Abstract
Lantibiotics are potent antimicrobial peptides characterized by the presence of dehydrated amino acids, dehydroalanine and dehydrobutyrine, and (methyl)lanthionine rings. In addition to these posttranslational modifications, some lantibiotics exhibit additional modifications that usually confer increased biological activity or stability on the peptide. LtnJ is a reductase responsible for the introduction of d-alanine in the lantibiotic lacticin 3147. The conversion of l-serine into d-alanine requires dehydroalanine as the substrate, which is produced in vivo by the dehydration of serine by a lantibiotic dehydratase, i.e., LanB or LanM. In this work, we probe the substrate specificity of LtnJ using a system that combines the nisin modification machinery (dehydratase, cyclase, and transporter) and the stereospecific reductase LtnJ in Lactococcus lactis. We also describe an improvement in the production yield of this system by inserting a putative attenuator from the nisin biosynthesis gene cluster in front of the ltnJ gene. In order to clarify the sequence selectivity of LtnJ, peptides composed of truncated nisin and different mutated C-terminal tails were designed and coexpressed with LtnJ and the nisin biosynthetic machinery. In these tails, serine was flanked by diverse amino acids to determine the influence of the surrounding residues in the reaction. LtnJ successfully hydrogenated peptides when hydrophobic residues (Leu, Ile, Phe, and Ala) were flanking the intermediate dehydroalanine, while those in which dehydroalanine was flanked by one or two polar residues (Ser, Thr, Glu, Lys, and Asn) or Gly were either less prone to be modified by LtnJ or not modified at all. Moreover, our results showed that dehydrobutyrine cannot serve as a substrate for LtnJ.
INTRODUCTION
Since the discovery of penicillin by Alexander Fleming in 1928, antibiotics have saved the lives of countless people. Regrettably, due to abuse and overuse, increasing resistance to antibiotics has been found among pathogenic bacteria, which has led to an urgent need for new antimicrobial compounds (1–4). Lanthipeptides, defined as posttranslationally modified peptides containing a lanthionine and/or methyllanthionine ring(s), are a type of ribosomal peptides produced by many Gram-positive bacteria (5, 6). They can be subdivided into 4 different classes based on the enzyme(s) that catalyze(s) the formation of lanthionine residues (6). Some of them (i.e., classes I and II) show antimicrobial activity and are referred to as lantibiotics (7).
The capability of lantibiotics to inhibit the growth of obstinate pathogens, including multidrug-resistant bacteria such as methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and oxacillin-resistant Gram-positive organisms, makes them very promising candidates for future antimicrobial development (7–9). So far, only a few lantibiotics have been commercially applied or are under development for medical use in spite of their promising properties (7, 10). Nisin, the model class I lantibiotic produced by the Gram-positive bacterium Lactococcus lactis, has been applied in industry as a food preservative for decades without triggering effective resistance in pathogens (11, 12); duramycin, a class II lantibiotic, is in phase II clinical trials and was proven to be safe and effective for the symptomatic treatment of cystic fibrosis by inhalation (13). Another class II lantibiotic, deoxyactagardine B (NVB302; Novacta Biosystems Limited), is undergoing a phase I clinical trial as a drug candidate for the treatment of Clostridium difficile infections (14). Furthermore, recent research has shown that some class III lanthipeptides have unexpected bioactivity to relieve neuropathic pain (15) and as antiviral compounds (16).
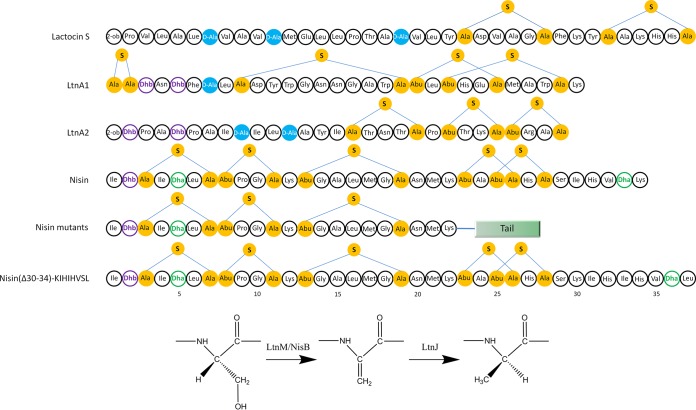
Besides the common (methyl)lanthionine, more than 15 extra structures have been unveiled in lanthipeptides playing a significant role in antimicrobial activity, resistance against proteases, and/or physicochemical resistance (12, 17–20). For instance, d-alanine was found within two lantibiotics: lactocin S and the two-component lantibiotic lacticin 3147 (Fig. 1) (21–23). According to the work of Cotter and his coworkers, the replacements of d-alanine by other residues (l-alanine, l-threonine, glycine, and l-valine) in lacticin 3147 caused a dramatic decrease in activity against L. lactis HP (24). Additionally, a gene designated ltnJ was predicted to encode a protein with significant similarity to zinc-dependent alcohol dehydrogenases and NAD(P)H-dependent quinone oxidoreductases of the zinc-containing alcohol dehydrogenase superfamily (24). LtnJ was shown to be responsible for the formation of d-alanines in lacticin 3147 with dehydroalanine (Dha) as an intermediate (24). This dehydroalanine reductase activity to generate d-alanine has been observed among some homologues of LtnJ, like SacJ from S. aureus C55 and PenN from Pediococcus pentosaceus FBB61 (25). In fact, d-alanine in lacticin 3147 can be formed by some homologues of LtnJ, although low efficiency was observed compared to LtnJ (25).
FIG 1.
Structures of lactocin S, lacticin 3147 (peptides LtnA1 and LtnA2), nisin, nisin with designed C-terminal tail, and nisin(insS29KIH; K34L). The residues involved in the formation of (methyl)lanthionine are in yellow, d-alanines are depicted in blue, and Dha and Dhb are colored green and purple, respectively. The pathway for d-alanine conversion from serine is depicted. 2-ob, 2-oxobutyryl group.
