Abstract
Pristionchus pacificus is a free-living nematode increasingly used as an organism for comparison to the more familiar model Caenorhabditis elegans. In this study, we examined the N-glycans of this organism isolated after serial release with peptide:N-glycosidases F and A; after fluorescent labelling with 2-aminopyridine, chromatographic fractionation by three types of reversed-phase HPLC (with either classical C18, fused core C18 or alkylamide bonded phases) followed by mass spectrometric analyses revealed key features of its N-glycome. In addition to paucimannosidic and oligomannosidic glycans typical of invertebrates, N-glycans with two core fucose residues were detected. Furthermore, a range of glycans carrying up to three phosphorylcholine residues was observed whereas, unlike C. elegans, no tetrafucosylated N-glycans were detected. Structures with three fucose residues, unusual methylation of core α1,3-fucose or with galactosylated fucose motifs were found in low amounts; these features may correlate with a different ensemble or expression of glycosyltransferase genes as compared to C. elegans. From an analytical perspective, both the alkylamide RP-amide and fused core C18 columns, as compared to a classical C18 material, offer advantages in terms of resolution and of elution properties, as some minor pyridylamino-labelled glycans (e.g., those carrying phosphorylcholine) appear in earlier fractions and so potential losses of such structures due to insufficient gradient length can be avoided.
Keywords: nematode, RP-HPLC, fucose, phosphorylcholine, pyridylamination, N-glycans
The range of asparagine-linked oligosaccharides (N-glycans) found in lower eukaryotes is indeed immense and glycobiologists have the multiple challenges of determining the structure and function of unusual glycan structures as well as defining glycan biosynthetic pathways (1). The concept of comparative glycomics as a tool to aid definition of the specificity of glycosylation-relevant enzymes is in its infancy, but with combined glycomic and genomic data on closely-related species it becomes a realistic option to help solve the aforementioned challenges. In the case of nematodes, an increasing number of genomes are becoming available, although the degree of assembly and annotation varies. In recent years, the Ascaris, Brugia, Haemonchus, Necator, Trichinella and Trichuris draft genomes have been sequenced as examples for animal parasites (2-7), Bursaphelenchus and Meloidogyne are examples for the plant parasites (8,9) and genomes of various Caenorhabditis species (10,11) as well as of free-living Panagrellus and Pristionchus (12,13) are now available. However, other than various animal parasites and Caenorhabditis elegans itself (14,15), there is little glycomic knowledge about nematodes.
A particularly highly-variable and complex set of glycan modifications in nematodes is fucosylation. N-glycans with up to four fucose residues have been detected in C. elegans, which is a remarkable number considering the relative lack of extended antennae in this species; indeed, these fucose residues are modifications of the mannosylchitobiosyl core region. Fucose can be α1,3-linked to either the reducing-terminal proximal and distal core N- acetylglucosamine (GlcNAc) residues, α1,6-linked to the proximal GlcNAc or α1,2-linked to galactose residues which are also associated with the core (16,17). At least some of these features have been found in parasitic nematodes such as Ascaris, Haemonchus and Oesophagostomum (17,18). Recently, we have shown that three C. elegans fucosyltransferases (FUT-1, FUT-6 and FUT-8) can be employed to generate trifucosylated N-glycan cores in vitro (19).
Another unusual aspect of nematode glycans is the presence of phosphorylcholine; this zwitterionic modification is otherwise known from some bacterial lipopolysaccharides, fungal cell walls and one cestode glycoprotein (Ag5 from Echinococcus granulosus) (20-22). The phosphorylcholine attached to the N-glycans of one nematode glycoprotein (ES-62 from Acanthocheilonema viteae) is apparently necessary for the immunomodulatory activity of this protein (23); on the other hand, a possibly contradictory aspect is that phosphorylcholine is recognised by mammalian C-reactive protein (24), a pentraxin whose levels in the serum increases during inflammation. Furthermore, galactosylation of core fucose and methylation have been observed on nematode N-glycans (14,15). Of these various N-glycan modifications, at least core α1,3-fucose and galactosylated core α1,6-fucose are potential anthelminthic targets as fungal lectins (specifically CCL2 and CGL2) recognising these elements are nematotoxic (25,26). To date, our information regarding the enzymatic basis for modifications of nematode N-glycans is restricted to identification of the three fucosyltransferases modifying the core region (FUT-1, FUT-6 and FUT-8) and the α1,6-fucose-modifying galactosyltransferase (GALT-1) (19,27-29) as well as conserved enzymes such as N-acetylglucosaminyltransferases I and II (30). The genetic basis for further modifications remains unknown.
Analysis of the various nematode glycans presents significant challenges as, in comparison to mammalian glycans, there are generally no commercially-available standards and little literature. However, a reliance of mass spectrometry alone may result in misleading interpretations. Therefore, methods which reliably separate glycans on the basis of structural type or isomeric status are highly valuable prior to mass spectrometric analyses. A gold standard in this respect may well be two-dimensional separations (e.g., hydrophilic interaction followed by reversed phase) in order to obtain fractions with only one or two glycan structures, but the low amounts of glycans obtained from many biological sources in combination with the multiplication of obtained fractions from two-dimensional HPLC mean that this approach may not be the most appropriate for a specific sample. Thus, a one-dimensional HPLC method which results in the greatest informational gain may be advantageous. This, though, may be dependent on the sample to be analysed: in previous studies, we have used normal-phase, reversed-phase (either in the classical C18 or the newer ‘RP-amide’ modes) and mixed mode columns (17,31,32). Although, reversed phase HPLC is probably the most robust method, there is the tendency that anionic glycans are poorly retained, which for instance when examining Dictyostelium discoideum N-glycans resulted in early-eluting fractions of some complexity (32). However, normal phase often offers little isomeric information.
