Abstract
The Chinese Han population, the largest population in the world, has traditionally been geographically divided into two parts, the Southern Han and Northern Han. In practice, however, these commonly used ethnic labels are both insufficient and inaccurate as descriptors of inferred genetic clustering, and can lead to the observation of “spurious association” as well as the concealment of real association. In this study, we attempted to address this problem by using 14 microsatellite markers to reconstruct the population genetic structure in 768 Han Chinese samples, including 384 Southern Han and 384 Northern Han, and in samples from Chinese minorities including 48 Yao and 48 BouYei subjects. Furthermore, with a dense set of markers around the region 5q34–35, we built fine-scale haplotype networks for each population/subpopulation and tested for association to schizophrenia susceptibility. We found that more variants in SLIT3 tend to associate with schizophrenia susceptibility in the genetically structured samples, compared to geographically structured samples and samples without identified population substructure. Our results imply that identifying the hidden genetic substructure adds power when detecting association, and suggest that SLIT3 or a nearby gene is associated with schizophrenia.
Schizophrenia is a common severe mental disorder that affects 1% of the population. As the largest population in the world (nearly 20% of all humankind), Chinese Han society also contains the largest number of schizophrenic patients on the globe. In general, Chinese samples have been considered to be a good genetic resource for studies of both genetic etiology and genetics-based drug target identification. However, due to the Han having an extremely long and complex demographic history (starting from the ancient Huaxia tribe during the 21st–8th centuries B.C., which then integrated with numerous tribes and ethnic groups), a precise classification of Han samples is required to reduce the effect of population stratification and so enhance the power of detection.
A geographical or ethnic label that is usually adequate for the overall classification of samples may only represent a certain proportion of the actual underlying population genetic structure, as the real information of human history is hidden in the genome. Even in “God-given isolated populations,” genetic heterogeneity is unavoidable (Devlin et al. 2002). Case-control studies based on statistical methods that use marker genotype data to infer the hidden population genetic structure have been proposed as a possible alternative to family-based designs (Pritchard et al. 2000b; Hoggart et al. 2003). In the present study, we attempted a genetic structuring of the Han Chinese by using a model-based clustering tool, STRUCTURE, which is able to demonstrate the presence of population structure, assign individuals to populations, study hybrid zones, and identify migrants and admixed individuals (Pritchard et al. 2000a).
As determined in previous studies (Chu et al. 1998; Su et al. 1999; Jin and Su 2000; YiDa and Cheng 2002), the Han Chinese population is generally thought to be naturally divided into two parts by the Yangtze River, which resulted in the formation of different founder populations with relatively isolated consanguinity. In addition, it is believed that the difference between the two parts of the Han Chinese is greater than that between a given subpopulation and ethnic minorities at the same location. However, several other studies have argued that a distinction between Southern and Northern Han could not be well supported (Ding et al. 2000; Oota et al. 2002).
Ignoring the question as to how homogeneous the Southern and Northern Chinese populations are, we focused on whether we could find a detectable cryptic borderline within the Han Chinese as a whole, and whether we could add power to an association study by taking account of any subdivision. Here, we used 14 microsatellite markers and 768 Han samples to test whether there is a detectable cryptic structure within the Han population. We wanted to understand what effect would be found if we brought such a cryptic subdivision into association tests.
SLIT3, a human homolog of the Drosophila `slit' gene, which has been shown to play a critical role in central nervous system midline formation and is located at 5q34–35, has been suggested to play possible roles in the formation and maintenance of the nervous system (Vargesson et al. 2001; Nguyen-Ba-Charvet and Chedotal 2002), and has also recently been shown to be involved in the embryological formation of the diaphragm (Yuan et al. 2003). However, we aimed to investigate SLIT3 as a candidate gene for schizophrenia.
RESULTS
Population Genetic Structures of the Han Chinese
To construct the population genetic structure of the Chinese populations, we selected a 14-microsatellite marker set from the ABI prism panels 1 and 8, where none of the markers have a reported linkage to schizophrenia. These markers were considered to be independent of each other because the minimum space between any two markers is larger than 1.0 cM, beyond the distance at which linkage disequilibrium is important. These markers were genotyped in four populations: SH (Southern Han) from Shanghai in the southeast of China (384), NH (Northern Han) from JiLin in northeast China (384), and Yao (48) and BouYei (48), each from the south of China. Subclusters were classified with the program STRUCTURE without knowing the original population information. To estimate the number of subclusters (K) present in the data, we used STRUCTURE to estimate the proportion of each individual's genome having ancestry in each subcluster and applied a prior probability of the data (Pr(X|K), where X represents the data). In this work, we estimated Pr(X|K) using a model allowing admixture for K from 1 to 4. When setting the prior probability, we estimated Pr(K|X) using Bayes' theorem (Table 1). Clearly, K=2 had the biggest posterior probability in Table 1 (∼1.00).
