Abstract
l-Fucose is a sugar present in human secretions as part of human milk oligosaccharides, mucins, and other glycoconjugates in the intestinal epithelium. The genome of the probiotic Lactobacillus rhamnosus GG (LGG) carries a gene cluster encoding a putative l-fucose permease (fucP), l-fucose catabolic pathway (fucI, fucK, fucU, and fucA), and a transcriptional regulator (fucR). The metabolism of l-fucose in LGG results in 1,2-propanediol production, and their fucI and fucP mutants displayed a severe and mild growth defect on l-fucose, respectively. Transcriptional analysis revealed that the fuc genes are induced by l-fucose and subject to a strong carbon catabolite repression effect. This induction was triggered by FucR, which acted as a transcriptional activator necessary for growth on l-fucose. LGG utilized fucosyl-α1,3-N-acetylglucosamine and contrarily to other lactobacilli, the presence of fuc genes allowed this strain to use the l-fucose moiety. In fucI and fucR mutants, but not in fucP mutant, l-fucose was not metabolized and it was excreted to the medium during growth on fucosyl-α1,3-N-acetylglucosamine. The fuc genes were induced by this fucosyl-disaccharide in the wild type and the fucP mutant but not in a fucI mutant, showing that FucP does not participate in the regulation of fuc genes and that l-fucose metabolism is needed for FucR activation. The l-fucose operon characterized here constitutes a new example of the many factors found in LGG that allow this strain to adapt to the gastrointestinal conditions.
INTRODUCTION
One of the few hexoses with “l” configuration found in nature is l-fucose (6-deoxy-l-galactose). It forms part of many glycans present at the surface of eukaryotic cells such as H and Lewis antigens, which are present not only in blood cells but also in epithelial cells at different mucosal sites (1). It is also present at the highly glycosylated mucin proteins of the intestinal mucosa and in a high proportion of the oligosaccharides present in human milk (2, 3). These facts make l-fucose an important sugar in the microbial ecology of the gastrointestinal tract. The importance of the fucosylation of mucosal glycoconjugates on the intestinal ecology is reflected by the fact that the intestinal microbiota composition is dependent on the secretor status of the individuals, which is defined by mutations in the FUT2 gene coding for an α(1,2)-fucosyltransferase that participates in the fucosylation of mucosal glycans (4, 5). Owing to its elevated concentration found in the intestinal niche, l-fucose can be used as a carbon source and its utilization has been identified as a key factor for intestinal colonization in some bacteria. Thus, the pathogen Campylobacter jejuni, primarily thought to be an asaccharolytic microorganism, was able to use l-fucose, and this provides it with a competitive advantage, as determined in a neonatal piglet infection model (6). Also, a ΔfucAO mutant of the probiotic Escherichia coli Nissle 1917 strain, unable to use l-fucose, showed 2 orders of magnitude lower colonization level in mouse intestine compared to the wild type (7).
l-Fucose metabolism has been extensively characterized in Escherichia coli (8–10). In this bacterium, l-fucose is transported by a permease and catabolized to fuculose-1-phosphate that is finally split into dihydroxyacetone-phosphate and l-lactaldehyde by a specific aldolase. l-Lactaldehyde can be further metabolized to 1,2-propanediol or lactate, and the dihydroxyacetone-phosphate enters glycolysis (10). A similar pathway has been described in Bacteroides (11). However, C. jejuni or intestinal commensals such as Bifidobacterium rely on a different route for its catabolism, which involves l-fucose dehydrogenase and l-fuconolactonase, although the corresponding genes and enzymes have not been fully characterized (6).
l-Fucose does not only serve as a carbon and energy source, but in some bacteria it acts as a signaling molecule. In enterohemorrhagic E. coli the two-component system FusKR senses extracellular l-fucose and activates virulence and metabolic genes (12). Streptococcus pneumoniae strains, although having all the genes which encode a complete l-fucose pathway are not able to use it. The S. pneumoniae fuc genes are clustered with genes encoding extracellular glycosidases that release fucosyl-oligosaccharides from Lewisy and type A and B histo-blood group antigens from the host cell surface. These fucosyl-oligosaccharides are taken up by specific transporters encoded in the same operon. The S. pneumoniae fuc genes are needed for virulence and are induced by l-fucose, but this sugar is not further metabolized (13).
Lactobacillus rhamnosus GG (LGG) is a bacterium marketed as probiotic (14). It was derived from the intestinal habitat of a healthy human and its health benefits as probiotic have been widely documented (14, 15). Genetic and biochemical studies have revealed that LGG possesses many mechanisms of survival and persistence at the gastrointestinal tract (16), which include bile salt resistance (17), proteic factors for mucosal attachment (e.g., mucus-binding pili [18]), glycosidases, sugar transporters, and catabolic enzymes needed to exploit the carbohydrate resources characteristic of the intestinal habitat (19). LGG is able to use l-fucose as a carbon source, and putative l-fucose utilization genes are annotated in its genome, but their functionality has never been proved, and no data on l-fucose utilization by intestinal lactobacilli are available. We establish here the involvement of LGG fuc genes in l-fucose utilization, revealing a new trait that may be important in the persistence and colonization of the intestinal habitat by LGG.
MATERIALS AND METHODS
Strains and growth conditions.
