Abstract
The duplication of DNA sequences contributes to genomic plasticity and is known to be one of the key factors responsible for evolution. The mechanisms underlying these rare events, which have been frequently mentioned by authors performing genomic analysis, have not yet been completely elucidated. These mechanisms were approached here in the yeast Saccharomyces cerevisiae, using a positive selection screen based on a particular mutated allele of the URA2 gene. Spontaneous revertants containing a duplication of the terminal part of the URA2 gene were selected and analyzed. Some important features of the duplicated regions, such as their chromosome location, size, and insertion sites, were characterized. The events selected correspond to a single inter- or intrachromosomal gene duplication process. The duplicated ATCase sequence is generally punctuated by a poly(A) tract and is always located in Ty1 sequences. In addition, the activation of a Ty1 transcription process increased the frequency of the duplication events. All in all, these data suggest that the duplication mechanism involves the reverse transcription of mRNA and the subsequent integration of the cDNA into a Ty1 area. The Ty1 elements and the retrotransposon-encoded function are key factors contributing to chromosomal reshaping. The genomic rearrangements described constitute experimental evidence for the recovery of a function involving duplication by retroposition.
Gene duplication can be said to be a key to molecular evolution (Ohno 1970). It results in redundancy and may be the first step in the emergence of new functions. Duplicated genes may have three types of fate. The most likely of these is a process called nonfunctionalization, in which one of the two copies of a duplicate pair may degenerate into a pseudogene or be lost from the genome as the result of the accumulation of several deleterious mutations followed by chromosomal rearrangements such as deletion (Lynch and Conery 2000; Lynch 2002). The second possible process, which is called subfunctionalization, consists of a division of the original functions of the ancestral gene between the two duplicates due to the differential accumulation of degenerative mutations in both copies (Force et al. 1999). The last and most important evolutionary gene duplication fate is known as neofunctionalization. One of the copies in a duplicate pair can acquire a new function by changing its coding or regulatory sequences, whereas the second copy keeps the original function (Zhang et al. 1998).
Based on experimental studies and data obtained in global genomic analyses, several hypotheses have been proposed to explain the presence of a duplicated copy. Gene duplication can involve either a single gene, segmental (Bailey et al. 2002; Koszul et al. 2004), single chromosome or aneuploidy (Hughes et al. 2000) or whole-genome duplication process (Wolfe and Shields 1997). In Saccharomyces cerevisiae, the only examples of in vivo single gene duplication described to date are those involving the ACP1 (Hansche et al. 1978), HIS4 (Greer and Fink 1979), ADH2 (Paquin et al. 1992), URA2 (Bach et al. 1995), and hexose transport genes (Brown et al. 1998). These duplication events have been observed at a very low frequency of 10–10 – 10–12 duplications per cell per generation, and the mechanisms responsible for these events are still poorly understood.
To understand the mechanisms involved in the duplication events at work in S. cerevisiae, we used a genetic screening method developed at our laboratory which can be used for the positive selection of spontaneous genomic rearrangements (Roelants et al. 1995). This screening method is based on the use of a mutated allele of the S. cerevisiae URA2 gene carrying the CPSase and ATCase functional domains (Fig. 1). In the ura2 15-30-72 mutant strain, the ATCase domain is inactive, resulting in a uracil auxotroph. The functional reactivation of the ATCase domain suffices to generate Ura+ prototrophs, but only at a low frequency of ∼10–10. Three types of reactivation events have been described previously: insertion of a Ty1 retrotransposon in the URA2 coding sequence upstream of the last mutation (Roelants et al. 1997), deletion of the sequence bearing the three mutations (Welcker et al. 2000), and duplication of the ATCase coding sequence and its insertion elsewhere in the genome under the dependence of a resident promoter (Bach et al. 1995).
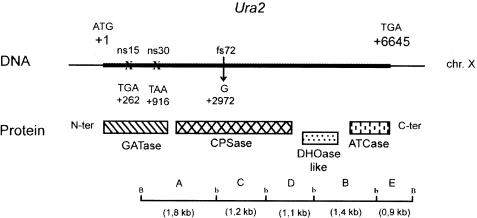
Figure 1.
Correspondence between the simplified restriction map of the URA2 locus and protein domains. The thick line indicates the URA2 coding sequence (restriction site: b, BglII; B, BamHI). A, C, D, B, and E denote the BamHI-BglII subfragments, ns15 and ns30 the positions of the nonsense mutations, and fs72 the position of the frameshift mutation (GATase, glutamine amidotransferase; CPSase, carbamoylphosphate synthetase; DHOase-like, dihydroorotase-like; ATCase, aspartate transcarbamylase).
