Abstract
The genome of the thermophilic bacterium Caldicellulosiruptor bescii encodes three multimodular enzymes with identical C-terminal domain organizations containing two consecutive CBM3b modules and one glycoside hydrolase (GH) family 48 (GH48) catalytic module. However, the three proteins differ much in their N termini. Among these proteins, CelA (or C. bescii Cel9A [CbCel9A]/Cel48A) with a GH9/CBM3c binary partner in the N terminus has been shown to use a novel strategy to degrade crystalline cellulose, which leads to its outstanding cellulose-cleaving activity. Here we show that C. bescii Xyn10C (CbXyn10C), the N-terminal GH10 domain from CbXyn10C/Cel48B, can also degrade crystalline cellulose, in addition to heterogeneous xylans and barley β-glucan. The data from substrate competition assays, mutational studies, molecular modeling, and docking point analyses point to the existence of only one catalytic center in the bifunctional xylanase/β-glucanase. The specific activities of the recombinant CbXyn10C on Avicel and filter paper were comparable to those of GH9/CBM3c of the robust CelA expressed in Escherichia coli. Appending one or two cellulose-binding CBM3bs enhanced the activities of CbXyn10C in degrading crystalline celluloses, which were again comparable to those of the GH9/CBM3c-CBM3b-CBM3b truncation mutant of CelA. Since CbXyn10C/Cel48B and CelA have similar domain organizations and high sequence homology, the endocellulase activity observed in CbXyn10C leads us to speculate that CbXyn10C/Cel48B may use the same strategy that CelA uses to hydrolyze crystalline cellulose, thus helping the excellent crystalline cellulose degrader C. bescii acquire energy from the environment. In addition, we also demonstrate that CbXyn10C may be an interesting candidate enzyme for biotechnology due to its versatility in hydrolyzing multiple substrates with different glycosidic linkages.
INTRODUCTION
Plant cell wall polysaccharides (PCWPs), composed mainly of cellulose and hemicellulose, are a promising rich resource for renewable biofuel development (1). The complete deconstruction of PCWPs into fermentable, simple mono- or oligosaccharides requires the concerted action of a complex array of glycoside hydrolases (GHs), including cellulases and hemicellulases (2, 3). The genomes of Gram-positive bacteria of the genus Caldicellulosiruptor encode an arsenal of thermophilic plant cell wall polysaccharide-degrading enzymes (3–5), which are appealing candidates in the design of novel, robust enzyme cocktails for PCWP deconstruction.
In the genome of Caldicellulosiruptor bescii, there is a gene cluster containing three genes which encode proteins harboring two tandemly linked CBM3b modules and a GH family 48 (GH48) cellobiohydrolase in the C terminus. These CBM3b and GH48 modules, as well as their linker sequences, are extremely similar at the level of the amino acid sequence (see Fig. S1 in the supplemental material). Notably, however, the three multimodular enzymes differ much in their N termini: CelA (or C. bescii Cel9A [CbCel9A]/Cel48A) has a GH9/CBM3c catalytic binary partner, C. bescii Xyn10C (CbXyn10C)/Cel48B (GenBank accession number ACM60945) has a GH10 catalytic domain, and the third protein (GenBank accession number ACM60948) has a GH74 domain. While the GH9 and GH48 catalytic domains in CelA have been extensively characterized (6, 7), no analysis of the GH10 and GH74 catalytic domains has been carried out.
Many characterized xylanases belong to families 10 and 11 of the glycoside hydrolases according to the classification of the Carbohydrate-Active Enzymes (CAZy) Database (http://www.cazy.org) (8). Members of GH11 were generally regarded to be true xylanases due to their stringent substrate specificity on xylans. In contrast, some members of the GH10 xylanases can hydrolyze other polysaccharides, such as tomatine (9) or barley β-glucan (10), in addition to xylans. A few GH10 xylanases even have perceivable activity on artificial cellulosic substrates, such as p-nitrophenol-β-cellobiose (pNPC) (11) or sodium carboxymethyl cellulose (CMC) (12). However, to the authors' knowledge, no GH10 xylanase has been reported to be able to hydrolyze both xylans and crystalline cellulose, the two major components of PCWPs, as feedstocks for biofuels.
In the present study, we focused on characterization of the GH10 module from the multimodular protein CbXyn10C/Cel48B. We cloned and functionally expressed the N-terminal GH10 domain in Escherichia coli. The recombinant protein, designated CbXyn10C, displayed activities on heterogeneous xylans, barley β-glucan, and, surprisingly, all tested cellulosic substrates, including CMC, phosphoric acid swollen cellulose (PASC), Avicel, and filter paper. The molecular mechanism by which CbXyn10C hydrolyzes xylan and cellulose was also explored using combinatorial methods.
MATERIALS AND METHODS
Gene identification and cloning.
The E. coli Trans1 strain (Transgen, Beijing, China) was used for plasmid construction and propagation throughout this study. The genome sequence of C. bescii was uploaded and analyzed on the RAST (Rapid Annotation Using Subsystem Technology; http://rast.nmpdr.org) server (13) as described previously (3). A gene encoding a multimodular protein (GenBank accession number ACM60945) which contained a GH10 domain at its N terminus, two CBM3b modules in the middle, and a GH48 domain at the C terminus was identified to be cb1944, and the protein that it encoded was designated CbXyn10C/Cel48B. The gene encoding the N-terminal GH10 domain (amino acids [aa] 38 to 376) was amplified from the genomic DNA of C. bescii DSMZ 6725 (purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) using PrimeSTAR HS DNA polymerase (TaKaRa, Shiga, Japan) and cloned into pET-46/Ek-LIC to obtain pET46-CbXyn10C according to the manual of the manufacturer (Merck KGaA, Darmstadt, Germany). Additionally, the genes encoding the N-terminal GH10 domain with one CBM3b module (aa 38 to 599) or two CBM3b modules (aa 38 to 852) and a truncation mutant of CelD (designated CelDTM1) (14) were amplified from the genomic DNA of C. bescii and subcloned into the EcoRI/XhoI-digested pET-28a(+) plasmid to obtain pET28-CbXyn10C-CBM3b, pET28-CbXyn10C-2CBM3b, and pET28-CelDTM1, which were used for expression of recombinant CbXyn10C-CBM3b, CbXyn10C-CBM3b-CBM3b, and CelDTM1, respectively. The primers used for cloning the genes are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequencea (5′–3′) | Usage |
|---|---|---|
| CbXyn10C-F | GACGACGACAAGATGCCTGACTGGAACATTCCAAGTTTATATG | Cloning of C. bescii xyn10C into pET46-Ek/LIC |
| CbXyn10C-R | GAGGAGAAGCCCGGTTATGATGTCCCTGATGCTTCTATTACTGC | |
| CbXyn10C-CBM3b-F | GGGAATTCCCTGACTGGAACATTCCAAGTTTATATG | Cloning of C. bescii xyn10C-CBM3b into pET-28a(+) |
| CbXyn10C-CBM3b-R | GGCTCGAGGGTGTCGGTGTCACTGTCGGTG | |
| CbXyn10C-2CBM3b-F | GGGAATTCCCTGACTGGAACATTCCAAGTTTATATG | Cloning of C. bescii xyn10C-CBM3b-CBM3b into pET-28a(+) |
| CbXyn10C-2CBM3b-R | GGCTCGAGGGTTGGAGTTGGTGCTGGTGTG | Cloning of celDTM1 into pET28a |
| CelDTM1-F | GGGAATTCCAGAGCATACTGTATGAAAAGG | |
| CelDTM1-R | GGCTCGAGTTACTCAAAAAGGATATTGGTAAATC | |
| Q94A-F | CTTTAGTGTGGCACAGCGCGACGCCTGATTGG | Mutation of Q94 to alanine |
| Q94A-R | CCAATCAGGCGTCGCGCTGTGCCACACTAAAG | |
| L306A-F | TTATTCATGGGCAAGATCTTTTAACG | Mutation of L306 to alanine |
| L306A-R | CGTTAAAAGATCTTGCCCATGAATAA | |
| E140A-F | ACGTTGTAAATGCAGCCATTGATGAGAATCAGTC | Mutation of E140 to alanine |
| E140A-R | TCATCAATGGCTGCATTTACAACGTCCCATGC | |
| E248A-F | ATAACAGCGCTTGATATGAGTTTATACAATTACGG | Mutation of E248 to alanine |
| E248A-R | CTCATATCAAGCGCTGTTATATGAATTTCTATGC |
The underlined nucleotides are the EcoRI (GAATTC) and XhoI (CTCGAG) restriction sites.