Lactobacillus strains (Table 1) were grown in MRS medium (Difco) or in basal MRS medium (10 g/liter Bacto peptone [Difco], 4 g/liter yeast extract [Pronadisa], 5 g/liter sodium acetate, 2 g/liter triammonium citrate, 0.2 g/liter magnesium sulfate 7-hydrate, 0.05 g/liter manganese sulfate monohydrate, and 1 ml/liter Tween 80) supplemented with 0.5% l-fucose (30 mM) or 4 mM fucosyl-α1,3-N-acetylglucosamine (Fuc-α1,3-GlcNAc; Carbosynth, Ltd., Compton, Berkshire, United Kingdom) at 37°C under static conditions. Bacterial growth was determined in microtiter plates in a Polarstar Omega plate reader at 37°C. Each well (100 μl of medium) was inoculated with bacteria grown in basal MRS without sugars at an initial optical density at 550 nm (OD550) of 0.1. The maximum growth rate (μ) was calculated by following the OD550 versus time. Three independent biological replicates for each growth curve were obtained. Anaerobic and aerobic growth of LGG was carried out in 10 ml of basal MRS medium supplemented with 20 mM l-fucose or 20 mM glucose in anaerobic jars (AnaeroGen; Oxoid) or in 50-ml Erlenmeyer flasks shaken at 200 rpm, respectively. The medium was inoculated at an initial OD550 of 0.1 and incubated at 37°C for 24 h. E. coli DH10B was used as cloning host, and it was grown in Luria-Bertani medium at 37°C under shaking at 200 rpm. The antibiotics used for plasmid selection were ampicillin at 100 μg/ml and chloramphenicol at 20 μg/ml for E. coli and erythromycin at 5 μg/ml for LGG.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| L. rhamnosus GG | Wild type | ATCC 53103 |
| L. rhamnosus BL394 | LGG with an 963-bp in-frame deletion in fucP | This study |
| L. rhamnosus BL395 | LGG with a frameshift at the fucI EcoRI site | This study |
| L. rhamnosus BL396 | LGG with an insertion of a pRV300-derivative at the fucR gene; Eryr | This study |
| L. casei ATCC 393 | Wild type | ATCC 393 |
| L. zeae KCTC3804 | Wild type; very similar to L. casei isolates | ATCC 15820 |
| E. coli DH10B | F− endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 ϕ80lacZΔM15 araD139 Δ(ara leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ− | Invitrogen |
| Plasmids | ||
| pRV300 | Nonreplicative plasmid for Lactobacillus; Ampr Eryr | 20 |
| pRΔFucP | pRV300 with a 1,221-bp fragment containing fused 5′ and 3′ fucP fragments | This study |
| pRFucI | pRV300 with a 997-bp fucI fragment containing a frameshift at the EcoRI site | This study |
| pRFucR | pRV300 with a 437-bp fucR internal fragment cloned at EcoRV site | This study |
Eryr, erythromycin resistance; Ampr, ampicillin resistance.
Construction of LGG mutants in fuc genes.
LGG mutants in fucP (LGG_02683) and fucI (LGG_02685) were constructed by replacing the wild-type genes with mutated variants. The LGG chromosomal DNA was isolated from 10-ml cultures with the DNA isolation kit for Cells and Tissues (Roche). The fucP gene was amplified by PCR with Pfx DNA polymerase (Invitrogen) and the oligonucleotide pair FucP1 (5′-GCCTTGCGATGGTCTATAG) and FucP2 (5′-GCTTCTTCTCAGTTGATCAAC) using the isolated chromosomal DNA as the template. The resulting 2,187-bp fragment was digested with AclI, and the 634- and 590-bp fragments corresponding to the 5′ and 3′ portions of fucP, respectively, were gel isolated. These fragments were ligated together with EcoRV-digested pRV300 (20), creating an integrative plasmid carrying a fucP gene with an internal 963-bp in-frame deletion (pRΔFucP). The fucI gene was amplified by PCR with oligonucleotides FucI1 (5′-ATTGGCGCGTTGAATGAAAG) and FucI2 (5′-ATAAGATCCGGCTGGTTTCC) and the obtained fragment was digested with PstI. The generated 997-bp fragment was gel isolated and ligated to pRV300 digested with EcoRV/PstI. The obtained plasmid was digested with EcoRI, treated with Klenow, and ligated, creating a frameshift in fucI as verified by sequencing (pRFucI plasmid). The plasmids containing the mutated fucP and fucI genes were transformed by electroporation in LGG with a GenePulser apparatus (Bio-Rad) as previously described (21). Integrants obtained by single crossover recombination were selected on MRS agar plates containing erythromycin. Clones were grown for about 200 generations in antibiotic-free MRS medium and plated on MRS medium. Colonies that had undergone a second recombination event leading to an erythromycin-sensitive phenotype were selected by replica plating and tested by PCR with appropriated oligonucleotides for the replacement of the wild-type genes by the mutated copy. After confirmation of the mutations by sequencing, two clones were selected and named BL394 (ΔfucP) and BL395 (fucI).
A 437-bp internal fragment of the fucR gene (LGG_02680) was amplified with oligonucleotides FucR1 (5′-GGCGTGGCTTTGGATATG) and FucR2 (5′-CATCATCGCTCGCTTGAC) and cloned into pRV300 digested with EcoRV. The resulting plasmid (pRFucR) was used to transform LGG selecting for erythromycin resistance. One strain with disrupted fucR was selected and named BL396.
Reverse transcription-quantitative PCR (RT-qPCR) analysis of fuc genes expression.
L. rhamnosus strains were grown in MRS basal medium containing 0.5% glucose, 0.5% l-fucose, or 0.5% l-fucose plus 0.2% glucose to an OD550 of 0.9 to 1. Bacterial cells from 9-ml cultures were recovered by centrifugation and washed with 9 ml of 50 mM EDTA (pH 8). The cell pellets were resuspended in 1 ml of TRIzol reagent (Gibco) and 1 g of glass beads (0.1 mm in diameter) were added. The bacteria were broken with a Mini-BeadBeater apparatus (Biospec Products, Bartlesville, OK), and total RNA was isolated according to the recommendations of the manufacturer of TRIzol. For experiments with Fuc-α1,3-GlcNAc, RNA was isolated from 1-ml cultures containing 4 mM sugar. Portions (100 ng) of RNA were digested with DNase I (RNase-free; Fermentas), and cDNA was obtained from 50 ng of DNase-treated RNA using a Maxima first-strand cDNA synthesis kit (Fermentas). qPCR was performed in a LightCycler 480 II system (Roche) with the LightCycler FastStart DNA Master SYBR green I mix (Roche). Primers were designed by using the Primer-BLAST service at the NCBI (http://www.ncbi.nlm.nih.gov/tools/primer-blast) in order to generate amplicons ranging from 70 to 100 bp in size. qPCR was performed for each cDNA sample with the following primers pairs: 5′-TGGCTAAGCATGGGTTCCTG and 5′-TCGCCATTGATCGTCTCTCG (fucI), 5′-CCGGAGAGCCAGTCAAAGTT and 5′-CTCAACAGGCCCAGCTACAA (fucK), 5′-TTTGTCGGCTGACACGATGA and 5′-GCCGGCGCTAATTCAGAAAG (fucP), 5′-GAGCAGATGGCCTGACAGTT and 5′-ACTGGCGGTTCACCATTAGG (fucU), 5′-GAAGTTGCTGGTGCAAAGGG and 5′-TCATCCGGGTATTCGCTGTG (fucA), and 5′-TCAGTTGTGCAGTGAGCAGT and 5′-TTCAACGCCTGCATCTGCTA (fucR). The reaction mixture (10 μl) contained 5 μl of 2× qPCR master mix, 0.5 μl of each primer (10 μM), and 1 μl of a 10-fold dilution of the cDNA synthesis reaction. Reaction mixtures without a template were run as controls. The cycling conditions were as follows: 95°C for 10 min, followed by 45 cycles of three steps consisting of denaturation at 95°C for 10 s, primer annealing at 60°C for 20 s, and primer extension at 72°C for 20 s. For each set of primers, the cycle threshold (crossing point [CP]) values were determined by the automated method implemented in the LightCycler software (version 4.0; Roche). The relative expressions were calculated using the software tool REST (relative expression software tool) (22) and three LGG reference genes (pyrG [LGG_02546], recG [LGG_01660], and leuS [LGG_00848]) were used simultaneously for the analysis. Linearity and amplification efficiency were determined for each primer pair, and every RT-qPCR analysis was performed at least in triplicate with two biologically independent samples.
