SUMMARY
The abundance of cell surface membrane proteins is regulated by internalization and delivery into intralumenal vesicles (ILVs) of multivesicular bodies (MVB). Many cargoes are ubiquitinated, allowing access to an ESCRT-dependent pathway into MVBs. Yet, how non-ubiquitinated proteins, such as Glycosylphosphatidylinisotol-anchored proteins, enter MVBs is unclear, supporting the possibility of mechanistically distinct ILV biogenesis pathways. Here we show a family of highly ubiquitinated tetraspan Cos proteins provide a Ub-signal in trans, allowing sorting of non-ubiquitinated MVB cargo into the canonical ESCRT- and Ub-dependent pathway. Cos proteins create discrete endosomal subdomains that concentrate Ub-cargo prior to their envelopment into ILVs and the activity of Cos proteins is required not only for efficient sorting of canonical Ub-cargo but is also essential for sorting non-ubiquitinated cargo into MVBs. Expression of these proteins increases during nutrient stress though a NAD+/Sir2-dpendent mechanism that in turn accelerates the down-regulation of a broad range of cell surface proteins.
INTRODUCTION
Lysosomal degradation of membrane proteins is mediated by their incorporation into intralumenal vesicles (ILVs), which reside within late-endosomes/multivesicular bodies (MVBs)(Piper and Katzmann, 2007). MVB sorting is conducted by the ESCRT (Endosomal Sorting Complex Required for Transport) machinery, through packaging of cargo into ILVs and scission of ILVs from the limiting endosomal membrane. The major sorting signal for this pathway is ubiquitin (Ub), which in yeast is typically ligated to MVB cargo by the HECT-type Ub-ligase Rsp5 that uses a host of adaptor proteins to associate with its substrates (Lin et al., 2008; Nikko and Pelham, 2009). A variety of ESCRTs (ESCRT-0, -I, -II) and ESCRT-associated factors such as Alix and Bro1, bind Ub to usher ubiquitinated membrane proteins into burgeoning ILVs (Piper et al., 2014). Before ILV formation is complete, most Ub is removed from cargo, catalyzed in yeast by the Doa4 deubiquitinating enzyme (Amerik et al., 2000). ESCRTs themselves are also largely removed from the nascent ILV.
Current models for the sorting process predict that cargo needs to be highly organized and immobilized to allow for proper ESCRT assembly, cargo deubiquitination and eventual ESCRT release in preparation for incorporation into ILVs (Hurley and Hanson, 2010; Nickerson et al., 2007). Yet, key aspects of how MVB cargo is captured, concentrated and packaged into ILVs have yet to be clarified. In vitro systems that successfully reconstitute ESCRT assembly and ILV formation lack the ability to sort cargo (Carlson and Hurley, 2012; Wollert et al., 2009), suggesting that auxiliary factors work in conjunction with ESCRTs. Even some ESCRT components are not strictly required for MVB sorting, as loss of Mvb12, Vta1, or the Alix-related Bro1 protein does not block all cargo sorting into yeast ILVs. Sorting independent of ESCRTs is more profound in animal cells, Plasmodium, and Dictyostelium, which generate MVBs when they lack, or are severely depleted of, various archetypal ESCRTs, suggesting auxiliary factors or alternate pathways may play prominent roles in MVB sorting (Blanc et al., 2009; Colombo et al., 2013; Edgar et al., 2014; Leung et al., 2008; Trajkovic et al., 2008).
Lipid-based mechanisms, such the generation of ceramide by sphingomyelinase or the accumulation of bis(monoacylglycero)phosphate / lysobisphosphatidic acid (BMP/LBPA) may contribute to MVB formation (Falguieres et al., 2008; Matsuo et al., 2004; Trajkovic et al., 2008). Also, tetraspanins (Tspans), many of which are major constituents of ILVs (Pols and Klumperman, 2009; Wubbolts et al., 2003), have been implicated in MVB sorting. Tspans are palmitoylated 4 transmembrane-spanning domain (TMD)-containing proteins with a characteristic large extracellular disulfide-containing loop. They cluster in membrane microdomains and form complexes with a wide variety of membrane proteins (Hemler, 2005; Stipp et al., 2003). A functional role for Tspans in endosomes was established by studies of CD63, which compensates for the depletion of ESCRTs during ILV formation in general, and appears tasked with sorting specific cargo such as the luminal protein Pmel17 into ILVs of melanosomes (Berson et al., 2001; Colombo et al., 2013; Edgar et al., 2014; Theos et al., 2006; van Niel et al., 2011). Yet, how Tspans integrate functionally with ESCRTs, and whether they and ESCRTs augment the same ILV formation pathway or control mechanistically distinct MVB biogenesis pathways, remains unclear.
Here we document a family of yeast MVB sorting factors called Cos proteins that functionally resemble Tspans found in mammalian ILVs. We show that nutrient-limiting conditions, which elevate COS expression, increase flux through the MVB pathway thereby causing down-regulation of various cell surface proteins. The metabolic link is NAD+ levels, which are sensed by the NAD+-dependent deaceytlase Sir2. We find that Cos proteins cluster in endosomal subdomains, which they help create, and trap cargo there to promote incorporation into ILVs. Among these cargoes are Glycosylphosphatidylinisotol (GPI)-anchored proteins that sort into MVB ILVs along an ESCRT-dependent pathway, yet have no cytosolic domain to mediate interaction with the Ub-binding ESCRT apparatus.
RESULTS
Sorting through the MVB pathway is stimulated in response to NAD+ depletion
Nutrient-stress caused by growing cells past mid-log phase induces many plasma membrane proteins to sort into the vacuole (Babst and Odorizzi, 2013; Beck et al., 1999; MacDonald et al., 2012b). This phenomenon is exemplified by the redistribution to the vacuole of the cell surface proteins Mup1-GFP (methionine transporter), Can1 (arginine transporter), Fur4 (uracil transporter), Snc1 (recycling v-SNARE), Ste3 (a-factor GPCR), and Yor1 (ABC transporter) (Figure 1 and S1a). This downregulation was also independent of many of the known Rsp5 arrestin-related E3 ligase adaptors that connect Rsp5 to its substrates (Lin et al., 2008; Nikko and Pelham, 2009). To understand this metabolic control we identified the key components of SD media that were being consumed or altered (Figure S1b). Interestingly, increased Yeast Nitrogen base, comprised mostly of B-vitamins and trace metals (Figure S1c), suppressed Mup1 vacuolar sorting. We traced the active ingredient to Niacin/Vitamin B3, a precursor for NAD+ also known as Nicotinic Acid (NA). NA is present in standard SD media at ~3 μM, but supplementing SD media to 100 µM NA stabilized Mup1 almost entirely at the cell surface in cells grown to late log-phase (Figure 1C). By contrast, in SD media without NA, Mup1-GFP was found in the vacuole even in cells grown to mid-log phase, and was found exclusively in the vacuole at late-log phase.
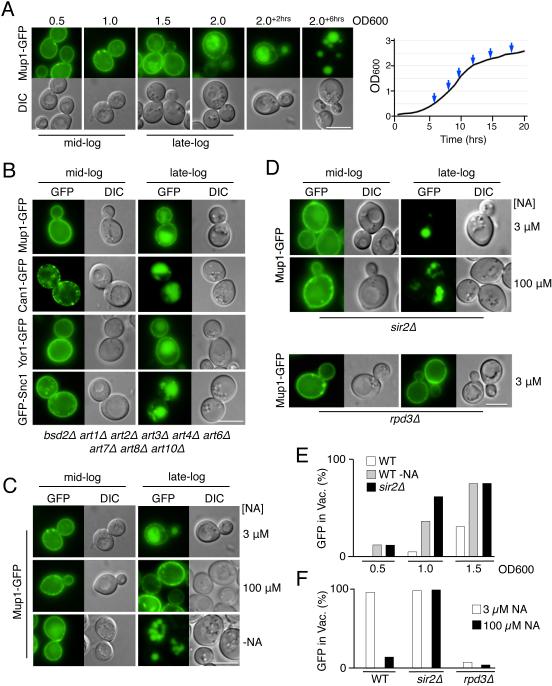
Figure 1. Niacin depletion at late-log phase induces down-regulation of cell surface proteins via transcriptional modulation by Sir2 and Rpd3.