Gene expression and protein purification.
The pET46-CbXyn10C, pET28-CbXyn10C-CBM3b, pET28-CbXyn10C-2CBM3b, and pET28-CelDTM1 plasmids were individually transformed into E. coli BL21(DE3) competent cells, and positive colonies on LB plates supplemented with 100 μg/ml of ampicillin were selected. Five to 10 colonies of the BL21(DE3) strains carrying one of the plasmids were inoculated into 10 ml of LB medium with 100 μg/ml of ampicillin and cultured at 37°C for 6 h. The preculture was then transferred to 1 liter LB medium with 100 μg/ml of ampicillin and cultured with shaking for 2.5 h. Then, 0.8 mM (final concentration) IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the medium and the culture was continued at 25°C for another 16 h. The cells were harvested by centrifugation at 12,000 × g for 10 min and then resuspended in a binding buffer (20 mM Tris-HCl, 500 mM NaCl, 10% glycerol, pH 7.6). The cell wall was disrupted by sonication, followed by centrifugation at 10,000 × g for 15 min. The supernatant was separated and loaded into a nickel-nitrilotriacetic acid (NTA) chelating column (GE Healthcare, Uppsala, Sweden). The bound protein was eluted from the resin with a linear imidazole gradient (40 to 400 mM) in the binding buffer. The eluted protein was analyzed on a 12% SDS-polyacrylamide gel, and the pure fractions were mixed. The purified protein was extensively dialyzed against a protein storage buffer containing 50 mM Tris-HCl and 150 mM NaCl, pH 7.5.
Substrate specificity.
The substrate specificity of CbXyn10C was determined by incubating 10 μM purified CbXyn10C in pH 6.5 McIlvaine buffer (200 mM sodium phosphate, 100 mM sodium citrate) at 75°C with different natural polysaccharides or artificial substrates, including 10 mg/ml of beech wood xylan, wheat arabinoxylan (WAX), barley β-glucan, lichenin, laminarin, CMC, Avicel, filter paper, arabic gum, debranched arabinan, sugar beet arabinan, and xyloglucan; 5 mg/ml of konjac glucomannan, carob bean gum, locust bean gum (LBG), and guar gum; 2.5 mg/ml of PASC and pectin; and 1 mM p-nitrophenyl β-d-glucoside (pNPG), p-nitrophenyl β-d-xylopyranoside (pNPX), and pNPC in a total volume of 1.0 ml. The amount of reducing sugars released into the reaction mixture was measured by using the 3,5-dinitrosalicylic acid (DNS) method (15). The release of p-nitrophenol (pNP) was measured on a Specord 205 spectrophotometer (Analytik, Jena, Germany) by monitoring the change in the absorbance (ΔA) at 405 nm.
Determination of optimal pH and temperature.
The optimal pH for CbXyn10C activity was determined in McIlvaine buffer (pH 3.0 to 8.0), 100 mM Tris-HCl (pH 7.0 to 9.0), and 100 mM glycine-NaOH (pH 9.0 to 12.0) in a total volume of 1.0 ml at 80°C for 10 min. To determine the optimal temperature for CbXyn10C activity, assays of activity were performed at temperatures ranging from 37°C to 95°C at pH 6.5. For analysis of pH stability, the protein was preincubated in 100 mM glycine-HCl (pH 1.0 to 3.0), McIlvaine buffer (pH 3.0 to 8.0), Tris-HCl (pH 7.0 to 9.0), or glycine-NaOH (pH 9.0 to 12.0) without substrate at 37°C for 1 h. Then, the residual activities were measured under optimal conditions (pH 6.5, 85°C, and 10 min). The thermal stability of CbXyn10C was determined by measuring its residual activity after incubation at 80°C, 85°C, or 90°C for different periods of time.
Assay of enzymatic activity.
To determine the specific activities of CbXyn10C on xylans, cellulose, and β-glucans, 100 μl of appropriately diluted enzyme was incubated with 900 μl of beech wood xylan, WAX, barley β-glucan, CMC, Avicel, filter paper, microcrystalline cellulose (MCC; catalog no. 310697; Sigma), PASC, and lichenin (10 mg/ml final) at the optimal temperature (or 75°C for Avicel, filter paper, and MCC) for certain periods of time. Similarly, the specific activity of CbXyn10C-CBM3b or CbXyn10C-CBM3b-CBM3b on 10 mg/ml of Avicel, MCC, or filter paper was also determined. The released reducing sugars were determined by using the DNS method. The kinetic parameters of the wild-type CbXyn10C as well as its site-directed mutants were determined in McIlvaine buffer containing 1 to 20 mg/ml of beech wood xylan, WAX, barley β-glucan, and CMC-Na or 0.1 to 4.0 mM pNPX and pNPG at the optimal pH and temperature. The Km and kcat values were estimated by fitting the data to the Michaelis-Menten equation using GraphPad Prism (version 5.01) software (GraphPad, San Diego, CA).
To study the effect of removing amorphous cellulose from crystalline cellulose on the hydrolyzing activity of CbXyn10C, 10 mg/ml of Avicel was first incubated with 10 μM CelDTM1 (14) in pH 6.5 McIlvaine buffer in a total volume of 1.0 ml at 70°C for 16 h. The ability of CelDTM1 to degrade amorphous cellulose has been reported by Velikodvorskaya et al. (14). As a control, Avicel was incubated with CelDTM1 that had been inactivated by boiling at 100°C for 10 min. After the pretreatment, the reaction mixture was heated at 100°C for 10 min to denature the CelDTM1. Then, 10 μM CbXyn10C was added to the pretreated Avicel and the reaction was carried out at 75°C. At different time points (30 min, 60 min, 90 min, 120 min, and 150 min), samples were taken out and the amounts of reducing sugars were measured using the DNS method.
Site-directed mutagenesis.
In total, four single mutants (the Q94A, L306A, E140A, and E248A mutants) and two double mutants (the Q94A/L306A and E140A/E248A mutants) were generated by site-directed mutagenesis, which was carried out by using a Fast mutagenesis system (Transgen, Beijing, China) according to the instructions of the manufacturer. Briefly, primer pairs (listed in Table 1) bearing the mutations were used to amplify the pET46-CbXyn10C plasmid. The PCR products were then transformed into DMT chemically competent E. coli cells (Transgen, Beijing, China). The mutant plasmids were verified for their integrity by sequencing and transformed into E. coli BL21(DE3) cells for protein expression. The procedure used for purification of the mutants was the same as that used for wild-type CbXyn10C.
Hydrolysis of Avicel, WAX, and xylo- and cellooligosaccharides for thin-layer chromatography (TLC) and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analyses.
One micromolar CbXyn10C was incubated with 20 mg/ml of Avicel in McIlvaine buffer in a total volume of 1.0 ml at pH 6.5 and 75°C for 16 h. Before analysis of the hydrolysis products, the enzyme was removed from the reaction system by using a Nanosep centrifugal 3K device (Pall, New York, NY). For oligosaccharides or WAX, the reactions were performed by incubating 1 μM CbXyn10C with 2.5 mg/ml each of the xylo- or cellooligosaccharides or 1 mg/ml of WAX in a total volume of 40 μl or 1 ml at pH 6.5 and 75°C for 16 h.
TLC.
TLC was used for analyses of the end products of hydrolysis of xylan, xylooligosaccharides, or cellooligosaccharides. Hydrolysis products were spotted on 10- by 20-cm Whatman 60-Å silica gel with a thickness of 0.25 mm (Silica gel 60 F254; Merck, Darmstadt, Germany). The gel was air dried at room temperature and then developed in a solution containing n-butanol–acetic acid–H2O with a volumetric ratio of 2:1:1. After drying at room temperature, the plate was soaked in ethanol-H2SO4 (19:1, vol/vol) and then incubated at 110°C for 5 min until clear spots could be visualized.
HPAEC-PAD.
