, this study demonstrates the potential contribution of flies to the dissemination of virulence and resistance genes into different ecological niches.
INTRODUCTION
Escherichia coli is a commensal bacterium commonly found in nature and in the lower intestine of warm-blooded organisms. However, some serotypes can cause enteric and extraintestinal infections in humans and animals (1). For instance, avian-pathogenic E. coli (APEC) is the major cause of colibacillosis in poultry production. It is a syndrome that causes respiratory infections associated with airsacculitis, pericarditis, and septicemia, resulting in a large economical burden for the poultry industry due to the loss of production and mortality (2). Hybridization studies detected APEC-specific DNA sequences presenting a high level of homology with DNA sequences of human extraintestinal pathogenic E. coli (ExPEC) strains (3). Both types have virulence determinants in common, suggesting that APEC could serve as a reservoir and a source of virulence for ExPEC (4). Moreover, it has been suggested that APEC strains are potential zoonotic pathogens (5).
Antimicrobials are the common treatment for avian colibacillosis caused by APEC. During the last years, increased resistance to frontline antimicrobials, such as fluoroquinolones and third-generation cephalosporins, has been reported for E. coli isolates from poultry (1). Additionally, the emergence of bacteria resistant to critically important antimicrobials is a major concern in human and veterinary medicine. The presence of isolates producing extended-spectrum beta-lactamases (ESBLs) and plasmid-mediated AmpC beta-lactamases among E. coli isolates from broilers has increased substantially in the last decade (6). Generally, the genes encoding beta-lactamases are located on plasmids, which can be transferred to other bacteria (7).
The persistence of E. coli in the environment has been described in the literature (8). However, the relevance of wildlife vectors in the persistence and spread of ESBL-producing E. coli in the farm environment has not been studied thoroughly (9, 10). In particular, flies are one of the most important vectors of human diseases worldwide (11). They have the capacity to horizontally transfer pathogens from different environments (12), posing a high risk to human health (13). Due to their movements, their capacity to fly long distances, and their attraction to decaying organic materials and food, houseflies amplify the risk of human exposure to foodborne pathogens (14–16). Moreover, they may also spread antibiotic resistance genes within microbial communities (17). The digestive tract of flies provides a suitable environment for the horizontal transfer of genes among bacteria, which contributes to the spread of resistance and virulence genes (18). Several studies have demonstrated that flies carry multidrug-resistant bacteria in hospital environments and have also demonstrated their role in the transmission of human pathogens within hospitals (17).
The objective of this study was to assess the potential contribution of flies to the spread of ESBL/AmpC-producing E. coli in the farm environment. For this purpose, isolates, resistant genes, and mobile genetic elements involved in the transmission of resistant genes were fully characterized. The genomic relationship among strains and the virulence content associated with APEC and ExPEC strains were also evaluated.
MATERIALS AND METHODS
Study design.
The study was conducted in five broiler farms (farms 1 to 5) each one with one or two houses, located in the Catalonia region (Spain). Broiler house capacities ranged from 15,000 to 46,000 birds per house. Only farm 2 presented cats at the premises. Minimum and maximum distances between farms were 15 km and 200 km, respectively. From May to November 2012, each broiler farm was visited twice per rearing cycle to capture flies (6 to 8 visits per farm in total). Overall, 23 broiler flocks were reared in the 5 study houses during the study period. Fly sampling was performed when chickens were ∼14 and 28 days old. At each visit, up to 50 flies were collected, always outside the same broiler houses (within a 10-m periphery). Overall, 682 flies were captured individually, placed into disposable sterile plastic bags, and transported refrigerated (to be kept alive) to the laboratory. Once at the laboratory, flies were anesthetized with CO2, identified to the genus or species level, and subsequently processed for Campylobacter isolation (our unpublished data) and thereafter for cephalosporin-resistant E. coli isolation. Each individual fly was aseptically macerated in plastic bags with 2.5 ml Bolton broth (CM0983 with selective supplement [SR0183] and laked horse blood [SR0048]; Oxoid, Basingstoke, United Kingdom) and incubated at 42°C for 24 h in 2-ml screw-cap tubes. A 10-μl loop of broth was plated onto MacConkey agar (Oxoid, Basingstoke, United Kingdom) supplemented with ceftriaxone (1 mg/liter). Three lactose-positive colonies from each plate were selected and confirmed to be E. coli by PCR (19). Subsequently, one representative was selected for further studies.
Phylogeny, pulsed-field gel electrophoresis, and multilocus sequence typing.
Isolates were discriminated into phylogenetic groups by PCR (phylogroup A, B1, B2, C, D, E, or F), as previously described by Clermont et al. (20, 21).
Pulsed-field gel electrophoresis (PFGE) was performed to analyze the genomic relatedness among E. coli isolates. PFGE of chromosomal DNA digested with the restriction enzyme XbaI was carried out according to PulseNet protocols (22). Salmonella enterica serovar Braenderup H9812 was used as a size marker. The results were analyzed by Fingerprinting II Informatix software (Applied Maths, Sint-Martens-Latem, Belgium). Isolates were considered to have a different pattern when at least one band difference was detected. The analysis of the bands generated was performed by using the Dice coefficient and unweighted-pair group method with arithmetic averages (optimization of 1.25% and position tolerance of 1.25%).