Sugar and metabolite analysis.
The concentration of l-fucose and Fuc-α1,3-GlcNAc in the supernatants was determined by high-pH anion-exchange chromatography with pulsed amperometric detection in an ICS3000 chromatographic system (Dionex) using a CarboPac PA100 column (Dionex). A combined gradient of 100 to 300 mM NaOH and 0 to 150 mM acetic acid was used for 20 min at a flow rate of 1 ml/min. 1,2-Propanediol and lactic acid were determined by high-pressure liquid chromatography with a Jasco PU-2080 Plus system coupled to refractive index (Jasco RI-2031Plus) or UV (210 nm) detectors with Rezex RCM-Monosaccharide and Rezex ROA-Organic Acid columns, respectively. The flow rates were 0.6 and 0.5 ml/min, respectively, in water or 5 mM H2SO4.
Sequence analysis.
The sequences of the LGG fuc genes were retrieved from the GenBank database (accession no. FM179322.1) and homology searches were performed with BLAST at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Genomic context analysis was performed on genomes deposited at the Microbial Genome Database for Comparative Analysis (http://mbgd.genome.ad.jp/) (23). Transcriptional terminators in the fuc gene cluster were searched with ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/).
Statistical analysis.
Statistical analysis was performed using GraphPad software (San Diego, CA). A Student t test was used to detect statistically significant differences between growth rates from the wild-type strain and the mutant strains. Statistical significance was accepted at P < 0.05.
RESULTS
LGG fuc operon.
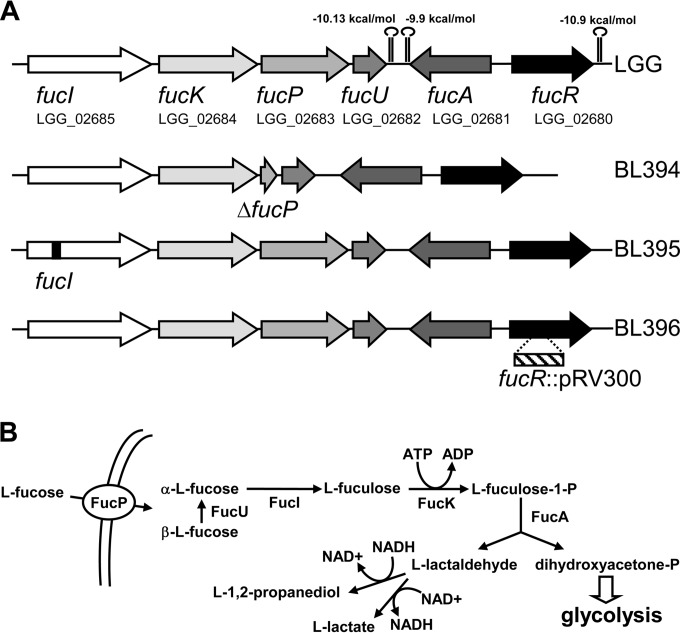
An l-fucose operon is annotated in the genome of LGG (LGG_02680 to LGG_02685) which codes for the enzymes that would comprise a complete pathway for l-fucose catabolism (Fig. 1). The genes fucI, fucK, fucP, and fucU would code for an l-fucose isomerase, l-fuculose kinase, an l-fucose/H+ symporter of the major facilitator superfamily (MFS), and l-fucose mutarotase, an enzyme that accelerates the conversion of the β anomer of l-fucose into the α anomer, the substrate for FucI (24). The divergently transcribed fucR gene codes for a transcriptional regulator of the DeoR family, whereas fucA, codes for an l-fuculose-1-phosphate aldolase that splits l-fuculose-1-phosphate into dihydroxyacetone-phosphate and l-lactaldehyde, and it is transcribed in the opposite direction to fucIKPU. In E. coli the l-lactaldehyde formed during l-fucose catabolism is detoxified by the action of an l-1,2-propanediol oxidoreductase encoded by a gene in the fuc cluster, fucO, with the concomitant production of 1,2-propanediol (10). No fucO homologue is found in the LGG fuc cluster, but two genes that encode proteins with 50% (LGG_00757) and 42% (LGG_02124) identity to E. coli FucO are present at another location in the LGG chromosome. Alternatively, in E. coli l-lactaldehyde can be transformed into lactic acid by the action of lactaldehyde dehydrogenase, encoded by the aldA gene (9). In LGG the gene LGG_02286 codes for a protein with 32% identity to E. coli AldA, which possesses the typical motifs of NAD(+)-dependent aldehyde dehydrogenases.
FIG 1.
l-Fucose metabolism in LGG. (A) Schematic representation of the fuc gene cluster in LGG. Stem-loops represent putative rho-independent transcriptional terminators, and their calculated ΔG values are given. The genetic structures of the different fuc mutants generated are shown. (B) Proposed catabolic pathway for l-fucose utilization in LGG. l-Lactaldehyde can follow two different routes, leading to the production of l-1,2-propanediol or l-lactate. In LGG, the route producing l-1,2-propanediol is more efficient (anaerobic growth; see the text).
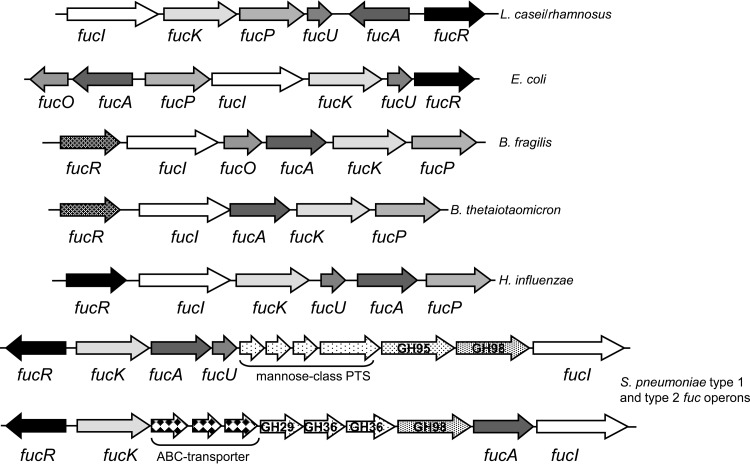
fuc clusters encoding similar catabolic pathways are found in the genomes of some intestinal commensals and pathogens, although the genetic structure varies and fucO and fucU genes are not always present (Fig. 2). A second type of transcriptional regulator, with the N-terminal DNA-binding domain of the GntR superfamily and a C-terminal ligand-binding domain of the sugar-binding domain of ABC transporters, replaces the DeoR family FucR in Bacteroides (11). Furthermore, the fuc operons from S. pneumoniae strains are more complex, and they lack the gene for an l-fucose/H+ symporter permease, which is replaced by genes encoding phosphoenolpyruvate/sugar phosphotransferase systems (PTS) of the mannose-class or sugar ABC-type transporters, and contain genes for intracellular and extracellular glycosyl hydrolases (Fig. 2) (13).