HPAEC-PAD was used for analysis of the hydrolysis products of Avicel. Briefly, 500 μl of appropriately diluted hydrolysates was analyzed on a Thermo Scientific Dionex ICS-5000 (Dionex Corporation, Sunnyvale, CA) high-performance liquid chromatography (HPLC) instrument equipped with a CarboPac PA100 guard column (4 by 50 mm) and an analytical column (4 by 250 mm). The flow rate was 1 ml/min, and the temperature was 22°C. The samples were resolved in a mobile phase of 100 mM NaOH using glucose and cellooligosaccharides (G2 to G6) as standards.
Amino acid sequence alignment.
ClustalW software (16) and the online BoxShade server (http://www.ch.embnet.org/software/BOX_form.html) were used to align the amino acid sequence of CbXyn10C with the amino acid sequences of selected GH10 enzymes from Cellulomonas fimi (GenBank accession number AAA56792.1), Streptomyces lividans (GenBank accession number WP_003978188.1), Paenibacillus sp. strain E18 (GenBank accession number ACY69972.1), Pseudomonas cellulosa (GenBank accession number WP_012488068), and Demequina sp. strain JK4 (GenBank accession number ACM41799.1).
Hydrolysis of mixed substrates.
To determine whether recombinant CbXyn10C utilizes the same or separate active centers to hydrolyze multiple substrates, substrate competition assays using xylan and barley β-glucan were performed as described previously (10). One hundred microliters of appropriately diluted CbXyn10C was incubated with mixtures of the substrates with a constant total concentration of 20 mg/ml but various concentrations of beech wood xylan (16 to 4 mg/ml) and barley β-glucan (4 to 16 mg/ml) in pH 6.5 McIlvaine buffer in a total volume of 1 ml at 85°C for 10 min. The observed overall activities of hydrolysis were compared with the theoretical values calculated on the basis of the assumption of the presence of a single or two catalytic centers (17).
Homology modeling of tertiary structure of CbXyn10C and molecular docking.
A homology model of CbXyn10C was built using the ModWeb server (https://modbase.compbio.ucsf.edu/modweb/) and the Bacillus alkali thermostable GH10 xylanase (PDB accession number 2F8Q), to which CbXyn10C has 45% identity, as a template. To perform docking studies, the coordinate files of cellopentaose and xylopentaose ligands were prepared in the PRODRG server (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg) (18). The online AutoDockVina server (http://vina.scripps.edu/) (19) was employed to calculate the relative binding free energy and generate ligand-protein complexes.
Viscosity reduction of mash.
Mash was prepared as described by Celestino et al. (20) with some modifications. Five grams of triturated and filtered malt was resuspended in 50 ml of McIlvaine buffer (pH 6.5). CbXyn10C (50 U, 80 U, 100 U, or 200 U) was incubated with 50 ml of mash at 60°C for 30 min, 70°C for 30 min, 80°C for 60 min, and 85°C for 30 min and then boiled for 5 min. After centrifugation and filtration, the viscosity of the mash supernatant (5 ml) was measured at 25°C by a capillary viscometer with a 0.6-mm diameter. The reduction of the viscosity was calculated according to the standard equation using the mash viscosity in the absence of enzyme as a control.
RESULTS
Cloning, expression, and purification of CbXyn10C.
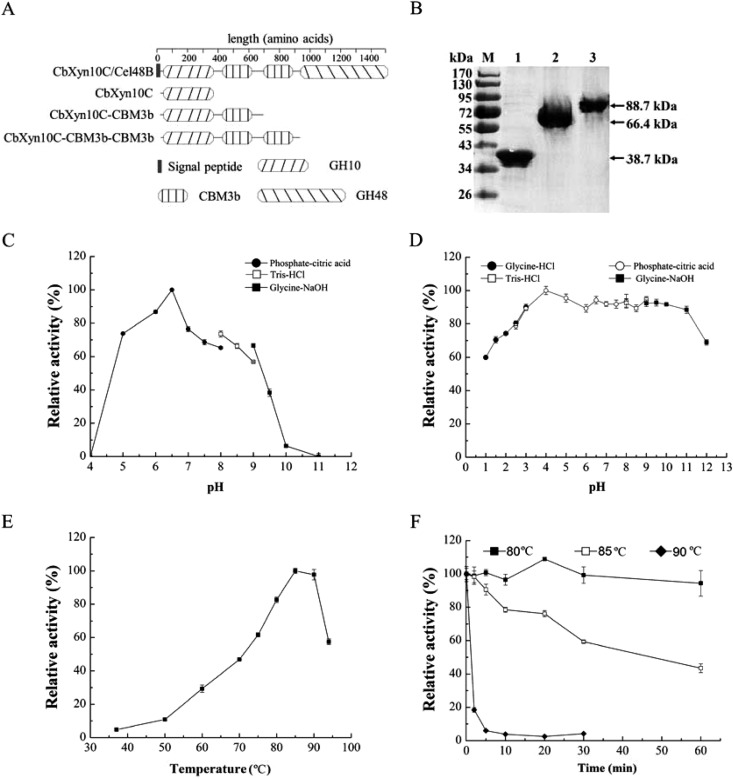
A gene (cb1944) encoding a putative multimodular enzyme was identified from the genome of C. bescii. Analysis of the Pfam database (http://pfam.xfam.org/) for the amino acid sequence of the protein indicated that this large protein (1,478 aa) contains one N-terminal GH10 domain, two nearly identical CBM3b domains in the middle, and one C-terminal GH48 domain (Fig. 1A). The CBM3b and GH48 domains are highly homologous to those in CbCel9B/Man5A (4), CbMan5B/Cel44A (21), and CbCel9A/Cel48A (6) or CelA (7, 22) from the same organism. Two GH family 10 xylanases (CbXyn10A and CbXyn10B) from C. bescii have been biochemically analyzed before. Thus, the N-terminal GH10 catalytic domain was designated CbXyn10C, and the full-length protein was designated CbXyn10C/Cel48B. Although CbXyn10C was computationally predicted to be a xylanase, no data regarding its biochemical properties have been reported. The genes encoding CbXyn10C, CbXyn10C-CBM3b, and CbXyn10C-CBM3b-CBM3b, which were devoid of a signal peptide, were thus cloned from the genomic DNA of C. bescii (Fig. 1A). The recombinant proteins were successfully expressed in a soluble form in E. coli and purified to near homogeneity, as resolved by SDS-PAGE (Fig. 1B).
FIG 1.
Gene cloning, expression, and purification of CbXyn10C, CbXyn10C-CBM3b, and CbXyn10C-CBM3b-CBM3b and the basic biochemical properties of CbXyn10C. (A) Domain organization of CbXyn10C/Cel48B. (B) Purification of CbXyn10C, CbXyn10C-CBM3b, and CbXyn10C-CBM3b-CBM3b. Lane M, molecular mass markers; lane 1, CbXyn10C; lane 2, CbXyn10C-CBM3b; lane 3, CbXyn10C-CBM3b-CBM3b. (C) Optimal pH for CbXyn10C activity. (D) pH stability of CbXyn10C. (E) Optimal temperature for CbXyn10C activity. (F) Thermostability of CbXyn10C. In panels C to F, we used the relative activities of CbXyn10C (in percent), with the maximal activities at the optimal pH (32,000 ± 120 μmol/min/μmol of enzyme) (C) and the optimal temperature (39,000 ± 160 μmol/min/μmol of enzyme) (E) or the activities before treatment (38,000 ± 130 μmol/min/μmol of enzyme [D]; 39,000 ± 130 μmol/min/μmol of enzyme [F]) being used as the reference activities (100%).
pH and temperature profile of CbXyn10C.
The optimal pH for CbXyn10C activity is 6.5 (Fig. 1C), which is close to the pHs of the characterized C. bescii xylan-degrading enzymes, including CbXyn10A, CbXyn10B, CbAra51A, CbAgu67A, and CbXyl3A (5). CbXyn10C was a thermophilic enzyme for which the optimal temperature was 85 to 90°C (Fig. 1E), as determined using beech wood xylan as the substrate. The thermophilic character of CbXyn10C is similar to that of other functionally characterized C. bescii glycoside hydrolases (4, 6, 22). Since C. bescii is a hyperthermophilic bacterium, this is not unexpected. CbXyn10C was stable under extremely acidic (pH 1.0) and alkaline (pH 12.0) conditions, retaining over 60% of its residual activity (Fig. 1D). When CbXyn10C was incubated in buffers with pHs ranging from 1.0 to 12.0 for 1 h, it retained over 80% of its activity at pH 2.5 to 11.0 (Fig. 1D). CbXyn10C did not lose any activity when it was incubated at 80°C for 1 h (Fig. 1F). At 85°C it gradually lost its activity, with a residual activity of 43% after 1 h. At 90°C, it rapidly lost all its activity within 20 min (Fig. 1F).