In the context of comparative nematode glycomics and the development of relevant methods, we have examined the N-glycans of Pristionchus pacificus, which is (like C. elegans) a free-living nematode used for some comparisons of development (33,34) and whose genome has been sequenced (13); an unusual feature is the dimorphism of its mouthparts and a pre-hatching moult. In the wild, some Pristionchus species appear to be associated with beetles (35). Our studies indicate that the N-glycome of P. pacificus is somewhat simpler than that of C. elegans, but still shows some features seen in various nematode species, such as mono- and difucosylation as well as modification by the zwitterion phosphorylcholine. As part of our study, we compared three different HPLC columns (classical C18, fused core C18 and RP-amide) to fractionate fluorescently-labelled N-glycans. On one hand, common trends in the elution of pyridylaminated forms of the standard paucimannosidic and oligomannosidic glycans were observed, whereas the order for glycans carrying core fucose and antennal phosphorylcholine differs. Probably the most superior resolution is attained with the fused core C18 column.
Experimental Procedures
Nematode cultivation
Pristionchus pacificus (PS312) was grown in liquid culture using E. coli OP50 as a food source, as previously done for C. elegans (27). Two independently-grown samples of mixed populations were analysed and 1-2 g worm pellets were homogenised and proteolysed with pepsin. N-glycans were then released from peptic peptides using peptide:N-glycosidase F (capable of removing most eukaryotic N-glycans other than those carrying α1,3-fucose on the reducing terminal GlcNAc), followed by peptide:N-glycosidase A (which is capable of removing core α1,3-fucosylated N-glycans), according to the procedures described previously. Free glycans were labelled with 2-aminopyridine (36,37) prior to MALDI-TOF MS and fractionation by reversed-phase HPLC (RP-HPLC).
Fractionation of pyridylaminated N-glycans
Separation of pyridylamino-labeled glycans was carried out on a Shimadzu HPLC system equipped with a fluorescence detector (RF 10 AXL; excitation at 320 nm and emission 400 nm). For standard RP-HPLC, a Hypersil ODS column (Agilent; a classical C18 column of the dimensions 250 × 4.6 mm) was used with 100 mM ammonium acetate, pH 4.0 (buffer A) and 30% (v/v) methanol (buffer B); a gradient of increasing buffer B (1% per minute) was programmed up to 30 minutes; a step up to 40% B for 5 minutes was followed by another step to 45% for 5 minutes followed by a return to starting conditions. A similar linear gradient (1% B per minute; 0.8 ml/min) for 45 minutes was used with a Kinetex™ 5μ XB-C18 column (250 × 4.6 mm; Phenomenex), which has iso-butyl side chains on fused core particles. For the Ascentis® Express 2.7 μ RP-Amide column (150 × 4.6 mm; Sigma-Aldrich) which has a fused core carrying alkyl amide chains as the bonded phase, a gradient of buffer B up to 35% over 34 minutes was applied at a flow rate of 0.8 ml/min as follows: 0-4 min, 0% B; 4-14 min, 0-5% B; 14-24 min, 5-15% B; 24-34 min, 15-35% B; 34-35 min, return to starting conditions. The recommended guard columns were used in all cases. The overall HPLC and MALDI-TOF MS results from two independently-grown nematode samples were similar, but only the data for one preparation are shown.
Mass spectrometry
Monoisotopic MALDI-TOF MS was performed using a Bruker Autoflex Speed (equipped with a 1000 Hz Smartbeam™-II laser) instruments in positive reflectron mode with 6-aza-2-thiothymine (ATT) as matrix. MS/MS was performed by laser-induced dissociation. Spectra were processed with the manufacturer’s software (Bruker Flexanalysis 3.3.80) using the SNAP algorithm with a signal/noise threshold of 6 for MS (unsmoothed) and 3 for MS/MS (four-times smoothed). Glycan spectra were manually interpreted on the basis of the masses of the predicted component monosaccharides, relative elution times as compared to previous studies, differences of mass in glycan series and fragmentation patterns.
Enzymatic and chemical treatments
Further analysis by MALDI-TOF MS was performed after treatment overnight with either β-galactosidase (recombinant Aspergillus niger lacA; prepared in-house (38)), α-fucosidase (bovine kidney from Sigma-Aldrich), α-mannosidase (Xanthomonas manihotis α1,2/3-specific from NEB) or β-N-acetylhexosaminidase (recombinant Apis mellifera FDL; prepared in-house, specific for the N-acetylglucosamine attached to core α1,3-mannose, i.e., the residue transferred by GlcNAc-TI (39)) in 25 mM ammonium acetate, pH 4.5, at 37 °C. For removal of α1,3-linked fucose or of phosphorylcholine, selected fractions were dried and incubated overnight at 0 °C with 3 μl 48% (v/v) hydrofluoric acid prior to evaporation; the samples were diluted in water and re-evaporated, prior to redissolving once again.