Multilocus sequence typing (MLST) was performed to determine the potential evolutionary relatedness between isolates. MLST was carried out by gene amplification and sequencing of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA), according to protocols and primers specified on the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli) and as previously described (23). Sequences were analyzed with Vector NTI Advance 11 (InforMax, Inc., Bethesda, MD). The allelic profiles of the gene sequences and the sequence types (STs) were obtained via the electronic database at the E. coli MLST website.
Antimicrobial susceptibility testing.
Disc diffusion was performed according to CLSI guidelines, using the following discs (Oxoid, Basingstoke, United Kingdom): cefoxitin at 30 mg; cefepime at 30 mg; ceftazidime at 30 mg; cefotaxime at 30 mg; cefotaxime-clavulanic acid at 30 and 10 mg, respectively; and ceftazidime-clavulanic acid at 30 and 10 mg, respectively. The synergies between cefotaxime and cefotaxime-clavulanic acid and between ceftazidime and ceftazidime-clavulanic acid were used as suggestive evidence of ESBL production; cefoxitin was used for the detection of AmpC-type beta-lactamase (24). Additionally, all isolates were tested for antimicrobial susceptibility using a MIC-based broth microdilution (VetMIC GN-mo; National Veterinary Institute, Uppsala, Sweden) for the following antimicrobial agents: ampicillin (1 to 128 mg/liter), cefotaxime (0.016 to 2 mg/liter), ceftazidime (0.25 to 16 mg/liter), nalidixic acid (1 to 128 mg/liter), ciprofloxacin (0.008 to 1 mg/liter), gentamicin (0.12 to 16 mg/liter), streptomycin (2 to 256 mg/liter), kanamycin (8 to 16 mg/liter), chloramphenicol (2 to 64 mg/liter), florfenicol (4 to 32 mg/liter), trimethoprim (1 to 128 mg/liter), sulfamethoxazole (8 to 1,024 mg/liter), tetracycline (1 to 128 mg/liter), and colistin (0.5 to 4 mg/liter). E. coli ATCC 25922 was used as a control strain. Isolates were considered to be wild-type (WT) or non-WT isolates based on epidemiological cutoff values according to EUCAST guidelines (http://www.eucast.org/) (25).
Resistance genes.
All strains were tested by PCR for the presence of the blaCTX-M, blaSHV, blaTEM, and blaCMY-2 genes as described previously by Hasman et al. (26). Sequencing of both strands of amplicons was performed. The presence of the aac(6′)-Ib-cr, qnrA, qnrB, qnrS, qepA, and oqxAB genes conferring resistance to fluoroquinolones was also assessed (27).
Plasmid DNA analysis.
One isolate from each PFGE clonal cluster was selected for plasmid characterization. The presence of plasmid replicons HI1, HI2, I1, X, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIA, and K was assessed by PCR-based replicon typing methods described previously (28, 29), including screening for the IncR replicon (30). The detection of plasmids and sizing were carried out on all the isolates by PFGE of total DNA digested with S1 nuclease (31). Restriction fragments from S1-PFGE gels (i.e., PFGE gels digested with S1 nuclease) were transferred onto positively charged nylon membranes and hybridized with specific probes for blaCTX-M-1 and blaCTX-M-9 and with specific probes for each previously identified replicon.
Detection of virulence-associated genes.
All 42 strains were tested for a pool of 35 virulence-associated genes (see Table 2), including 7 adhesins, 4 siderophores, 9 toxins, 8 capsule synthesis-associated genes or protectins, and 7 miscellaneous genes, by PCR using primers described previously (32, 33). The five virulence factors for ExPEC detection (pap, sfa/foc, afa/dra, iutA, and kpsM II) (34) together with the five potential APEC virulence genes (iutA, hlyF, iss, iroN, and ompT) (35) were included in the PCR analysis. Virulence scores were calculated as the sum of all virulence genes detected for each isolate; pap, sfa/foc, clbB-clbN, and kpsM II were counted only once, regardless of the number of elements or subunits identified (maximum possible score of 27).
TABLE 2.