FIG 2.
l-Fucose catabolic gene clusters in bacteria. A schematic representation of different gene clusters for l-fucose catabolism is shown. In S. pneumoniae strains, two different fuc clusters (type 1 and type 2) have been described. Genes encoding glycosyl hydrolases (GH) are shown with the GH family number.
Genome inspection in the rest of Lactobacillus species revealed that fuc clusters are exclusively found in some L. rhamnosus isolates (e.g., GG, HN001, LRHMDP2, and LRHMDP3 strains), Lactobacillus casei and Lactobacillus zeae (only one sequenced representative strain is available for these species: strains ATCC 393 and ATCC 15820, respectively). According to this, the ATCC 15820 strain was able to ferment l-fucose (data not shown). In contrast, the ATCC 393 strain did not form acid from l-fucose (data not shown). This inability is probably due to the presence of a frameshift in the fucK gene of this strain (resulting in the two different annotated genes LBCZ_2453 and LBCZ_2454) that gives truncated l-fuculose kinases lacking the N-terminal 328 and C-terminal 181 amino acids, respectively. Among the rest of lactobacilli, homologues to some fuc genes were only found in the draft genomes of newly described species: Lactobacillus shenzhenensis LY-73T, Lactobacillus composti JCM14202, and Lactobacillus herbinensis DSM16991 (data not shown). However, they were not grouped in an identifiable fuc cluster. It seems therefore that within Lactobacillus the presence of a complete set of fuc genes organized in an operon structure is exclusive for L. rhamnosus and for the L. casei/L. zeae group. Despite of the elevated number of sequenced strains of Lactobacillus paracasei (25), no fuc genes were found in this species, which forms part of the phylogenetically related L. casei/L. paracasei/L. rhamnosus group.
Another remarkable feature of the LGG fuc region was the presence of an adjacent set of genes (LGG_02687 to LGG_02692) encoding an l-rhamnulose-1-phosphate aldolase, l-rhamnose isomerase, l-rhamnose mutarotase, l-rhamnulokinase, a MFS permease, and a transcriptional regulator of the AraC family, which would constitute a complete pathway for l-rhamnose (6-deoxy-l-mannose) utilization, analogous to the l-fucose catabolic pathway. Therefore, it appears that this chromosomal region in LGG consist of genes for the catabolism of l-deoxy sugars.
The mutation in fuc catabolic genes impairs growth on l-fucose.
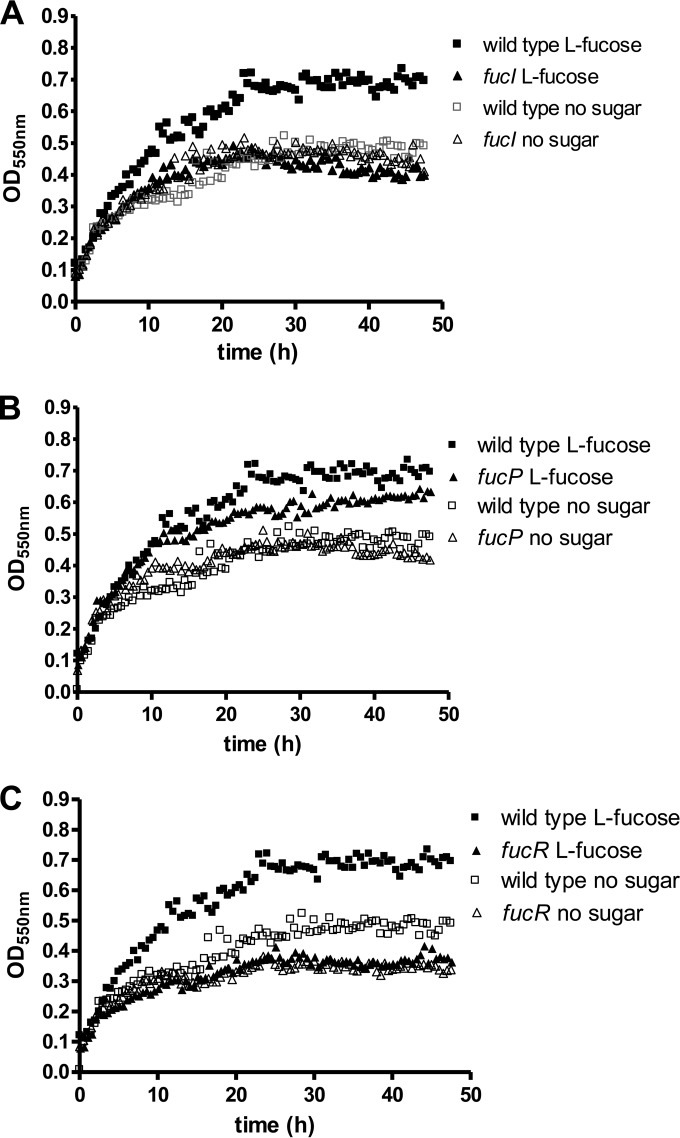
In order to prove the involvement of fuc genes in l-fucose catabolism, we constructed mutants in fucI and fucP (Fig. 1A). The fucI strain failed to acidify the growth medium when it was supplemented with l-fucose, whereas in the fucP mutant acidification was slower compared to the wild type (data not shown). Under our experimental conditions, LGG showed slow growth in unsupplemented basal MRS medium, probably due to the consumption of residual carbon sources present in this complex medium, as has been described earlier (26). When the medium was supplemented with l-fucose, the wild-type strain showed a growth rate (0.114 ± 0.022 h−1) compatible with l-fucose utilization, whereas the fucI mutant strain displayed a growth rate significantly lower than that of the wild type (0.077 ± 0.007 h−1, P = 0.048) and lower ODs were reached (Fig. 3A). In the fucP mutant, l-fucose was only able to sustain a reduced growth rate (0.078 ± 0.006 h−1, P = 0.048), but the attained ODs were higher compared to the fucI strain (Fig. 3B), suggesting that, although FucP is the main l-fucose transporter in LGG, in the absence of this permease l-fucose is probably entering the cells by alternative and less efficient transporter(s). In order to determine whether, as with E. coli (10), the fate of l-fucose in LGG depends on the oxygen availability, the wild-type supernatants were analyzed after growth under anaerobic or aerobic conditions. The analysis evidenced the production of 1,2-propanediol when cells were grown with l-fucose under anaerobiosis (Table 2). This compound was not detected in the supernatants of glucose-grown cells. This confirmed that in LGG the l-lactaldehyde formed by the action of the l-fuculose-1-phosphate aldolase on l-fuculose-1-phosphate can be metabolized by an l-1,2-propanediol oxidoreductase as occurs in E. coli. Lactate production from l-fucose under anaerobic conditions was lower in cells grown with glucose. Aerobic conditions did not have a strong impact on lactic acid production or growth with glucose. However, these conditions did not favor l-fucose utilization, which was very inefficient and incomplete after 24 h (Table 2).