A) Wild-type cells expressing Mup1-GFP were grown in minimal media lacking methionine (SD-Met) and imaged at different stages of growth. The final time points (+2 and +6) were 2 or 6 hrs after the cell culture had reached OD600 2.0. Right, corresponding growth curve of the culture with sampled time points indicated.
B) Localization of GFP-tagged cargo proteins in mutant cells lacking 9 known Rsp5 adaptor proteins grown to mid- and late-log phase.
C) Cells expressing Mup1-GFP were grown to mid- and late-log phase in SD-Met media with 3 µM NA, 100 µM NA, or SD-Met media without NA.
D) Mup1-GFP localization in sir2Δ and rpd3Δ null mutant cells in SD-Met media containing indicated levels of NA.
E) Quantitation of intravacuolar Mup1-GFP localization from microscopy shown in C and D. Quantified is the proportion of cells (n = >150) showing GFP fluorescence in the vacuole.
F) Percentage of Mup1-GFP expressing cells (n = >130) showing vacuolar GFP in WT, sir2Δ, and rpd3Δ cells grown in standard SD-Met media with 3 µM or 100 µM NA. Bar = 5 µm.
One prominent class of NAD+-sensors is the family of NAD+-dependent histone deacytelase enzymes (HDACs) knows as sirtuins (Hekimi and Guarente, 2003; Wood et al., 2004). The yeast sirtuin, Sir2, mediates gene silencing at the mating-type, ribosomal DNA and the telomeric loci, and is inhibited when NAD+ levels are low (Bryk et al., 1997; Gottschling et al., 1990; Landry et al., 2000; Loo et al., 1995). The effect of NA-depletion on MVB sorting of Mup1-GFP was mimicked by deleting SIR2, which caused cells at mid-log phase to accumulate Mup1 in the vacuole and caused exclusive localization of Mup1-GFP to the vacuole in cells grown to late-log phase (Figure 1D,E). Increasing NA levels to 100 µM did not suppress Mup1-GFP delivery to the vacuole in sir2Δ cells, suggesting the effects of high NA levels or NA-depletion are mediated through Sir2. Previous studies have shown that some of the effects of Sir2 on transcription are opposed by the activity of the Rdp3-Sin3 Class I HDAC complex (Bernstein et al., 2000; Rundlett et al., 1996). We found that Rpd3 had opposing effects on Mup1-GFP down-regulation in response to nutrient stress (Figure 1D,F and S1d) suggesting that the opposing effects of Rpd3 and Sir2 comprise reciprocal control over the transcription of factors that induce MVB sorting of a broad range of cell surface proteins.
A novel family of yeast MVB sorting factors
To find the genes responsible for nutrient-stress induced MVB sorting, we cross-referenced existing microarray data for those that were repressed in rpd3Δ mutants and induced when cells were grown toward stationary phase (Bernstein et al., 2000; Gasch et al., 2000). Of those 25, 15 also had substantially increased transcription in sir2Δ null mutants (Bedalov et al., 2001) (Figure 2A and S2a). Eleven belonged to the family of conserved sequence (COS) genes that are found in subtelomeric regions within 9 of the 16 yeast chromosomes (Figure S2b,e); regions known to be repressed by Sir2 and activated by Rdp3 (Bernstein et al., 2000). The 11 COS genes, COS1 – COS10 and COS12, are remarkably similar at both the protein and nucleotide level (Figure S2d) (Spode et al., 2002). Cos proteins are comprised of ~380 residues, predicted to form 4 membrane spanning segments that bridge 2 small extracellular loops and larger cytosolic N and C-terminal tails (Figure 2B). We found that GFP fusions of different Cos proteins sort to the vacuole lumen, consistent with previous high-throughput studies (Huh et al., 2003), and do so along an ESCRT-dependent route (Figure 2C and S4a,d).
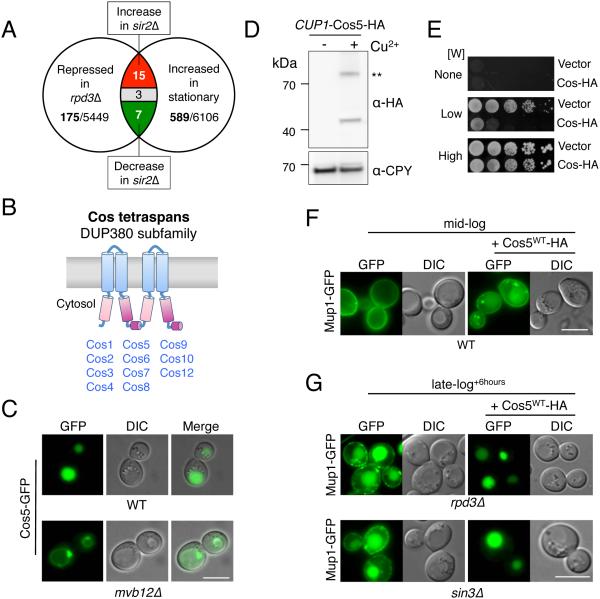
Figure 2. COS genes encode MVB sorting accelerators.
A) Venn diagram summarizing microarray mRNA expression data mined for genes that increase expression upon entry towards stationary phase and upon SIR2 deletion, and decrease expression upon loss of RPD3.
B) A cartoon of the predicted secondary structure for Cos proteins, which contain 4 transmembrane-spanning domains (TMDs). Pink and purple regions indicate internal regions sharing primary structural homology.
C) Localization of Cos5-GFP in wild-type cells and in mvb12Δ cells, which are defective for MVB sorting.
D) Immunoblot of WT cells carrying a low-copy plasmid encoding C-terminally HA-tagged Cos5 expressed from the inducible CUP1 promoter. Cells were gown in either 50 µM copper chelator (BCSA: bathocuproinedisulfonate) or 50 µM CuCl2 (− + Cu2+, respectively) prior to lysis, SDS-PAGE, and immunoblotting. A dimeric form of Cos5-HA is indicated (**).
E) Wild-type cells carrying CUP1-Cos5-HA or vector plasmid were serially diluted and grown on SD plates containing 50 µM CuCl2 and either no Trp (None), 4 µg/ml Trp (Low), or 20 µg/ ml Trp (High).
F) Localization of Mup1-GFP in wild-type cells grown to mid-log phase in SD-Met media containing 50 µM CuCl2. Cells were carrying vector plasmid or the CUP1-Cos5-HA plasmid.
G) Localization of Mup1-GFP in rpd3Δ and sin3Δ null cells carrying vector alone or the CUP1-Cos5-HA plasmid that were grown to late-log phase + 6 hours in SD-Met media containing 50 µM CuCl2. Bar = 5 µm.
Overexpressing a representative of the Cos family, Cos5, in wild-type cells caused Mup1-GFP to be shunted from its exclusive localization at the cell surface to endosomes and the vacuole interior of cells grown to mid-log phase (Figure 2D,F). Similarly, Mup1-GFP retained at the cell surface of Cos-depleted rpd3Δ or sin3Δ cells grown past late-log phase can be directed to the vacuole by overexpressing Cos5-HA (Figure 2G). Overexpressing Cos5-HA also compromised the ability of Trp− auxotrophic cells to grow on low levels of tryptophan (Figure 2E), consistent with down-regulation of the tryptophan transporter Tat2 from the cell surface (Schmidt et al., 1994).