Distribution of virulence genes among the 42 isolates, the largest phylogenetic groups, and relevant ST clonal complexesb
| Virulence gene or E. coli type | Product(s) | No. (%) of isolates |
P valuea for phylogroup A/ST10 clonal complex vs phylogroup A/non-ST10 clonal complex | |||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 42) | Phylogroup A (n = 15) | Phylogroup B1 (n = 18) | Phylogroup C (n = 7) | Phylogroup A/ST10 clonal complex (n = 8) | Phylogroup A/non-ST10 clonal complex (n = 7) | |||
| Virulence genes | ||||||||
| Adhesins | ||||||||
| fimH | d-Mannose-specific adhesin of type 1 fimbriae | 42 (100) | 15 (100) | 18 (100) | 7 (100) | 8 (100) | 7 (100) | |
| papEF | Pilus associated with pyelonephritis (P fimbriae) | 11 (26) | 5 (33) | 4 (22) | 0 | 5 (63) | 0 | 0.0256 |
| papG | P fimbriae carrying Gal(α1-4) Gal-specific PapG adhesin at its distal end | 3 (7) | 0 | 2 (11) | 0 | 0 | 0 | |
| papA | Major structural subunit of the P fimbrial shaft | 3 (7) | 0 | 3 (17) | 0 | 0 | 0 | |
| papC | Pilus assembly; central region of the pap operon | 4 (10) | 0 | 3 (17) | 0 | 0 | 0 | |
| sfa/focDE | Central region of the sfa and foc operons | 1 (2) | 0 | 1 (6) | 0 | 0 | 0 | |
| afa/draBC | Dr antigen-specific adhesin operons (AFA, Dr, F1845) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Toxins | ||||||||
| cnf1 | Cytotoxic necrotizing factor 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| cdtB | Cytolethal distending toxin | 0 | 0 | 0 | 0 | 0 | 0 | |
| sat | Secreted autotransporter | 0 | 0 | 0 | 0 | 0 | 0 | |
| hlyD | Alpha-hemolysin | 0 | 0 | 0 | 0 | 0 | 0 | |
| hlyF | Hemolysin F | 37 (88) | 12 (80) | 16 (89) | 7 (100) | 8 (100) | 4 (57) | |
| astA | Enteroaggregative E. coli heat-stable toxin (EAST1) | 12 (29) | 7 (47) | 4 (22) | 0 | 7 (86) | 0 | 0.0014 |
| tsh | Temp-sensitive hemagglutinin-serine protease | 12 (29) | 2 (13) | 4 (22) | 6 (86) | 2 (25) | 0 | |
| stx1 | Shiga toxin 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stx2 | Shiga toxin 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Siderophores | ||||||||
| fyuA | Yersinia siderophore receptor (ferric yersiniabactin uptake) | 6 (14) | 0 | 3 (17) | 3 (43) | 0 | 0 | |
| iutA | Ferric aerobactin receptor (iron uptake; transport) | 32 (76) | 9 (60) | 15 (83) | 6 (86) | 8 (100) | 1 (14) | 0.0014 |
| iroN | Novel catecholate siderophore receptor | 33 (79) | 9 (60) | 15 (83) | 7 (100) | 8 (100) | 1 (14) | 0.0014 |
| ireA | Iron-regulated element (novel siderophore receptor) | 6 (14) | 0 | 5 (28) | 0 | 0 | 0 | |
| Protectins | ||||||||
| kpsM II | Group II capsule | 2 (5) | 1 (7) | 0 | 0 | 0 | 1 (14) | |
| kpsM II-K2 | K2 subgroup II capsule | 1 (2) | 1 (7) | 0 | 0 | 0 | 1 (14) | |
| kpsM II-K5 | K5 subgroup II capsule | 1 (2) | 0 | 1 (6) | 0 | 0 | 0 | |
| kpsM II-K1 | K1 subgroup II capsule | 0 | 0 | 0 | 0 | 0 | 0 | |
| kpsM III | Group III capsule | 10 (24) | 7 (47) | 2 (11) | 0 | 7 (86) | 0 | 0.0014 |
| cvaC | Colicin V from serum resistance-associated plasmids | 20 (48) | 1 (7) | 12 (67) | 6 (86) | 1 (13) | 0 | |
| iss | Increased serum survival (outer membrane protein) | 36 (86) | 10 (67) | 17 (94) | 7 (100) | 8 (100) | 2 (29) | 0.0070 |
| traT | Surface exclusion, serum survival (outer membrane protein) | 37 (88) | 12 (80) | 17 (94) | 6 (86) | 8 (100) | 4 (57) | |
| Miscellaneous | ||||||||
| ompT | Outer membrane protein (protease) T | 37 (88) | 12 (80) | 16 (89) | 7 (100) | 8 (100) | 4 (57) | |
| ibeA | Invasion of brain endothelium | 1 (2) | 1 (7) | 0 | 0 | 0 | 1 (14) | |
| malX | Pathogenicity-associated island marker | 1 (2) | 1 (7) | 0 | 0 | 1 (13) | 0 | |
| usp | Uropathogenic-specific protein (bacteriocin) | 1 (2) | 1 (7) | 0 | 0 | 0 | 1 (14) | |
| clbB | Hybrid peptide-polyketide synthase (colibactin) | 32 (76) | 12 (80) | 13 (72) | 6 (86) | 6 (75) | 6 (86) | |
| clbN | Nonribosomal synthetase (colibactin) | 0 | 0 | 0 | 0 | 0 | 0 | |
| fliCH7 | H7 flagellin variant | 1 (2) | 0 | 1 (6) | 0 | 0 | 0 | |
| E. coli types | ||||||||
| APEC | 34 (81) | 9 (60) | 17 (94) | 7 (100) | 8 (100) | 1 (14) | 0.0014 | |
| ExPEC | 12 (29) | 6 (40) | 4 (22) | 0 | 5 (63) | 1 (14) | ||
P values (by Fisher's exact test) are shown where the P value was <0.05.
The mean virulence scores (number of virulence genes detected), adjusted for multiple detections of the pap, sfa/foc, clbB-clbN, and kpsM II operons, were 8.8 (range, 1 to 12) for the total number of isolates and 7.8 (1 to 12) for phylogroup A, 9.1 (4 to 12) for phylogroup B1, 9.7 (6 to 11) for phylogroup C, 10.6 (10 to 12) for phylogroup A/ST10 clonal complex, and 4.6 (1 to 7) for phylogroup A/non-ST10 clonal complex isolates. Virulence scores were compared by use of the Mann-Whitney U test (P < 0.0001).
Statistical analysis.
Differences in the prevalences of phylogroups and STs between the distinct groups were determined by Fisher's exact test. The associations between groups were assessed by calculation of the odds ratio (OR) with 95% confidence intervals (CIs). The null hypothesis was rejected for data with P values of <0.05. Statistical analyses were performed by using GraphPad Prism, version 3.1, software (GraphPad Software, Inc., San Diego, CA). Virulence scores were compared by the Mann-Whitney U test.