FIG 3.
Growth of LGG on l-fucose. (A) fucI mutant; (B) fucP mutant; (C) fucR mutant. In all graphs, the growth profile of the wild-type strain is represented for a better comparison. The l-fucose concentration was 0.5% (30 mM). The data presented are the means from three determinations. The standard deviation did not exceed 15%.
TABLE 2.
Product formation and growth characteristics of LGG with l-fucose or d-glucose under different conditions
| Parameter | Mean ± SDa |
|||
|---|---|---|---|---|
| Anaerobic |
Aerobic |
|||
| l-Fucose | d-Glucose | l-Fucose | d-Glucose | |
| Concn (mM) | ||||
| 1,2-Propanediol | 10.0 ± 0.1 | ND | 1.3 ± 0.1 | ND |
| Lactate | 10.0 ± 1.3 | 33.8 ± 1.3 | 0.7 ± 0.1 | 35.7 ± 6.7 |
| Residual sugar | 0.10 ± 0.06 | ND | 12.73 ± 0.63 | ND |
| Final OD550 | 1.20 ± 0.14 | 1.75 ± 0.07 | 0.75 ± 0.06 | 1.65 ± 0.07 |
| Final pH | 5.36 ± 0.01 | 5.02 ± 0.01 | 7.09 ± 0.14 | 5.09 ± 0.01 |
Growth occurred in basal MRS medium supplemented with 20 mM l-fucose or 20 mM d-glucose for 24 h at 37°C. ND, not detected.
The fucR gene codes for a transcriptional activator of the fuc cluster.
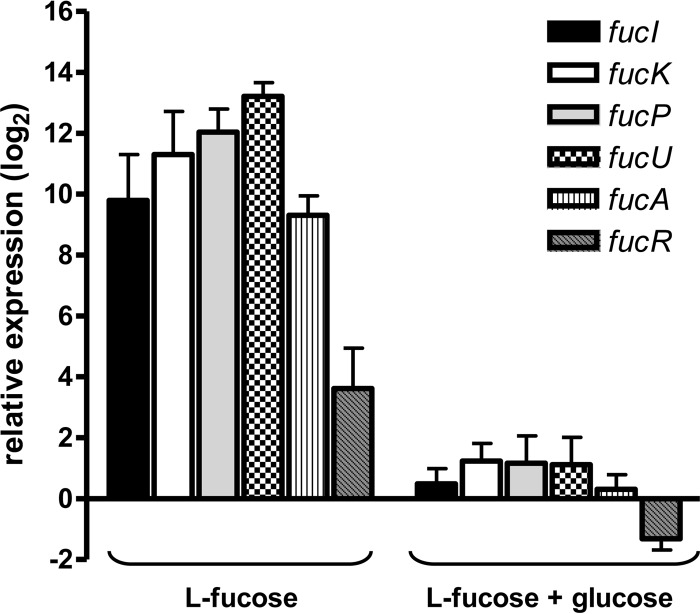
To ascertain the role of the product of fucR in the regulation of the fuc operon a disrupted mutant was constructed (Fig. 1A). Similar to the fucI mutant, the fucR mutant strain displayed a growth rate (0.067 ± 0.013 h−1, P = 0.032) significantly lower than that of the wild type. Thus, fucR mutant failed to grown with l-fucose as a carbon source (Fig. 3C), supporting the role of FucR as a transcriptional activator of fuc genes. Gene expression analysis of the fuc operon revealed that all fuc genes were induced by the presence of l-fucose by a factor ranging from 12 (fucR) to 9,400 (fucU) compared to growth on glucose (Fig. 4). According to the gene structure of the LGG fuc cluster (Fig. 1A), at least three different promoters are necessary for the transcription of all genes. Our results show that the three promoters are responsive to l-fucose and suggested that FucR autoregulates its own transcription, although fucR displayed the lower level of induction compared to the rest of fuc genes.
FIG 4.
Expression of fuc genes in LGG. The LGG wild-type strain was grown with 0.5% l-fucose or 0.5% l-fucose plus 0.2% glucose, and the expression of fuc genes was determined by RT-qPCR. The relative expression is referred to bacterial cells grown with 0.5% glucose. The LGG pyrG (LGG_02546), recG (LGG_01660), and leuS (LGG_00848) genes were used as a reference.
The presence of glucose in addition to l-fucose caused a strong carbon catabolite repression of all fuc genes, whose expression was restored to levels close to those found during growth on glucose, with a factor of repression (i.e., the fold change in l-fucose/fold change in l-fucose plus glucose) ranging from 30- to 4,400-fold. In agreement with this observation catabolite responsive element (cre) sites which fit the consensus sequence defined for Lactococcus lactis (WGWAARCGYTWWMA) (27) were found upstream of fucI (−86TGAAAGCGCTTATT, two mismatches) or fucA and fucR (−57TGCAAGCGCTTACG, one mismatch; the numbering is relative to fucR). These sites are the target for binding of the global transcriptional regulator CcpA, which controls carbon catabolite repression in low-G+C Gram-positive bacteria (28).
Growth of LGG with a fucose-containing disaccharide.