Results
Overall N-glycomic profile
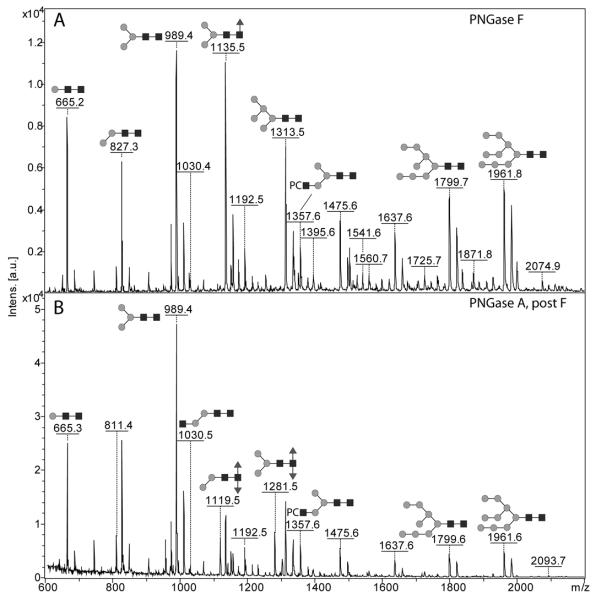
An initial perusal of the N-glycomes of Pristionchus pacificus released with PNGase F and PNGase A and analysed by MALDI-TOF MS would suggest similar profiles as compared to Caenorhabditis elegans with a set of probable paucimannosidic and oligomannosidic N-glycans (Hex5-9HexNAc2 and Hex1-3HexNAc2Fuc0-1) being the major N-glycans with some amounts of phosphorylcholine-modified glycans (addition of 165 Da) and, in the glycans released with PNGase A, some difucosylated species (Figure 1). It is known from other studies on glycans from plants, insects, nematodes and other non-mammalian sources that core α1,3-fucose, either on its own or in combination with core α1,6-fucose, prevents removal of such modified N-glycans by PNGase F (40); therefore, the sequential use of both enzymes to release the N-glycans was performed. As judged by the fluorescence of the labeled glycan pools, the amount of glycan in the pool released by PNGase A after PNGase F was some ten-fold lower than the pool released by PNGase F. As seemingly the PNGase F digestion was incomplete, many glycans in the PNGase A pool are lower-intensity ‘remainders’ of glycans in PNGase F pool (Table 1); as only a minority of glycans are found solely in the PNGase A pool, the proportion of core α1,3-fucosylated N-glycans in this species is probably under 5% of the total.
Figure 1. MALDI-TOF mass spectra of the complete N-glycan pools from Pristionchus pacificus.
The N-glycans released by (A) PNGase F or (B) by PNGase A after PNGase F were pyridylaminated (i.e., fluorescently-labelled with 2-aminopyridine) and are annotated with selected major glycan structures; the annotations are for the [M+H]+ forms, but sodiated ions were also detected. The example glycans are depicted according to the symbolic nomenclature of the Consortium for Functional Glycomics; circles, hexose; squares, N- acetylhexosamine; triangles, fucose; PC, phosphorylcholine.
Table I. List of predicted N-glycans for Pristionchus pacificus.
Glycans released by either PNGase F (F) or PNGase A after F (A), whose structures were proven on the basis of retention time (in glucose units for the three columns used), MS/MS fragmentation pattern and sensitivity/resistance to chemical and enzymatic treatments, are shown according to the nomenclature of the Consortium for Functional Glycomics; calculated m/z for pyridylamino-labelled glycans (as [M+H]+) are presented as well as, for some structures, the ‘Schachter-style’ abbreviation.
| Structure | m/z | Retention time | ||
|---|---|---|---|---|
| RPC18 | RP-amide | Kinetex | ||
|
|
665.28 | A/F 6.4 | A/F 6.2 | F 7.8 |
|
|
811.35 | A 4.3 | A 4.3 | A 4.9 |
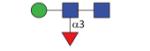
|
811.35 | A/F 7.7 | F 7.5 | A/F 11.5 |
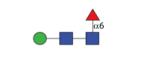
|
811.35 | A/F 10.3 | A/F 8.2 | A/F 13.5 |
|
|
827.34 (0M) | A/F 5.9 | A/F 6.1 | A/F 7.5 |
|
|
827.34 (M0) | A/F 7.7 | A/F 7.2 | A/F 9.5 |
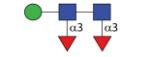
|
957.40 | A 5.9 | n.d. | A 6.8 |
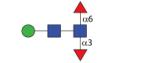
|
957.40 | A 7.0 | A 6.3 | A 8.3 |
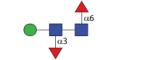
|
957.40 | n.d. | A 8.7 | F > 25.0 |
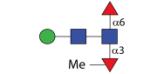
|
971.42 | A 19.0 | A 9.5 | A > 25 |
|
|
973.40 (0MF3) | A 4.2 | A 4.5 | A 4.7 |
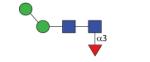
|
973.40 (M0F3) | A 5.0 | A 5.0 | A 5.5 |
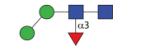
|
973.40 | A/F 7.7 | F 7.2 | A/F 10.2 |
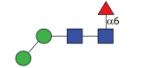
|
973.40 (0MF6) | A/F 9.8 | A/F 8.2 | A/F 13 |
|
|
973.40 (M0F6) | A/F 13.0 | A/F 9.0 | A/F 16 |
|
|
989.39 (MM) | A/F 7.2 | A/F 7.2 | A/F 9.5 |
|
|
1030.42 (0Gn) | A/F 6.5 | A/F 6.1 | A/F 7.5 |
|
|
1119.46 | A 5.2 | A 5.4 | A 6.8 |
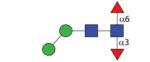
|
1119.46 | A 7.0 | A 6.3 | A 8.3 |
|
|
1119.46 | F 13.0 | n.d. | F > 25.0 |
|
|
1135.45 (MMF3) | A 5.0 | A 5.1 | A 5.5 |
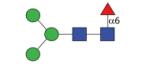
|
1135.45 (MMF6) | A/F 11.5 | A/F 9.5 | A/F 16.0 |