Among these three reactivation events, we focused here on the duplication events which lead to the generation of a Ura+ prototroph. In this case, duplication is synonymous with recovery of gene function which is associated with a change in the regulatory sequences involved. We sought to determine the chromosome location, size, and insertion sites of the duplicate regions in a set of eight spontaneous revertants isolated in a previous work (Roelants et al. 1995), with a view to establishing the underlying gene duplication mechanism. We focused here on four strains in which single ATCase sequence duplication occurs. In these revertants, the fact that the duplicated segment is punctuated at the 3′ end by a poly(A) tract and is located between Ty1 sequences strongly suggests that the Ty1 retrotransposon contributes to this process. To test the role of the Ty1 retrotransposon in this process, 27 revertants containing a duplication event were selected, starting with a strain in which the Ty1-mediated transposition was enhanced and analyzed: The enhancement of the Ty1 transcription increased the frequency of the genomic rearrangements, and that of the duplication events in particular (Roelants 1996).
The genetic screening method based on the URA2 gene provides a powerful tool for understanding the mechanisms involved in gene duplication in S. cerevisiae. The results indicate that mRNA may mediate spontaneous duplication events and that Ty elements are involved in the formation of the duplicates. After its integration, the duplicated sequence becomes part of a chimerical gene characterized by a new pattern of expression. In light of these new molecular data, we discuss the possible role of the Ty1 elements in the chromosomal reshaping process. The genomic rearrangements described constitute an example of duplication by retroposition.
RESULTS
Characterization and Chromosome Location of the Duplication Events
Starting with an initial ura2 15-30-72 mutant strain, four spontaneous revertants (Rev 25, Rev 37, Rev 45, and Rev 56) carrying a duplication of the ATCase sequence were isolated (Roelants et al. 1995). In each revertant, the BamHI restriction pattern detected by Southern blot hybridization using an ATCase coding sequence probe consists of two bands, the one corresponding to the ura2 15-30-72 resident copy, which is preserved at its locus, and the other to a duplicated copy inserted into a novel chromosomal region.
The chromosome location of each duplicated copy was detected by pulse field gel electrophoresis (PFGE), followed by Southern hybridization with an ATCase probe. In the Rev 25, Rev 37, and Rev 45 strains, chromosome X carrying the initial mutated allele was detected, as well as a second chromosome containing the duplicated copy. These strains result from an interchromosomal duplication event in which another chromosome (IV–XII, III, and VII–V, respectively) acts as a recipient sequence for the duplicated ATCase segment. The double chromosome numbers (IV–XII and VII–V) are due to the fact that the gel-shifted chromosomes had similar lengths. With Rev 56, the fact that only one chromosome was detected suggests that either an intrachromosomal duplication occurred on chromosome X or the duplicated sequence is localized on a chromosome which comigrates with the chromosome X.
Estimation of the Size of the Duplicated Region
There are at least two different ways in which duplication can arise: via a single gene duplication or a segmental duplication process (Sankoff 2001). To determine the type of events which occurred in the four revertants, we estimated the size of the duplicated segment by Southern hybridization using several probes corresponding to regions adjacent to the URA2 gene. By such a walking procedure, if there is a single gene duplication, only the ATCase probe will show the chromosome carrying the duplication, whereas a segmental duplication process will result in one or several 3′ adjacent genes of the URA2 locus being coduplicated with the ATCase sequence, and the corresponding probe used on these genes will also show the chromosome carrying the duplicated segment.
In the three strains containing interchromosomal duplications (Rev 25, Rev 37, and Rev 45), the walking procedure was performed directly on the chromosome by hybridizing the PFGE. The hybridization of the karyotypes of the strains, using probes specific to the YJL131c and YJL132w (MRS3) located at 0.375 kb and 4.16 kb downstream of the URA2 (YJL130w) locus, respectively, yielded only chromosome X, which indicates that these interchromosomal duplication events correspond only to the duplication of the AT-Case sequence.
In Rev 56, this procedure could not be used because the resident and duplicated copies comigrate. In this case, the walking procedure was performed on genomic DNA by analyzing the BamHI patterns with two probes, one corresponding to the ATCase coding region and the other to the intergenic region between the ATCase sequence and the YJL131c open reading frame (ORF). The restriction pattern observed using the ATCase probe consisted of two bands, whereas the intergenic probe yielded only one band, which indicates that only the ATCase coding sequence was duplicated.
Sequence Analysis of the Insertion Sites of the Duplicated Regions
The insertion sites of the duplicated copy were identified by sequencing the 5′ and 3′ flanking regions. The presence of specific motifs might help to gain further insights into the mechanisms involved in the occurrence of duplications.