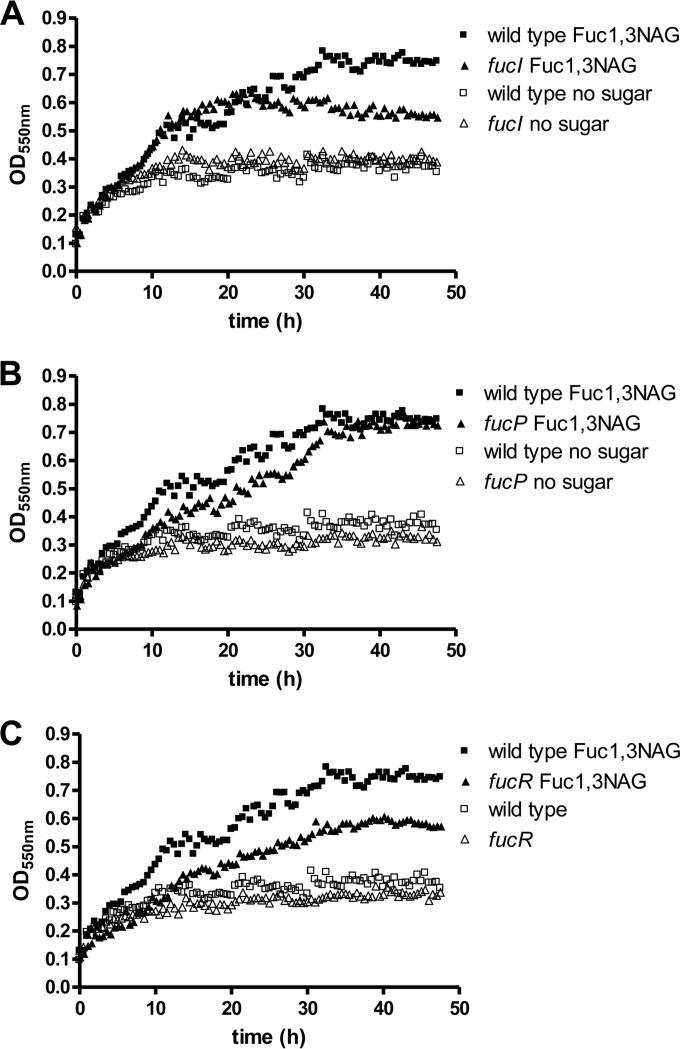
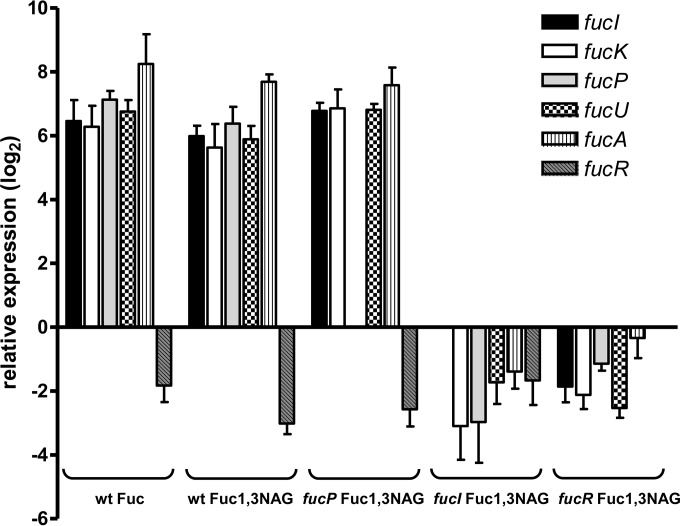
A previous work has shown that Lactobacillus casei BL23 is able to utilize the fucosyl-disaccharide fucosyl-α1,3-N-acetylglucosamine (Fuc-α1,3-GlcNAc), which is transported by a specific permease of the PTS class and split into l-fucose and N-acetylglucosamine by the α-l-fucosidase AlfB (29). However, this strain is not able to use l-fucose and only the N-acetylglucosamine moiety of Fuc-α1,3-GlcNAc is catabolized, while l-fucose is quantitatively expelled to the growth medium. LGG was also able to grow with 4 mM Fuc-α1,3-GlcNAc (Fig. 5) and, in agreement with the presence of an l-fucose catabolic pathway, no l-fucose was found in the supernatants. In contrast, compared to the wild type, the LGG fucI and fucR mutants reached a lower final OD on Fuc-α1,3-GlcNAc (Fig. 6). In these experiments 110% ± 6.1% (fucI mutant) and 70.8% ± 1.4% (fucR mutant) of the theoretical l-fucose from Fuc-α1,3-GlcNAc (4 mM) was detected in the bacterial supernatants at the stationary phase. This was consistent with the fact that only the N-acetylglucosamine moiety of the disaccharide was efficiently metabolized in these mutants. The fucP mutant, deficient in the l-fucose permease, showed a growth pattern on Fuc-α1,3-GlcNAc similar to the wild type (Fig. 5) and only 3.1% ± 0.3% of the theoretical l-fucose generated from Fuc-α1,3-GlcNAc was detected in supernatants. These data indicated that the l-fucose intracellularly generated, possibly by the action of an AlfB homologue (coded by LGG_02652), was metabolized, and that FucP was not fully necessary for this process. In agreement with this, growth on Fuc-α1,3-GlcNAc induced expression of all fuc genes, although the use of lower concentrations of disaccharide (4 mM) led to lower induction levels (Fig. 6) compared to growth with 0.5% (30 mM) l-fucose (Fig. 4). Also, under these conditions the fucR gene showed a downregulation compared to cells grown with glucose. In the ΔfucP strain expression of the fuc cluster after growth with fuc-α1,3-GlcNAc was comparable to that found in the wild type, further supporting the fact that fucP is dispensable for Fuc-α1,3-GlcNAc metabolism. In the fucI and fucR mutants no induction of fuc genes was observed after growth on Fuc-α1,3-GlcNAc (Fig. 6), which was in agreement with the observed excretion of l-fucose in these strains when cells were grown on the fucosyl-disaccharide.
FIG 5.
Growth of LGG with fucosyl-α-1,3-N-acetylglucosamine. (A) fucI mutant; (B) fucP mutant; (C) fucR mutant. In all graphs, the growth pattern of the wild-type strain is represented for a better comparison. Fucosyl-α-1,3-N-acetylglucosamine was used at 4 mM. The data presented are means from three determinations. The standard deviations did not exceed 15%.
FIG 6.
Expression of fuc genes in LGG grown in fucosyl-α-1,3-N-acetylglucosamine. LGG wild-type, fucP, fucI, and fucR strains were grown in the presence of 4 mM fucosyl-α-1,3-N-acetylglucosamine, and the expression of fuc genes was monitored by RT-qPCR. For each bacterial strain, the relative expression is referred to the expression in the same strain grown with 4 mM glucose. The expression in wild type in the presence of 4 mM l-fucose is also shown. The LGG pyrG (LGG_02546), recG (LGG_01660), and leuS (LGG_00848) genes were used as a reference.