It has been shown that the nisin biosynthetic machinery possesses high substrate tolerance and can modify diverse peptides fused to the nisin leader peptide, not only those related to lanthipeptides but also unrelated peptide sequences (26–30). Previously, we successfully introduced d-alanine into nisin by expressing LtnJ together with the nisin biosynthetic machinery in a dual-plasmid system in L. lactis (18). The Dha at position 5 of nisin has been demonstrated to be modified by LtnJ, while the other Dha, at position 33, is most likely not (18).
In this study, nisin was used as a model peptide to investigate if the surrounding residues of Dha affect the conversion of Dha into d-alanine. Therefore, a mutant oligopeptide tail (AAIS26LALTIK) was fused at the C terminus of truncated nisin [i.e., NisA(Δ23–34)], generating a peptide designated NisAtail (Fig. 1). Another 18 variants were designed to probe the influence of flanking residues in the conversion of Dha into d-alanine by LtnJ. We show that the polar or hydrophobic nature of flanking amino acids in the substrate plays a vital role in this reaction.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and vectors used in this work are listed in Table 1. L. lactis strains were cultured in M17 medium supplemented with 0.5% glucose (GM17) at 30°C for genetic manipulation or in a minimal expression medium (MEM) for protein expression and purification assays (29). Chloramphenicol and/or erythromycin was used at 5 μg/ml when necessary.
TABLE 1.
Strains and vectors used in this worka
| Strain or vector | Characteristics | Reference |
|---|---|---|
| Strains | ||
| L. lactis NZ9000 | pepN::nisRK; expression host strain | 41 |
| L. lactis NZ9700 | Nisin-producing transconjugant containing Tn5276; used for cloning of the attenuator | 42 |
| Vectors | ||
| pIL3EryBTC | Eryr nisBTC; modification and transport of lantibiotics | 18 |
| pNZ-nisA | Cmr nisA; expression of nisin | 18 |
| pNZ-nisA-ltnJ | Cmr nisA ltnJ; expression of nisin and LtnJ | 18 |
| pNZ-nisA-T-ltnJ | Derivative of pNZ-nisA-ltnJ; attenuator between nisA and ltnJ | This work |
| pNZ-nisAtail | Derivative of pNZ-nisA; ΔnisA; NisAtail | This work |
| pNZ-S26insS | Derivative of pNZ-nisA; ΔnisA; S26insS | This work |
| pNZ-S26insSS | Derivative of pNZ-nisA; ΔnisA; S26insSS | This work |
| pNZ-S26insT | Derivative of pNZ-nisA; ΔnisA; S26insT | This work |
| pNZ-L27K | Derivative of pNZ-nisA; ΔnisA; L27K | This work |
| pNZ-I25K | Derivative of pNZ-nisA; ΔnisA; I25K | This work |
| pNZ-L27E | Derivative of pNZ-nisA; ΔnisA; L27E | This work |
| pNZ-I25E | Derivative of pNZ-nisA; ΔnisA; I25E | This work |
| pNZ-L27F | Derivative of pNZ-nisA; ΔnisA; L27F | This work |
| pNZ-I25F | Derivative of pNZ-nisA; ΔnisA; I25F | This work |
| pNZ-L27N | Derivative of pNZ-nisA; ΔnisA; L27N | This work |
| pNZ-I25N | Derivative of pNZ-nisA; ΔnisA; I25N | This work |
| pNZ-L27G | Derivative of pNZ-nisA; ΔnisA; L27G | This work |
| pNZ-I25G-L27G | Derivative of pNZ-nisA; ΔnisA; I25G-L27G | This work |
| pNZ-I25G | Derivative of pNZ-nisA; ΔnisA; I25G | This work |
| pNZ-L27A | Derivative of pNZ-nisA; ΔnisA; L27A | This work |
| pNZ-I25A-L27A | Derivative of pNZ-nisA; ΔnisA; I25A-L27A | This work |
| pNZ-I25A | Derivative of pNZ-nisA; ΔnisA; I25A | This work |
| pNZ-S26T | Derivative of pNZ-nisA; ΔnisA; S26T | This work |
| pNZ-nisin(Δ30–34)-KIHIHVSL | Derivative of pNZ-nisA; ΔnisA; nisin(Δ30–34)-KIHIHVSL | This work |
| pNZ-nisAtail-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; NisAtail | This work |
| pNZ-S26insS-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; S26insS | This work |
| pNZ-S26insSS-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; S26insSS | This work |
| pNZ-S26insT-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; S26insT | This work |
| pNZ-L27K-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27K | This work |
| pNZ-I25K-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25K | This work |
| pNZ-L27E-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27E | This work |
| pNZ-I25E-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25E | This work |
| pNZ-L27F-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27F | This work |
| pNZ-I25F-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25F | This work |
| pNZ-L27N-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27N | This work |
| pNZ-I25N-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25N | This work |
| pNZ-L27G-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27G | This work |
| pNZ-I25G-L27G-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25G-L27G | This work |
| pNZ-I25G-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25G | This work |
| pNZ-L27A-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; L27A | This work |
| pNZ-I25A-L27A-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25A-L27A | This work |
| pNZ-I25A-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; I25A | This work |
| pNZ-S26T-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; S26T | This work |
| pNZ-nisin(Δ30–34)-KIHIHVSL-T-ltnJ | Derivative of pNZ-nisA-T-ltnJ; ΔnisA; nisin(Δ30–34)-KIHIHVSL | This work |
Cmr, chloramphenicol resistance; Eryr, erythromycin resistance.
Molecular cloning.
Molecular cloning techniques were performed as described by Sambrook and Russell (31). Preparation of competent cells and transformation were performed as described previously (32). Fast digest restriction enzymes and ligase were supplied by Fermentas and used according to the manufacturer's instructions.
Construction of recombinant vectors.