RESULTS
During the course of the study, a total of 682 flies were captured from the environments surrounding five different broiler farms. The 42 ESBL producers were collected from farm 1 (9%; n = 193), farm 2 (3%; n = 138), farm 3 (15%; n = 109), and farm 4 (4%; n = 134). Finally, all flies collected from farm 5 (n = 108) were negative for ESBL-producing E. coli. Most of the fly species were classified as Musca domestica (n = 615), followed by Ophyra spp. (n = 33), Stomoxys calcitrans (n = 15), Muscina stabulans (n = 7), Fannia canicularis (n = 6), and others (n = 6). A total of 42 ESBL-producing E. coli strains were isolated mainly from M. domestica (n = 41), and 1 was isolated from Muscina stabulans.
PFGE, phylogeny, and MLST.
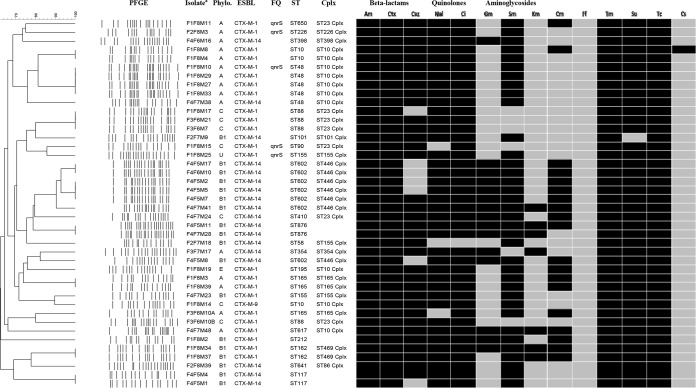
XbaI PFGE analysis revealed a total of 29 different PFGE restriction profiles among the 42 E. coli isolates (Fig. 1). The number of fragments generated ranged from 14 to 21, and their sizes varied from 20 to 1,135 kb. In almost all cases, epidemiologically related isolates belonged to the same farm, except for three isolates (F1F8M17, F3F6M21, and F3F6M7) from farms 1 and 3 that presented identical fingerprints (Fig. 1).
FIG 1.
PFGE dendrogram illustrating the phenotypic and genotypic relationships of the strains, phylogenies, and cephalosporin resistance genes. PFGE, pulsed-field gel electrophoresis; Phylo., phylogroup; ESBL, extended-spectrum beta-lactamase gene; FQ, fluoroquinolone resistance genes; ST, sequence type; Cplx, clonal complex; U, unknown; Am, ampicillin (WT, ≤8 mg/liter); Ctx, cefotaxime (WT, ≤0.25 mg/liter); Caz, ceftazidime (WT, ≤0.5 mg/liter); Nal, nalidixic acid (WT, ≤16 mg/liter); Ci, ciprofloxacin (WT, ≤0.064 mg/liter); Gm, gentamicin (WT, ≤2 mg/liter); Sm, streptomycin (WT, ≤16 mg/liter); Km, kanamycin (WT, ≤8 mg/liter); Cm, chloramphenicol (WT, ≤16 mg/liter); Ff, florfenicol (WT, ≤16 mg/liter); Tm, trimethoprim (WT, ≤2 mg/liter); Su, sulfamethoxazole (WT, ≤64 mg/liter); Tc, tetracycline (WT, ≤8 mg/liter); Cs, colistin (WT, ≤2 mg/liter). a, isolates were named based on the numbers assigned to the farm (F), flock (F), and fly (M).
Four different phylogroups were represented among the 42 isolates. Of these, 15 were of group A (36%), 18 were of group B1 (43%), 7 were of group C (17%), 1 was of group E (2%), and 1 was of an unknown group (2%) (Fig. 1).
MLST analyses identified 21 STs belonging to 11 different clonal complexes (Fig. 1). The most common clonal complex was the ST10 clonal complex (n = 10), containing four different ST types (ST10, ST48, ST195, and ST617), followed by the ST446 clonal complex (n = 7; all ST602) and the ST23 clonal complex (n = 7), comprising four different ST types (ST88, ST90, ST410, and ST650). Only two different complexes were represented by more than one phylogenetic group (phylogroups A and E for ST10 and phylogroups C and A for ST23).
Antimicrobial susceptibility testing and resistance genes.
Disc diffusion demonstrated that all isolates presented the ESBL phenotype. All the strains were multiresistant (resistant to >3 antimicrobial families). Furthermore, 79% of the isolates had a non-WT phenotype for more than eight antimicrobials. MIC determination confirmed that all strains had a non-WT phenotype for cephalosporins (100% resistance to cefotaxime and 83% resistance to ceftazidime), with 23 isolates yielding amplicons for blaCTX-M-1, 18 yielding amplicons for blaCTX-M-14, and 1 yielding amplicons for blaCTX-M-9. Out of the 42 isolates, 33 harbored the blaTEM gene. None of the isolates were positive for blaSHV or blaCMY-2. In addition, 93% of the isolates had a non-WT phenotype for nalidixic acid, and 98% had a non-WT phenotype for ciprofloxacin. The presence of qnrS genes was detected in five isolates obtained from farms 1 and 2. The genes aac(6′)-Ib-cr, qnrA, qnrB, qepA, and oqxAB were not found in this collection. Additionally, 100% of the strains had a non-WT phenotype for ampicillin, trimethoprim, and tetracycline; 98% had a non-WT phenotype for sulfamethoxazole; 86% had a non-WT phenotype for streptomycin; 45% had a non-WT phenotype for chloramphenicol; 36% had a non-WT phenotype for gentamicin; 17% had a non-WT phenotype for kanamycin; and 2% had a non-WT phenotype for colistin. All isolates had a non-WT phenotype for florfenicol.