Table 1.
Inferring the Number of Clusters in the Mixed Han Population
| K | In Pr (X|K) | Pr (K|X) |
|---|---|---|
| 1 | -34021.5 | ∼0 |
| 2 | -33585.7 | 1.00 |
| 3 | -34973.8 | ∼0 |
| 4 | -35899.9 | ∼0 |
Very interestingly, the allocation of individuals by STRUCTURE showed that a cryptic borderline was observable between the SH and NH samples, in which 88.4% of the SH and 38.8% of the NH fell into cluster A, and 11.6% of the SH and 61.2% of the NH fell into cluster B (Table 2). Then, we defined cluster A as the New Southern Han Cluster (NSHC) and cluster B as the New Northern Han Cluster (NNHC). Each individual was associated with two probabilities, which indicate the degree to which its genome was classified into each cluster. The criterion for allocation was set such that when an individual's probability of being in one cluster was more than 0.75, it was classified into this cluster. In other words, an individual with more than a 3/4 proportion of genetic background in the cluster should be allocated into the corresponding population, and one with less than 3/4 background in either of the two clusters should be treated as an ambiguous class member. Ambiguously classified members were not used in subsequent analyses. In total, 483 individuals were allocated into NSHC, 237 individuals into NNHC, and 48 individuals were categorized as ambiguous (Table 3).
Table 2.
Proportion of Membership of Both South Han and North Han Populations in STRUCTURE-Defined Subclusters
| Population | NSHC | NNHC |
|---|---|---|
| SH | 0.884 | 0.116 |
| NH | 0.388 | 0.612 |
Table 3.
Sample Reallocation From the Original Population to the STRUCTURE-Defined New Clusters, Including Case and Control Information
|
NSHC
|
NNHC
|
Ambiguous Class
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Original Population | Control | Case | Control | Case | Control | Case | |||
| SH | 172 | 173 | 6 | 6 | 14 | 13 | |||
| NH | 67 | 71 | 115 | 110 | 10 | 11 | |||
| Sum | 239 | 244 | 121 | 116 | 24 | 24 | |||
| Total | 483 | 237 | 48 | ||||||
Schizophrenia case and unaffected control samples were both randomly collected, and so they should be evenly distributed in each newly generated cluster if the new clustering is reliable. We obtained confirmation of this in the final clustering data (Table 3).
Using the same marker set, we succeeded in distinguishing the Yao, one of the Chinese southern minorities, from the NNHC, but failed in the other three cases (Table 4). This indicates that the marker set is powerful enough to identify the borderline between NSHC and NNHC, and the borderline between NNHC and some of the southern minorities (such as Yao), respectively, but is not able to separate NSHC from the adjacent southern minorities (such as Yao and BouYei). So, NSHC seems to be more tightly linked with Yao and BouYei than with NNHC. This result supports the viewpoints of Chu et al. (1998), Su et al. (1999), Jin and Su (2000), and YiDa and Cheng (2002), in the light of which we consider NSHC as representative of the genetically structured Chinese Southern Han population.
Table 4.