DISCUSSION
LGG is widely used as a probiotic, and this strain has the ability to survive and transiently colonize the gastrointestinal tract in animal models and humans (16, 19). In this context, the presence in LGG of genes involved in the utilization of host-derived glycans constitutes a competitive advantage for its persistence in the gut. LGG codes for about 40 putative glycosidases, many carbohydrate transporters, and catabolic enzymes that would allow it to take advantage of the carbohydrate resources of the mucosa (19, 30). Thus, LGG is able to grow with mucin as a carbon source (31), and it possesses extracellular factors that upregulate the production of mucin in the colonic epithelium (32). Some authors have reported that LGG, although being able to use mucus-derived glycans, was not capable of fermenting l-fucose (31). However, results obtained by others (30) and those presented here showed that LGG ferments l-fucose. The explanation for this may derive from differences in isolates of LGG from different laboratories. Thus, it has been reported that LGG isolates from diverse commercial probiotic products present heterogeneity in their genome sequences, which include point mutations and deletions of big portions of the chromosome (33).
We have established here that the fuc genes present in the LGG genome are indeed responsible for the observed l-fucose fermenting capacity of this strain. The presence of specific fuc genes, mutant analysis and the fact that 1,2-propanediol was detected in culture supernatants of LGG grown with l-fucose supports the idea that the utilization of this sugar in LGG follows the same pathway to that described in E. coli. Depending on the growth conditions (anaerobic or aerobic), E. coli directs the lactaldehyde resulting from l-fucose catabolism toward 1,2-propanediol (FucO activity) or lactate (AldA activity) (10). Although the LGG genes responsible for lactaldehyde metabolism have not yet been identified, it is possible that the redox status dictates its metabolism toward these two compounds in L. rhamnosus. However, we showed that l-fucose catabolism in LGG was only favored under anaerobic conditions, which suggests that lactaldehyde conversion to l-lactate is not efficient in this strain.
The utilization of carbohydrate resources typically found in the gastrointestinal niche is a characteristic of members of the intestinal microbiota, such as Bacteroides or Bifidobacterium but also of intestinal lactobacilli. Consequently, the genomes of species such as Lactobacillus acidophilus, Lactobacillus johnsonii, or Lactobacillus gasseri carry genes encoding many carbohydrate catabolic pathways, although they lack sialidases, α-fucosidases, or N-acetylglucosaminidases, which are typical in other intestinal commensals or pathogens (34–36). In this respect, the L. casei/L. paracasei/L. rhamnosus group present unique features, as these species are the only lactobacilli where α-fucosidades and genes for the utilization of l-fucose are present. A recent analysis of 100 L. rhamnosus strains isolated from several sources showed that they can be clustered on the basis of their sugar fermenting capacity (30). Strains with an l-fucose-fermenting phenotype were derived from the oral and intestinal habitat, whereas strains from dairy origin are l-fucose negative. This is in agreement with the hypothesis that lactobacilli evolved by gene loss and acquisition driven by the particular niches they inhabit. In some cases, this resulted in genome reduction after adaptation to less complex environments such as milk (25). Whether the L. rhamnosus fuc cluster was present in the ancestor of the L. casei/L. paracasei/L. rhamnosus group, and it was most recently lost in L. paracasei and in some strains of L. rhamnosus or it represents a recent acquisition remains to be elucidated. Interestingly, the fuc locus of LGG is located in a chromosomal region which seems dedicated to the catabolism of l-deoxy sugars, since it also contains a cluster with genes (rha) putatively involved in l-rhamnose catabolism, the sugar from which the species name is derived. However, previous data and our own results (data not shown) showed that LGG was not able to utilize l-rhamnose (19, 30).
The LGG fuc genes are subject to a dual regulation: induction by growth on l-fucose triggered by FucR and catabolite repression probably dependent on the CcpA transcriptional regulator. In B. thetaiotaomicron l-fucose is the inducer of the fuc genes through its binding to a GntR-superfamily transcriptional repressor (11). Transcriptional analysis in LGG grown with Fuc-α1,3-GlcNAc, which leads to the intracellular generation of l-fucose, excludes the fact that l-fucose itself could be the effector of FucR, as it failed to induce the fuc genes in a mutant deficient in the first step of the catabolic route (fucI). Furthermore, FucP does not participate in the regulation of fuc genes. Genetic studies in E. coli pointed to fuculose-1-phosphate as the inducer molecule of the fuc operon via FucR. The same situation probably exists for L. rhamnosus, since the regulators of both species belong to the same DeoR class. FucR DNA-binding sites have not been experimentally established for E. coli or other bacteria. In enterobacteria a consensus sequence for binding can be derived from the alignment of several fuc promoters, resulting in a 36-bp imperfect inverted repeated sequence. Inspection of the LGG fuc promoters did not reveal similar sequences, but two tandem repetitions of the sequence TGAAGAAAA separated by 14 bp are present in the fucI promoter, and another similar sequence is present in the fucA-fucR intergenic region. Whether these sequences are the target for FucR has to be verified.
L. casei BL23 and LGG are the only lactobacillus strains described thus far that are able to use fucosylated oligosaccharides (29, 37). Both strains share the same set of α-l-fucosidases (AlfA, AlfB, and AlfC), which can act on specific oligosaccharides derived from glycoconjugates present at mucosal surfaces, such as the LewisX antigen core, but also on human milk oligosaccharides (37). In addition, the alfRB-EFG operon, responsible for the uptake and hydrolysis of Fuc-α1,3-GlcNAc, is present in both strains (29). However, BL23 does not possess a pathway for l-fucose and, similar to LGG mutants in fucI and fucR, expels the l-fucose moiety of Fuc-α1,3-GlcNAc. A minimal amount of the l-fucose derived from Fuc-α1,3-GlcNAc was detected in the supernatants of the LGG fucP mutant (3%), whereas this sugar was totally absent in the supernatants from the wild type under the same conditions. This suggests that part of the intracellularly generated l-fucose is diffusing to the extracellular medium and that, in the absence of FucP, this released sugar cannot re-enter the cell being metabolized. In accordance with the activating role of FucR, no induction of the fuc genes was detected in the fucR mutant when it was grown with Fuc-α1,3-GlcNAc. In this mutant, only 30% of the intracellular l-fucose from the disaccharide was utilized, which suggests that the basal transcription of the fuc catabolic genes in the absence of activator allows a limited l-fucose metabolism when this sugar is produced intracellularly.