CbXyn10C hydrolyzes multiple glycosidic linkages.
CbXyn10C was incubated with a variety of model polysaccharide substrates, including heterogeneous xylans (beech wood xylan and WAX), β-glucans (barley β-glucan, lichenin, laminarin, and celluloses, including CMC, PASC, Avicel, and filter paper), pectin, mannans (carob bean gum, guar gum, locust bean gum), konjac flour, arabinans (arabic gum, debranched arabinan, sugar beet arabinan), and xyloglucan. On the basis of the findings of the reducing sugar assay, CbXyn10C was most active on the xylan substrates (Fig. 2), with specific activities of 39,000 ± 160 and 51,000 ± 240 μmol/min/μmol of enzyme on beech wood xylan and WAX, respectively (Table 2). It also had activities on barley β-glucan (3,400 ± 15 μmol/min/μmol of enzyme) and lichenin (180 ± 3.8 μmol/min/μmol of enzyme), two substrates with a mixed linkage of β-1,3/1,4-glucosidic bonds, but no activity on laminarin, which has β-1,3/1,6-glucosidic linkages (Fig. 2; Table 2) or laminarihexaose (a glucose hexamer with a β-1,3-glucosidic linkage; data not shown). Unexpectedly, CbXyn10C displayed activities not only on soluble CMC (39 ± 3.3 μmol/min/μmol of enzyme) but also on the amorphous cellulose PASC (1.2 ± 0.4 μmol/min/μmol of enzyme) and even on the crystalline cellulosic substrates Avicel (3.4 ± 0.8 μmol/min/μmol of enzyme) and filter paper (5.4 ± 0.3 μmol/min/μmol of enzyme) (Fig. 2; Table 2). Note that the activities of CbXyn10C on four different forms of cellulose (CMC, PASC, Avicel, and filter paper) did not differ much. The cellulose-binding CBM3bs have been demonstrated to enhance the hydrolysis of crystalline cellulose by GH9/CBM3c in CbCel9B/Man5A and CelA (4, 6). Since CbXyn10C was able to hydrolyze Avicel and filter paper, two commonly used model crystalline cellulosic substrates, we also determined if the two CBM3bs in CbXyn10C/Cel48B could affect the hydrolysis of these two substrates by CbXyn10C. Not surprisingly, appending one or two CBM3bs significantly increased the activity of CbXyn10C on the two substrates by 2.1- to 2.6-fold (Table 3).
FIG 2.
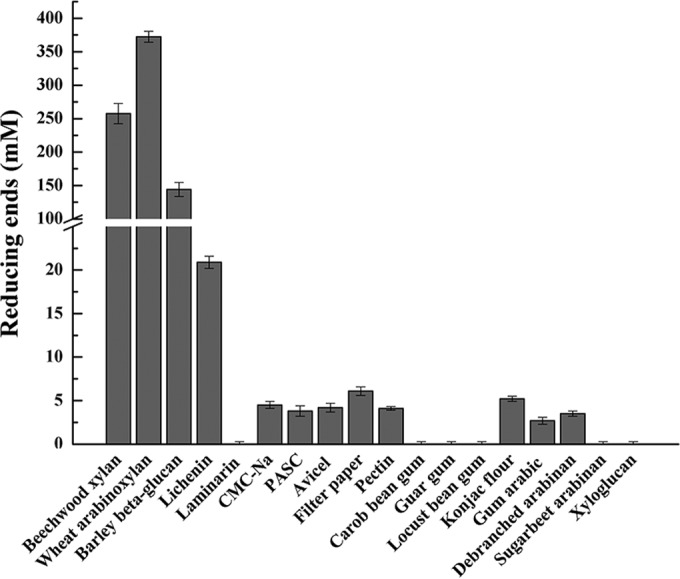
Screening of activity of CbXyn10C on various polysaccharides. The reaction mixture contained 10 μM CbXyn10C in pH 6.5 McIlvaine buffer with each of a series of polysaccharides, including 10 mg/ml of beech wood xylan, WAX, barley β-glucan, lichenin, laminarin, CMC, Avicel, filter paper, gum arabic, debranched arabinan, sugar beet arabinan, and xyloglucan; 5 mg/ml of konjac glucomannan, carob bean gum, locust bean gum (LBG), and guar gum; and 2.5 mg/ml of PASC and pectin. The release of reducing sugars in the reaction mixture was measured by using the DNS method.
TABLE 2.
Substrate specificity and kinetic parameters of CbXyn10Ca
| Substrate | Sp act (μmol/min/μmol of enzyme) | Km | kcat (s−1) | kcat/Km (ml s−1 mg−1) |
|---|---|---|---|---|
| Beech wood xylan | 39,000 ± 160 | 0.3 ± 0.1 mg ml−1 | 630 ± 16 | 2,100 |
| WAX | 51,000 ± 240 | 0.3 ± 0.0 mg ml−1 | 1,100 ± 15 | 3,700 |
| Barley β-glucan | 3,400 ± 15 | 2.7 ± 0.2 mg ml−1 | 85 ± 5.6 | 31 |
| CMC | 39 ± 3.3 | 4.3 ± 0.0 mg ml−1 | 1.8 ± 0.0 | 0.42 |
| pNPX | 19 ± 0.4 | 47 ± 0.1 μM | 0.1 ± 0.0 | 0.002 |
| pNPG | 0.5 ± 0.0 | 1.7 ± 0.0 μM | 0.02 ± 0.00 | 0.011 |
| Avicel | 3.4 ± 0.8 | ND | ND | ND |
| Filter paper | 5.4 ± 0.3 | ND | ND | ND |
| MCC | 2.3 ± 0.0 | ND | ND | ND |
| PASC | 1.2 ± 0.4 | ND | ND | ND |
| Lichenin | 180 ± 3.8 | ND | ND | ND |
The specific activities and kinetic parameters of CbXyn10C on konjac flour, arabic gum, and debranched arabinan were not determined in this study. No activity was detected on laminarin, pectin, carob bean gum, guar gum, locust bean gum, sugar beet arabinan, or xylogluan. The concentrations used for specific activity determination were 10 mg/ml for all polysaccharide substrates, while a series of concentrations (1 to 20 mg/ml) was used for estimation of the kinetic parameters of CbXyn10C on these substrates. The concentrations of pNPX and -pNPG used for specific activity measurement were 1 mM, while those used for kinetic studies were 0.1 to 4.0 mM. The released reducing sugars were determined by using the DNS method, and pNP was monitored spectrophotometrically at 405 nm. ND, not determined.
TABLE 3.
Enzymatic activities of wild-type and mutant CbXyn10C on xylan and cellulosesa
| CbXyn10C enzyme | Sp act (μmol/min/μmol of enzyme) |
CMC |
Xylan |
(kcat/Km for CMC)/(kcat/Km for xylan) (10−4) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Avicel | Filter paper | MCC | kcat (s−1) | Km (mg ml−1) | kcat/Km (ml s−1 mg−1) | kcat (s−1) | Km (mg ml−1) | kcat/Km (ml s−1 mg−1) | ||
| Wild type | 3.4 ± 0.8 | 5.4 ± 0.3 | 2.3 ± 0.0 | 1.8 ± 0.0 | 4.3 ± 0.0 | 0.42 | 630 ± 16 | 0.3 ± 0.1 | 2,100 | 2.0 |
| Q94A | 2.4 ± 0.2 | 6.2 ± 0.4 | ND | 1.3 ± 0.1 | 4.0 ± 0.3 | 0.33 | 310 ± 12 | 0.3 ± 0.1 | 1,000 | 3.3 |
| L306A | 2.9 ± 0.1 | 6.5 ± 0.3 | ND | 1.0 ± 0.0 | 2.6 ± 0.0 | 0.38 | 390 ± 25 | 0.5 ± 0.1 | 780 | 4.9 |
| Q94A/L306A | 3.4 ± 0.1 | 6.2 ± 0.1 | ND | 0.8 ± 0.0 | 3.3 ± 0.0 | 0.24 | 360 ± 17 | 0.6 ± 0.1 | 600 | 4.0 |
| E140A | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| E248A | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| E140A/E248A | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| CbXyn10C-CBM3b | 7.3 ± 0.0 | 12 ± 0.1 | 4.9 ± 0.0 | ND | ND | ND | ND | ND | ND | ND |
| CbXyn10C-CBM3b-CBM3b | 8.4 ± 0.0 | 14 ± 0.1 | 7.2 ± 0.0 | ND | ND | ND | ND | ND | ND | ND |
ND, not determined; MCC, microcrystalline cellulose with the amorphous region removed (Sigma).