Determination of the 5′ Boundaries
The 5′ junction of the duplicated regions was cloned using a plasmid insertion and excision strategy. The first step consisted of integrating a plasmid downstream of the starting point of the duplicated region. After this integration step, HindIII digestion and ligation of the digestion product were carried out in order to recover a plasmid containing the unknown junction via an Escherichia coli transformation and plasmid extraction procedure. The unknown junction was then sequenced and used to perform BLAST queries against the Saccharomyces Genome Database (http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd).
The analysis of the junction indicates that the starting point of the duplicated copy is located downstream of the three mutations, either at the end of the CPSase or in the DHOase-like domain (Fig. 2). In the four revertants, the 5′ of the duplicated region is attached to a Ty1 sequence with the following structure: Ty δ LTR followed by a variable part of a TyA sequence, which is fused in frame with the duplicated ATCase sequence, leading to a chimerical TyA-ATCase gene (Fig. 2). In addition, the four junctions showed the existence of short regions of homology (2–11 bp) between the TyA sequence and the 5′ duplicated part of the URA2 gene (Fig. 3).
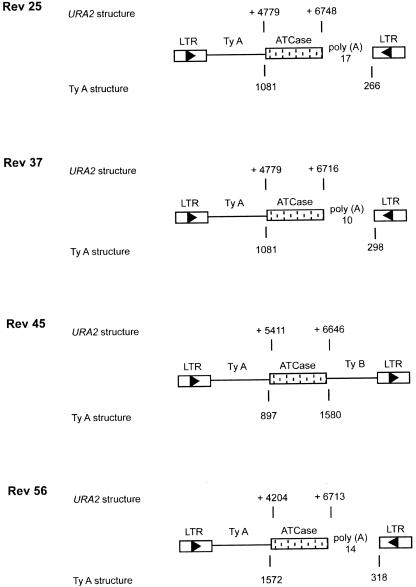
Figure 2.
The 5′ and 3′ insertion sites of the four spontaneous revertants were analyzed. The boxed arrowheads indicate Ty LTRs and the direction of the Ty transcription. In the URA2 structure, the nucleotide locations of the beginning and the end of the duplicated region are given, position +1 being the A nucleotide of the ATG initiator codon of the URA2 gene. In the Ty1 structure, the numbers given below are the coordinates observed in the Ty1 part of the various revertants.
Figure 3.
The 5′ junction is attached to a LTR-TyA structure. The boldface type indicates homologies existing between the TyA sequence of the Ty1 retrotransposon and the 5′ duplicated part of the URA2 gene.
In order to establish whether these sequences belong to resident or newly transposed Ty1 elements, we used a long-range asymmetric PCR technique (thermal asymmetric interlaced PCR; Szabados et al. 2002). This technique was used to determine the 5′ region situated upstream of the Ty structure in two revertants (Rev 25 and Rev 56). In both revertants, the sequences correspond to resident Ty1 structures located on chromosome IV downstream of a tRNA-Ile gene and on chromosome XVI down-stream of a tRNA-Ala gene (Fig. 4).
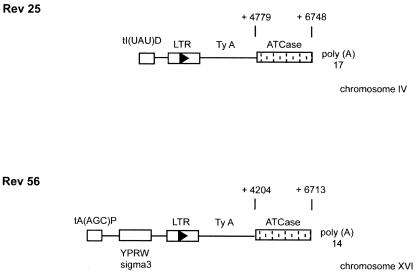
Figure 4.
The 5′ region situated downstream the Ty sequences of the Rev 25 and Rev 56 spontaneous revertants were analyzed. The boxed arrowheads indicate Ty LTRs and the direction of the Ty transcription. In the URA2 structure, the nucleotide locations of the beginning and the end of the duplicated region are given, position +1 being the A nucleotide of the ATG initiator codon of the URA2 gene.
Determination of the 3′ Insertion Site of the Duplicated Region
The asymmetric PCR technique (thermal asymmetric interlaced PCR) described by Liu et al. (1995) was used to determine the 3′ boundaries of the single ATCase sequence duplication. The results show that the duplicated sequence is punctuated by a poly(A) tract ranging from 10 to 14 bases in length in three of the four strains (Rev 25, Rev 37, Rev 56). Moreover, analysis of the sequences shows that the ATCase coding sequence is complete in all of the strains investigated, but that the stop codon adjacent region varies from 68 nt to 103 nt, which suggests that the polyadenylation site differs among the three strains. The presence of a poly(A) tail yielded additional information and provided evidence of the presence of an RNA intermediate.