The Cos proteins are required for efficiently sorting MVB cargo
We next generated a strain lacking all COS genes and COS pseudogenes (Figure S3c,d). Unlike wild-type cells, cosΔ cells no longer efficiently delivered Mup1-GFP to the vacuole at late-log phase; rather, Mup1-GFP was localized to the cell surface and intracellular endosomal puncta (Figure 3A). In wild-type cells, an inframe fusion of Ub to Mup1-GFP serves as a sufficient signal for MVB sorting in the absence of methionine (Figure S3a). However Mup1-GFP-Ub sorting was largely blocked in cosΔ cells lacking methionine but could be restored by addition of methionine or an additional Ub moiety to the fusion protein (di-Ub) (Figure 3B), implying that greater ubiquitination of cargo surmounts the cosΔ defect. Defects were also observed for Ste3-GFP that is constitutively ubiquitinated and sorted into the MVB pathway (Davis et al., 1993; Roth et al., 1998). In mid-log phase grown wild-type cells, Ste3-GFP was largely in the vacuole, however, in cosΔ cells, a significant proportion of Ste3-GFP was also observed within endosomes, accompanied by a sizeable loss of a vacuolar processed GFP detected by immunoblot (Figure S3b). This defect was also suppressed by an inframe fusion of Ub. We next monitored the cellular pool of Ub using GFP-Ub. Wild-type cells accumulate some GFP-Ub in the vacuole due to the MVB pathway (Ren et al., 2008), yet this was largely blocked in cosΔ cells (Figure 3C).
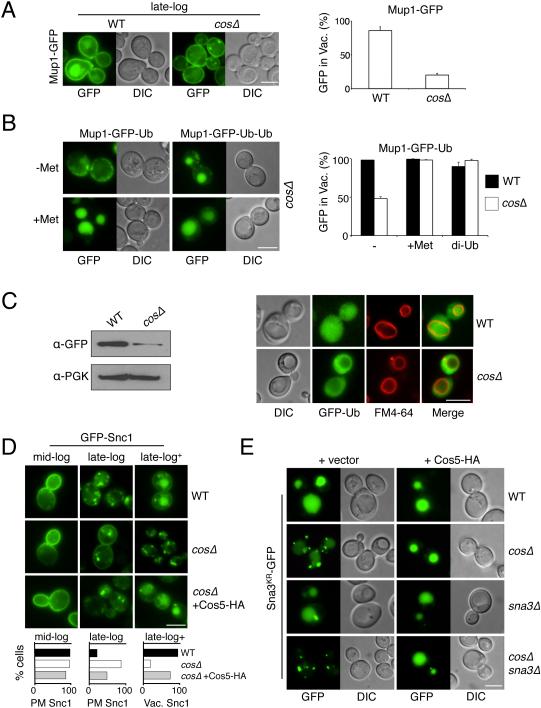
Figure 3. Cos proteins are required for efficient MVB sorting.
A) Localization of Mup1-GFP in WT and cosΔ cells grown to late-log phase in standard SD-Met media. Right, quantitation of the percentage of cells (n=>200) with vacuolar GFP (mean and SD of 3 experiments).
B) Sorting defects of Mup1-GFP-Ub in cosΔ cells were overcome by addition of 20 μg/ml methionine to the media or by expressing a Mup1-GFP-Ub-Ub fusion. Quantitation of cells (n=>50) with intravacuolar GFP is shown (mean and SD of 2 experiments).
C) Localization of GFP-Ub in WT and cosΔ mutant cells labeled with FM4-64 (red) to reveal the limiting membrane of the vacuole. Left, immunoblot of PGK and vacuolar processed GFP from WT and cosΔ cells expressing GFP-Ub.
D) Localization of GFP-Snc1 in WT, cosΔ cells, and cosΔ cells expressing Cos5-HA from the CUP1-Cos5-HA plasmid that were grown in SD media to mid-log (OD600 1.0), late-log (OD600 2.0), or 6 hrs past late-log (late-log+). Below, quantitation of the percentage of cells (n=>100) grown to mid-log and late-log that have GFP-Snc1 on the plasma membrane (PM), and cells grown to late-log+ that have vacuolar GFP-Snc1.
E) Localization of Sna3KR-GFP (in which the lysines of Sna3 are changed to arginines) in WT, cosΔ, sna3Δ and cosΔsna3Δ mutant cells. Cells also carried either vector alone or the CUP1-Cos5-HA plasmid and were grown in 50 µM CuCl2. Bar = 5 µm.
We found the delivery of GFP-Snc1 to the vacuole, which is only observed at late-log phase, was dramatically reduced in cosΔ null mutants (Figure 3D). Instead, GFP-Snc1 accumulated in endosomal compartments suggesting that Cos proteins function at endosomes to convey proteins delivered there further into the MVB ILVs. Re-expressing Cos5-HA restored sorting of GFP-Snc1 into the vacuole of cosΔ null cells demonstrating that the defects observed were due to the specific loss of the COS genes.
We next tested MVB sorting of a synthetic Ub-independent cargo, Sna3KR-GFP, a version of the small polytopic membrane Rsp5 adaptor protein Sna3 in which ubiquitinatable lysines are substituted with arginines (Reggiori and Pelham, 2001). In cosΔ cells, Sna3KR-GFP was found primarily in endosomes and blocked from entry into the vacuole, most strikingly when endogenous SNA3 was also deleted (Figure 3E). This was corrected by overexpressing Cos5-HA from a plasmid. These data suggest Cos proteins mediate efficient sorting of a wide number of MVB cargoes.
The Cos proteins are ubiquitinated trafficking effectors
Previous mass-spectrometry experiments indicated that the Cos proteins are ubiquitinated (Hitchcock et al., 2003; Peng et al., 2003; Reggiori and Pelham, 2001). We found that ubiquitination of Cos proteins is required for their MVB sorting (Figure 4A). A Cos5KR-GFP fusion protein, in which all 33 lysines of Cos5 were altered to arginine, was unable to sort into the vacuole of wild-type cells. Pulse/chase experiments (Figure 4B) showed that degradation of Cos5-GFP had a half-time of ~20 min and was dependent on vacuolar proteases. The Cos5KR-GFP mutant, by contrast, was very stable. Sorting into the vacuole was restored by reintroducing a single lysine at positions 2 or 7 within the N-terminus and was correlated with the overall level of Cos5 ubiquitination (Figure S4b,c). Sorting of Cos5 into the vacuole was also blocked in cells compromised for Rsp5 function (Figure 4A and S4e), and blocked by fusing Cos5 to the catalytic domain of the deubiquitinating enzyme UL36 (Figure 4A), a manipulation that removes Ub on the host protein (Stringer and Piper, 2011). Ubiquitination was also important for Cos5 to have an effect on the sorting of other cargoes. Overexpressing Cos5-HA in mid-log grown cells caused Mup1-GFP to accumulate in endosomes and the vacuole (Figure 4C). Yet, overexpressing Cos5KR-HA to similar levels had no effect on Mup1-GFP sorting. Importantly, the defective Cos5KR-HA could be converted to a functional protein by an inframe fusion of Ub, Cos5KR-HA-Ub, which drove sorting of Mup1-GFP into endosomes and the vacuole to an extent on par with the wild-type Cos5-HA (Figure 4C,D).
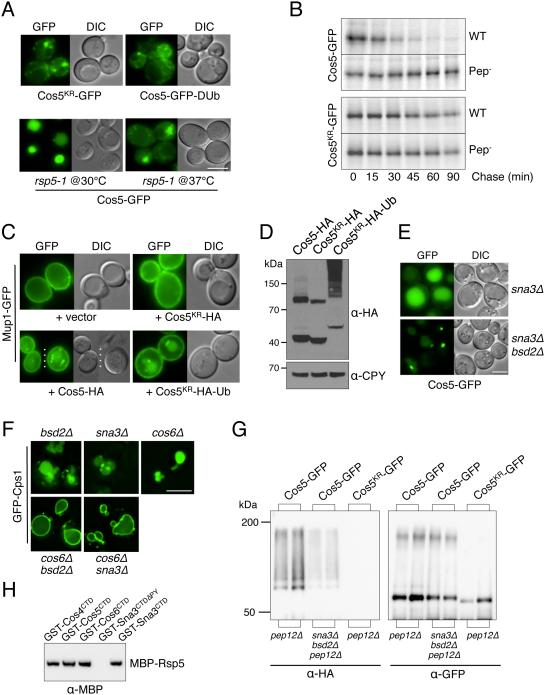
Figure 4. Cos5 requires ubiquitination for its MVB sorting and its function in facilitating MVB sorting of other cargo.