Crystalline celluloses, including Avicel and filter paper, are not completely crystalline but, rather, have larger or smaller amounts of amorphous cellulose. The degree of crystallinity is indicated by the crystalline index (23). To obtain more insight into how CbXyn10C acts on crystalline cellulose, microcrystalline cellulose (MCC), which has the amorphous regions removed and is commercially available, was used as the substrate. CbXyn10C showed a specific activity of 2.3 ± 0.0 μmol/min/μmol of enzyme on MCC (Tables 2 and 3). Similarly, appending one or two CBM3b modules enhanced its activity on MCC to 4.9 ± 0.0 and 7.2 ± 0.0 μmol/min/μmol of enzyme, respectively (Table 3). In addition, the GH5-CBM28 truncation mutant (designated CelDTM1) of C. bescii CelD, an enzyme specifically hydrolyzing amorphous cellulose (14), was used to pretreat Avicel. On pretreated, more crystalline Avicel, CbXyn10C displayed activity similar to that on Avicel incubated with inactivated CelDTM1 (Fig. 3), as reflected by the slopes of the curves of the amount of released reducing sugars against time. The results from the two experiments collectively suggested that CbXyn10C can act on highly crystalline celluloses.
FIG 3.
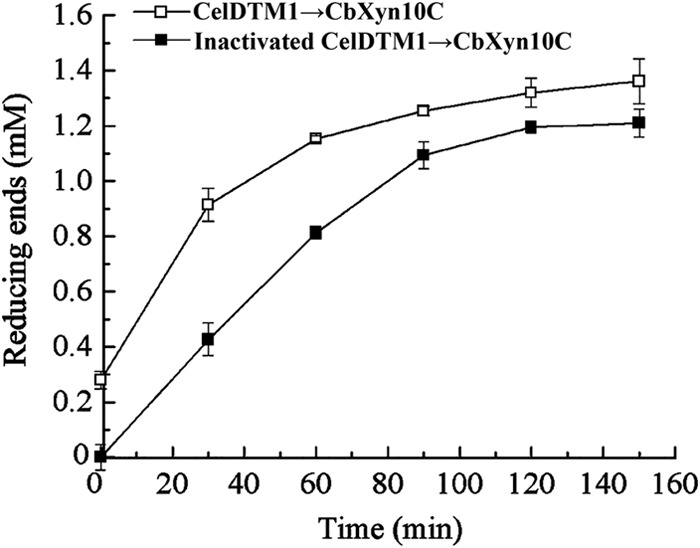
Effect of removing the amorphous region from Avicel on the activity of CbXyn10C. Avicel (10 mg/ml) was first incubated with 10 μM CelDTM1 in pH 6.5 McIlvaine buffer at 70°C for 16 h. As a control, Avicel was incubated with CelDTM1 that had been inactivated by boiling at 100°C for 10 min. Then, the samples were boiled at 100°C for 10 min to degrade the CelDTM1. Ten micromolar CbXyn10C was added to the pretreated Avicel, and the reaction was carried out at 75°C. At different time points (30 min, 60 min, 90 min, 120 min, and 150 min), samples were taken out and the reducing sugars were measured using the DNS method.
CbXyn10C was also active on konjac flour, which has a backbone of mixed β-1,4-glucosidic/mannosidic linkages (Fig. 2). Arabic gum and debranched arabinan, but not other substrates (carob been gum, guar gum, locust bean gum, and xylogluan), were also substrates of CbXyn10C (Fig. 2). CbXyn10C was also active on pNPX and pNPG (Table 2) but not on pNP-cellobiose. Taken together, CbXyn10C is a versatile glycoside hydrolase that can degrade a variety of natural plant cell wall polysaccharides and artificial substrates.
CbXyn10C hydrolyzes xylan and cellulose in different modes.
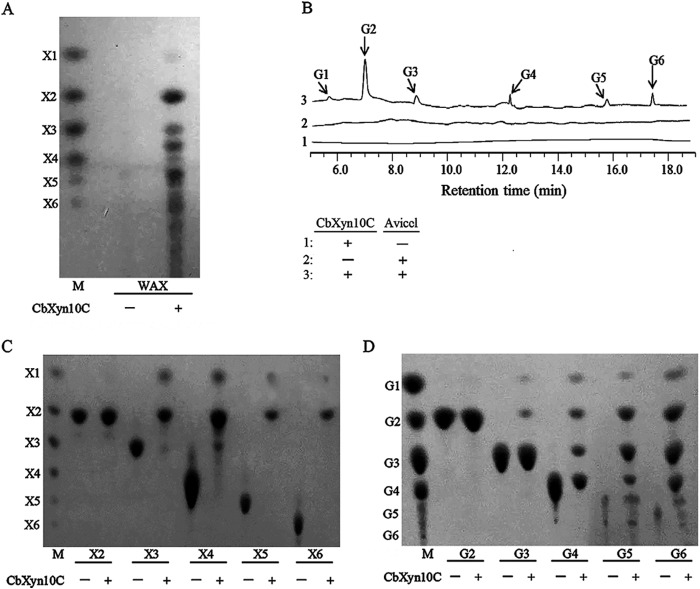
To gain insights into how CbXyn10C reacts with xylan and cellulose, the two major plant cell wall polysaccharides preferred for biofuel production, the end reaction products of WAX and Avicel hydrolysis by CbXyn10C were analyzed by TLC (for WAX) or HPLC (for Avicel). Xylobiose was the most prominent product of WAX hydrolysis, with a slight amount of xylose and minor amounts of xylooligosaccharides being produced (Fig. 4A). Some products could not be aligned with the xylooligosaccharide standards (Fig. 4A). Since WAX is composed of a β-1,4-xylosidic backbone with arabinose side chains, the unaligned spots might be xylooligosaccharides with arabinose side chains. However, the nature of these spots needs further studies. A dominant amount of cellobiose with minor amounts of glucose, cellotriose, cellotetraose, cellopentaose, and cellohexaose was detected from the end product of Avicel hydrolysis (Fig. 4B). A further hydrolysis using xylooligosaccharides (xylobiose [X2] to xylohexaose [X6]) as the substrates revealed that CbXyn10C was inactive on xylobiose but hydrolyzed xylotriose, xylotetraose, xylopentaose, and xylohexaose into xylose and xylobiose (Fig. 4C). In contrast, the end products of cellooligosaccharide hydrolysis were a series of shorter oligosaccharides and glucose (Fig. 4D). The results clearly demonstrate that CbXyn10C binds xylan and cellulose substrates differently and hydrolyzes these substrates via different modes of action.
FIG 4.
Hydrolysis products of WAX (A), Avicel (B), xylooligosaccharides (C), and cellooligosaccharides (D) by CbXyn10C analyzed by thin-layer chromatography (A, C, and D) or HPAEC-PAD (B). One micromolar CbXyn10C was incubated with 20 mg/ml of Avicel, 2.5 mg/ml each xylo-/cellooligosaccharides, or 1 mg/ml of WAX in McIlvaine buffer at pH 6.5 and 75°C for 16 h. X1, xylose; X2 to X6, xylobiose to xylohexaose; G1, glucose; G2 to G6, cellobiose to cellohexaose. Lanes M, molecular mass markers.
CbXyn10C has one catalytic center.