Plasmid isolation was performed with the plasmid DNA extraction kit (Roche). The transcriptional attenuator region between nisA and nisB was amplified from genomic DNA of L. lactis NZ9700 using primers P-for-nisA-T and P-rev-nisA-T-2nd. After digestion using BglII and KpnI, the region was ligated into pNZ-nisA-ltnJ, which was amplified using the primers P-for-kpnI-ltnJ and P-rev-bglII-Cmr to insert the KpnI site, resulting in the plasmid pNZ-nisA-T-ltnJ. Primers P-for-ALTIK and P-rev(01–19) (Table 2) were designed for the construction of the 19 nisin analogues (Table 3; Fig. 1). Round PCR was performed with pNZ-nisA as the template, primer P-for-ALTIK as the forward primer, and P-rev(01–19) as the reverse primer to create 19 different derivatives of pNZ-nisA (Table 1). These resulting pNZ-nisA derivatives were digested by HindIII and XhoI and ligated with a fragment containing the transcriptional attenuator and ltnJ to generate 19 different derivatives of pNZ-nisA-T-ltnJ (Table 1). This fragment was amplified with the primers P-for-HindIII-T-ltnJ and P-ltnJ-rev-XhoI (Table 2) using pNZ-nisA-T-ltnJ (18). By applying two sets of round PCR on pNZ-nisA with two pairs of primers, P-for-K34L/P-rev-K34L and P-for-S29insKIH/P-rev-S29insKIH (Table 2), pNZ-nisin(Δ30–34)-KIHIHVSL was created (Table 1). Similarly, pNZ-nisin(Δ30–34)-KIHIHVSL-T-ltnJ was obtained with pNZ-nisA-T-ltnJ as the template (Table 1).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Characteristic/function |
|---|---|---|
| P-for-nisA-T | GGAAGATCTAGTCTTATAACTATAC | BglII cleavage site |
| P-rev-nisA-T-2nd | CGGGGTACCTGTTTTTTCCTCTC | KpnI cleavage site |
| P-for-KpnI-ltnJ | CGGGGTACCCTGTAAGGAGAAAAATTATG | KpnI cleavage site |
| P-rev-BglII-Cmr | GGAAGATCTTGGAGCTGTAATATAAAAAC | BglII cleavage site |
| P-for-HindIII-T-ltnJ | CCCAAGCTTGTAAGCAAATAACCAAATC | HindIII cleavage site |
| P-ltnJ-rev-XhoI | CCGCTCGAGTTAGTGGTGGTGGTGGTGGTGTGTATCATAAGAAGTATCATATCTC | XhoI cleavage site |
| P-for-ALTIK | GCGTTAACAATTAAATAAGCTTTCTTTGAACC | General forward primer; 5′ phosphorylation |
| P-rev01 | AAGAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of NisAtail |
| P-rev02 | AAGAGAAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of S26insS |
| P-rev03 | AAGAGAAGAAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of S26insSS |
| P-rev04 | AAGTGTAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of S26insT |
| P-rev05 | TTTAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27K |
| P-rev06 | AAGAGATTTAGCTGCTTTCATGTTACAACCCATC | Construction of I25K |
| P-rev07 | TTCAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27E |
| P-rev08 | AAGAGATTCAGCTGCTTTCATGTTACAACCCATC | Construction of I25E |
| P-rev09 | AAAAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27F |
| P-rev10 | AAGAGAAAAAGCTGCTTTCATGTTACAACCCATC | Construction of I25F |
| P-rev11 | ATTAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27N |
| P-rev12 | AAGAGAATTAGCTGCTTTCATGTTACAACCCATC | Construction of I25N |
| P-rev13 | ACCAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27G |
| P-rev14 | ACCAGAACCAGCTGCTTTCATGTTACAACCCATC | Construction of I25G-L27G |
| P-rev15 | AAGAGAACCAGCTGCTTTCATGTTACAACCCATC | Construction of I25G |
| P-rev16 | TGCAGATATAGCTGCTTTCATGTTACAACCCATC | Construction of L27A |
| P-rev17 | TGCAGATGCAGCTGCTTTCATGTTACAACCCATC | Construction of I25A-L27A |
| P-rev18 | AAGAGATGCAGCTGCTTTCATGTTACAACCCATC | Construction of I25A |
| P-rev19 | AAGTGTTATAGCTGCTTTCATGTTACAACCCATC | Construction of S26T |
| P-for-K34L | TTTTAAGCTTTCTTTGAACC | Construction of nisin(Δ30–34)-KIHIHVSL |
| P-rev-K34L | GCTTACGTGAATACTACAATG | Construction of nisin(Δ30–34)-KIHIHVSL; 5′ phosphorylation |
| P-for-S29insKIH | AAAATTCACATTCACGTAAGCTTTTAAGC | Construction of nisin(Δ30–34)-KIHIHVSL |
| P-rev-S29insKIH | ACTACAATGACAAGTTGCTG | Construction of nisin(Δ30–34)-KIHIHVSL; 5′ phosphorylation |
Restriction sites engineered in the primers are underlined.
TABLE 3.