A) Top, sorting of GFP fused to Cos5 lacking its lysine residues (Cos5KR-GFP) and Cos5-GFP fused to the UL36 deubiquitinating (DUb) catalytic domain in WT cells. Bottom, Cos5-GFP localization in temperature-sensitive rsp5-1 mutant cells grown at 30°C and 37°C for 4 hrs.
B) Pulse/chase immunoprecipitation of Cos5-GFP and Cos5KR-GFP in WT (Pep+) and Pep- cells. Cells were labeled with 35S~Met for 10 min followed by the indicated chase times. Cos5-GFP and Cos5KR-GFP were immunopreciptated from cell lysates with α-GFP antibodies prior to SDS-PAGE and autoradiography.
C) Localization of Mup1-GFP in cells co-transformed with a vector control or plasmids over-expressing Cos5-HA, Cos5KR-HA and Cos5KR-HA-Ub from the CUP1 promoter. Cells were grown to mid-log phase in SD-Met media containing 50 µM CuCl2.
D) Corresponding immunoblot analysis of cells in C) expressing Cos5-HA, Cos5KR-HA and Cos5KR-HA-Ub with α-HA and α-CPY, provided as a loading control.
E) Localization of Cos5-GFP in sna3Δ and sna3Δ bsd2Δ double null mutant cells.
F) Synthetic effects of deleting COS6 and SNA3 or BSD2. Localization of GFP-Cps1 in the indicated single and double null mutants is shown.
G) Cos5-GFP and Cos5KR-GFP were immunoprecipitated from lysates of cells also expressing HA-Ub using α-GFP antibodies. Immunoprecipitates were immunoblotted with α-GFP and α-HA. Cells carried the pep12Δ mutation alone or in combination with sna3Δ and bsd2Δ mutations.
H) Binding of MBP-Rsp5 to fusion proteins between GST and the C-terminal domains (CTD) of Cos4, Cos5, Cos6, Sna3, and the Sna3-CTD lacking its Rsp5-binding PY motif (ΔPY). GST fusion proteins were immobilized on GSH-agarose. Beads were incubated with recombinant MBP-Rsp5, washed, and immunoblotted with α-MBP antibodies. Bar = 5 µm.
We found that sorting of Cos5-GFP to the vacuole was slightly perturbed in cells lacking Sna3 or Bsd2, two polytopic membrane proteins that act as Rsp5 substrate adaptors (Hettema et al., 2004; MacDonald et al., 2012b) and was significantly compromised in a sna3Δ bsd2Δ double mutant (Figure 4E). We also found that loss of Sna3 or Bsd2 caused synthetic MVB sorting defects when combined with a single deletion of a COS gene (COS6 in Figure 4F and COS1, COS2, COS4, or COS5 in Figure S4g). Together these data indicate that Sna3 and Bsd2 collaborate with Cos proteins to facilitate MVB sorting, possibly by mediating ubiquitination of the Cos proteins themselves. To test this model, Cos5-GFP and Cos5KR-GFP were immunoprecipitated from cells co-expressing HA-Ub (Figure 4G). Cells also carried a pep12Δ mutation that stabilizes Ub-modified MVB cargo (Katzmann et al., 2001). Ubiquitination of Cos5-GFP was diminished by loss of Bsd2 and Sna3 and no ubiquitination could be found on Cos5KR-GFP. These data demonstrate that Sna3 and Bsd2 contribute to the ubiquitination of Cos proteins, however, residual ubiquitination of Cos proteins remained in bsd2Δ sna3Δ cells. We also found that Rsp5 could bind directly to Cos proteins (Figure 4H). GST fusions to the C-terminal domains (CTD) of Cos4, Cos5, and Cos6 bound a recombinant MBP-Rsp5 fusion protein at levels similar to those attained by a GST fusion protein containing the CTD of Sna3, which has a PPxY motif that binds strongly to Rsp5 (McNatt et al., 2007; Oestreich et al., 2007; Stawiecka-Mirota et al., 2007; Watson and Bonifacino, 2007). Together these results show that Cos proteins are major targets of Rsp5-dependent ubiquitination, that multiple mechanisms are used by Rsp5 to ubiquitinate Cos proteins, and that ubiquitination of Cos proteins in turn is critical for their ability to sort themselves and other proteins into the MVB pathway.
Cos proteins form endosomal microdomains that concentrate ubiquitinated cargo
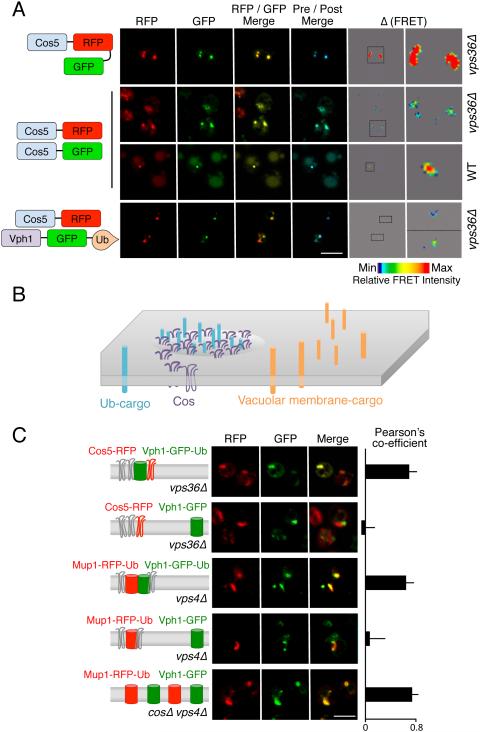
Immunoblot analysis of Cos5-HA suggests Cos proteins can self associate, as indicated by dimeric and oligomeric species detectable at higher exposures (Figure 2D, 4D and S5a). To explore this in vivo we examined the localization of Cos proteins within endosomes with respect to other Ub-cargo. FRET (Fluorescence Resonance Energy Transfer) microscopy showed that Cos proteins interact with each other in endosomal compartments (Figure 5A). A strong FRET signal between Cos5-GFP and Cos5-mCherry was only found at peri-vacuolar / late endosomal structures of wild-type cells or within class E endosomes of vps36Δ cells. We also found that Cos5 interacted with Ub-cargo by FRET using a fusion of Vph1-GFP to Ub (Figure 5A). Vph1 is a membrane protein subunit of the V-ATPase, which normally localizes to the limiting membrane of the vacuole but is redirected into the MVB pathway when translationally fused to Ub (Figure S5b) (Urbanowski and Piper, 2001). We also found that GFP-Ub, which accumulates on MVB cargo on endosomes of ESCRT null mutants (Macdonald et al., 2012a; Ren et al., 2008), co-localized with Cos proteins in endosomal structures of vps36Δ and vps4Δ cells (Figure S5c).
Figure 5. Cos proteins associate in endosomal subdomains they help create.
A) Mutant vps36Δ cells containing an enlarged endosomal class E compartment were used to localize a Cos5 fusion protein containing both mCherry (RFP) and GFP or to localize Cos5-GFP and Cos5-RFP co-expressed from different plasmids. FRET was measured by increase in donor (GFP) fluorescence after acceptor (RFP) photobleaching. Maximal FRET was set at the level displayed for Cos5-RFP-GFP tandem fusion protein. Shown is RFP fluorescence, GFP fluorescence, the merge between RFP and GFP, the merge of the GFP signal before and after (blue) photobleaching, and the scaled FRET signal. The indicated inset is also magnified. Also shown is the co-localization and FRET of Cos5-GFP and Cos5-RFP in WT cells. Below, co-localization of Cos5-RFP and Vph1-GFP-Ub to the same endosomal subdomain within vps36Δ cells.
B) Schematic model of endosomal membrane sub-compartments where Ub-cargo and Cos proteins cluster in subdomains that are distinct from subdomains containing other membrane proteins.