Clustering Analysis of Genetically Structured Han and Minorities
|
Yao & NSHC
|
BouYei & NSHC
|
||||
|---|---|---|---|---|---|
| Population | 1 | 2 | Population | 1 | 2 |
| NSHC | 0.499 | 0.501 | NSHC | 0.500 | 0.500 |
| Yao | 0.514 | 0.486 | BouYei | 0.502 | 0.498 |
| Yao & NNHC | BouYei & NNHC | ||||
| Population | 1 | 2 | Population | 1 | 2 |
| NNHC | 0.071 | 0.929 | NNHC | 0.500 | 0.500 |
| Yao | 0.966 | 0.034 | BouYei | 0.501 | 0.499 |
Association Between SLIT3 Variations and Schizophrenia
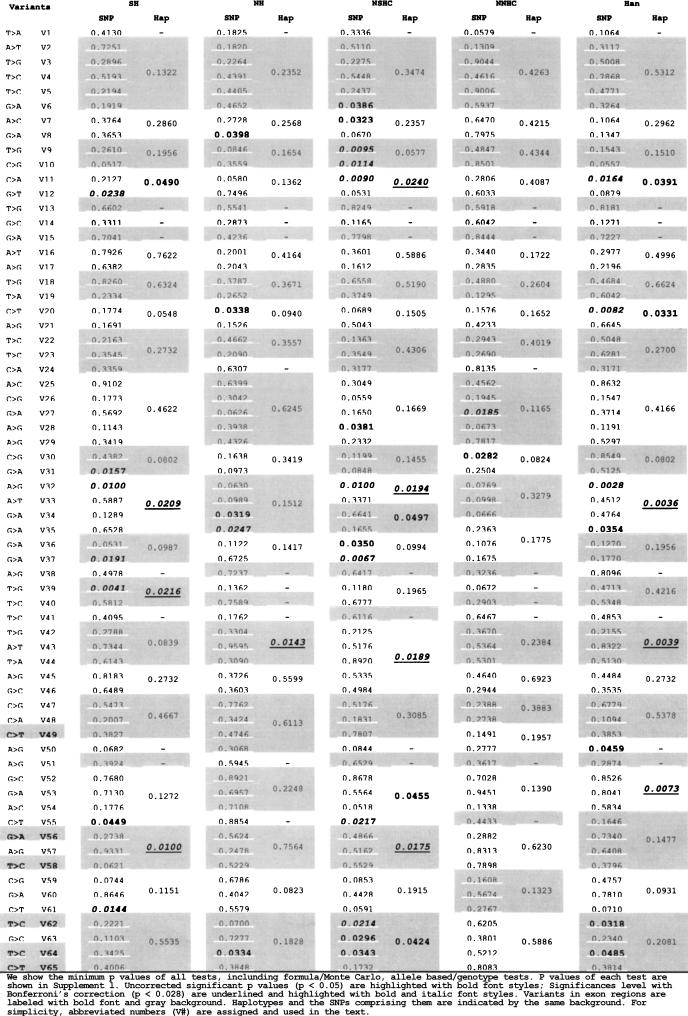
We extensively tested the association of the gene SLIT3 within the 5q34–35 region to schizophrenia in 768 case-control samples of the Han Chinese (384 cases and 384 controls) by genotyping individual single-nucleotide polymorphisms (SNPs) and constructing and testing haplotypes and individual SNPs for association. After dividing these samples into geographic clusters and genetic clusters, we obtained four subclusters. Each of these subclusters contained part of the cohort, including SH (192 cases, 192 controls), NH (192 cases, 192 controls), NSHC (244 cases, 239 controls), and NNHC (116 cases, 121 controls). Association with susceptibility was tested in each subcluster using both Monte Carlo tests and formula-based tests. The relevant results can be found in the Supplement 1. We show the minimum P-values of all tests in Table 5.
Table 5.
The Minimum P-Values of All Tests, Including Formula/Monte Carlo, Allele-Based/Genotype-Based Tests
Considering that multiple tests were carried out for each marker in five sample clusters, we should include a correction of the significance level. As SH, NH, NSHC, NNHC, and Han were not independent groups, we calculated the correlation between them (Suppl. 4). After Bonferroni's multiple test correction in light of this correlation, the adjusted significant threshold (α) is 0.028.
After correction, a number of markers, including coding variants and blocks containing coding variants, still showed significance (Tables 5, 6, 7) below this threshold. SLIT3 is thought to be involved in the development of the nervous system, but there are no reports of schizophrenia susceptibility at this locus. Our result, with significant SNPs and haplotypes in introns and exons, suggests that SLIT3 may confer moderate schizophrenia susceptibility, especially in the genetically structured NSHC population.
Table 6.
Two cSNPs Showing Significant Results in Monte Carlo Tests
|
NSHC
|
Han
|
|||
|---|---|---|---|---|
| T1 | T4 | T1 | T4 | |
| V62 |
0.0214
|
0.0284 | 0.0318 | 0.0338 |
| V64 | 0.0343 | 0.0393 | 0.0485 | 0.0542 |
T1 and T4 are the output P-values of CLUMP software. Uncorrected significant P-values (P < 0.05) are in bold font. The significance level with Bonferroni's correction (P < 0.028) is underlined bold italic font.
Table 7.