Core peptide sequences of NisA mutant peptidesa
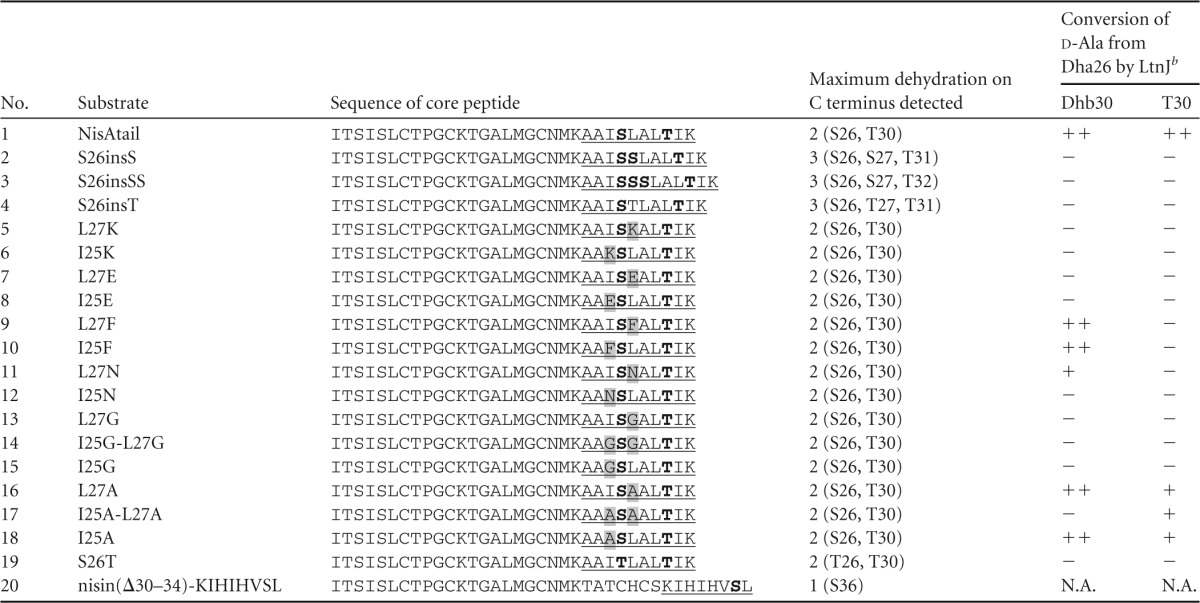
C-terminal extensions in nisin are underlined, with the dehydratable residues serine and threonine in bold. The replacements of I25 and L27 flanking serine residues are shaded.
Symbols: ++, >50%; +, 10 to 50%; −, no conversion. N.A., not applicable.
All the constructs were verified by sequencing.
Protein expression and purification.
The pNZ-derivative vectors containing the mutant structural gene with or without ltnJ were transformed into NZ9000(pIL3EryBTC). The expression assays were performed as described previously (29). Briefly, MEM was inoculated at 2% from an overnight culture of the producer strain grown in GM17. When the fresh culture reached an optical density (OD; 600 nm) of 0.4 to 0.6, nisin was added at a final concentration of 5 ng/ml. Cells were harvested after 2 h of induction by centrifugation at 4°C for 10 min at 5,000 rpm, and the supernatant was kept for the isolation of the peptides. The cell-free supernatant was mixed at a 1:1 ratio with a 100 mM lactic acid solution and applied to a 5-ml HiTrap SP-Sepharose (GE Healthcare) column for cationic exchange chromatography. Bound peptides were washed with 50 mM lactic acid (pH 4.0) and eluted with 50 mM lactic acid, 1 M NaCl, pH 4.0. Subsequently, a PD-10 desalting column (GE Healthcare) was used to desalt the sample according to the provider's instructions. The production was evaluated by Tricine-SDS-PAGE as described before (33).
The peptides were purified to homogeneity by reverse-phase high-performance liquid chromatography (RP-HPLC). A Jupiter 4-μm Proteo 90-Å column, 250 by 4.6 mm (Phenomenex), was used. Solvents for RP-HPLC were solvent A (0.1% trifluoroacetic acid [TFA] in water) and solvent B (0.1% TFA in acetonitrile). Following a 10-min washing step with 20% solvent B, a gradient of 27.5 to 47.5% solvent B over 35 min was executed at a flow rate of 1 ml/min. Peptides were detected by measuring the absorbance at 205 nm. The production levels of the different mutants were assessed by determining the areas of the peaks.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) and LC-MS.
The PD-10-desalted peptide was digested with 1 μl of 1-mg/ml sequencing-grade trypsin in 5 mM CaCl2 and 50 mM Tris (pH 6.8) solution at 37°C for 16 h.
After trypsin digestion, the proteolytic mix was injected into an Ultimate 3000 nano-LC-MS/MS system (Dionex, Amsterdam, The Netherlands) in line connected to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The sample mixture was loaded on a trapping column (Acclaim PepMap; C18; 5-mm length by 300-μm inside diameter [i.d.]; 5-μm particle size; 100-Å porosity; Dionex) and washed. After 3 min, the mixture was separated using a 42-min linear gradient from 95% solvent C (0.1% formic acid) to 90% solvent D (0.1% formic acid in acetonitrile) at a flow rate of 250 nl/min. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the five most abundant doubly and triply charged ions with a minimal signal of 2,500 in a given MS spectrum. Full-scan MS spectra were acquired from m/z 300 to 1,300 in the Orbitrap spectrometer at a target value of 1E6 with a resolution of 60,000. The five most intense ions which met the set criteria were then isolated for fragmentation in the linear ion trap, with a dynamic exclusion of 10 s. Peptides were fragmented after filling the ion trap at a target value of 1E4 ion counts. Data were analyzed using Peak6 software (Bioinformatics Solutions Inc., Waterloo, Ontario, Canada).
The intact peptides were analyzed using the same setup with the following modifications: the column temperature was 50°C, and after a 5-min wash, a 44-min gradient from 10% to 40% of solvent D at a flow rate of 300 nl/min was performed. The mass range was set to 400 to 1,900 Da. Data were processed using Xtract software (Thermo). Arbitrary abundance of peptide fragments of interest was calculated on the basis of proportional relationship between the area of the chromatographic peak and the corresponding peptide. The conversion rate of LtnJ was obtained by dividing the arbitrary abundance of tail peptide containing d-alanine-26 by the summed arbitrary abundance of tail peptides containing d-alanine-26 and Dha26, respectively.
RESULTS
Improvement of the production of nisin and its derivatives.