C) Confocal microscopy of subdomains in the class E compartments of vps4Δ and vps36Δ cells. Cos5-RFP, Mup1-RFP-Ub, and Vph1-GFP-Ub are used as indicated (left) to mark localization of ubiquitinated proteins. Vph1-GFP is used as indicated as a cargo protein that does not undergo MVB sorting but still accumulates within enlarged endosomal “class E” compartments. Right, the Pearson’s coefficient was determined for >30 cells to quantify co-localization of proteins in each condition (average and SD). Bar = 5 µm.
In contrast to Ub-cargo, Vph1-GFP that also accumulates within class E endosomes of ESCRT mutants, did not co-localize with Cos proteins and instead occupied adjacent subdomains (Figure 5B & C). This separation of subdomains, one marked by Cos proteins and Ub-cargo and another marked by the non-MVB cargo Vph1-GFP, were found not only in vps36Δ cells, but also in vps23Δ, vps4Δ, and escrt-IIIΔ cells (Figure S5d). We found that the Cos/Ub-cargo domains could also be visualized with Mup1-RFP-Ub, which segregates with Vph1-GFP-Ub and not Vph1-GFP within class E compartment/endosomes. Using Mup1-RFP-Ub together with Vph1-GFP revealed that segregation of Ub-cargo within class E endosomal compartments required Cos proteins, as these markers were not segregated in endosomes of cosΔ vps4Δ cells (Figure 5C).
The Cos protein network traps cargo at endosomes
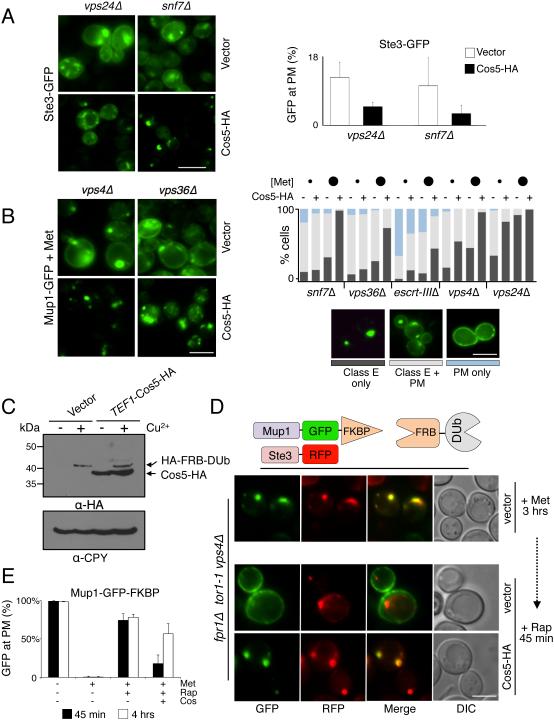
The effects of Cos overexpression on GFP-Snc1 and Mup1-GFP localization suggested that Cos proteins immobilize cargo in endosomes prior to sorting into ILVs (Figure 2F, 3E and 4C). To test the idea that Cos proteins retain cargo on endosomes, we compared the distribution of Ste3-GFP in vps24Δ and snf7Δ with Cos5-HA overexpression (Figure 6A). In control cells Ste3-GFP was found within late-endosomal/class E compartments and also at the cell surface; overexpressing Cos5-HA shifted Ste3 more to endosomal compartments. Previous studies compared the steady-state distribution of Mup1-GFP on the plasma membrane with endosomes in various ESCRT null mutants as a measure for how well cargo is trapped on endosomes (Teis et al., 2008). Those studies found differences in the degree of Mup1 localization at the cell surface depending on which ESCRT subunit was deleted, implying that exaggerated ESCRT-III polymers might trap cargo on endosomes. We found that localization of Mup1-GFP to the cell surface in all ESCRT mutants was also modulated by how much extracellular methionine was present, likely reflecting how strongly Mup1 is ubiquitinated. Therefore, we assessed Mup1-GFP distribution at low [0.5 μg/ml] and modest [4.0 μg/ml] concentrations of methionine and quantified the distribution of Mup1-GFP between the cell surface and class E/endosomal compartments (Figure 6B). We found that in all ESCRT mutants tested, overexpression of Cos5-HA shifted the distribution of Mup1-GFP from the cell surface to class E/endosomal compartments. Notably, this was found in vps36Δ mutants, snf7Δ mutants, and escrt-IIIΔ mutants (lacking all ESCRT-III subunits) indicating that the ability of Cos proteins to trap cargo did not strictly require ESCRT-III polymers.
Figure 6. Cos expression sequesters MVB cargo in endosomes.
A) Localization of Ste3-GFP in vps24Δ and snf7Δ mutants. Cells carrying vector alone or the CUP1-Cos5-HA plasmid were grown in SD-Met containing 50 µM CuCl2. Right, quantitation of percentage of cells (n=>200 over 2 experiments) showing Ste3-GFP at the cell surface (average + SD).
B) Localization of Mup1-GFP in vps4Δ and vps36Δ mutants. Cells carrying vector alone or the CUP1-Cos5-HA plasmid were grown in SD containing 50 µM CuCl2 and 4 µg/ml Methionine. Right, quantitation of the percentage of cells (n=200) showing different distributions of Mup1 in the ESCRT null strains indicated. For quantitation, cells were grown in low (small circles, 0.5 µg/ml) or modest (large circles, 4 µg/ml) concentrations of methionine. Cells were classified as having Mup1-GFP localized to only class E compartments (dark grey bars), to a mix between class E compartments and plasma membrane (light grey), or to the plasma membrane exclusively (blue).
C) Immunoblot of HA-FRB-M48 induced by addition of 50 μM Copper Chloride co-expressed with Cos5-HA driven from the constitutive TEF1 promoter.
D) Triggered recycling to the plasma membrane from class E endosomal compartments in the presence and absence of Cos5-HA overexpression. Mutant vps4Δ cells (carrying fpr1Δ and tor1-1 mutations) expressing Ste3-RFP, Mup1-GFP-FKBP, and FRB-DUb, were grown in SD+Met media for 3 hrs followed by addition of 50 µM CuCl2 for an additional 15 min. Rapamycin (Rap: 10µM) was then added for 45 min allowing Mup1-GFP to relocalize to the cell surface.
E) Quantitation of experiment from D for proportion of cells (n=>150) with Mup1-GFP-FKBP at the cell surface. Controls lacking Rapamycin show Mup1-GFP-FKBP is internalized by addition of 40 μg/ml methionine and does not return to the surface after 4 hrs. Surface recycling was measured at 45min (black bars) and 4hrs (white bars) after addition of 10 µM Rapamycin (mean + SD over 3 experiments). Bar = 5 µm.
We next developed a kinetic assay to follow recycling of Mup1-GFP back to the cell surface (Figure 6). We took advantage of the ability of Rapamycin to heterodimerize chimeric proteins containing FK506-binding protein (FKBP-12) and the FRB domain of mTOR (Belshaw et al., 1996; Liberles et al., 1997; Spencer et al., 1993). Here we fused Mup1-GFP to FKBP-12 and co-expressed it with FRB fused to the deubiquitinating enzyme M48 and tested feasibility with a set of control experiments (Figure S6). To assess endosomal trapping, Mup1-GFP-FKBP was expressed in vps36Δ cells grown for 16 hrs in SD media lacking methionine. High concentrations of methionine [40 μg/ml] were then added for 3 hrs to drive all of the Mup1-GFP-FKBP into endosomal/class E compartments (Figure 6D,E). These high levels of methionine also repress the MUP1 promoter ensuring that only a pre-existing population of Mup1-GFP-FKBP was followed (Menant et al., 2006). Rapamycin triggered a dramatic redistribution of Mup1-GFP-FKBP to the cell surface within 45 min (Figure 6D,E), while having no effect on a co-expressed Ste3-mCherry that remained localized to endosomal compartments. By contrast, overexpressing Cos5-HA caused Mup1-GFP-FKBP to remain in endosomal compartments (Figure 6C,D,E). Extending the treatment of Rapamycin to 4 hr did allow recycling of more Mup1-GFP-FKBP to the cell surface of Cos5-HA overexpressing cells underscoring the dramatic delay in the efflux of cargo from endosomes (Figure S6c).