A Risk Haplotype Comprised of Coding Region SNPs
|
NSHC
|
Han
|
|||||
|---|---|---|---|---|---|---|
| V62-V64 | Control | Case | Control | Case | ||
| T-T | 260 | 240 | 439 | 409 | ||
| T-C | 34 | 30 | 52 | 52 | ||
| C-T | 1 | 1 | 2 | 1 | ||
| C-C | 83
|
123
|
147 | 186 | ||
| P = 0.0124 | P = 0.0551 | |||||
DISCUSSION
One problem is that there must be a correlation between sample clusters because NSHC and NNHC are reallocated from SH and NH. We calculated the correlation level in Supplement 4. Additionally, in order to study the correlation effect on detection power, we employed a simulation method (see Methods). Based on the allele frequencies of the studied SNPs (Suppl. 3), this program uses simulations to test the detection power within SH, NH, NSHC, and NNHC under random distribution. Therefore, this method can estimate the probability of correlation-affected power change at each locus under the definite sample correlation. We summarize the simulation results in Supplement 2.
In summary, we found that:
Thirteen SNPs showed significance in NSHC, whereas only two SNPs were detected as associating in NNHC. This indicates that after identifying the cryptic structure, more genetic homogeneity, which can add to the power of detecting of schizophrenia susceptibility, is present in the new NSHC subcluster.
Ten SNPs (V6, 7, 9, 10, 11, 28, 36, 62, 63, 64) which did not show significance in SH or NH were detected as associating in NSHC; three significant SNPs of SH (V32, 37, 55) also showed significance in NSHC, and their significance levels in NSHC were no less than those observed in SH. Based on the simulation results (Suppl. 2), four of them (V9, 11, 62, 64), were likely to be caused by sample correlation. Additionally, correlation significantly lowered the detection power at V28, 36, 37, 55, and 63, although the five SNPs still showed significance in adding SH to NSHC. The association significance values of another four SNPs (V6, 7, 10, 32) were not significantly affected by the correlation. By the identification of cryptic subdivisions, obscured associations may be explored and the genetic homogeneity increased in subpopulations, which adds to the detection power.
Four of the significant SNPs in SH (V12, 31, 39, 61) did not show significance in NSHC and NNHC; another four significant SNPs (V8, 20, 34, 35) in NH did not show significance in NSHC and NNHC; and one significant SNP (V50) in Han did not show significance in any of the subclusters. The correlation of samples could cause this tendency at V12, 31, 34, 35, 39, and 61. The association at these loci could also be caused by population stratification, especially at V8, 20, and 50. After identifying the cryptic structure, `spurious association' is removed and stratification can be considered to be minimized.
Another interesting phenomenon is that six of the seven SNPs that were significant in the whole Han population, all except V50, also showed significance in at least one subpopulation as mentioned above. Although the stratification will tend to “conceal” many potentially significant loci, the association reports in whole Han samples seem to be highly repeatable.
Ding et al. (2000) and Oota et al. (2002) suggested that populations in East Asia are relatively homogeneous. If this is true, the effect of stratification should be smaller in our samples. However, we still observed an increase in power. We think this increase in power is due to two advantages of cryptic subdivision analysis: stratification is removed, and the genetically structured subpopulations' homogeneity is increased.
We propose to construct an LD map in a larger scale around this region in the Han Chinese population in order to find the real disease gene, or to confirm the role that SLIT3 may play in the etiology of schizophrenia.
METHODS
Subjects
All 864 of the individuals studied are unrelated local Chinese from different races. We collected 768 Han Chinese including 384 from the South (Shanghai) and 384 from the North (Jilin), and two minority races with 48 individuals from Yao and 48 from BouYei, both of which are located in the South of China. There were a total of 384 schizophrenics, 185 males and 198 females, 192 from the South (Shanghai) and 192 from the North (Jilin). Subjects studied had an average age of 41.58 yrs.
Subjects with schizophrenia were strictly diagnosed according to the criteria of DSM-III-R (DSM-III-R; American Psychiatric Association, 1987). Written informed consent was obtained from either the patient or the patient's relatives after the procedure had been fully explained.