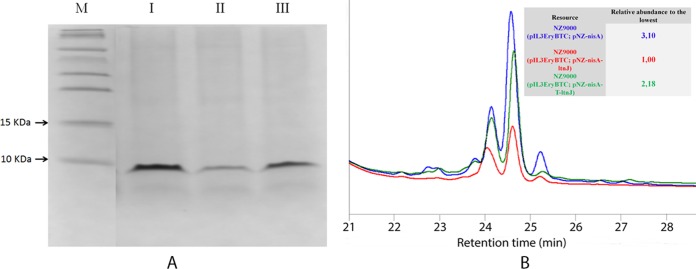
The production level of nisin decreased 3-fold when coexpressed with LtnJ using L. lactis NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ) as a host strain compared to use of the strain L. lactis NZ9000(pIL3EryBTC, pNZ-nisA) (Fig. 2). An attenuator structure located downstream of the nisA gene in the nisin biosynthesis cluster (11) was cloned into pNZ-nisA-ltnJ between the genes nisA and ltnJ, resulting in pNZ-nisA-T-ltnJ. This regulatory element ensures high transcription of the structural gene but a reduced transcription level of ltnJ from the nisA promoter. The peptides produced in the culture supernatant of NZ9000(pIL3EryBTC, pNZ-nisA), NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ), and NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ) were purified by cationic exchange chromatography. Tricine-SDS-PAGE was used to visualize the semipurified peptide. The peptides from the three strains migrated as a band of approximately 6 kDa, which is in line with the theoretic mass of the nisin precursor (Fig. 2A). Peptides from NZ9000(pIL3EryBTC, pNZ-nisA), NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ), and NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ) were further purified via RP-HPLC. An intense peak with a retention time of 24.6 min was observed in all the strains (Fig. 2B). The purified peptide from NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ) was collected for a more accurate characterization by LC-MS. We could observe a peak corresponding to a peptide with a positive mass shift of 2 Da compared to the peptide from NZ9000(pIL3EryBTC, pNZ-nisA), indicating that one Dha was converted into d-alanine by LtnJ (see Fig. S1 in the supplemental material). These results are consistent with the previous results when the attenuator was not present in the construction (18). Comparing the areas under the curves of the different peaks from nisin and its derivatives on the HPLC profile, we observed that the level of nisin produced by NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ) was 2-fold higher than that of NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ), although it was still lower than the level of nisin without the addition of LtnJ to the system (Fig. 2B).
FIG 2.
(A) Tricine-SDS-PAGE analysis after cationic exchange chromatographic purification of nisin with/without d-alanine. M, molecular mass marker; I, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA); II, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ); III, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ). (B) HPLC profile of nisin with/without d-alanine. Blue, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA); red, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA-ltnJ); green, protein purified from NZ9000(pIL3EryBTC, pNZ-nisA-T-ltnJ).
LtnJ can modify the C terminus of peptides.
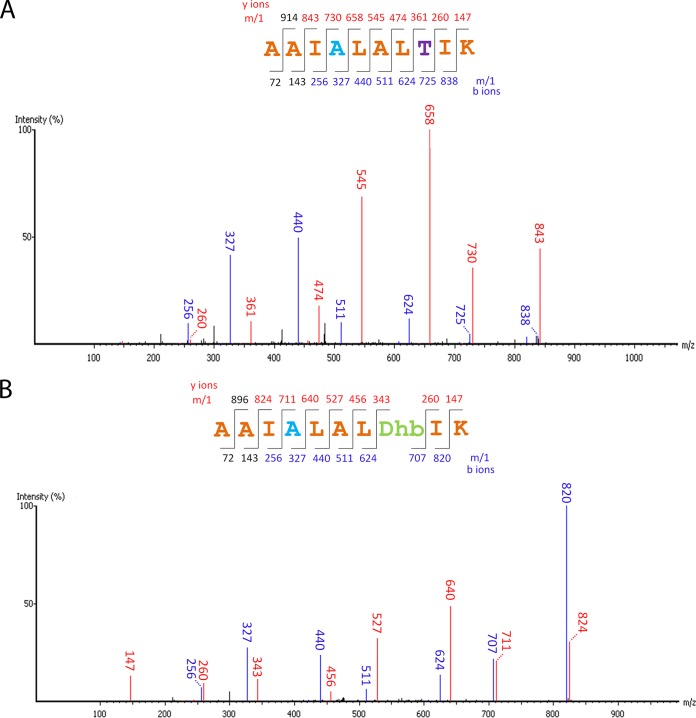
d-Alanine has been observed at the N-terminal part of the two peptides, LtnA1 and LtnA2, that compose lacticin 3147 (22). Recently, van Heel et al. showed that when LtnJ was expressed with the nisin machinery, the Dha at position 5 in nisin could be modified into d-alanine while Dha at position 33 was most likely unmodified (18). To investigate if the function of LtnJ is position dependent, plasmids pNZ-NisAtail and pNZ-NisAtail-T-ltnJ, producing mutants of nisin in the absence or presence of LtnJ, respectively, were constructed (Table 1). In these mutants, the C-terminal region of nisin (residues 23 to 34) was replaced by a linear peptide sequence (AAIS26LALTIK) (Table 3) containing a serine that could be dehydrated more effectively than Ser33 in nisin (34). Additionally, the lack of cysteine residues in this fragment (and therefore the absence of lanthionine rings) facilitates MS/MS fragmentation and characterization. Peptides purified by cationic exchange chromatography from both NZ9000(pIL3EryBTC, pNZ-NisAtail) and NZ9000(pIL3EryBTC, pNZ-NisAtail-T-ltnJ) were digested by trypsin and analyzed by LC-MS/MS. The conversion of Dha to d-alanine was detected only in the case of NZ9000(pIL3EryBTC, pNZ-NisAtail-T-ltnJ) compared to the control strain NZ9000(pIL3EryBTC, pNZ-NisAtail), as expected (Fig. 3). De novo sequencing data for the C terminus of NisAtail obtained following trypsin digestion showed that when LtnJ was expressed, 70% of Dha26 was hydrogenated into d-alanine in the fully dehydrated peptide. This percentage is 57% for peptides where Thr30 was not dehydrated (Fig. 4). Our results prove that LtnJ can modify Dha when it is located at the C-terminal part of the precursor peptide.