Cos proteins act as cargo adaptors for GPI-anchored proteins
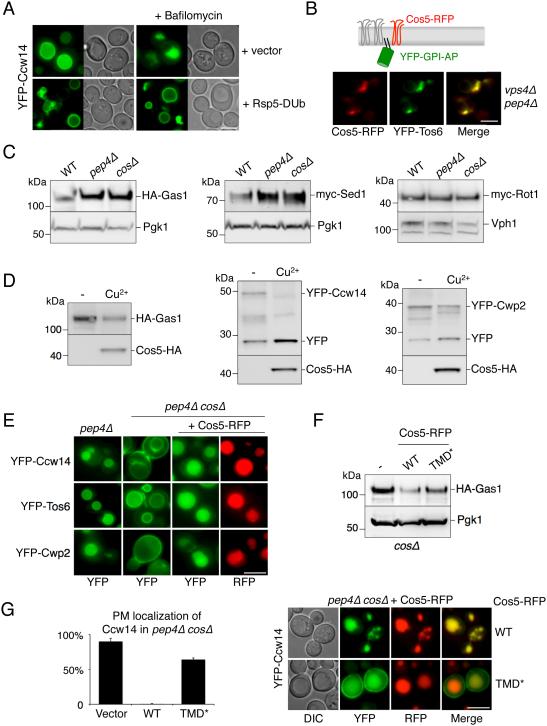
We hypothesized a model in which the functional role of Cos proteins is to create an endosomal environment, or subdomain (Figure 5), conducive to MVB sorting. This could explain how cargo that has no Ub signal of its own might still access the canonical ESCRT- and Ub-dependent MVB pathway for degradation. To explore this possibility we examined the MVB sorting of an unequivocal Ub-independent class of cargo: GPI anchored proteins (GPI-APs), which do not have cytosolic domain on which to attach Ub or otherwise interact directly with ESCRT components (Mayor and Riezman, 2004) (Figure S7a). Previous experiments demonstrate that many Fluorescent Protein (FP)-tagged GPI-anchored proteins are sorted to the vacuole (Ast et al., 2013; Bagnat and Simons, 2002; Castillon et al., 2009; Cohen et al., 2013; Rodriguez-Pena et al., 2002). Analysis in pep4Δ cells that lack active vacuolar hydrolases to cleave YFP from lumenally exposed GPI-APs clearly showed that GPI-APs enter the vacuole via intralumenal vesicles (Figure S7a,b,c). We also found that MVB sorting of GPI-APs relied on proper Rsp5 and ESCRT function (Figure 7A,B). In addition, GPI-APs strongly co-localized with Cos proteins within endosomal class E compartments of vps4Δ pep4Δ cells demonstrating that like canonical Ub-cargo, they too can localize to discrete Cos-containing endosomal subdomains (Figure 7B).
Figure 7. MVB sorting of GPI-anchored proteins is Cos-dependent.
A) Localization of YFP-Ccw14 in wild-type cells carrying a vector control or expressing Rsp5-DUb. Cells were also imaged 3 hrs after the addition of 1 µM Bafilomycin.
B) Co-localization of Cos5-RFP and YFP-Tos6 in vps4Δ pep4Δ cells.
C) Cells (WT, pep4Δ, cosΔ) expressing HA-Gas1, myc-Sed1 or myc-Rot1 were grown to log phase, prior to immunoblot analysis with α-HA or α myc antibodies and α-PGK or α-Vph1 blots for loading control.
D) The levels of GPI-APs (HA-Gas1, YFP-Ccw14 or YFP-Cwp2) in cells co-expressing Cos5-HA were measured by immunoblotting. Cultures were grown in the presence of copper chelator (−) or 50 µM copper chloride (Cu2+). In the case of YFP-tagged proteins, the vacuolar-processed form of the protein was also detectable (YFP).
E) Sorting of YFP tagged GPI-APs was assessed by fluorescence microscopy in pep4Δ and pep4Δ cosΔ cells. Defective sorting observed in cosΔ cells was restored upon expression of Cos5-RFP from a CUP1-Cos5-RFP plasmid.
F) Levels of HA-Gas1 are shown from wild-type cells co-expressing vector (−), Cos5-RFP (WT) and Cos5TMD*-RFP (TMD*). PGK immunoblot was used as a loading control.
G) Mutant cosΔ cells co-expressing YFP-Ccw14 and either WT or TMD* versions of Cos5-RFP. Left, the number of cells with YFP signal detected at the cell surface was quantified for each condition (average + SD). Bar = 5 µm.
In cosΔ cells, MVB sorting of GPI-APs was completely blocked, yet was restored upon overexpressing Cos5-RFP from a plasmid (Figure 7E). This was consistent with the effect of deleting Cos proteins on the overall levels of two GPI-APs, Gas1 and Sed1 assessed by immunoblotting (Figure 7C). Loss of Cos or Pep4 function did not alter levels of the ER-localized myc-Rot1 GPI-AP (Takeuchi et al., 2006), confirming the specificity of Cos proteins with regard to vacuole-dependent degradation. Finally, we found that overexpressing Cos5-HA in otherwise wild-type cells resulted in more GPI-APs directed to the vacuole for degradation (Figure 7D). These results were corroborated in pep4Δ cells grown in media containing high levels of NA, which activates Sir2 to silence subtelomerically-localized COS genes (Figure S7d). Here, sorting of YFP-Ccw14, YFP-Tos6, and YFP-Cwp2 into the vacuole was modest but was strongly increased upon overexpression of Cos5-RFP from a plasmid.
These data demonstrate that Cos proteins mediate entry of GPI-APs into the Ub-dependent and ESCRT-dependent MVB sorting pathway. The simplest explanation is that GPI-APs congregate with Cos proteins, which supply a Ub sorting signal in trans. It is unlikely that such a mechanism is mediated by direct interaction between GPI-APs and Cos proteins because GPI-APs are entirely luminal proteins with small lipid anchors and Cos proteins only have 4-6 predicted residues on their luminal side, providing no basis for direct interaction. Rather, Cos proteins might form a membrane subcompartment into which GPI-APs are segregated and held, a model consistent with the co-localization of GPI-APs within Cos-containing endosomal subdomains (Figure 7B). A prediction of this model is that residues within the membrane spanning domains are critical for the ability of Cos proteins to mediate MVB sorting of GPI-APs. Indeed, Cos proteins have many residues within their TMDs that are atypical for such hydrophobic stretches and these in turn might create a local environment that promotes trapping of cargoes before their incorporation into ILVs. To test this idea, we created a version of Cos5 with TMDs composed entirely of conventional hydrophobic residues (TMD*; Figure S7e). Despite these changes, Cos5TMD*-RFP still sorted into the vacuole lumen, however, its ability to confer sorting on GPI-APs was greatly reduced. This was reflected by the inability of overexpressed Cos5TMD*-RFP to decrease steady-state levels of HA-Gas1 (Figure 7F) or to completely shift YFP-Ccw14 from the cell surface into the vacuole lumen (Figure 7G).
DISCUSSION
Cos proteins contribute to the MVB sorting of a variety of cargo and are used as a broad mechanism to drive down-regulation of cell surface proteins when nutrients, in particular Niacin, are depleted. This mechanism allows cells to integrate protein turnover with the overall metabolic activity and requirements of the cell whilst reducing the abundance of transporters, receptors, and other proteins at the cell surface. Such a response may also provide an additional mechanism to generate amino acids during mild nutrient stress as a complementary form of autophagy (Babst and Odorizzi, 2013).