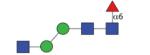
|
1176.48 | n.d. | F 8.2 | F 13.0 |
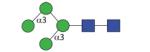
|
1151.45 | A/F 7.2 | A/F 7.2 | A/F 9.5 |
|
|
1192.47 (MGn) | A/F 7.0 | A/F 6.8 | A/F 8.4 |
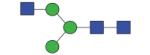
|
1192.47 (GnM) | A/F 10.3 | A/F 9.0 | F/A 16.0 |
|
|
1195.47 | A/F 7.7 | A/F 5.5 | A/F 6.9 |
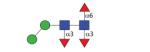
|
1265.51 | A 8.0 | A 6.8 | A 9.2 |
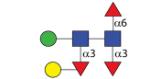
|
1265.51 | A 9.8 | A 8.0 | A 11.2 |
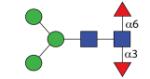
|
1281.51 (MMF3F6) | A 7.2 | A 6.8 | A 8.5 |
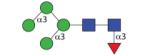
|
1297.50 | A 5.0 | A 5.0 | A 5.8 |
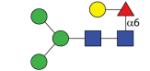
|
1297.50 | n.d. | F 11.0 | A/F > 20.0 |
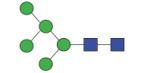
|
1313.50 (Man5) | A/F 7.0 | A/F 7.2 | A/F 8.7 |
|
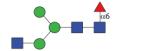
1338.53 (MGnF6) | A/F 10.3 | A/F 8.6 | A/F 13.0 |
|
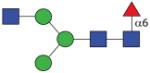
1338.53 (GnMF6) | F 17.0 | F 12.0 | A/F > 25.0 |
|
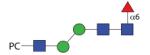
1341.53 | F 13.0 | F/A 7.7 | A/F 11.5 |
|
|
1357.53 | A/F 8.0 | A/F 6.2 | A/F 7.5 |
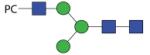
|
1357.53 | A/F 11.5 | A/F 8.17 | A/F 14.5 |
|
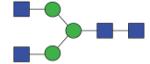
1395.55 (GnGn) | A/F 9.0 | A/F 8.10 | A/F 11.5 |
|
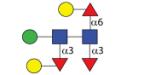
1427.56 | A 11.5 | A 8.2 | A 12.7 |
|
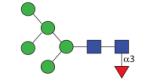
1459.56 | A 4.7 | A 5.0 | A 5.4 |
|
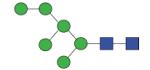
1475.55 | A/F 5.5 | A/F 6.1 | A/F 6.5 |
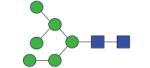
|
1475.55 | A/F 5.9 | A/F 6.3 | A/F 6.9 |
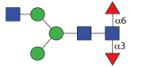
|
1484.59 | A 9.0 | A 7.8 | A 11.2 |
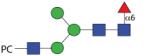
|
1503.59 | A/F 13.0 | A/F 8.0 | A/F 11.5 |
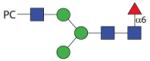
|
1503.59 | A/F 19.0 | A/F 11.0 | F > 25.0 |
|
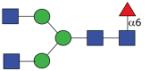
1541.61 (GnGnF6) | F 14.0 | F 10.5 | F 21.0 |
|
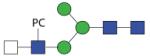
1560.61 | F 8.0 | F 6.8 | F 8.8 |
|
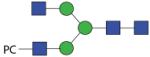
1560.61 | F 10.3 | F 7.5 | F 9.5 |
|
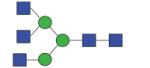
1598.63 | A/F 6.5 | F 6.3 | F 7.5 |
|
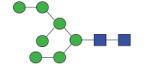
1637.60 | A/F 5.0 | A/F 5.5 | A/F 5.3 |
|
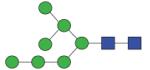
1637.60 | A/F 5.7 | A/F 6.1 | A/F 6.2 |
|
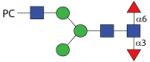
1649.64 | A/F 9.8 | A 7.2 | A 10.0 |
|
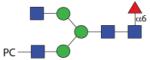
1706.67 | F 17.0 | F 9.5 | F 16.0 |
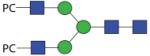
|
1725.66 | A/F 15.0 | A/F 6.8 | A/F 9.5 |
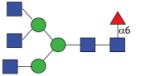
|
1744.69 | n.d. | F 8.2 | F 11.5 |
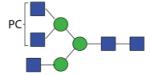
|
1763.69 | F 7.2 | F 5.5 | F 6.3 |
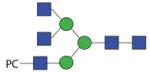
|
1763.69 | n.d. | F 5.9 | F 6.9 |
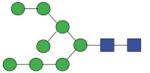
|
1799.66 (Man8B) | A/F 4.6 | A/F 5.2 | A/F 5.0 |

|
1799.66 (Man8A) | A/F 5.3 | F 5.8 | A/F 6.0 |
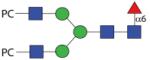
|
1871.72 | A/F > 20 | F 8.6 | F 14.5 |
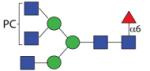
|
1909.75 | n.d. | F 6.8 | F 8.8 |
|
|
1928.74 | F 17.0 | F 4.0 | F 5.0 |
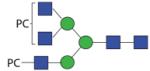
|
1928.74 | n.d. | n.d. | F 5.7 |
|
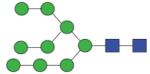
1961.71 (Man9) | A/F 4.9 | A/F 5.5 | A/F 5.3 |
|
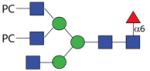
2074.80 | F > 20.0 | n.d. | F 6.9 |
|
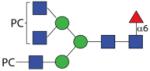
2074.80 | n.d. | F 6.3 | F 8.0 |
|
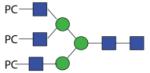
2093.80 | A/F > 20.0 | A/F 4.8 | F 4.8 |
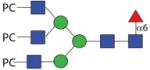
|
2239.86 | F > 20.0 | F 4.5 | F 6.7 |
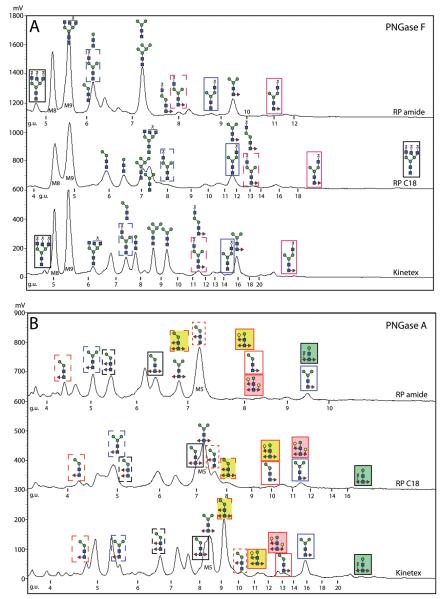