Localization of blaCTX-M.
The replicons IncFIB and IncI1 were detected in the majority of the isolates (90% and 83%, respectively), being associated or not with any of the CTX-M genes. However, IncP, IncK, IncY, Inc FIA, IncHI1, IncH12, and IncN were also detected (Table 1).
TABLE 1.
Identification and characterization of the plasmid locations of blaCTX-M-1, blaCTX-M-14, and blaCTX-M-9 among 29 CTX-M-producing E. coli isolatese
| Gene and isolatea | ST | Clonal complex | Replicon(s)b | Plasmidc | Inc type(s)d | Plasmid size (kb) |
|---|---|---|---|---|---|---|
| blaCTX-M-1 | ||||||
| F1F8M8 | 10 | 10 | I1, FIB | pST10-1 | I1 | 110 |
| F1F8M10 | 48 | 10 | I1, FIB | pST48-1 | I1 | 110 |
| F1F8M17 | 88 | 23 | I1, FIB, P | pST88 | I1 | 110 |
| F1F8M15 | 90 | 23 | I1, FIB | pST90 | I1, FIB | 120 |
| F1F8M25 | 155 | 155 | I1, FIB, P | pST155-1 | I1, FIB | 120 |
| pST155-2 | I1, FIB | 190 | ||||
| F1F8M34 | 162 | 469 | I1, FIB | pST162 | I1 | 110 |
| F1F8M3 | 165 | 165 | I1, FIB, P | pST165-1 | I1 | 110 |
| F1F8M19 | 195 | 10 | I1, FIB, P, K | pST195 | I1 | 110 |
| F1F8M2 | 212 | I1, FIB, Y, P | pST212 | I1 | 110 | |
| F1F8M11 | 650 | 23 | I1, FIB, P | pST650 | I1 | 110 |
| F3F6M10B | 88 | 23 | I1, FIB, P | pST88 | I1 | 110 |
| F3F6M10A | 165 | 165 | I1 | pST165-2 | I1 | 120 |
| F4F7M23 | 155 | 155 | I1, FIB | pST155-3 | I1, FIB | 110 |
| F4F7M48 | 617 | 10 | I1, FIA, FIB | pST617-1 | 50 | |
| pST617-2 | 300 | |||||
| F2F8M3 | 226 | 226 | pST226 | 50 | ||
| blaCTX-M-14 | ||||||
| F4F7M38 | 48 | 10 | I1, FIB | pST48-2 | I1 | 90 |
| F4F5M4 | 117 | FIB | pST117 | FIB | 145 | |
| F4F6M16 | 398 | 398 | FIB | pST398 | 75 | |
| F4F7M24 | 410 | 23 | HI1, HI2, I1, FIB, P | pST410 | I1 | 110 |
| F4F5M8 | 602 | 446 | HI1, HI2, I1, FIB, P | pST602-1 | I1 | 110 |
| F4F5M2 | 602 | 446 | HI1, HI2, I1, FIB, P | pST602-1 | I1 | 110 |
| F4F5M17 | 602 | 446 | HI1, HI2, I1, FIB, P | pST602-1 | I1 | 110 |
| F4F7M41 | 602 | 446 | HI1, HI2, I1, FIB, P | pST602-2 | I1 | 90 |
| F4F5M11 | 876 | HI1, HI2, I1, FIB, P | pST876 | 90 | ||
| F2F7M18 | 58 | 155 | I1, FIB | pST58 | 90 | |
| F2F7M9 | 101 | 101 | FIB | pST101 | 90 | |
| F2F8M39 | 641 | 86 | I1, N | pST641 | I1 | 100 |
| F3F7M17 | 354 | 354 | FIA, FIB | pST602-2 | FIB | 90 |
| blaCTX-M-9 | ||||||
| F1F8M14 | 10 | 10 | I1, FIB, P | pST10-2 | I1, FIB, P | 120 |
Isolates were named based on the numbers assigned to the farm (F), flock (F), and fly (M).
Replicon identifications are based on positive amplifications from the PCR-based replicon typing method.
Plasmids were named based on the source strain sequence type and plasmid size.
In all E. coli isolates, replicons from plasmids containing the different bla genes were identified by PCR-positive amplification and by Southern hybridization of the S1-digested fragments.
One representative for each PFGE cluster is shown. p(ST number), plasmid location; Inc, identified replicon.
All blaCTX-M-1 isolates hybridized with a plasmid of ∼110 kb containing an IncI1 replicon, except for the following exceptions: two isolates (F1F8M15 and F3F7M23) contained both IncI1 and IncFIB in a 120-kb plasmid, and one extra isolate (F1F8M25) contained IncI1 together with IncFIB in two plasmids of 120 and 190 kb (Table 1). Additionally, isolate F4F7M48 carried a second copy of the gene on a large plasmid of 300 kb (Table 1). The IncI1 and IncFIB replicons were identified on blaCTX-M-14-carrying plasmids of different sizes. The isolate carrying blaCTX-M-9 exhibited three different replicons (IncI1, IncFIB, and IncP) on the same plasmid (Table 1).