These 3′ sequences were then used as BLAST queries against the Saccharomyces Genome Database (http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd). The results showed that in all duplications, the 3′ junction is also attached to Ty1 sequences. However, these sequences are not in the same orientation as those located at the 5′ junction, and therefore belong to another Tyl structure (Fig. 2).
Duplication of the ATCase Sequence Is Facilitated by Expression of the Ty Retrotransposon
The presence of a poly(A) tract at the end of the duplicated copy in three spontaneous revertants strongly suggests that a reverse transcription of the corresponding mRNA has occurred. One obvious source of reverse transcriptase activity in yeast is the Ty1 retrotransposon. A set of selections starting with the FRJ1 (MATa, ura2 15-30-72, trp1-4) strain, which in addition to the starting mutant, contains the pFJ2 plasmid carrying a Ty1 element under a GAL1 promoter, was therefore undertaken. The transcription of the plasmidic Ty1 element was induced in galactose-containing medium for 72 h, and the resulting Ura+ prototrophs were then selected. Among the 46 independent revertants selected, 23 strains containing a duplication were selected and further analyzed as described above.
The chromosome location shows that the active ATCase sequence can be situated on any of the 16 chromosomes of S. cerevisiae (Table 1). Nineteen events correspond to interchromosomal duplications, whereas four are intrachromosomal duplications. Due to the walking procedure based on Southern hybridization, it was established that in all of the cases studied, single ATCase sequence duplication occurred.
Table 1.
Chromosome Location of the ATCase Duplicated Sequence in the Set of Strains in Which Tyl-Mediated Transposition Was Increased
| Chromosomes | Revertants |
|---|---|
| IV—XII | 72-74-154-160 |
| VII—XV | 16-69-153-156 |
| XIII—XVI | 63-68 |
| II—XIV | 36-67 |
| X | 24-38-116-119 |
| XI | 152-158 |
| V—VIII | 22-30-149 |
| III | 114 |
| I—VI | 40 |
The results of the analysis of the 5′ junction indicate that the starting point of the duplicated region varies from one revertant to another and is always located downstream of mutation 72. The 5′ insertion sites of 19 revertants have a structure similar to those observed in the spontaneous revertants: a δ LTR-TyA sequence in frame with the duplicated ATCase sequence (Fig. 5A). In the four other revertants, the ATCase sequence is attached to a complete Ty1 element in an orientation opposite to that of the ATCase sequence (Fig. 5B).
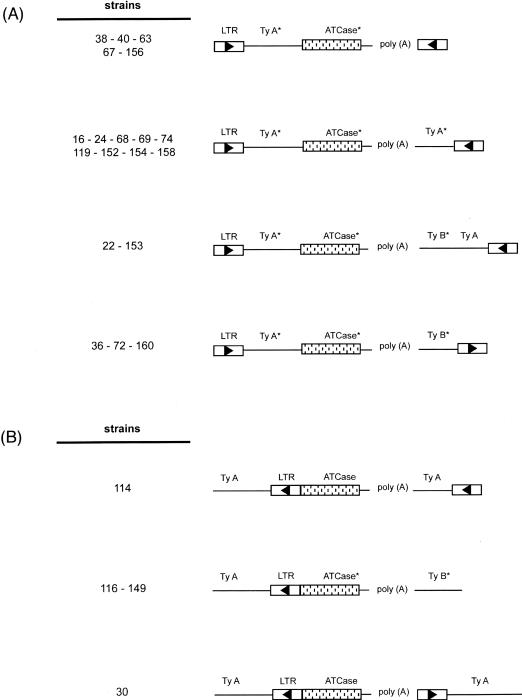
Figure 5.
Diagram of 5′ and 3′ chromosomal insertion of the single ATCase duplication detected in the revertants selected in a Ty1 overexpression context. The boxed arrowheads indicate Ty LTRs and the direction of Ty transcription. TyA and TyB form the Ty ORF. The symbol * has been added to sequences which are variable in length from one strain to another. Revertants 16, 72, and 74 lack the poly(A) tract. (A) In frame insertion of the ATCase duplicated region in a δ LTR Ty1. (B) Attachment of the duplicated ATCase region to a complete Ty1 element.
The 3′ junction of 20 revertants is also punctuated by a poly(A) tract ranging from 11 to 73 nt in length. In three revertants (16, 72, and 74), this poly(A) track was found to be absent. As in the spontaneous revertants, the ATCase coding sequence is complete in all of the strains studied here, but the poly(A) tail is located in a region ranging from 56 to 122 nt after the stop, which suggests that the polyadenylation site of the URA2 messenger is not unique. All of the 3′ junctions are also Ty sequences, but they can differ from one strain to another. The sequences can consist of a δ LTR, a δ LTR followed by a variable part of a TyA sequence, or a TyB sequence (Fig. 5A,B). These sequences are mostly in orientations opposite to those located at the 5′ junction, and therefore belong to another Ty1 element.