FIG 3.
MS/MS spectra of C-terminal tryptic fragment of NisAtail when LtnJ was present, showing the occurrence of d-alanine conversion. Expected masses for y and b ions are listed above and below the peptide sequence, respectively. Ions that were positively identified in the MS/MS spectrum are highlighted in blue (b ions) or red (y ions). d-Alanine converted from serine is colored blue; threonine and dehydrated threonine are labeled in purple and green, respectively. (A) Partly dehydrated peptide; (B) fully dehydrated peptide.
FIG 4.
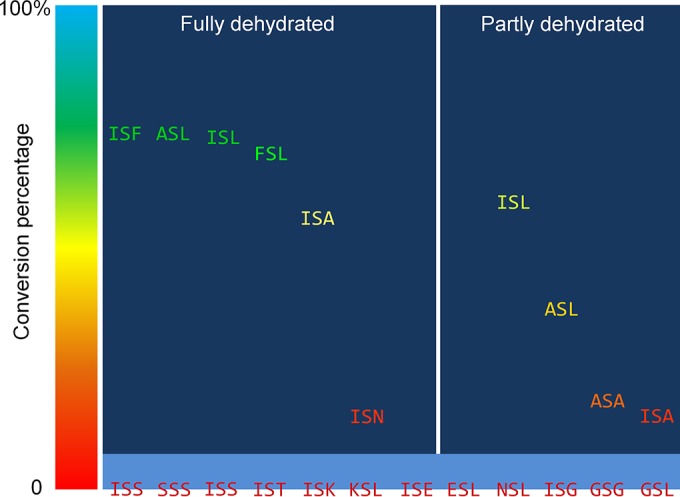
Graphical representation of the conversion percentage of Ser26 flanked by different residues into d-alanine by LtnJ. “Fully dehydrated” indicates that both Ser26 and Thr30 were dehydrated. “Partly dehydrated” indicates that only Ser26 was dehydrated. A descending tendency of conversion percentage (XSF > XSL > XSA) shows a preference for larger hydrophobic amino acids at the C terminus of Ser26.
Flanking residues of the substrate Dha affect the modification by LtnJ.
To explore the role of flanking residues of Dha during biosynthesis, 17 nisin mutant peptides (peptides 2 to 18) (Fig. 1; Table 3) were designed and constructed similarly to the mutant NisAtail described above. Thus, we replaced the C terminus of nisin with a peptide sequence (AAIS26LALTIK) in which flanking amino acids of Ser26 (i.e., I25 and L27) were mutated into serine, threonine, lysine, glutamic acid, phenylalanine, asparagine, glycine, or alanine (Table 3). The modification extent of this polypeptide tail in nisin was thoroughly studied by LC-MS/MS for all these mutants. According to the LC-MS/MS profile, conversion into d-alanine from Ser26 was observed in L27F, I25F, L27N, L27A, I25A-L27A, and I25A (Table 3; see also Fig. S2 to S7 in the supplemental material). The conversion rate of Dha into d-alanine is 71% (L27F), 67% (I25F), 13% (L27N), 54% (L27A), and 71% (I25A) compared to the fully dehydrated peptide (Fig. 4; see also Fig. S3 in the supplemental material). In the cases where the threonine at position 30 is not dehydrated, the conversion of Dha into d-alanine is not detected for L27F or I25F, is hardly detected for L27N, and drops to 13% for L27A and 35% for I25A (Fig. 4; see also Fig. S8 in the supplemental material for L27A). An exception is that I25A-L27A was modified by LtnJ only when threonine was not dehydrated with a conversion rate of 16%.
In mutant peptides (peptides 2 to 8 and 13 to 15) where Ser26 was neighbored by small polar amino acids (Ser/Thr), charged residues (Lys/Glu), or glycine, hydrogenation was not detected (Table 3), although serine was dehydrated in all cases. A similar result is obtained when asparagine is present at the N terminus of Ser26. On the other hand, a relatively low rate of conversion by LtnJ was detected when asparagine was located C terminally to Ser26 (Table 3; Fig. 4). The presence of charges, either positive or negative, did not allow the conversion into d-alanine in spite of Dha being present. Similarly, the presence of dehydrated residues Dha and dehydrobutyrine (Dhb) inhibited the hydrogenation of the adjacent Dha (Table 3).
In order to study the modification of Ser33 in nisin, a mutant nisin(Δ30–34)-KIHIHVSL (Table 3; Fig. 1) was designed in which the last residue, Lys34, was mutated into Leu to favor the dehydration by NisB and a Lys-Ile-His sequence was inserted downstream of Ser29, offering a cleavage site for trypsin digestion. A mass shift of 2 Da was not observed when LtnJ was coexpressed with nisin(Δ30–34)-KIHIHVSL (Table 3).
Dhb is not a substrate for LtnJ.
The mutant S26T was designed to mimic the conditions in which Dha is converted into d-alanine and therefore provide optimal conditions under which to investigate the conversion of Dhb into d-aminobutyrate (Table 3). Although threonines at positions 26 and 30 were dehydrated by NisB, providing a possible substrate for LtnJ (i.e., Dhb), d-aminobutyrate was not detected in any case (Table 3). This result indicated that the additional methyl group in Dhb prevents the hydrogenation catalyzed by LtnJ.
DISCUSSION
The combination of different posttranslational modification enzymes in peptide design and production is a promising approach (35). The success of these efforts depends to a great extent on the correct characterization of the enzymes that can be used (e.g., dependence on leader peptide recognition, cofactors, target sequence, substrate tolerance, etc.). In a previous study, we demonstrated that the modification machinery of nisin can be expanded with additional posttranslational modification enzymes, in this case, LtnJ and GdmD, which do not require their original leader peptide sequence for target recognition (18). However, we observed a reduced production of nisin in the presence of LtnJ (18). Therefore, we attempted to improve the production of the structural peptide. In diverse lantibiotic operons and other bacteriocin systems, an inverted repeat sequence is observed between the structural gene and the next open reading frame (ORF) within the same operon (36–38). By introducing an attenuator between nisA and ltnJ in our plug-and-play production system (18), the production level of the structural gene was improved more than 2-fold. These sequences may act as processing sites for the mRNA or as transcriptional attenuators which, in either case, ensure an appropriate ratio of enzymes and structural peptide. Additionally, the inverted repeats can affect the stability of the transcript as in the case of enterocin AS-48 (38) or Pep5 (39).