Despite the impression that the N-glycomes of C. elegans and P. pacificus are similar, a more exact examination of the overall mass spectra indicated a lack of tetrafucosylated glycans from the glycome of the latter. For a closer inspection of the pools of pyridylamino-labelled N-glycans, HPLC was employed and each fraction from the PNGase F and PNGase A pools was subject to MALDI-TOF MS and MS/MS. Due to the excellent experience with the Ascentis Express RP-amide column for fractionation of N-glycans from oyster plasma and haemocytes (31), we first used this column to fractionate the pyridylaminated P. pacificus N-glycans; however, to explore the use of other columns, we also supplemented this fractionation by comparing the RP-amide column with classical and fused core RP-HPLC materials (Agilent Hypersil C18 RP-HPLC, as previously used by us with C. elegans N-glycans, and Kinetex XB-C18; Figure 2). Our structural propositions (Table 1) are corroborated by a range of MS/MS before and after chemical and enzymatic digestion; in total some 1300 MS and MS/MS spectra were recorded, manually interpreted and annotated. For simplicity, only a selection of these data, indicative of each type of structural motif, is presented in the main text; a summary of evidence for each proposed glycan structure is given in the Supplementary Table.
Figure 2. Comparisons of RP-HPLC fractionation of P. pacificus pyridylamino-labelled N-glycans.
Chromatograms obtained with pyridylaminated N-glycans released either with PNGase F alone (A) or with PNGase A after PNGase F (B) are shown for the Ascentis® Express RP-amide, Agilent Hypersil ODS C18 and Phenomenex Kinetex™ XB-C18 columns; the chromatograms are normalised with respect to the elution of the Man8,9GlcNAc2 and Man3GlcNAc2Fuc1 glycans and the calibration in terms of glucose units is shown. The chromatograms are also annotated with the structures of selected N-glycans with either similar or contrasting relative elution times with boxes in the same form (colour, shading or dashes) to highlight the elution position of the same glycan in different chromatograms.
Oligo- and paucimannosidic glycans
The major oligomannosidic glycans are Man5GlcNAc2 (7.0 g.u. on the Agilent C18 RP-HPLC column) and Man8-9GlcNAc2 (4.6 and 4.9 g.u.) with also a significant amount of Man3GlcNAc2 (7.2 g.u.), which are in line with literature values (41). On the Kinetex XB-C18 and RP-amide columns, the elution times of Man3,5,8,9GlcNAc2 (respectively 9.5/8.7/5.0/5.3 g.u. and 7.2/7.2/5.2/5.5 g.u.) are not directly comparable to those on the standard C18 column, although the tendency is similar; it can be concluded that all three columns can be used to effectively separate N-glycan isomers, whose structures can then be verified by MS/MS or exoglycosidase digestion. The Man3,5,6GlcNAc2 glycans lose, as expected, one or two mannose residues upon α1,2/3-mannosidase digestion (Figure 3B and J).
Figure 3. Diagnostic digestion of pyridylamino-labelled paucimannosidic N-glycans with α 1,2/3-mannosidase.
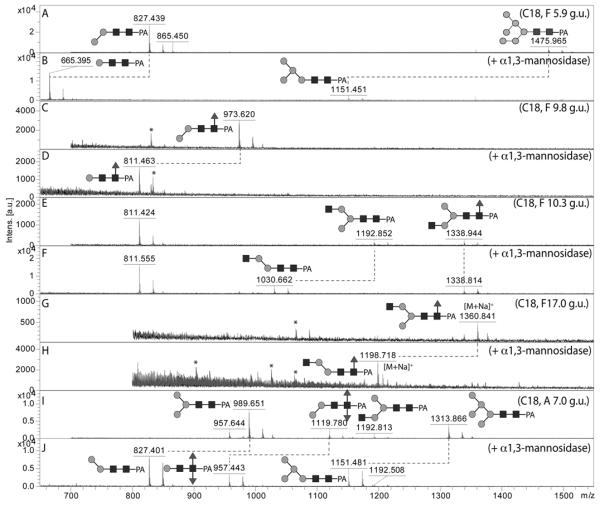
The effects of α1,2/3-mannosidase on five different RP-HPLC (C18) fractions from either the PNGase F or PNGase A pools was monitored by MALDI-TOF MS: (A, B) spectra of the PNGase F C18 fraction of 5.9 g.u. (hence annotated as F 5.9 g.u.) before and after α1,2/3-mannosidase digestion, demonstrating the isomeric forms of Man2GlcNAc2 and Man6GlcNAc2; (C, D) spectra of the PNGase F C18 fraction of 9.8 g.u. before and after α1,2/3-mannosidase digestion, demonstrating the major isomeric form of Man2GlcNAc2Fuc1; (E,F) spectra of the PNGase F C18 fraction of 10.3 g.u. before and after α1,2/3-mannosidase digestion, demonstrating two isomeric forms of Man3GlcNAc3Fuc0-1; (G,H) spectra of the PNGase F C18 fraction of 17 g.u. before and after α1,2/3-mannosidase digestion, demonstrating a minor isomeric form of Man3GlcNAc3Fuc1; (I,J) spectra of the PNGase A C18 fraction of 7.0 g.u. before and after α1,2/3-mannosidase digestion, demonstrating the isomeric status of Man2GlcNAc2Fuc2 as well as co-eluting Man3,5GlcNAc2. Note that the α1,3-mannose attached to the core α1,6-mannose is resistant to this enzyme. Asterisks indicate non-glycan contaminants in some low-intensity fractions; glycans are annotated with the m/z values for the [M+H]+ ions, unless otherwise indicated.