DISCUSSION
New functions can arise via gene duplication processes followed by changes in the coding or regulatory sequence. In S. cerevisiae, a few in vivo gene duplication events have been described (Welch et al. 1990; Dorsey et al. 1993; Brown et al. 1998), but the mechanisms responsible for these events are still poorly understood. Here we describe a single gene duplication process occurring in four spontaneous revertants selected using a positive screening method based on a mutated allele of the URA2 gene. The genetic background of the strain used differs from the wild type in terms of a single mutated allele containing three point mutations. The chromosomal rearrangements observed therefore reflect what may have happened during the evolution of the S. cerevisiae genome. The uracil prototrophy of the revertants selected is recovered because of the duplication of the ATCase sequence and its expression after being integrated into a novel chromosomal region, which was consistently found to a Ty1 area.
Several hypotheses can be put forward to explain the contribution of the Ty1 sequences to the process of duplication. One of these hypotheses involves a process of homologous recombination between two repeated Tyl sequences. In all of the cases of chromosomal rearrangement such as translocations (Fischer et al. 2000; Dunham et al. 2002) described thus far, the events are bounded by transposon-related sequences at the breakpoints. The present data suggest the occurrence of genomic rearrangements mediated by Ty1 sequences which simply serve to mediate homologous recombination processes between the host and donor chromosomes. In our system, a Ty1 element inserted into the second half of the CPSase coding region of the URA2 gene may have recombined with another Ty1 or δ sequence located elsewhere in the genome, leading to the duplication of the region between these two sequences. If the transposition-recombination hypothesis is correct, one would find similar sequences near both the original and duplicated ATCase regions. However, in the present study, all of the duplicated sequences were located between Ty1 sequences, but no Ty sequences were observed in the vicinity of the original URA2 sequence. The above hypothesis therefore does not seem to provide a valid explanation for the duplication events observed in this study.
The fact that duplicated segments are punctuated by a poly(A) tract in three revertants suggests that these copies may arise from the reverse transcription of the URA2 mRNA. Because Ty1 is one possible source of reverse transcriptase, a set of Ura+ selections was performed in a Ty1 overexpression context. In this setting, the frequency of reversion was found to be 10-fold higher than in a strain with no Ty1 overexpression. Twenty-three revertants, corresponding to ∼50% of all the selected strains, were duplication events, whereas in the spontaneously selected revertants, only 17% correspond to these events (Roelants et al. 1995). The analysis of these 23 strains shows that they are similar to the four spontaneous Ura+ revertants. They are all single-gene duplications located in the vicinity of Ty1, and 20 of the duplicated segments contain a poly(A) tract. The absence of the poly(A) tract in the other three strains as well as in the spontaneous Rev 45 might be attributable to a deletion event, as described in the case of pseudogenes processed via the reverse transcriptase of LINE retrotransposons in higher eukaryotes (Esnault et al. 2000).
All these results strongly suggest that the duplication events selected here result from a process of retroposition (Derr et al. 1991; Zhang 2003). One possible mechanism might be the formation of a cDNA as the result of the reverse transcription of the mRNA corresponding to the part of the URA2 sequence containing the ATCase sequence, which might be located down-stream of mutation 72. Xu and Boeke (1990) reported that TRP1 and HIS3 RNAs were associated with the Ty virus-like particles (VLPs). Our own data suggest that the URA2 mRNA is packed in the Ty VLPs and reverse transcribed, because the Ty1 reverse transcriptase is located in the VLPs (Mellor et al. 1985).
The reverse transcription is then followed by the integration of this cDNA into the genome on any of the chromosomes present. However, the integration of the cDNA is not a random occurrence, because the presence of the Ty1 sequence systematically observed downstream and upstream of the duplicated ATCase sequence clearly shows that Ty1 elements provide more than the reverse transcriptase function. The presence of short regions of homology ranging from 3–11 nucleotides in both the TyA and 5′ duplicated ATCase sequences might provide a clue to the mechanism resulting in the integration of the duplicated copy into a Ty1 resident copy. Reverse transcription would in this case be initiated in the poly(A) region via the ATCase sequence and switch at the level of the 5′ junction observed on a Ty element template, leading to the formation of the chimeric structure observed: a δ LTR TyA segment in frame with the duplicated ATCase sequence. This possibility is consistent with the hypothesis that the hybrid Ty1/Ty2 LTRs may have been generated by a reverse transcription-mediated template switching event (Jordan and McDonald 1998).