A bifunctional glycoside hydrolase may use one (4) or two (24) centers for catalyzing the hydrolysis of different substrates. By comparing the enzyme activities observed on mixed substrates with those obtained by calculation, the number of catalytic centers in bifunctional glycoside hydrolases can be estimated (4, 25). If one enzyme has one active center, the two substrates will compete for the same center, leading to activity on mixed substrates lower than that obtained for the enzyme if it is assumed that it has two separate, noncompeting centers. On the basis of the observation that β-1,4-xylosidic and β-1,3/1,4-glucosidic bonds were the two preferred linkages cleaved by CbXyn10C, we determined the activity of CbXyn10C on mixed substrates containing various concentrations of beech wood xylan and barley β-glucan, which contain β-1,4-xylosidic and β-1,3/1,4-glucosidic linkages, respectively. As shown in Table 4, in the presence of decreasing concentrations of beech wood xylan (16 to 4 mg/ml) with increasing concentrations of barley β-glucan (4 to 16 mg/ml), the measured activities of CbXyn10C were 0.20 to 0.22 ΔA/min/mg, which were much more similar to those calculated for enzymes with a single catalytic center (0.16 to 0.18 ΔA/min/mg) than to those calculated for enzymes with two different active centers (0.27 to 0.29 ΔA/min/mg) (Table 4). This suggests that CbXyn10C may have one rather than two catalytic centers for multiple substrates. Moreover, mutating either or both of the two glutamate catalytic residues (E140 and E248, where E is glutamate) (Fig. 5) to alanine simultaneously abolished the catalytic ability of CbXyn10C on xylan, barley β-glucan, and Avicel, further supporting the suggestion that CbXyn10C has one catalytic center (Table 3).
TABLE 4.
Hydrolysis of mixed substrates by CbXyn10C
| Reaction no. | Substrate concn in reaction mixture (mg/ml) |
Observed value (ΔAa/min/mg) | Total calculated velocity of hydrolysis (ΔA/min/mg) with: |
||
|---|---|---|---|---|---|
| Beech wood xylan | Barley β-glucan | Same active center | Different active center | ||
| 1 | 16 | 4 | 0.20 ± 0.00 | 0.18 | 0.27 |
| 2 | 12 | 8 | 0.21 ± 0.00 | 0.18 | 0.29 |
| 3 | 10 | 10 | 0.21 ± 0.00 | 0.18 | 0.29 |
| 4 | 8 | 12 | 0.22 ± 0.00 | 0.17 | 0.29 |
| 5 | 4 | 16 | 0.21 ± 0.00 | 0.16 | 0.29 |
ΔA, change in absorbance at 540 nm.
FIG 5.
Amino acid sequence alignment of CbXyn10C with selected GH10 xylanase sequences. The organisms (GenBank accession numbers) used as sources for the sequences were C. bescii (ACM60945) for CbXyn10C, Paenibacillus sp. strain E18 (ACY69972) for PsXynBE18, C. fimi (AAA56792) for CfCex, S. lividans (WP_003978188.1) for SlXyl10A, Demequina sp. strain JK4, (ACM41799) for DsMxyn10, and P. cellulosa WP_012488068 for PcXyl10A. Empty triangles, conserved catalytic residues; filled triangles, Q94 and L306, the residues for discriminating xylose- and glucose-configured substrates; hyphens, gaps in protein sequences.
Mutation of the xylan/cellulose-discriminating residues increased the substrate selectivity on cellulose over xylan.
Some GH10 xylanases are also able to cleave β-1,4-glucosidic bonds, in addition to the β-1,4-xylosidic linkage (9, 10). The amino acid sequences of such proteins were aligned with the amino acid sequence of CbXyn10C (Fig. 5). Among these proteins, Cex of C. fimi (CfCex) can degrade fluoro- and aryl-β-cellobiosides (11), Xyl10A of P. cellulosa (PcXyl10A) can degrade pNPC and 4-methylumbelliferyl-β-cellobioside (MUG) (26), Xyl10A of S. lividans (SlXyl10A) can degrade pNPC (27), Mxyn10 of Demequina sp. JK4 (DsMxyn10) can degrade CMC (12), and XynBE18 of Paenibacillus sp. E18 (PsXynBE18) can degrade barley β-glucan and lichenan (10), in addition to xylans. The two Glu catalytic residues E140 and E248 are well conserved among all of these proteins (Fig. 5). In PcXyl10A, Y87 and L314 are responsible for discriminating xylan and cellulosic substrates (26). Y87 has a counterpart of Gln in all five other proteins, including CbXyn10C, while L314 is conserved in CbXyn10C and PsXynBE18 but replaced by a valine in CfCex or an arginine in SlXyl10A and DsMxyn10 (Fig. 5). The two corresponding residues in CbXyn10C (Q94 and L306) were thus mutated to alanine individually or in combination. The kinetic parameters of the mutants on xylan and cellulosic substrates were compared to those of the wild type. Mutation of L306 to alanine decreased both the kcat of CbXyn10C on CMC from 1.8 ± 0.0 s−1 to 1.0 ± 0.0 s−1 and the Km from 4.3 ± 0.0 mg/ml to 2.6 ± 0.0 mg/ml, while the kcat and Km values of the Q94A mutant and the wild type were comparable. Mutation of both residues further decreased the kcat to 0.8 ± 0.0 s−1. The mutant with the Q94A mutation had a decreased kcat of 310 ± 12 s−1 on xylan, whereas that for the wild type was 630 ± 16 s−1, while the Km did not change significantly. The L306A mutant had a decreased kcat of 390 ± 25 s−1 but an increased Km of 0.5 ± 0.1 mg/ml on xylan, leading to a further decrease of catalytic efficiency of 780 ml s−1 mg−1. The Q94A/L306A double mutant had a much lower catalytic efficiency of 600 ml s−1 mg−1 due to a decreased kcat and an increased Km. Notably, the substrate preference for cellulose over xylan upon single or double mutation significantly increased, as indicated by the change of the ratio of the kcat/Km for CMC to the kcat/Km for xylan from 2.0 × 10−4 (for the wild type) to 3.3 × 10−4 (for the Q94A mutant), 4.9 × 10−4 (for the L306A mutant), and 4.0 × 10−4 (for the Q94A/L306A double mutant) (Table 3). The specific activities of the single and double mutants on the crystalline celluloses Avicel and filter paper were comparable to those of CbXyn10C (Table 3).
Application potential of CbXyn10C.
The ability of CbXyn10C to hydrolyze multiple types of plant cell wall polysaccharides makes it an interesting candidate enzyme for potential industrial usages. The raw materials barley and malt in the brewing industry have considerable amounts of xylan and β-1,3/1,4-glucan, which increase wort viscosity and decrease yields. The xylan- and β-glucan-degrading enzymes are, hence, commonly used to decrease the viscosity of mash. Bearing both xylanase and β-1,3/1,4-glucanase activities, CbXyn10C is regarded to be potentially useful in brewing. Its ability to reduce the viscosity of mash was thus determined using an increasing amount of the enzyme. Addition of 50 U, 80 U, 100 U, and 200 U CbXyn10C increasingly reduced the viscosity of mash by 16%, 20%, 22%, and 24%, respectively (data not shown).
DISCUSSION
The genomes of members of the genus Caldicellulosiruptor encode an arsenal of glycoside hydrolases attacking a variety of plant cell wall polysaccharides (28). Within the Caldicellulosiruptor genus, C. bescii is one of the best-characterized species regarding the presence of PCWP-degrading enzymes. The multimodular protein C. bescii CelA is appealing due to its superior crystalline cellulose-degrading ability, which results from a unique domain organization representing a newly discovered strategy of cellulose hydrolysis (7). The gene structure of CelA entails an N-terminal GH9/CBM3c endocellulase and a C-terminal GH48 exocellulase separated by two consecutive CBM3b domains. The specific cellulase activity of CbXyn10C on Avicel is 3.4 ± 0.8 μmol/min/μmol of enzyme, which is lower than but still in the same order of magnitude of that of CelA′ (18 μmol/min/μmol of enzyme) (21), which was purified from the culture broth of C. bescii and roughly contains the native GH9/CBM3c binary partner. This specific activity of CbXyn10C is, however, very similar and comparable to that of GH9/CBM3c module of CelA recombinantly expressed in E. coli (6). As has been observed by Zverlov et al., CelA has posttranslational modifications (22) which were regarded to be responsible for the observed difference in the specific activities between natural and recombinant CelA proteins and its truncation mutants (6).