Microsatellite Marker Selection and Structure Inference
Microsatellite markers genotyped included D1S206, D1S484, D1S2726, D1S2797, D1S2800, D1S2842, D1S2878, D5S406, D5S422, D5S433, D6S289, D6S309, D6S1581, and D6S1610. All markers used were believed to be neutral and to have no correlation with schizophrenia in Han Chinese. The chromosome 1 markers were from the ABI Prism linkage mapping panel 1 and the chromosome 5 and 6 markers from ABI Prism panel 8. All of them were amplified according to the manufacturer's instructions. We assigned individuals into clusters using the admixture model in the program STRUCTURE, with no correlation in allele frequencies among the populations and a burn-in time of at least 1 million steps, followed by another 1 million steps of the Markov Chain for data collection. We carried out multiple runs for each set of conditions to ensure that the chain had converged.
Candidate SNP Discovery
We searched for SNPs in the promoter region (–2000 bp), exons, and additional ∼40-kb intron regions of SLIT3 by sequencing DNA pools consisting of 200 Han Chinese schizophrenic and 60 European Caucasian nonaffected samples, respectively. All the samples were sequenced in both forward and reverse directions to validate SNP discovery. We discovered 65 candidate SNPs, which were then confirmed using another pool of the same size from the Han Chinese samples. We also selected 47 SNPs from dbSNP, using those reported by at least two sources. In total, we used 112 candidate SNPs for genotyping.
Genotyping of SNPs
All 112 candidate SNPs were genotyped using ABI's TaqMan technology (Assay-by-Design) on an ABI7900 system. All probes and primers were designed by the Assay-by-Design service of ABI. The standard PCR reactions of 5 μL were carried out by using TaqMan Universal PCR Master Mix reagent kits as described by the manufacturer. During assay development, 11 SNPs failed at the primer and probe design stage, 15 SNPs failed in amplification or showed ambiguous genotyping results, and 21 SNPs were removed due to low frequency of the minor allele (< 3%). We finally successfully genotyped 65 SNPs for the 1150 samples using ∼116,150 reactions. All genotypes were collected using the SDS software provided by ABI.
Constructing Haplotype Networks
We calculated the pairwise D′ value of each pair of the 65 SNPs using the EHPLUS program (Xie and Ott 1993; Terwilliger and Ott 1994) and the program 2LD (Zapata et al. 2001). Then we constructed a haplotype block by considering adjacent markers where |D′| values between two SNPs was greater than 0.80. This construction was carried out in each sample cluster individually. By using a new statistical model-based program, PHASE (Stephens et al. 2001), which is considered to be an improvement on traditional Expectation Maximization algorithm-based methods, we obtained the estimated genotypes of haplotype blocks for every individual. Only common haplotypes (frequency >3%) were passed for association study.
Statistical Analysis
Haplotype probabilities for each individual were obtained using PHASE (Stephens et al. 2001). We carried out a series of nonfamily-based association studies in each population cluster using both single SNPs and the haplotypes. Formula-based standard χ2 test and Monte Carlo tests were both used. We used the software CLUMP (Sham and Curtis 1995) for Monte Carlo tests of those variations to report four P-values in each test, from which we chose two, normal χ2 result (T1) and the χ2 for the “clumped” 2×2 table (T4), which is suggested to have enough power of detecting association, as mentioned in the manual. The results of tests for all variants are listed in Supplement 1. Table 5 summarizes each marker's minimum P-value. When considering the correlation between tests for each marker, we adjusted the significance level to 0.028 by Bonferroni's multiple test correction. Significant results as determined by the new threshold are distinguished by different style and color in Table 5 from results with the uncorrected threshold P < 0.05. To verify our statistical results, we also did cross-validation by subdividing each of the sample sets into two random subsets of roughly equal sizes for independent tests. Those results fully support our inferences. The details of cross-validation can be found in Supplements 5–8.
Simulation
We built a simulation program to estimate how correlation among samples may influence the detection power of subsequent analysis. (1) Reading the input of allele frequencies in SH's control, SH's case, NH's control, and NH's case, the program randomly generates the genotypes of each individual in a single simulation. (2) Based on the given correlated sampling structure of SH, NH, NSHC, and NNHC (Table 3), the program then calculates the χ2 values and compared them among different clusters. Repeating steps (1) and (2) 1,000,000 times (the number of repeats can be defined in the input file), we obtained the frequencies of occurrences “χ2 in SH > χ2 in NSHC,” “χ2 in NH > χ2 in NSHC,” “χ2 in SH > χ2 in NNHC,” and “χ2 in NH > χ2 in NNHC.” These frequencies should reflect how same-sample reallocation affected the susceptibility detection power. The simulation result for each SNP, program source code, and README file can be found in supplementary files (“README for simulate.txt” and “simulate.c”).