At present, an impressive number of different posttranslational modifications have been unveiled in lantibiotics (20). For example, aminovinyl-cysteine in gallidermin and epidermin, lysinoalanine in cinnamycin, or N-terminal lactate formation in epilancin 15X increases the biological activity and/or constrains the structure of the lantibiotics in which it naturally appears. d-Alanine, present in lacticin 3147, is one of the modifications that has been further investigated. Previous studies demonstrated the importance of d-alanine for the activity of lacticin 3147 and identified LtnJ as the responsible reductase producing d-alanine by deletion-complementation experiments (24). As reported previously, when LtnJ is coexpressed with the model lantibiotic nisin, Dha33 within nisin is likely not modified by LtnJ, whereas Dha5 is partially reduced by LtnJ to form d-alanine (18). In the same article, the hypothesis that amino acids flanking Dha may affect the function of LtnJ was raised. In order to clarify the specificity of LtnJ and to extend the applicability of this enzyme as a general tool for the designed modification of peptides, its selectivity was investigated in this study. For this purpose, we designed a set of mutants of nisin where the C terminus was replaced by a linear polypeptide tail (AAIS26LALTIK) that facilitates de novo sequencing by LC-MS/MS due to the absence of lanthionine rings in this region. In this tail, the substrate Dha (i.e., Ser26) is flanked by different residues. In our experiment, the serine in position 26 was dehydrated by NisB in all cases, rendering Dha26 (Dha27 for S26insS and S26insSS). However, only in some specific cases, Dha26 was converted into the hydrogenated form d-alanine. The high ratio of hydrogenated peptide compared to the corresponding mutant in NisAtail, L27F, I25F, L27A, and I25A emphasizes the importance of hydrophobic residues flanking the intermediate Dha for the function of LtnJ. The conversion observed in L27N (although in small amounts) and not detectable in I25N indicates a clear preference for an N-terminal hydrophobic residue flanking Dha. This phenomenon suggests that the active site in LtnJ is most likely surrounded by nonpolar residues. Additionally, the presence of d-alanine at the C terminus of the designed mutants demonstrates that the activity of LtnJ is not restricted to the N-terminal part of the peptide, although in its natural substrates d-alanine occurs only in that part of the peptide (Fig. 1). Recently published research showed that the reductase CrnJ from Carnobacterium maltaromaticum C2 could modify dehydroamino acids into d-amino acids at very different locations in the core peptide of carnolysin (40). Whether LtnJ can do so as well remains to be proved. The failure of modification by LtnJ in the tail of nisin(Δ30–34)-KIHIHVSL was probably due to the negative charge of the carboxyl group of the C-terminal leucine.
In both peptide LtnA1 and peptide LtnA2, the natural substrates of LtnJ, there are Dhb residues that are not converted into a reduced form (24). This is also the case for lactocin S (21). In order to determine whether this effect is caused by the different location and flanking residues or whether Dhb cannot serve as a substrate for LtnJ at all, we engineered a mutant where Dha was replaced by Dhb at the same location and surrounded by the same environment. Our data also exclude Dhb as a substrate for LtnJ and indicate that LtnJ is a Dha-specific hydrogenase, as previously suggested (24). Nevertheless, the reductase CrnJ from the carnolysin gene cluster can reduce Dhb flanked by hydrophobic amino acids and render d-aminobutyrate, which points at a broader substrate tolerance for CrnJ than for LtnJ (40).
An interesting observation is that among these modified mutants (NisAtail, L27F, I25F, L27A, I25A-L27A, and I25A) where Ser26 was flanked by hydrophobic amino acids, LtnJ preferred to modify Dha with a larger residue (Leu and Phe) on its C-terminal side rather than a smaller one (Ala).
In this study, we shed light on the specificity of LtnJ in terms of peptide sequence. We show the promiscuity of this leader-independent reductase when hydrophobic residues are neighboring Dha, which is its only substrate, as Dhb is never found to be modified by LtnJ. We also show that the reductase action can also take place at the C-terminal region of lantibiotics, although this has not been found in nature. Our data provide a deeper insight into the use of LtnJ as a tool to engineer designed peptides in future research.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Marcel de Vries from the Mass Spectrometry Core Facility of the University of Groningen for the assistance with LC-MS/MS.
D. Mu was supported by the China Scholarship Council (no. 2010605032). M. Montalbán-López was supported by the ALW-NWO project SynMod (project number 855.01.162) and the FW7 program SynPeptide. J. Deng was supported by the China Scholarship Council (no. 201406220171).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00475-15.