The integration of this structure into the genome may involve either of two distinct processes. It may be integrated by the Ty integrase into a novel chromosomal site. However, the Ty sequences which surround the ATCase duplicated sequences are not in the same orientation and do not therefore form an entire Ty element which could be inserted via the integrase. Furthermore, in the case of integration by integrase, one might expect to find the duplicated sequence in regions containing genes transcribed by RNA polIII (Devine and Boeke 1996). This is not the case, because in all of the revertants isolated here, the integration was always found to occur within the Ty1 structure. In addition, the exact insertion sites were determined in two spontaneous revertants (Rev 25 and Rev 56), and the duplicated ATCase sequence was found to be inserted into a resident Ty1 and not into a mobilized component. The second process which might lead to the integration of this structure is a homologous recombination event resulting from gene conversion between preexisting chromosomal Ty elements and the 5′ end of the cDNA.
Moreover, in order to be transcribed, the duplicated ATCase segment needs to have a promoter sequence, which is not the case in the initial duplicated ATCase sequence, but only in the 5′ Ty attached region. In 23 revertants, the ATCase is fused in frame to a variable part of the TyA sequence. In these strains, the promoter located in the δ LTR can activate the transcription of the chimeric gene. Because the diploids obtained by crossing the parental strain with the revertants are auxotrophic or bradytrophic with respect to uracil, the chimeric gene shows the same pattern of expression as Ty, which is repressed in the diploid context. In contrast, in four revertants obtained in the Ty overexpression context, an entire Ty1 element is inserted in the orientation opposite to that of the ATCase segment. The expression of the ATCase in the latter revertants is probably initiated in the δ LTR, as described by Roelants et al. (1997).
Another relevant point worth mentioning is the finding by Levine et al. (1990) that the insertion of cDNA is often associated with large-scale chromosomal rearrangements. The cloning of the junction in the Rev 56 revertant shows that the duplicated copy is situated on chromosome XVI. The migration of this chromosome differs between the Rev 56 strain and the initial ura2 15-30-72, as it is much smaller in the former case. This result suggests that the gene duplication event is also associated with other chromosomal rearrangements, which can be detected by PFGE if they lead to changes in chromosome size.
The present results support the idea that yeast genes may result from the reverse transcription of mRNA, which would explain the paucity of introns observed in S. cerevisiae (Fink 1987). Moreover, the general process occurring in our model seems to be be similar to that postulated for the formation of pseudogenes in higher eukaryotes (Moran et al. 1999; Esnault et al. 2000). These studies showed that the LINEs retrotransposon can mobilize transcribed DNA not associated with a LINE sequence. The corresponding mRNA is then retroposed, that is, then reverse transcribed due to the LINE machinery and integrated into the genome. The results of this study provide experimental evidence that spontaneous duplications can occur in S. cerevisiae via an LTR Ty1 retroposition mechanism.
METHODS
Yeast Strains and Plasmids
Yeast strain ura2 15-30-72 is an isogenic derivative of the laboratory strain FL100 (ATCC 28583; Welcker et al. 2000). Strain FRJ1 was obtained by crossing ura2 15-30-72 (Gal–) with a trp1–4 (Gal+) strain followed by selection of the ura2 trp1 strains able to catabolize galactose. The pFJ2 plasmid was constructed by inserting the TRP1 marker into the unique BamHI site of the plasmid pJEF1271 (Eichinger and Boeke 1988) and was introduced into the FRJ1 strain. The integrative pFLB, pFLC, and pFLD plasmids were constructed by inserting the corresponding BglII-BglII subfragment into the unique BamHI site of the pFL35 plasmid (Bonneaud et al. 1991).
Selection of Ura+ Revertants From the ura2 15-30-72 Mutant Strain
Ura+ revertants were isolated as described by Roelants et al. (1995) from the ura2 15-30-72 and FRJ1 [pFJ2] strains.
Determination of the 5′ Insertion Site by Plasmid Integration and Excision
The 5′ junction of the duplicate regions was cloned using a plasmid integration/excision strategy. For this purpose we first located the beginning of the duplicated fragment, using the BglII-BamHI restriction map (Fig. 1). Depending on the beginning of the duplicated segment, one integrative plasmid (pFLB, pFLC, or pFLD) was introduced downstream of the modified subfragment (B, C, or D). If the starting point of the duplicated region is situated in the C subfragment, the pFLD plasmid containing the D fragment will be integrated (Fig. 1). After the transformation step, HindIII digestion and ligation were carried out in order to recover a plasmid containing the unknown junction, which was sequenced.