The substrate competition experiment suggested that CbXyn10C most likely harbors only one catalytic center. Two facts further support the existence of one active center: first, replacement of the two catalytic residues (E140 and E248) with alanine simultaneously abolished the catalytic ability of CbXyn10C on xylan, cellulose, and barley β-glucan; second, single or double mutations of the two discriminating residues Q94 and L306 to alanine increased the substrate preference of CbXyn10A for cellulose rather than for xylan, although at the cost of enzyme activity, which is consistent with the findings for other GH10 xylanases with substrate promiscuity on xylan and cellulose (29). A computationally modeled tertiary structure of CbXyn10C revealed that the protein has an open cleft (Fig. 6), which is usually observed in the endo-acting glycoside hydrolases and complies with the endo faction of CbXyn10C on both xylose- and glucose-configured substrates. In addition, xylopentaose and cellopentaose can easily be docked into this cleft (Fig. 6), further confirming that CbXyn10C uses one catalytic center for hydrolyzing multiple substrates.
FIG 6.
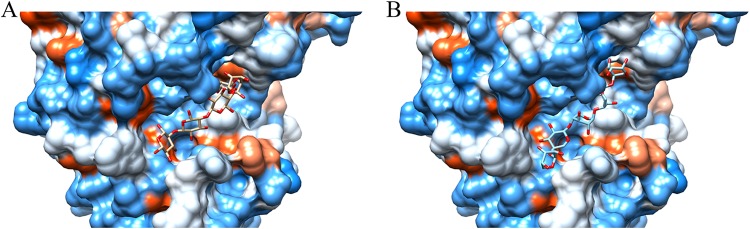
Modeled structure of CbXyn10C, in which cellopentaose (A) and xylopentaose (B) are docked into the substrate binding cleft. The three-dimensional structure of CbXyn10C was modeled by using the online ModWeb server (https://modbase.compbio.ucsf.edu/modweb/). Molecular docking was done by using the PRODRG server (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg).
The modes of recognizing xylose- or glucose-configured substrates by the same catalytic center of CbXyn10C are apparently different, determined from analyses of the hydrolysis products of xylo- or cellooligosaccharides. While xylose and xylobiose were the only two end products of xylooligosaccharide hydrolysis, cellooligosaccharides with higher degrees of polymerization, in addition to glucose and cellobiose, were present in the hydrolysis of glucose oligomers. This may suggest that the substrate binding cleft of CbXyn10C interacts with glucose-configured substrates less stably than xylose-configured substrates, thereby requiring more glucose units. While the catalytic efficiency of pNPX and pNPG differs by only 5-fold, the relative activities of CbXyn10 on celluloses were ∼105 less than those on xylans (Table 2), similar to the findings observed for PcXyl10A (30), suggesting that the binding of glucose-configured substrates by the distal regions of the substrate binding cleft is not optimized. Note that the activities of CbXyn10C on crystalline, amorphous, and soluble cellulose do not differ much. CMC is often a much better substrate for many endoglucanases. However, the manually introduced carboxymethyl groups on certain hydroxyl groups of the glucopyranose monomers may become a steric hindrance for the binding cleft of CbXyn10C, which is not optimized for cellulosic substrates. The similar activities of CbXyn10C on three different forms of cellulose are not unique. For example, a GH131 endoglucanase from Podospora anserina has activities of 2.0 ± 0.4, 2.0 ± 0.1, 6.3 ± 0.3, and 15 ± 2.7 U/mg on Avicel, PASC, hydroxyethyl cellulose, and CMC, respectively (31).
In comparison with the activities of the xylanases, many cellulose-degrading enzymes have activities on the β-1,4-xylosidic linkage. For example, the GH3 CdxA (cellodextrinase A) from Prevotella bryantii B14 is highly active on cellohexaose, but it can also degrade xylohexaose to some extent (32). The GH7 endoglucanase I of Trichoderma reesei can degrade birch wood xylan, in addition to cellulosic substrates (33). Substrate promiscuity is rarely observed for xylanases, particularly for crystalline cellulose. One possible reason may be that the glucose unit has a larger group (—CH2OH) than xylose (—H2) attached to the sugar ring. The modeled CbXyn10C-cellopentoase complex structure revealed many direct hydrogen bonds between cellopentaose and the binding cleft of CbXyn10C. The amino acid residues that were predicted to be involved in binding were N186, Q216, H218, W223, E248, N255, and G257. A hydrophobic interaction between Y185 and the glucose unit at the −2 subsite was also found. Cellopentaose displayed an extended conformation. An anomeric carbon on cellopentaose was 3.7 Å away from the nucleophile E248, suggestive of the formation of a stable enzyme-substrate complex. Further mutational studies and biochemical assays are, however, required to provide experimental evidence on the roles of key residues in hydrolyzing cellulose.
CbXyn10C/Cel48B (Athe_1857) is the third most abundant glycoside hydrolase secreted by C. bescii grown on the crystalline cellulose Avicel (34). As a GH10 xylanase, CbXyn10C has evolved to be able to hydrolyze multiple substrates with different glycosidic linkages. Based on our biochemical data, we hypothesize that CbXyn10C/Cel48B may help C. bescii acquire energy from various plant cell wall polysaccharides. In CelA, the GH48 module is weaker than GH9/CBM3c in degrading crystalline cellulose (6), raising the question of why an N-terminal xylanase but not an endocellulase, like that in CelA, exists in the large multimodular protein CbXyn10C/Cel48B. As the endocellulase activity of CbXyn10C has been discovered to be comparable to that of GH9/CBM3c expressed in E. coli, it is noteworthy that addition of one or two cellulose-binding CBM3bs enhanced the hydrolyzing activities of both CbXyn10C and GH9/CBM3c on crystalline celluloses. These improved activities were again comparable to each other. For CbXyn10C-CBM3b-CBM3b, the activities on Avicel and filter paper were 8.4 ± 0.0 and 14 ± 0.1 μmol/min/μmol of enzyme, respectively, while those for GH9/CBM3c-CBM3b-CBM3b of CelA were 10 ± 1.0 and 7.0 ± 1.8 μmol/min/μmol of enzyme, respectively. As the C-terminal tandem CBM3b-CBM3b-GH48 in CbXyn10C/Cel48A both is structurally identical to CelA and has a sequence highly homologous to that of CelA (see Fig. S1 in the supplemental material), it is now reasonable to speculate that the entire CbXyn10C/Cel48B protein may also utilize the same strategy that CelA uses to hydrolyze crystalline cellulose and thus harbor good cellulose-degrading activity. This, however, remains to be elucidated when the entire recombinant or natural purified protein becomes available in the future. Note that no colonies could be obtained when the pET plasmids bearing any of the three C. bescii gh48-containing full-length genes were transformed into BL21(DE3). The third (and last) multimodular protein with a GH48 exocellulase in C. bescii contains a GH74 domain (GenBank accession number ACM60948) in the N terminus, followed also by the highly similar CBM3b-CBM3b-GH48 in the same order. The GH74 enzymes have substrate promiscuities for cellulose (35) and xyloglucan (36). Therefore, it would be interesting to investigate if the GH74 domain also harbors a cellulase activity.
CbXyn10C is able to effectively reduce the viscosity of mash, which contains xylan and β-1,3/1,4-glucan as major components. From the experiments using MCC or the more crystalline Avicel with the amorphous regions removed by CelDTM1, it was further demonstrated that CbXyn10C is able to act on highly crystalline cellulose. This character would be appealing to the biofuel and biorefinery industry. In conclusion, the versatility of CbXyn10C in hydrolyzing multiple substrates, including crystalline cellulose, makes CbXyn10C an interesting enzyme for understanding how C. bescii utilizes various plant polysaccharides and the molecular mechanism underlying the substrate promiscuity of GH10 enzymes and a candidate enzyme with potential applications in biotechnology.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (31400067), the Fundamental Research Funds for Central Public-Interest Institutes of the Chinese Academy of Agricultural Sciences (2015ZL045), the National High Technology Research and Development Program of China (2012AA022208), and the National Science Foundation for Distinguished Young Scholars of China (31225026).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00432-15.