REFERENCES
- 1.D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knerr PJ, van der Donk WA. 2012. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem 81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 7.Dischinger J, Basi Chipalu S, Bierbaum G. 2014. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 304:51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 9.Montalbán-López M, Sánchez-Hidalgo M, Valdivia E, Martínez-Bueno M, Maqueda M. 2011. Are bacteriocins underexploited? Novel applications for old antimicrobials. Curr Pharm Biotechnol 12:1205–1220. doi: 10.2174/138920111796117364. [DOI] [PubMed] [Google Scholar]
- 10.van Heel AJ, Montalban-Lopez M, Kuipers OP. 2011. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol 7:675–680. doi: 10.1517/17425255.2011.573478. [DOI] [PubMed] [Google Scholar]
- 11.Lubelski J, Rink R, Khusainov R, Moll G, Kuipers OP. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 13.Grasemann H, Stehling F, Brunar H, Widmann R, Laliberte TW, Molina L, Doring G, Ratjen F. 2007. Inhalation of Moli1901 in patients with cystic fibrosis. Chest 131:1461–1466. doi: 10.1378/chest.06-2085. [DOI] [PubMed] [Google Scholar]
- 14.Crowther GS, Baines SD, Todhunter SL, Freeman J, Chilton CH, Wilcox MH. 2013. Evaluation of NVB302 versus vancomycin activity in an in vitro human gut model of Clostridium difficile infection. J Antimicrob Chemother 68:168–176. doi: 10.1093/jac/dks359. [DOI] [PubMed] [Google Scholar]
- 15.Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Gühring H, Vértesy L, Wink J, Hoffmann H, Brönstrup M, Sheldrick GM, Süssmuth RD. 2010. Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew Chem Int Ed Engl 49:1151–1154. doi: 10.1002/anie.200905773. [DOI] [PubMed] [Google Scholar]
- 16.Férir G, Petrova MI, Andrei G, Huskens D, Hoorelbeke B, Snoeck R, Vanderleyden J, Balzarini J, Bartoschek S, Brönstrup M, Süssmuth RD, Schols D. 2013. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. PLoS One 8:e64010. doi: 10.1371/journal.pone.0064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupke T, Götz F. 1997. In vivo reaction of affinity-tag-labelled epidermin precursor peptide with flavoenzyme EpiD. FEMS Microbiol Lett 153:25–32. doi: 10.1111/j.1574-6968.1997.tb10459.x. [DOI] [PubMed] [Google Scholar]
- 18.van Heel AJ, Mu D, Montalban-Lopez M, Hendriks D, Kuipers OP. 2013. Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth Biol 2:397–404. doi: 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- 19.Velásquez JE, Zhang X, van der Donk WA. 2011. Biosynthesis of the antimicrobial peptide epilancin 15X and its N-terminal lactate. Chem Biol 18:857–867. doi: 10.1016/j.chembiol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 21.Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Inger C, Abildgaard M, Nes IF. 1994. In vivo conversion of l-serine to d-alanine in a ribosomally synthesized polypeptide. J Biol Chem 269:27183–27185. [PubMed] [Google Scholar]
- 22.Ryan MP, Jack RW, Josten M, Sahl HG, Jung G, Ross RP, Hill C. 1999. Extensive post-translational modification, including serine to d-alanine conversion, in the two-component lantibiotic, lacticin 3147. J Biol Chem 274:37544–37550. doi: 10.1074/jbc.274.53.37544. [DOI] [PubMed] [Google Scholar]
- 23.Martin NI, Sprules T, Carpenter MR, Cotter PD, Hill C, Ross RP, Vederas JC. 2004. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43:3049–3056. doi: 10.1021/bi0362065. [DOI] [PubMed] [Google Scholar]
- 24.Cotter PD, O'Connor PM, Draper LA, Lawton EM, Deegan LH, Hill C, Ross RP. 2005. Posttranslational conversion of l-serines to d-alanines is vital for optimal production and activity of the lantibiotic lacticin 3147. Proc Natl Acad Sci U S A 102:18584–18589. doi: 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suda S, Lawton EM, Wistuba D, Cotter PD, Hill C, Ross RP. 2012. Homologues and bioengineered derivatives of LtnJ vary in ability to form d-alanine in the lantibiotic lacticin 3147. J Bacteriol 194:708–714. doi: 10.1128/JB.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJ, Kuipers OP, Moll GN. 2007. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl Environ Microbiol 73:1792–1796. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majchrzykiewicz JA, Lubelski J, Moll GN, Kuipers A, Bijlsma JJ, Kuipers OP, Rink R. 2010. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother 54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kluskens LD, Kuipers A, Rink R, de Boef E, Fekken S, Driessen AJ, Kuipers OP, Moll GN. 2005. Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry 44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 29.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJ, Moll GN, Kuipers OP. 2005. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 30.Rink R, Kluskens LD, Kuipers A, Driessen AJ, Kuipers OP, Moll GN. 2007. NisC, the cyclase of the lantibiotic nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry 46:13179–13189. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Holo H, Nes I. 1995. Transformation of Lactococcus by electroporation. Methods Mol Biol 47:195–199. [DOI] [PubMed] [Google Scholar]
- 33.Schägger H. 2006. Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 34.Karakas Sen A, Narbad A, Horn N, Dodd HM, Parr AJ, Colquhoun I, Gasson MJ. 1999. Post-translational modification of nisin. Eur J Biochem 261:524–532. doi: 10.1046/j.1432-1327.1999.00303.x. [DOI] [PubMed] [Google Scholar]
- 35.Montalbán-López M, Zhou L, Buivydas A, van Heel AJ, Kuipers OP. 2012. Increasing the success rate of lantibiotic drug discovery by synthetic biology. Expert Opin Drug Discov 7:695–709. doi: 10.1517/17460441.2012.693476. [DOI] [PubMed] [Google Scholar]
- 36.Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 37.McAuliffe O, O'Keeffe T, Hill C, Ross RP. 2001. Regulation of immunity to the two-component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Mol Microbiol 39:982–993. doi: 10.1046/j.1365-2958.2001.02290.x. [DOI] [PubMed] [Google Scholar]
- 38.Fernández M, Sánchez-Hidalgo M, García-Quintáns N, Martínez-Bueno M, Valdivia E, López P, Maqueda M. 2008. Processing of as-48ABC RNA in AS-48 enterocin production by Enterococcus faecalis. J Bacteriol 190:240–250. doi: 10.1128/JB.01528-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pag U, Heidrich C, Bierbaum G, Sahl HG. 1999. Molecular analysis of expression of the lantibiotic Pep5 immunity phenotype. Appl Environ Microbiol 65:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohans CT, Li JL, Vederas JC. 2014. Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with d-amino acids. J Am Chem Soc 136:13150–13153. doi: 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 41.Kuipers OP, De Ruyter PG, Kleerebezem M, De Vos WM. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol 15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 42.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Eur J Biochem 216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.