An example of the isomeric designations is described for Man8GlcNAc2: the major form, eluting at 4.6 g.u. on the C18 column is the Man8B isomer (a product of ‘endoplasmic reticulum’ mannosidase I), whereas only a minor amount of the Man8A isomer is present (5.3 g.u.). Compatible with this assumption are the MS/MS data, which show relatively intense m/z 989 and 1151 signals respectively for the Man8A and Man8B structures (Supplementary Figure 1), which may result from the loss of the entire α1,6-antenna during fragmentation (see also our previous study (32)). There is also a remarkable level of small ‘paucimannosidic’ glycans with m/z 665 and 827, whose presence is suggestive of high mannosidase activity in vivo; specific α1,2/3-mannosidase digestion enabled us to confirm the major Man2GlcNAc2 isomer (5.9 g.u.) as being Manα1,3Manβ1,4GlcNAcβ1,4GlcNAc (‘0M’ according to a nomenclature based on that of Schachter (42); Figure 3A and B).
Mono-, di- and trifucosylated glycans
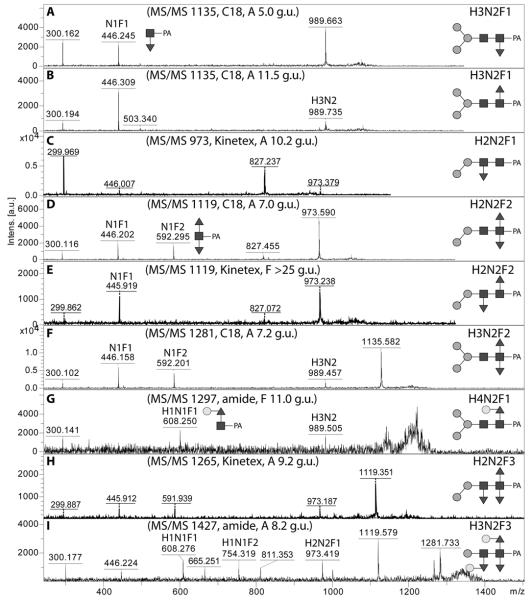
As in many invertebrates, the major fucosylated N-glycan in Pristionchus has the composition Hex3HexNAc2Fuc1 (m/z 1135). However, there were two isomeric glycans with this composition, both which displayed a typical core fucose MS/MS fragment of m/z 446 (Figure 4A and B). The early-eluting form present only in the PNGase A digest (5.0-5.5 g.u. on all columns) carries α1,3-fucose on the reducing-terminal GlcNAc, whereas core α1,6-fucose results in late elution (43), a conclusion corroborated by the respective sensitivities towards hydrofluoric acid and bovine α-fucosidase (Supplementary Figure 2B and D). For other simple monofucosylated glycans (Hex1-2HexNAc2Fuc1; m/z 811 and 973), there was surprisingly a greater range of structural diversity with respectively three and five different elution positions (see also Supplementary Figure 3). In addition to the typical ‘isomeric splitting’ due to α1,3- and α1,6-fucosylation of the reducing terminal GlcNAc and two different linkages of the single α-mannose in Hex2HexNAc2Fuc1 (α1,3 or α1,6, resulting for the former in earlier elution and α1,2/3mannosidase sensitivity; see Figure 3C and D), yet another position for fucose was apparent. For instance, a ‘fifth’ isomer of Hex2HexNAc2Fuc1 (7.7 g.u. or 10.2 g.u., respectively on the C18 and Kinetex columns) was present in both the PNGase F and A digests; this glycan, sensitive to hydrofluoric acid, showed only a very weak m/z 446 fragment (see Figure 4C and Supplementary Figures 2G and 3I), which we find to be a hallmark for glycans with fucose on the second (distal) core GlcNAc and which is consistent with a low-level ‘rearrangement’ of fucose during fragmentation (44). For fucosylated N-glycans with one non-reducing terminal GlcNAc (Hex3HexNAc3Fuc1; m/z 1338), there are two variations possible: so-called MGnF6 and GnMF6 (analogous to the MGn and GnM discussed below) eluting at 10.3 and 17 g.u.. These two glycans had contrasting sensitivities to α1,2/3-mannosidase in keeping with their proposed structures (Figure 3E-H).
Figure 4. MALDI-TOF MS/MS of pyridylamino-labelled mono-, di- and tri-fucosylated N-glycans.