Transformation of Yeast and Bacteria
Yeasts were transformed using the method developed by Becker and Guarente (1991). E. coli transformation was performed as described by Dower et al. (1988).
Southern Blot Analysis
Total DNA from S. cerevisiae was prepared as described by Hoffman and Winston (1987). Restriction endonuclease digestion steps were carried out as described by the manufacturers. DNA blots were prepared from PFGE and conventional agarose gels by transferring DNA to Hybond N+ membrane (Amersham). DIG-labeled DNA probes were prepared using the DNA labeling and detection kit (Roche).
Tail-PCR
Thermal asymmetric interlaced PCR was performed as described by Liu et al. (1995) using the same PCR conditions as well as the same arbitrary degenerate primers. The nested primers located at the 3′ end of the ATCase sequence were NF1 (TCGTGCTGCT TACTTCAGACA), NF2 (TGGTTTGTTCGTCAGAATGG), and NF3 (GCCATGGTCATGGGTGT).
Long-Range Tail-PCR
Long-range thermal asymmetric interlaced PCR was performed as described by Szabados et al. (2002), using the same PCR conditions and arbitrary degenerate primers. The nested primers located at the 5′ end of the ATCase sequence were D1.2 (GGGAG GTTAACAATAAAACCG), D1.5 (CGATGCCAAATCTGCAGTTT), and D1.6 (GCTACTGCTGTGATCTTGTTC).
DNA Sequencing and Sequence Analysis
Double-strand DNA fragments obtained after PCR amplification were purified using a MicroSpin Column S-400 (Amersham). DNA sequencing was performed on the PCR-purified fragments using the method described by Sanger et al. (1977) and a Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham). We performed BLAST analysis after sequencing the PCR product to determine the exact boundaries of the duplication events. This was done with the SGD database.
PFGE
Chromosomal DNA was prepared as described by Carle and Olson (1985). Chromosomes were separated on a 1% agarose gel (Pharmacia) in a 0.5X TBE buffer at 7V/cm for 22 h with a pulse time of 45 sec and an angle of 120°, using a Bio-Rad CHEF-DIII mapper apparatus. The gel was stained with ethidium bromide to identify the chromosomal pattern specific for each strain.
Acknowledgments
We thank Philippe Hammann and Malek Alioua for their help with DNA sequencing in the Strasbourg CNRS/IBMP department facilities. This work was supported in part by EU Grant Comprehensive Yeast Genome Database CYGD (QLRI CT 1999 01333) and by the Génolevure-2 sequencing consortium GDR CNRS 2354. J.S. is supported by a grant from the French Ministère de l'Enseignement Supérieur et de la Recherche.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2363004.
References
- Bach, M.L., Roelants, F., de Montigny, J., Huang, M., Potier, S., and Souciet, J.L. 1995. Recovery of gene function by gene duplication in Saccharomyces cerevisiae. Yeast 11: 169–177. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Becker, D.M. and Guarente, L. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol 194: 182–187. [DOI] [PubMed] [Google Scholar]
- Bonneaud, N., Ozier-Kalogeropoulos, O., Li, G.Y., Labouesse, M., Minvielle-Sebastia, L., and Lacroute, F. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7: 609–615. [DOI] [PubMed] [Google Scholar]
- Brown, C.J., Todd, K.M., and Rosenzweig, R.F. 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15: 931–942. [DOI] [PubMed] [Google Scholar]
- Carle, G.F. and Olson, M.V. 1985. An electrophoretic karyotype for yeast. Proc. Natl. Acad. Sci. 82: 3756–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr, L.K., Strathern, J.N., and Garfinkel, D.J. 1991. RNA-mediated recombination in S. cerevisiae. Cell 18: 355–364. [DOI] [PubMed] [Google Scholar]
- Devine, S.E. and Boeke, J.D. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes & Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Dorsey, M.J., Hoeh, P., and Paquin, C.E. 1993. Phenotypic identification of amplifications of the ADH4 and CUP1 genes of Saccharomyces cerevisiae. Curr. Genet. 12: 392–396. [DOI] [PubMed] [Google Scholar]
- Dower, W.J., Miller J.F., and Ragsdale, C.W. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16: 6127–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, M.J., Badrane, H., Ferea, T., Adams, J., Brown, P.O., Rosenzweig, F., and Botstein, D. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, D.J. and Boeke, J.D. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: Cell-free Ty1 transposition. Cell 54: 955–966. [DOI] [PubMed] [Google Scholar]
- Esnault, C., Maestre, J., and Heidmann, T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367. [DOI] [PubMed] [Google Scholar]