Intestinal bacteria belonging to Bifidobacterium and Bacteroides are well adapted to exploit the intestinal carbohydrate resources, and they possess α-l-fucosidases that allow them to scavenge l-fucose from the mucosa (38, 39). However, intestinal commensals, such as E. coli, that do not express α-l-fucosidases depend on other bacterial groups for l-fucose release from fucose-containing glycans. The LGG fuc genes studied here probably represent an adaptation of this strain to dwell in the particular niche of the gastrointestinal tract. However, since LGG α-l-fucosidases are intracellular enzymes, this strain must probably rely on the fucosidase and glycosidase activities from other members of the intestinal microbiota for the cross-feeding of l-fucose and fucosyl-oligosaccharides.
ACKNOWLEDGMENTS
This study was supported by the Spanish Ministry of Economy and Competitiveness (MINECO)/FEDER through projects AGL2010-18696 and AGL2013-40657-R and by the Valencian Government through project ACOMP/2012/030. J.E.B. received a Santiago Grisolía Fellowship from the Generalitat Valenciana.
REFERENCES
- 1.Becker DJ, Lowe JB. 2003. Fucose: biosynthesis and biological function in mammals. Glycobiology 13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 2.Bode L. 2009. Human milk oligosaccharides: prebiotics and beyond. Nutr Rev 67(Suppl 2):S183–S191. doi: 10.1111/j.1753-4887.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- 3.Coyne MJ, Reinap B, Lee MM, Comstock LE. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 4.Wacklin P, Makivuokko H, Alakulppi N, Nikkila J, Tenkanen H, Rabina J, Partanen J, Aranko K, Matto J. 2011. Secretor genotype (FUT2 gene) is strongly associated with the composition of bifidobacteria in the human intestine. PLoS One 6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. 2013. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. 2011. l-Fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc Natl Acad Sci U S A 108:7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. 2007. l-Fucose stimulates utilization of d-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun 75:5465–5475. doi: 10.1128/IAI.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YM, Zhu Y, Lin EC. 1987. The organization of the fuc regulon specifying l-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet 210:331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- 9.Chen YM, Zhu Y, Lin EC. 1987. NAD-linked aldehyde dehydrogenase for aerobic utilization of l-fucose and l-rhamnose by Escherichia coli. J Bacteriol 169:3289–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldoma L, Aguilar J. 1988. Metabolism of l-fucose and l-rhamnose in Escherichia coli: aerobic-anaerobic regulation of l-lactaldehyde dissimilation. J Bacteriol 170:416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A 96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins MA, Suits MD, Marsters C, Boraston AB. 2014. Structural and functional analysis of fucose-processing enzymes from Streptococcus pneumoniae. J Mol Biol 426:1469–1482. doi: 10.1016/j.jmb.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Saxelin M, Tynkkynen S, Mattila-Sandholm T, de Vos WM. 2005. Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol 16:204–211. doi: 10.1016/j.copbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Yan F, Polk DB. 2012. GG: an updated strategy to use microbial products to promote health. Funct Food Rev 4:77–84. [PMC free article] [PubMed] [Google Scholar]
- 16.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Adaptation factors of the probiotic Lactobacillus rhamnosus GG. Benef Microbes 1:335–342. doi: 10.3920/BM2010.0032. [DOI] [PubMed] [Google Scholar]
- 17.Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, Savijoki K, Nyman TA, Surakka A, Salusjarvi T, de Vos WM, Tynkkynen S, Kalkkinen N, Varmanen P. 2011. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics 10:M110.002741. doi: 10.1074/mcp.M110.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reunanen J, von Ossowski I, Hendrickx AP, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78:2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leloup L, Ehrlich SD, Zagorec M, Morel-Deville F. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol 63:2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Keersmaecker SC, Braeken K, Verhoeven TL, Perea Velez M, Lebeer S, Vanderleyden J, Hols P. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl Environ Microbiol 72:4923–4930. doi: 10.1128/AEM.02605-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchiyama I, Higuchi T, Kawai M. 2010. MBGD update 2010: toward a comprehensive resource for exploring microbial genome diversity. Nucleic Acids Res 38:D361–D365. doi: 10.1093/nar/gkp948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins MA, Boraston AB. 2011. Structure of the fucose mutarotase from Streptococcus pneumoniae in complex with l-fucose. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1524–1530. doi: 10.1107/S1744309111046343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, Horvath P, Heidenreich J, Perna NT, Barrangou R, Steele JL. 2012. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533. doi: 10.1186/1471-2164-13-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landete JM, Ferrer S, Monedero V, Zuñiga M. 2013. Malic enzyme and malolactic enzyme pathways are functionally linked but independently regulated in Lactobacillus casei BL23. Appl Environ Microbiol 79:5509–5518. doi: 10.1128/AEM.01177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:1366–1381. doi: 10.1128/JB.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutscher J, Ake FM, Derkaoui M, Zebre AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Diaz J, Rubio-del-Campo A, Yebra MJ. 2012. Lactobacillus casei ferments the N-acetylglucosamine moiety of fucosyl-α-1,3-N-acetylglucosamine and excretes l-fucose. Appl Environ Microbiol 78:4613–4619. doi: 10.1128/AEM.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douillard FP, Ribbera A, Kant R, Pietila TE, Jarvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lahteinen T, Brouns SJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez B, Saad N, Schmitter JM, Bressollier P, Urdaci MC. 2010. Adhesive properties, extracellular protein production, and metabolism in the Lactobacillus rhamnosus GG strain when grown in the presence of mucin. J Microbiol Biotechnol 20:978–984. doi: 10.4014/jmb.0911.11007. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, Yan F. 2014. Activation of EGF receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J Biol Chem 289:20234–20244. doi: 10.1074/jbc.M114.553800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sybesma W, Molenaar D, van IJcken W, Venema K, Kort R. 2013. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol 79:2233–2239. doi: 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl Environ Microbiol 74:4610–4625. doi: 10.1128/AEM.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci U S A 101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Diaz J, Monedero V, Yebra MJ. 2011. Utilization of natural fucosylated oligosaccharides by three novel α-l-fucosidases from a probiotic Lactobacillus casei strain. Appl Environ Microbiol 77:703–705. doi: 10.1128/AEM.01906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakurama H, Tsutsumi E, Ashida H, Katayama T, Yamamoto K, Kumagai H. 2012. Differences in the substrate specificities and active-site structures of two α-l-fucosidases (glycoside hydrolase family 29) from Bacteroides thetaiotaomicron. Biosci Biotechnol Biochem 76:1022–1024. doi: 10.1271/bbb.111004. [DOI] [PubMed] [Google Scholar]