Examples of MS/MS spectra for glycans monofucosylated on the reducing-terminal GlcNAc (A,B), monofucosylated on the distal GlcNAc (C), difucosylated on the reducing-terminal GlcNAc (D,F), fucosylated on both the reducing-terminal and distal GlcNAc (E), galactosylated on core fucose (G) or trifucosylated on the core (H,I) are shown for glycans identified in various RP-HPLC fractions. Key fragments are annotated with abbreviations of the form HxNyFz where H is hexose, N N-acetylhexosamine, and F fucose or with diagrammatic depictions, in which the α1,3- and α1,6-fucose residues are respectively shown in the ‘down’ or ‘up’ positions.
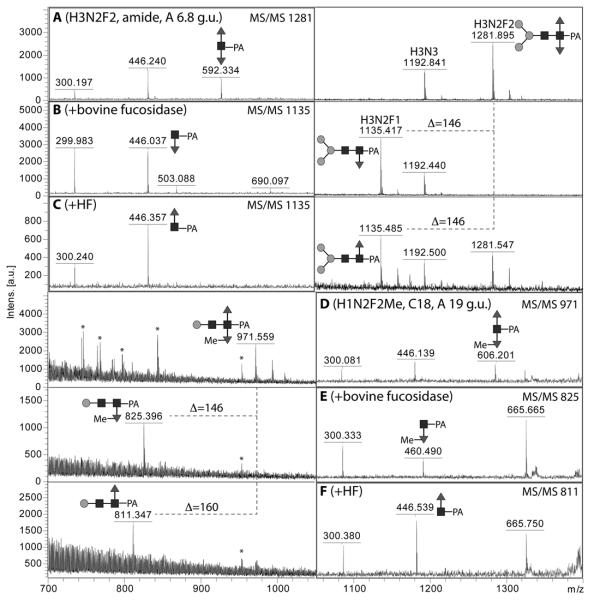
In the PNGase A digest, some difucosylated N-glycans with compositions Hex1-3HexNAc2-3Fuc2 were detected which elute between 7 and 9 g.u. on the C18 column; these g.u. values are similar to those previously reported for difucosylated insect N-glycans (45). MS/MS showed the presence of m/z 446 and 592 as major fragments for one isomer with m/z 1119 as well as for the single glycan with m/z 1281 (Figure 4D and F). Selected digests were also performed (see Figure 5A-C) and showed that (i) bovine α-fucosidase only removed one fucose residue from the putative Man3GlcNAc2Fuc2 glycan (m/z 1281) with the product eluting at 5 g.u. (like MMF3) when rechromatographed on the Hypersil C18 column, (ii) hydrofluoric acid also removed one fucose from the same Man3GlcNAc2Fuc2 glycan with the product eluting, as for MMF6, at 11.5 g.u. when rechromatographed and (iii) α1,2/3-mannosidase removed one mannose from the putative Man2GlcNAc2Fuc2 (m/z 1119; Figure 3I and J). Two further isomers of Man2GlcNAc2Fuc2 were also observed (5.2 and 13 g.u. on the C18 column), but do not display difucosylation of the proximal GlcNAc (see below). One further difucosylated glycan (m/z 971) also lost one fucose upon bovine α-fucosidase treatment, but lost 160 Da when treated with hydrofluoric acid; thereby, the m/z 606 fragment was replaced, respectively, by ones of m/z 460 and 446 (Figure 5D-F). These data indicate that this glycan carries a methyl residue on a core α1,3-fucose and so a structure of Manβ1,4GlcNAcβ1,4(Fucα1,6)(MeFucα1,3)GlcNAc is proposed.
Figure 5. Chemical and enzymatic defucosylation of pyridylamino-labelled difucosylated N-glycans.
Selected fractions (either RP-amide or C18; A,D) of PNGase A release N-glycans were subject to either bovine α-fucosidase (B,E) or hydrofluoric acid treatment (C,F). Examples are shown here for Man3GlcNAc2Fuc2 and Man1GlcNAc2Fuc2Me1; the corresponding MS/MS spectra are shown on the left (A-C) or right (D-F) and are annotated to indicate the key fragments. Losses of fucose or methylfucose are indicated by the Δ values. Asterisks indicate non-glycan contaminants in the low-intensity 19 g.u. fraction.