REFERENCES
- 1.Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2010. Feedstocks for lignocellulosic biofuels. Science 329:790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 2.Moon YH, Iakiviak M, Bauer S, Mackie RI, Cann IK. 2011. Biochemical analyses of multiple endoxylanases from the rumen bacterium Ruminococcus albus 8 and their synergistic activities with accessory hemicellulose-degrading enzymes. Appl Environ Microbiol 77:5157–5169. doi: 10.1128/AEM.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su X, Han Y, Dodd D, Moon YH, Yoshida S, Mackie RI, Cann IK. 2013. Reconstitution of a thermostable xylan-degrading enzyme mixture from the bacterium Caldicellulosiruptor bescii. Appl Environ Microbiol 79:1481–1490. doi: 10.1128/AEM.03265-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su X, Mackie RI, Cann IK. 2012. Biochemical and mutational analyses of a multidomain cellulase/mannanase from Caldicellulosiruptor bescii. Appl Environ Microbiol 78:2230–2240. doi: 10.1128/AEM.06814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su X, Zhang J, Mackie RI, Cann IK. 2012. Supplementing with non-glycoside hydrolase proteins enhances enzymatic deconstruction of plant biomass. PLoS One 7:e43828. doi: 10.1371/journal.pone.0043828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi Z, Su X, Revindran V, Mackie RI, Cann I. 2013. Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS One 8:e84172. doi: 10.1371/journal.pone.0084172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MW, Lunin VV, Himmel ME, Bomble YJ. 2013. Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 8.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roldan-Arjona T, Perez-Espinosa A, Ruiz-Rubio M. 1999. Tomatinase from Fusarium oxysporum f. sp. lycopersici defines a new class of saponinases. Mol Plant Microbe Interact 12:852–861. doi: 10.1094/MPMI.1999.12.10.852. [DOI] [PubMed] [Google Scholar]
- 10.Shi P, Tian J, Yuan T, Liu X, Huang H, Bai Y, Yang P, Chen X, Wu N, Yao B. 2010. Paenibacillus sp. strain E18 bifunctional xylanase-glucanase with a single catalytic domain. Appl Environ Microbiol 76:3620–3624. doi: 10.1128/AEM.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notenboom V, Birsan C, Warren RA, Withers SG, Rose DR. 1998. Exploring the cellulose/xylan specificity of the β-1,4-glycanase Cex from Cellulomonas fimi through crystallography and mutation. Biochemistry 37:4751–4758. doi: 10.1021/bi9729211. [DOI] [PubMed] [Google Scholar]
- 12.Meng X, Shao Z, Hong Y, Lin L, Li C, Liu Z. 2009. A novel pH-stable, bifunctional xylanase isolated from a deep-sea microorganism, Demequina sp. JK4. J Microbiol Biotechnol 19:1077–1084. [PubMed] [Google Scholar]
- 13.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velikodvorskaya GA, Chekanovskaya LA, Lunina NA, Sergienko OV, Lunin VG, Dvortsov IA, Zverlov VV. 2013. Family 28 carbohydrate-binding module of the thermostable endo-1,4-β-glucanase CelD from Caldicellulosiruptor bescii maximizes enzyme activity and irreversibly binds to amorphous cellulose. Mol Biol 47:581–586. doi: 10.1134/S0026893313040158. [DOI] [PubMed] [Google Scholar]
- 15.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 17.Chen HG, Yan X, Liu XY, Wang MD, Huang HM, Jia XC, Wang JA. 2006. Purification and characterization of novel bifunctional xylanase, XynIII, isolated from Aspergillus niger A-25. J Microbiol Biotechnol 16:1132–1138. [Google Scholar]
- 18.Schuttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 19.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celestino KRS, Cunha RB, Felix CR. 2006. Characterization of a β-glucanase produced by Rhizopus microsporus var. microsporus, and its potential for application in the brewing industry. BMC Biochem 7:23. doi: 10.1186/1471-2091-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L, Su X, Schmitz GE, Moon YH, Zhang J, Mackie RI, Cann IK. 2012. Molecular and biochemical analyses of the GH44 module of CbMan5B/Cel44A, a bifunctional enzyme from the hyperthermophilic bacterium Caldicellulosiruptor bescii. Appl Environ Microbiol 78:7048–7059. doi: 10.1128/AEM.02009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zverlov V, Mahr S, Riedel K, Bronnenmeier K. 1998. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144(Pt 2):457–465. doi: 10.1099/00221287-144-2-457. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Baker J, Himmel M, Parilla P, Johnson D. 2010. Cellulose crystallinity index: measurement techniques and their impact on interpreting cellulase performance. Biotechnol Biofuels 3:10. doi: 10.1186/1754-6834-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodd D, Kocherginskaya SA, Spies MA, Beery KE, Abbas CA, Mackie RI, Cann IK. 2009. Biochemical analysis of a β-d-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23. J Bacteriol 191:3328–3338. doi: 10.1128/JB.01628-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chinchetru MA, Cabezas JA, Calvo P. 1989. Purification and characterization of a broad specificity β-glucosidase from sheep liver. Int J Biochem 21:469–476. doi: 10.1016/0020-711X(89)90126-2. [DOI] [PubMed] [Google Scholar]
- 26.Andrews SR, Charnock SJ, Lakey JH, Davies GJ, Claeyssens M, Nerinckx W, Underwood M, Sinnott ML, Warren RA, Gilbert HJ. 2000. Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in discriminating between glucose and xylose binding in the proximal active site of Pseudomonas cellulosa xylanase 10A. J Biol Chem 275:23027–23033. doi: 10.1074/jbc.M000128200. [DOI] [PubMed] [Google Scholar]
- 27.Ducros V, Charnock SJ, Derewenda U, Derewenda ZS, Dauter Z, Dupont C, Shareck F, Morosoli R, Kluepfel D, Davies GJ. 2000. Substrate specificity in glycoside hydrolase family 10: structural and kinetic analysis of the Streptomyces lividans xylanase 10A. J Biol Chem 275:23020–23026. doi: 10.1074/jbc.275.30.23020. [DOI] [PubMed] [Google Scholar]
- 28.Blumer-Schuette SE, Lewis DL, Kelly RM. 2010. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl Environ Microbiol 76:8084–8092. doi: 10.1128/AEM.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichinose H, Diertavitian S, Fujimoto Z, Kuno A, Lo Leggio L, Kaneko S. 2012. Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86. Process Biochem 47:358–365. doi: 10.1016/j.procbio.2011.06.002. [DOI] [Google Scholar]
- 30.Andersson S, Wikberg H, Pesonen E, Maunu S, Serimaa R. 2004. Studies of crystallinity of Scots pine and Norway spruce cellulose. Trees 18:346–353. doi: 10.1007/s00468-003-0312-9. [DOI] [Google Scholar]
- 31.Lafond M, Navarro D, Haon M, Couturier M, Berrin J-G. 2012. Characterization of a broad-specificity β-glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol 78:8540–8546. doi: 10.1128/AEM.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodd D, Kiyonari S, Mackie RI, Cann IK. 2010. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J Bacteriol 192:2335–2345. doi: 10.1128/JB.01654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakazawa H, Okada K, Kobayashi R, Kubota T, Onodera T, Ochiai N, Omata N, Ogasawara W, Okada H, Morikawa Y. 2008. Characterization of the catalytic domains of Trichoderma reesei endoglucanase I, II, and III, expressed in Escherichia coli. Appl Microbiol Biotechnol 81:681–689. doi: 10.1007/s00253-008-1667-z. [DOI] [PubMed] [Google Scholar]
- 34.Lochner A, Giannone RJ, Rodriguez M Jr, Shah MB, Mielenz JR, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl Environ Microbiol 77:4042–4054. doi: 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takada G, Kawagushi T, Yoneda T, Kawasaki M, Sumitani J-I, Arai M. 1998. Molecular cloning and expression of the cellulolytic system of Aspergillus aculeatus, p 364–373. In Ohminya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T (ed), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co, Ltd, Tokyo, Japan. [Google Scholar]
- 36.Martinez-Fleites C, Guerreiro CI, Baumann MJ, Taylor EJ, Prates JA, Ferreira LM, Fontes CM, Brumer H, Davies GJ. 2006. Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem 281:24922–24933. doi: 10.1074/jbc.M603583200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.