Acknowledgments
This work was supported by grants from the National 973 and 863 programs (2002AA223021; 2002BA711A07-14; 2001CB510304), the National Natural Science Foundation of China, the Ministry of Education, and the Shanghai Municipal Commission for Science and Technology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1758204.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Chu, J.Y., Huang, W., Kuang, S.Q., Wang, J.M., Xu, J.J., Chu, Z.T., Yang, Z.Q., Lin, K.Q., Li, P., Wu, M., et al. 1998. Genetic relationship of populations in China. Proc. Natl. Acad. Sci. 95: 11763–11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, B., Bacanu, S.A., Roeder, K., Reimherr, F., Wender, P., Galke, B., Novasad, D., Chu, A., Cuenco, K.T., Tiobek, S., et al. 2002. Genome-wide multipoint linkage analyses of multiplex schizophrenia pedigrees from the oceanic nation of Palau. Mol. Psychiatry 7: 689–694. [DOI] [PubMed] [Google Scholar]
- Ding, Y.C., Wooding, S., Harpending, H.C., Chi, H.C., Li, H.P., Fu, Y.X., Pang, J.F., Yao, Y.G., Yu, J.G., Moyzis, R., et al. 2000. Population structure and history in East Asia. Proc. Natl. Acad. Sci. 97: 14003–14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart, C.J., Parra, E.J., Shriver, M.D., Bonilla, C., Kittles, R.A., Clayton, D.G., and McKeigue, P.M. 2003. Control of confounding of genetic associations in stratified populations. Am. J. Hum. Genet. 72: 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. and Su, B. 2000. Natives or immigrants: Modern human origin in East Asia. Nat. Rev. Genet. 1: 126–133. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet, K.T. and Chedotal, A. 2002. Role of Slit proteins in the vertebrate brain. J. Physiol. Paris 96: 91–98. [DOI] [PubMed] [Google Scholar]
- Oota, H., Kitano, T., Jin, F., Yuasa, I., Wang, L., Ueda, S., Saitou, N., and Stoneking, M. 2002. Extreme mtDNA homogeneity in continental Asian populations. Am. J. Phys. Anthropol. 118: 146–153. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., and Donnelly, P. 2000a. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., Rosenberg, N.A., and Donnelly, P. 2000b. Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, P.C. and Curtis, D. 1995. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Annu. Hum. Genet. 59: 97–105. [DOI] [PubMed] [Google Scholar]
- Stephens, M., Smith, N., and Donnelly, P. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978—989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, B., Xiao, J., Underhill, P., Deka, R., Zhang, W., Akey, J., Huang, W., Shen, D., Lu, D., Luo, J., et al. 1999. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am. J. Hum. Genet. 65: 1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, J. and Ott, J. 1994. Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore.
- van den Oord, E.J.C.G. 2002. Association studies in psychiatric genetics: What are we doing? Mol. Psychiatry 7: 827–828. [DOI] [PubMed] [Google Scholar]
- Vargesson, N., Luria, V., Messina, I., Erskine, L., and Laufer, E. 2001. Expression patterns of Slit and Robo family members during vertebrate limb development. Mech. Dev. 106: 175–180. [DOI] [PubMed] [Google Scholar]
- Xie, X. and Ott, J. 1993. Testing linkage disequilibrium between a disease gene and marker loci. Am. J. Hum. Genet. 53: 1107.8105690 [Google Scholar]
- YiDa, Y. and Cheng, Z. 2002. Chinese surnames: Colony genetics and population distribution. East China Normal University Press, Shanghai.
- Yuan, W., Rao, Y., Babiuk, R.P., Greer, J.J., Wu, J.Y., and Ornitz, D.M. 2003. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc. Natl. Acad. Sci. 100: 5217–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata, C., Carollo, C., and Rodriguez, S. 2001. Sampling variance and distribution of the D′ measure of overall gametic disequilibrium between multiallelic loci. Ann. Hum. Genet. 65: 395–406. [DOI] [PubMed] [Google Scholar]
- Zhao, T., Zhang, G., Zhu, Y., Zheng, S., Liu, D., Chen, Q., and Zhang, X. 1986. The distribution of immunoglobulin Gm allotypes in forty Chinese populations (in Chinese). Acta. Anthropology Sin. 6: 1–8. [Google Scholar]