Detection of virulence genes.
The prevalences of 35 virulence-associated genes, including the genes associated with APEC and ExPEC, are illustrated in Table 2. The virulence genes detected with the highest prevalences were fimH (100%), traT (88%), clbB (76%), and cvaC (48%). The presence of astA (29%), tsh (29%), papEF (26%), and kpsM III (24%) was confirmed for an intermediate percentage of the isolates. In contrast, the presence of fyuA (14%), ireA (14%), papC (10%), papA (7%), papG (7%), kpsM II (5%), sfa/focDE (2%), kpsM II-K2 (2%), kpsM II-K5 (2%), ibeA (2%), malX (2%), usp (2%), and fliCH7 (2%) was confirmed for a lower number of strains. None of the isolates were positive for afa/draBC, cnf1, cdtB, sat, hlyD, stx1, stx2, kpsM II-K1, and clbN. A total of 12 (29%) isolates from this study were identified as ExPEC according to the ExPEC definition. Additionally, 79%, 88%, 88%, 76%, and 86% of the strains yielded amplicons for iroN, ompT, hlyF, iutA, and iss, respectively; these genes have been described as the minimal predictors of APEC virulence. A total of 34 (81%) isolates were considered APEC, since they harbored between 4 and 5 of these genes. Moreover, 11 (26%) of the isolates were considered ExPEC and APEC at the same time.
Statistical analysis.
No significant differences in the numbers of virulence genes found between phylogroups were observed. Phylogroups A, B1, and C exhibited virulence scores of between 7.8 and 9.7 (Table 2). Phylogroup C/ST23 clonal complex (mean, 9.8; range, 6 to 11), phylogroup A/ST10 clonal complex (mean, 10.6; range, 10 to 12), and phylogroup B1/ST446 complex (mean, 9.6; range, 9 to 12) isolates exhibited similar virulence scores but different gene contents (data not shown). Significant differences in virulence scores between phylogroup A/ST10 clonal complex isolates and phylogroup A/non-ST10 clonal complex isolates were observed (P < 0.0001). The virulence factors that were significantly different were characteristic of APEC (iss, iutA, iroN, and astA) and ExPEC (papEF and kpsM III) (Table 2).
DISCUSSION
M. domestica is an arthropod distributed worldwide and the most abundant fly species in animal production and food at homes and restaurants. Flies are suspected reservoirs and vectors for human and animal pathogens due to their contact with animal manure, food, and humans. They can pick up bacteria present in those sites and transport them to the kitchen (36). Some studies have suggested that flies can also play an important role in the dissemination of antimicrobial resistance genes within the bacterial community (37, 38). In our study, the presence of multidrug-resistant E. coli isolated from flies, and in particular ESBL-producing E. coli, demonstrates the capacity of houseflies to disseminate and transport resistance genes located in mobile genetic elements. Additionally, five of the isolates also harbored plasmid-mediated quinolone resistance genes. qnrS genes were previously associated with the same plasmids harboring ESBL genes (39). The continuous increase in the prevalence of antimicrobial-resistant bacteria has been associated with the use of these drugs to treat human and animal infections, and the presence of ESBL-producing E. coli in flies suggests that animals and the farm environment are colonized and inhabited by these microorganisms. Flies are a reservoir of resistant bacteria and can contribute to the spread of resistance genes between different ecological niches.
Some studies have suggested that there is a relationship between different E. coli phylogenetic groups and the virulence capabilities of the strains (38, 40). Commensal isolates belong mainly to phylogenetic groups A, B1, and C (41, 42). In contrast, the most virulent phylogroups described in the literature are phylogroup B2 followed by phylogroup D, which are mainly responsible for extraintestinal infections (38, 40). None of the ESBL-producing isolates from this study belonged to the B2 and D phylogroups; most of them belonged to phylogroups A, B1, and C and possessed quite high virulence scores. Furthermore, the ESBL-producing E. coli A/ST10 clonal complex isolates from this study had significantly higher virulence scores than isolates of other STs from the same phylogroup. Similar results were obtained in other studies, where phylogroup A/ST10 isolates of APEC and ExPEC origins were described as emerging pathogens, suggesting that this ST complex may have relevant zoonotic potential (43, 44).
PFGE results demonstrated the same clonal groups in the same farms, suggesting dissemination of epidemiologically related clones within farm environments. An exception was the three strains from farm 1 and 3 belonging to the same PFGE. These farms were about 25 km apart. This observation would reinforce what has been previously reported, that flies can travel long distances, spreading resistant bacteria (14–16). Additionally, identical fingerprints have been recovered from different flies belonging to the same farm at different time points, including different broiler cycles, demonstrating the capacity of these bacteria to survive and persist in the environment for long periods of time.
This study also demonstrated the presence of E. coli isolates with virulence-associated genes characteristic of both APEC (81%) and ExPEC (29%) and the capacity of flies to transport them. Some of these virulence genes are also associated with mobile genetic elements, highlighting the relevance of flies in the transmission of virulence determinants in broiler farms and hospital settings (17).