- Fink, G.R. 1987. Pseudogenes in yeast? Cell 49: 5–6. [DOI] [PubMed] [Google Scholar]
- Fischer, G., James, S.A., Roberts, I.N., Oliver, S.G., and Louis, E.J. 2000. Chromosomal evolution in Saccharomyces. Nature 405: 451–454. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, H. and Fink, G.R. 1979. Unstable transpositions of his4 in yeast. Proc. Natl. Acad. Sci. 76: 4006–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansche, P.E., Beres, V., and Lange, P. 1978. Gene duplication in Saccharomyces cerevisiae. Genetics 88: 673–687. [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C.S. and Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., Roberts, C.J., Dai, H., Jones, A.R., Meyer, M.R., Slade, D., Burchard, J., Dow, S., Ward, T.R., Kidd, M.J., et al. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25: 333–337. [DOI] [PubMed] [Google Scholar]
- Jordan, I.K. and McDonald, J.F. 1998. Evidence for the role of recombination in the regulatory evolution of Saccharomyces cerevisiae Ty elements. J. Mol Evol. 47: 14–20. [DOI] [PubMed] [Google Scholar]
- Koszul, R., Caburet, S., Dujon, B., and Fischer, G. 2004. Eucaryotic genome evolution though the spontaneous duplication of large chromosomal segments. EMBO J. 23: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, K.L., Steiner, B., Johnson, K., Aronoff, R., Quinton, T.J., and Linial, M.L. 1990. Unusual features of integrated cDNAs generated by infection with genome-free retroviruses. Mol. Cell Biol. 10: 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Lynch, M. 2002. Genomics: Gene duplication and evolution. Science 297: 945–947. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Mellor, J., Malim, M.H., Gull, K., Tuite, M.F., McCready, S., Dibbayawan, T., Kingsman, S.M., and Kingsman, A.J. 1985. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature 318: 583–586. [DOI] [PubMed] [Google Scholar]
- Moran, J.V., DeBerardinis, R.J., and Kazazian, H.H. 1999. Exon shuffling by L1 retrotransposition. Science 283: 1530–1544. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer, Berlin.
- Paquin, C.E., Dorsey, M., Crable, S., Sprinkel, K., Sondej, M., and Williamson, V.M. 1992. A spontaneous chromosomal amplification of the ADH2 gene in Saccharomyces cerevisiae. Genetics 130: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants, F. 1996. Rôle du transposon Ty1 dans la réactivation fonctionnelle d'un allèle mutant du gène URA2 et dans les remaniements chromosomiques associés chez Saccharomyces cerevisiae. Thesis, pp 130–136. Université Louis Pasteur, Strasbourg I, France.
- Roelants, F., Potier, S., Souciet, J.L., and de Montigny, J. 1995. Reactivation of the ATCase domain of the URA2 gene complex: A positive selection method for Ty insertions and chromosomal rearrangements in Saccharomyces cerevisiae. Mol. Gen. Genet. 246: 767–773. [DOI] [PubMed] [Google Scholar]
- Roelants, F., Potier, S., Souciet, J.L., and de Montigny, J. 1997. Delta sequence of Ty1 transposon can initiate transcription of the distal part of the URA2 gene complex in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 148: 69–74. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff, D. 2001. Gene and genome duplication. Curr. Opin. Genet. Dev. 11: 681–684. [DOI] [PubMed] [Google Scholar]
- Szabados, L., Kovacs, I., Oberschall, A., Abraham, E., Kerekes, I., Zsigmond, L., Nagy, R., Alvarado, M., Krasovskaja, I., Gal, M., et al. 2002. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 32: 233–242 [DOI] [PubMed] [Google Scholar]
- Welch, J.W., Maloney, D.H., and Fogel, S. 1990. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol. Gen. Genet. 222: 304–310. [DOI] [PubMed] [Google Scholar]
- Welcker, A.J., de Montigny, J., Potier, S., and Souciet, J.L. 2000. Involvement of very short DNA tandem repeats and the influence of the RAD52 gene on the occurrence of deletions in Saccharomyces cerevisiae. Genetics 156: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H. and Shields, D.C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Xu, H. and Boeke, J.D. 1990. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: Analysis of mini-Ty1 elements. Mol. Cell Biol. 10: 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. 2003. Evolution by gene duplication: An update. Trends Ecol. Evol. 18: 292–298. [Google Scholar]
- Zhang, J., Rosenberg, H.F., and Nei, M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd; Saccharomyces Genome Database.