While the efficiency of MVB sorting was compromised for many Ub-cargoes in the absence of Cos proteins, sorting defects were most severe for cargoes that contain no Ub signal (e.g. Sna3KR and GPI-APs). Our data indicate that Cos proteins form oligomers that establish specialized regions of endosomes that trap Ub-cargo as well as GPI-APs, supporting a “cargo corral” model whereby immobilization leads to more efficient incorporation into forming ILVs. The Cos proteins within this “corral” are highly ubiquitinated, allowing them to provide Ub sorting signals in trans to cargo proteins with which they associate. This ubiquitinated conglomerate would provide assistance to a wide variety of conventional membrane proteins that might otherwise escape incorporation into ILVs as they transit through endosomes. In the case of GPI-APs, the Cos corral provides a way to cluster GPI-anchored proteins and other non-ubiquitinated proteins and surround them with multiple Ub sorting signals allowing their entry into ILVs along an ESCRT-dependent route. Formation of a “Cos corral” would also organize canonical Ub-cargo in a manner that allows for its deubiquitination prior to ILV incorporation while preventing such deubiquitinated cargoes from escaping their MVB fate.
This model explains how luminal sided GPI-APs rely on the E3 ligase Rsp5, ESCRTs and Cos proteins to enter the MVB pathway. Yet, exactly how Cos proteins segregate cargo remains to be determined. Part of the answer may lie in the unusual composition of the TMDs of the Cos proteins that is required for Cos protein function and that might establish a particular lipid architecture that favors coalescence of membrane proteins (Epand and Epand, 2011). In particular, the presence of arginines within the TMDs may induce thinning or other changes in the membrane bilayer (Krepkiy et al., 2009) that could affect diffusion of proximal cargo proteins. This model can explain how luminal sided GPI-APs rely on the E3 ligase Rsp5, ESCRTs and Cos proteins to enter the MVB pathway (Figure 7). In support of this model, mutations within the TMD of Cos5 greatly reduce its ability to sort GPI-APs. Future structure/function studies will determine the biophysical characteristics of Cos proteins that allow them to operate.
Ubiquitination of Cos proteins is a critical requirement for their function, and multiple mechanisms appear to contribute to their ubiquitination. Cos proteins may associate with Rsp5 indirectly via the Rsp5-binding membrane proteins Sna3 and Bsd2 as well as bind directly through their C-terminal domains (Figure 4). Cos proteins also have a very high proportion of lysines (9% of all residues), and preliminary mass-spectrometry analysis from our lab (data not shown) and others (Hitchcock et al., 2003; Peng et al., 2003) indicates that 10 or more ubiquitination sites are distributed throughout the cytosolic regions. These qualities would ensure that Cos proteins are replete with the Ub-sorting signals they provide to their associated cohort of cargo. In addition, they might allow Cos proteins to retain at least some Ub following cargo deubiquitination by Doa4, thus providing a means to keep its associated cargo engaged with the ESCRT apparatus. Indeed, the delivery of the intravacuolar pool of Ub along the MVB pathway requires the Cos proteins, implying that they may themselves be the major carriers of vacuolar localized Ub (Figure 3).
This study raises the question as to what the functional analogs of Cos proteins are in animal cells, where Ub-independent sorting and sorting of GPI-APs has been clearly documented (Dores et al., 2012; Satpute-Krishnan et al., 2014). Our model suggests that the role of Cos proteins in the MVB sorting process may be at least partly analogous to those of the Tspans in mammals. Tspans are enriched in membrane subdomains that can capture or contain a variety of other polytopic membrane proteins (Hemler, 2003). Some Tspans, such as CD63/LAMP3, are major components of the ILVs of mammalian MVBs (Pols and Klumperman, 2009) and contribute to the formation of MVBs and the sorting of particular cargo (Colombo et al., 2013; Edgar et al., 2014; van Niel et al., 2011). Whether Tspan-dependent MVB cargo is ushered into an ILV pathway that is distinct from that for Ub-dependent cargo has yet to be fully resolved. Ubiquitination of some Tspans has been described, and the MARCH Ub ligases have been proposed to regulate Tspan turnover (Bartee et al., 2010; Lineberry et al., 2008). Also, Tspans have been implicated in the MVB sorting of the EGF receptor, a prototypical Ub-dependent MVB cargo (Danglot et al., 2010; Odintsova et al., 2000; Odintsova et al., 2013). Cos proteins bear some resemblance to Tspans with respect to their molecular structures, localization, and proposed function. Cos proteins have polar residues within their 4 TMDs, associate with one another, and congregate in membrane subdomains. In addition, Cos proteins and a subset of Tspans are efficiently sorted into MVB ILVs. Tspans are widely expressed throughout Eukaryota yet are conspicuously absent from yeast (Garcia-España et al., 2008). Some of the defining sequence motifs of Tspans that distinguish them from other 4-TMD-contianing proteins are absent in Cos proteins. These include a large cysteine-rich extracellular domain between TMDs 3 and 4, and juxtamembrane domain-localized cysteine residues that serve as sites for palmitoylation. These features may simply be incompatible for function within the yeast secretory pathway, obliging their loss from a common ancestor. Alternatively, Cos proteins and Tspans may have evolved convergently to meet a related functional need in the MVB sorting process. One of the central observations here is that Cos proteins were required for sorting GPI-APs into ILVs, and this process in animal cells has been hypothesized to be mediated by Tspans (Sobo et al., 2007; van Niel et al., 2006). This extends the possibility that yeast Cos proteins are the functional analogs of Tspans or that other cell types possess a different analog that serves this central purpose.
EXPERIMENTAL PROCEDURES
General
Yeast strains, plasmids, and where they were used in this study are described in Tables S1 and S2. Details on deleting all COS genes are provided in supplemental data as are procedures for pulse chase analysis, production of recombinant proteins and in vitro binding studies.
Cell Culture
Standard yeast minimal and rich media were used as previously described (MacDonald et al., 2012b; Norgan et al., 2013). Synthetic defined (SD) 1x media containing 2% glucose, 1x yeast nitrogen base (Research Products International, Mount Prospect, IL) or 1x yeast nitrogen base lacking NA (Formedium, Norfolk, UK) was supplemented with amino acid mixtures that allow appropriate selection (Formedium, Norfolk, UK). Expression from the CUP1 promoter was induced with 50 μM CuCl2 or repressed with batho-cuproine disulphonate. Optical density at 600 nm was used to measure yeast growth: Mid-log was OD600 = 0.5 - 1.0; late-log phase was OD600 = 1.5 - 2.0, and late-log+ was 2-6 hrs after cells had reached OD600 = 2.0.
Microscopy
Conventional fluorescence microscopy of cells resuspended in 100 mM Tris.HCl pH 8.0, 0.2% (w/v) NaN3 and NaF3 was performed as previously described (Macdonald et al., 2012a). For deconvolved fluorescence images, micrographs of cells imaged with an Olympus IX70 and Coolsnap HQ camera (Photometrics, Tuscon, AZ) were processed using Delta Vision software (Applied Precision Issaquah, WA). Confocal images and FRET data were collected on live cells using a Zeiss 710 laser confocal microscope. Data were processed using Fiji software and change in GFP fluorescence upon bleaching was converted to a color-coded heat map calibrated across all measurements and images.
Immunoblot analysis
Sources of antibodies included: Monoclonal α-HA (Covance research products Inc. Berkely, CA); Polyclonal α-PGK1 (Tom Stevens, University of Oregon, OR); Polyclonal α-GFP (Urbanowski and Piper, 1999). Polyclonal α-Rsp5 (Stamenova et al., 2004); α-MBP (Lee et al., 2009); and Polyclonal anti-CPY (Tom Stevens, University of Oregon, OR). For immunoblot analysis, cells were grown to mid-log phase, resuspended in 0.2 M NaOH for 2 min, lysed at 25°C in 50 mM Tris.HCl pH 6.8, 5% SDS, 10% glycerol and 8 M urea and immediately subjected to SDS-PAGE.
Immunoprecipitations
Immunoprecipitations were described in (Katzmann et al., 2001). Yeast expressing the Cos5 constructs (10 OD) and (HA)-ubiquitin were TCA precipitated processed and lysed with glass beads. Immunoprecipitations were performed using GFP Rabbit Serum Polyclonal Antibody (Life Technologies). Samples were subjected to SDS-PAGE and Western blotting. Cos5 was detected with monoclonal anti-GFP AV-JL8 (Clontech) and the Ubiquitination status was determined with monoclonal anti-HA.11 (Covance) to recognize HA-ubiquitin.