In the present study, we found that blaCTX-M-1 and blaCTX-M-14 are the most prevalent ESBL genes detected in E. coli isolates obtained from flies captured in the areas surrounding broiler farms (55% and 43%, respectively). This result is in agreement with data from previous studies, which demonstrated that blaCTX-M-1 is one of the most prevalent ESBL genes detected in Enterobacteriaceae of broiler origin (45–48). Also in line with studies performed on broiler farms, the most common replicons encountered in this study were IncI1 and IncFIB (49, 50). Moreover, we have found five isolates with the same ESBL gene harbored in two different plasmids. Having two or more copies of a resistance mechanism in different locations would ensure the maintenance and persistence of these genes even if selective pressure enforces the loss of one of these copies.
In conclusion, this study has demonstrated a very diverse population of multidrug-resistant E. coli recovered from flies at different broiler farms. ESBL-producing E. coli in flies reflects the colonization status of the farm environment. Flies are probably not the source but the result of the colonization of animals. These isolates contained a high number of virulence-associated genes and ESBL genes, which could be easily introduced and disseminated into farms through the flies and subsequently could potentially colonize animals. Additional biosecurity measures, aimed at blocking or reducing the entrance of flies into broiler houses, should be undertaken. Otherwise, zoonosis control and antimicrobial resistance reduction may be frustrated. Flies are also contributing to pathogen evolution since the transfer of resistance- and virulence-associated genes between different strains could be facilitated through flies.
ACKNOWLEDGMENTS
This work was partially supported by projects AGL2011-28836 from the Ministerio de Economia y Competitividad of Spain, RTA2009-00117-00-00 from INIA (Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Spain), and CamCon (Campylobacter Control—Novel Approaches in Primary Poultry Production; funded by the European Community's Seventh Framework Programme, FP7/2007-2013, under grant agreement no. 244547).
We acknowledge the support received from farmers and poultry companies participating in the study as well as Marta Verdun and Sandra Talavera (CReSA) for their technical assistance in the identification of flies to the species level.
M.S.-G. is a Ph.D. student registered with the Universidad Autónoma de Barcelona.
REFERENCES
- 1.Yang H, Chen S, White DG, Zhao S, McDermott P, Walker R, Meng J. 2004. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J Clin Microbiol 42:3483–3489. doi: 10.1128/JCM.42.8.3483-3489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schouler C, Schaeffer B, Bree A, Mora A, Dahbi G, Biet F, Oswald E, Mainil J, Blanco J, Moulin-Schouleur M. 2012. Diagnostic strategy for identifying avian pathogenic Escherichia coli based on four patterns of virulence genes. J Clin Microbiol 50:1673–1678. doi: 10.1128/JCM.05057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Campos TA, Lago JC, Nakazato G, Stehling EG, Brocchi M, de Castro FP, da Silveira WD. 2008. Occurrence of virulence-related sequences and phylogenetic analysis of commensal and pathogenic avian Escherichia coli strain (APEC). Pesq Vet Bras 28:533–540. doi: 10.1590/S0100-736X2008001000015. [DOI] [Google Scholar]
- 4.Mokady D, Gophna U, Ron EZ. 2005. Virulence factors of septicemic Escherichia coli strains. Int J Med Microbiol 295:455–462. doi: 10.1016/j.ijmm.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Ewers C, Janssen T, Kiessling S, Philipp HC, Wieler LH. 2004. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet Microbiol 104:91–101. doi: 10.1016/j.vetmic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. 2008. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect 14(Suppl 1):144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 7.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 8.Brennan FP, Abram F, Chinalia FA, Richards KG, O'Flaherty V. 2010. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl Environ Microbiol 76:2175–2180. doi: 10.1128/AEM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaak H, Hamidjaja RA, van Hoek AH, de Heer L, de Roda Husman AM, Schets FM. 2014. Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl Environ Microbiol 80:239–246. doi: 10.1128/AEM.02616-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usui M, Iwasa T, Fukuda A, Sato T, Okubo T, Tamura Y. 2013. The role of flies in spreading the extended-spectrum beta-lactamase gene from cattle. Microb Drug Resist 19:415–420. doi: 10.1089/mdr.2012.0251. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg B. 1973. Flies and disease, vol 2 Biology and disease transmission. Princeton University Press, Princeton, NJ. [Google Scholar]
- 12.Wales AD, Carrique-Mas JJ, Rankin M, Bell B, Thind BB, Davies RH. 2010. Review of the carriage of zoonotic bacteria by arthropods, with special reference to Salmonella in mites, flies and litter beetles. Zoonoses Public Health 57:299–314. doi: 10.1111/j.1863-2378.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- 13.Olsen AR. 1998. Regulatory action criteria for filth and other extraneous materials. III. Review of flies and foodborne enteric disease. Regul Toxicol Pharmacol 28:199–211. [DOI] [PubMed] [Google Scholar]
- 14.Barreiro C, Albano H, Silva J, Teixeira P. 2013. Role of flies as vectors of foodborne pathogens in rural areas. ISRN Microbiol 2013:718780. doi: 10.1155/2013/718780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murvosh CM, Thaggard CW. 1966. Ecological studies of the house fly. Ann Entomol Soc Am 59:533–547. doi: 10.1093/aesa/59.3.533. [DOI] [PubMed] [Google Scholar]