Steady state recycling assay
For Ste3-GFP, cells were grown to mid-log phase in SD media and the number of cells exhibiting any cell surface Ste3-GFP were compared to the number of cells with only intracellular Ste3-GFP. For Mup1-GFP, cells were grown to early-log phase in minimal media lacking methionine before methionine (0.5 μg/ml or 4 μg/ml) was added for 1 hr.
Triggered Recycling Assay
The CUP1-HA-FRB-M48 gene was integrated into cells carrying the tor1-1 and fpr1Δ mutations (Haruki et al., 2008). The chromosomal copy of MUP1 was modified to express Mup1-GFP-FKBP as the sole copy of Mup1 under its endogenous promoter; and VPS4 was deleted. Cells grown to mid-log phase were treated with 40 μg/ml methionine for 3 hrs, washed 3x and resuspended in fresh methionine-containing media with 50 μM CuCl2 for 15 min before a 45 min or 4 hr treatment with 10 µM Rapamycin.
Supplementary Material
HIGHLIGHTS.
Niacin depletion down-regulates cell surface proteins by elevating Cos protein levels
Cos proteins create endosomal subdomains that trap MVB cargo
Various ubiquitinated cargoes rely on Cos proteins for efficient MVB sorting
Cos proteins provide ubiquitin in trans to convey GPI-APs into the ESCRT pathway
ACKNOWLEDGEMENTS
The work was funded by AHA postdoctoral fellowship award to CM, NIHRO1-GM058202 to RP, NIHRO1-GM073024 to DK. The authors wish to thank: Mark Stamnes for help with genome sequence analysis; Tom Moninger at the University of Iowa Central Microscopy Facility for help with microscopy; Staff of the Iowa Institute of Human Genetics for help with Illumina genome sequencing; Ling Song for help with generating the escrt-IIIΔ strain; Stanley Winistorfer for assistance; the laboratory of Maya Schuldiner for providing of GPI-AP plasmids, and Chris Stipp and Charles Brenner for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T, Cohen G, Schuldiner M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013;152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G. The balance of protein expression and degradation: an ESCRTs point of view. Curr Opin Cell Biol. 2013;25:489–494. doi: 10.1016/j.ceb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Simons K. Cell surface polarization during yeast mating. Proc Natl Acad Sci U S A. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E, Eyster CA, Viswanathan K, Mansouri M, Donaldson JG, Früh K. Membrane-Associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS ONE. 2010;5:e15132. doi: 10.1371/journal.pone.0015132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci USA. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci U S A. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic. 2009;10:161–171. doi: 10.1111/j.1600-0854.2008.00855.x. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci U S A. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Megyeri M, Chen OC, Condomitti G, Riezman I, Loizides-Mangold U, Abdul-Sada A, Rimon N, Riezman H, Platt FM, et al. The yeast p5 type ATPase, spf1, regulates manganese transport into the endoplasmic reticulum. PLoS ONE. 2013;8:e85519. doi: 10.1371/journal.pone.0085519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- Danglot L, Chaineau M, Dahan M, Gendron MC, Boggetto N, Perez F, Galli T. Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J Cell Sci. 2010;123:723–735. doi: 10.1242/jcs.062497. [DOI] [PubMed] [Google Scholar]
- Davis NG, Horecka JL, Sprague GF., Jr. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol. 2012;197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JR, Eden ER, Futter CE. Hrs- and CD63-Dependent Competing Mechanisms Make Different Sized Endosomal Intraluminal Vesicles. Traffic. 2014;15:197–211. doi: 10.1111/tra.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. Journal of peptide science : an official publication of the European Peptide Society. 2011;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- Falguieres T, Luyet PP, Bissig C, Scott CC, Velluz MC, Gruenberg J. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol Biol Cell. 2008;19:4942–4955. doi: 10.1091/mbc.E08-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-España A, Chung P-J, Sarkar IN, Stiner E, Sun T-T, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc Natl Acad Sci U S A. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JRE, Oestreich AJ, Payne JA, Gunawan MS, Norgan AP, Katzmann DJ. The HECT domain of the ubiquitin ligase Rsp5 contributes to substrate recognition. J Biol Chem. 2009;284:32126–32137. doi: 10.1074/jbc.M109.048629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci U S A. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Lineberry N, Su L, Soares L, Fathman CG. The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. The Journal of biological chemistry. 2008;283:28497–28505. doi: 10.1074/jbc.M805092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 2012a;13:331–338. doi: 10.1038/embor.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Stringer DK, Piper RC. Sna3 Is an Rsp5 Adaptor Protein that Relies on Ubiquitination for Its MVB Sorting. Traffic. 2012b doi: 10.1111/j.1600-0854.2011.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- McNatt MW, McKittrick I, West M, Odorizzi G. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol Biol Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menant A, Barbey R, Thomas D. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 2006;25:4436–4447. doi: 10.1038/sj.emboj.7601330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, Russell MR, Odorizzi G. A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 2007;8:644–650. doi: 10.1038/sj.embor.7401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgan AP, Davies BA, Azmi IF, Schroeder AS, Payne JA, Lynch GM, Xu Z, Katzmann DJ. Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J Biol Chem. 2013;288:26147–26156. doi: 10.1074/jbc.M113.494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Current biology : CB. 2000;10:1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- Odintsova E, van Niel G, Conjeaud H, Raposo G, Iwamoto R, Mekada E, Berditchevski F. Metastasis suppressor tetraspanin CD82/KAI1 regulates ubiquitylation of epidermal growth factor receptor. J Biol Chem. 2013;288:26323–26334. doi: 10.1074/jbc.M112.439380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007;18:707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nature biotechnology. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Piper RC, Dikic I, Lukacs GL. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Pashkova N, Winistorfer S, Piper RC. DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J Biol Chem. 2008;283:21599–21611. doi: 10.1074/jbc.M802982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pena JM, Rodriguez C, Alvarez A, Nombela C, Arroyo J. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J Cell Sci. 2002;115:2549–2558. doi: 10.1242/jcs.115.12.2549. [DOI] [PubMed] [Google Scholar]
- Roth AF, Sullivan DM, Davis NG. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Ajinkya M, Bhat S, Itakura E, Hegde RS, Lippincott-Schwartz J. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 2014;158:522–533. doi: 10.1016/j.cell.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Hall MN, Koller A. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol. 1994;14:6597–6606. doi: 10.1128/mcb.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobo K, Chevallier J, Parton RG, Gruenberg J, van der Goot FG. Diversity of raft-like domains in late endosomes. PLoS ONE. 2007;2:e391. doi: 10.1371/journal.pone.0000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Spode I, Maiwald D, Hollenberg CP, Suckow M. ATF/CREB sites present in sub-telomeric regions of Saccharomyces cerevisiae chromosomes are part of promoters and act as UAS/URS of highly conserved COS genes. J Mol Biol. 2002;319:407–420. doi: 10.1016/S0022-2836(02)00322-4. [DOI] [PubMed] [Google Scholar]
- Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. The Journal of biological chemistry. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–1296. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol. 2011;192:229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Kimata Y, Hirata A, Oka M, Kohno K. Saccharomyces cerevisiae Rot1p is an ER-localized membrane protein that may function with BiP/Kar2p in protein folding. Journal of biochemistry. 2006;139:597–605. doi: 10.1093/jb/mvj063. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, Berson JF, Thomas PC, Raposo G, Marks MS. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem. 1999;274:38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic. 2001;2:622–630. doi: 10.1034/j.1600-0854.2001.20905.x. [DOI] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. Journal of biochemistry. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Watson H, Bonifacino JS. Direct binding to Rsp5p regulates ubiquitination-independent vacuolar transport of Sna3p. Mol Biol Cell. 2007;18:1781–1789. doi: 10.1091/mbc.E06-10-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.