- 16.Nazni WA, Luke H, Wan Rozita WM, Abdullah AG, Sa'diyah I, Azahari AH, Zamree I, Tan SB, Lee HL, Sofian MA. 2005. Determination of the flight range and dispersal of the house fly, Musca domestica (L) using mark release recapture technique. Trop Biomed 22:53–61. [PubMed] [Google Scholar]
- 17.Boulesteix G, Le Dantec P, Chevalier B, Dieng M, Niang B, Diatta B. 2005. Role of Musca domestica in the transmission of multiresistant bacteria in the centres of intensive care setting in sub-Saharan Africa. Ann Fr Anesth Reanim 24:361–365. (In French.) doi: 10.1016/j.annfar.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Petridis M, Bagdasarian M, Waldor MK, Walker E. 2006. Horizontal transfer of Shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J Med Entomol 43:288–295. doi: 10.1093/jmedent/43.2.288. [DOI] [PubMed] [Google Scholar]
- 19.Heininger A, Binder M, Schmidt S, Unertl K, Botzenhart K, Doring G. 1999. PCR and blood culture for detection of Escherichia coli bacteremia in rats. J Clin Microbiol 37:2479–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 22.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 23.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI. 2008. Clinical and Laboratory Standards Institute performance standards for antimicrobial susceptibility testing: eighteenth informational supplement M100-S18. CLSI, Wayne, PA. [Google Scholar]
- 25.Schwarz S, Silley P, Simjee S, Woodford N, van Duijkeren E, Johnson AP, Gaastra W. 2010. Assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother 65:601–604. doi: 10.1093/jac/dkq037. [DOI] [PubMed] [Google Scholar]
- 26.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother 56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 27.Coelho A, Mirelis B, Alonso-Tarrés C, Nieves Larrosa M, Miró E, Cliville Abad R, Bartolomé RM, Castaner M, Prats G, Johnson JR, Navarro F, González-López JJ. 2009. Detection of three stable genetic clones of CTX-M-15-producing Klebsiella pneumoniae in the Barcelona metropolitan area, Spain. J Antimicrob Chemother 64:862–864. doi: 10.1093/jac/dkp264. [DOI] [PubMed] [Google Scholar]
- 28.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 30.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 31.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JR, O'Bryan TT, Low DA, Ling G, Delavari P, Fasching C, Russo TA, Carlino U, Stell AL. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun 68:3327–3336. doi: 10.1128/IAI.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol 46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forster M, Sievert K, Messler S, Klimpel S, Pfeffer K. 2009. Comprehensive study on the occurrence and distribution of pathogenic microorganisms carried by synanthropic flies caught at different rural locations in Germany. J Med Entomol 46:1164–1166. doi: 10.1603/033.046.0526. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Yang Y, Zhao F, Fan X, Zhong W, Qiao D, Cao Y. 2013. Multi-drug resistant gram-negative enteric bacteria isolated from flies at Chengdu Airport, China. Southeast Asian J Trop Med Public Health 44:988–996. [PubMed] [Google Scholar]
- 38.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 39.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 40.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Merens A, Mitanchez D, Vu-Thien H. 2010. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum beta-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J Clin Microbiol 48:2459–2463. doi: 10.1128/JCM.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol 11:654–662. doi: 10.1016/j.meegid.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Dissanayake DR, Octavia S, Lan R. 2014. Population structure and virulence content of avian pathogenic Escherichia coli isolated from outbreaks in Sri Lanka. Vet Microbiol 168:403–412. doi: 10.1016/j.vetmic.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 44.Maluta RP, Logue CM, Casas MR, Meng T, Guastalli EA, Rojas TC, Montelli AC, Sadatsune T, de Carvalho Ramos M, Nolan LK, da Silveira WD. 2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. doi: 10.1371/journal.pone.0105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Accogli M, Fortini D, Giufre M, Graziani C, Dolejska M, Carattoli A, Cerquetti M. 2013. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin Microbiol Infect 19:E238–E240. doi: 10.1111/1469-0691.12128. [DOI] [PubMed] [Google Scholar]
- 46.Belmar Campos C, Fenner I, Wiese N, Lensing C, Christner M, Rohde H, Aepfelbacher M, Fenner T, Hentschke M. 2014. Prevalence and genotypes of extended spectrum beta-lactamases in Enterobacteriaceae isolated from human stool and chicken meat in Hamburg, Germany. Int J Med Microbiol 304:678–684. doi: 10.1016/j.ijmm.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Ben Sallem R, Ben Slama K, Rojo-Bezares B, Porres-Osante N, Jouini A, Klibi N, Boudabous A, Saenz Y, Torres C. 2014. IncI1 plasmids carrying blaCTX-M-1 or blaCMY-2 genes in Escherichia coli from healthy humans and animals in Tunisia. Microb Drug Resist 20:495–500. doi: 10.1089/mdr.2013.0224. [DOI] [PubMed] [Google Scholar]
- 48.Zurfluh K, Wang J, Klumpp J, Nuesch-Inderbinen M, Fanning S, Stephan R. 2014. Vertical transmission of highly similar bla CTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol 5:519. doi: 10.3389/fmicb.2014.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bortolaia V, Guardabassi L, Trevisani M, Bisgaard M, Venturi L, Bojesen AM. 2010. High diversity of extended-spectrum beta-lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob Agents Chemother 54:1623–1626. doi: 10.1128/AAC.01361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Stephan R, Karczmarczyk M, Yan Q, Hachler H, Fanning S. 2013. Molecular characterization of bla ESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol 4:188. doi: 10.3389/fmicb.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]