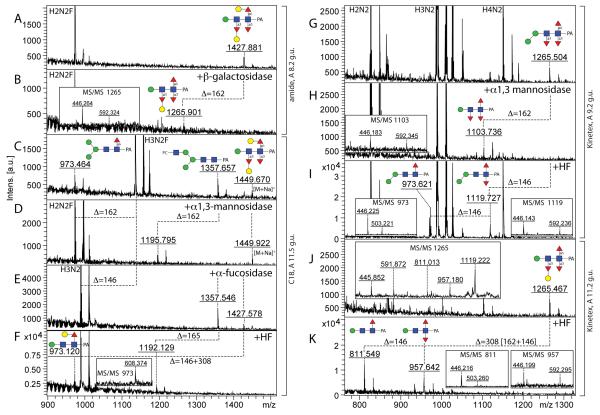
MS/MS of some low intensity glycans (e.g., m/z 1297 and 1427; Hex4HexNAc2Fuc1 and Hex3HexNAc2Fuc3) was found to result in m/z 608 or 754 fragments (Figure 4 G and I), which is reminiscent of glycans from C. elegans with a Galβ1,4Fucα1,6 modification of the reducing-terminal GlcNAc (17); incubation overnight with a diluted recombinant Aspergillus β-galactosidase resulted in a shift of these glycans to ones with a m/z 446 fragment (Figure 6 A and B; compare also Supplementary Figure 2 H and I). The m/z 1427 glycan is one of three trifucosylated glycans detected in our study and appeared insensitive to α1,2/3-mannosidase and bovine fucosidase (Figure 6 D and E). Treatment with hydrofluoric acid of the fucosidase-treated fraction resulted in loss of two fucoses and one hexose; MS/MS of the product of m/z 973 showed a fragment of m/z 608 indicative of galactosylation of core fucose (Figure 6F). It can be proposed that the m/z 1427 glycan is Man1GlcNAc2 decorated with Galβ1,4Fucα1,6 and α1,3-fucose on the reducing terminus and a hexosylated α1,3-fucose on the distal (second) core GlcNAc residue; by comparison, capping of distal fucose by α1,2-galactose (which explains the loss of a unit of 308 Da upon hydrofluoric acid treatment) and the presence of β1,4-galactose on the core α1,6-fucose (which explains the bovine fucosidase insensitivity) are akin to glycans previously found in C. elegans (17). Fucosylation of the distal GlcNAc is also concluded for minor isomers of Hex1-2HexNAc2Fuc1 (m/z 811 and 973; see above) as well as of two forms each of Hex1HexNAc2Fuc2 and Hex2HexNAc2Fuc2-3 (m/z 957, 1119 and 1265; see Table 1 and example MS/MS in Figure 4E and H as well as in Supplementary Figure 3). The two isomeric trifucosylated glycans with m/z 1265 were both sensitive to hydrofluoric acid, but differed regarding the position of the second hexose residue, which was either removed with α1,2/3-mannosidase or along with a fucose when treated with hydrofluoric acid (see Figure 6 H, I and K),
Figure 6. Chemical and enzymatic treatments of pyridylamino-labelled trifucosylated N-glycans.
Selected fractions (either RP-amide, C18 or Kinetex) of trifucosylated N-glycans were analysed by MALDI-TOF MS with the same m/z 1427 glycan being found in both the amide 8.2 g.u and C18 11.5 g.u. fractions (A,C; see MS/MS in Figure 4I), whereas two isomers with m/z 1265 were detected in two different Kinetex fractions (9.2 and 11.2 g.u.; G and J; MS/MS respectively in Figure 4H or inset in panel J). The fractions were treated with Aspergillus β1,4-specific galactosidase (B), Xanthomonas α1,2/3-mannosidase (D,H), bovine α-fucosidase (E), hydrofluoric acid (I,K) or hydrofluoric acid after α-fucosidase (F). Major glycan species in the relevant fractions are ‘off-scale’ in order to highlight the low-intensity, but complex structures. Losses of 146, 162 and 165 (i.e., of fucose, hexose and phosphorylcholine) are indicated and most m/z values are for [M+H]+ ions. Insets show regions of relevant MS/MS spectra for digestion products.
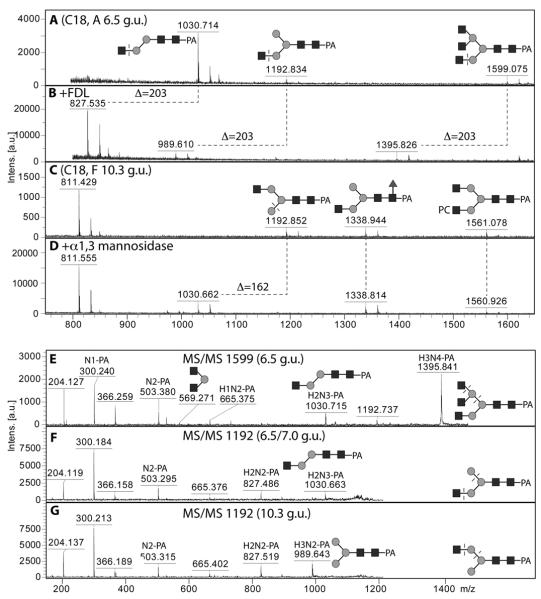
Glycans with free terminal N-acetylglucosamine
A number of glycans with three or more N-acetylhexosamine residues, but without phosphorylcholine, were detected in individual HPLC fractions. Two differently-eluting forms of Hex3HexNAc3 (m/z 1192) can be detected (7.0 and 10.3 g.u.; C18) and these correspond to the two variations of N-glycans with one non-reducing terminal GlcNAc (so-called MGn and GnM). Whereas MGn, as well as the Hex2HexNAc3 glycan (m/z 1030), were sensitive towards the arm-specific Apis mellifera FDL hexosaminidase (Figure 7 A and B), the GnM loses one hexose when incubated with α1,2/3-mannosidase (Figure 7 C and D); these data verify that the GlcNAc is respectively on the ‘lower’ or ‘upper’ antenna (i.e., α1,3- or α1,6-antenna). The two isomers of Hex3HexNAc3 also fragment differently, with MS/MS of MGn resulting in a significant m/z 1030 fragment, whereas the GnM has dominant m/z 827 and 989 fragments (Figure 7 F and G); as for the oligomannosidic glycans, we consider that this is due to higher lability of the ‘core’ α1,6-linked mannose during fragmentation. A Hex3HexNAc5 glycan, also sensitive to the FDL hexosaminidase (Figure 7B), is proposed to be triantennary due to the m/z 569 fragment (Hex1HexNAc2; Figure 7E); considering the repertoire of glycosyltransferases in nematodes (especially GlcNAc-TI, TII and TV (30,46)) and the presence of the m/z 1030 fragment, the m/z 569 is proposed to be derived from the α1,6-arm.
Figure 7. MALDI-TOF MS analysis of isomers of pyridylamino-labelled N-glycans with non-reducing N-acetylglucosamine residues.
Selected C18 RP-HPLC fractions (A, C) were treated with either the specific FDL β-hexosaminidase (B) or with α1,2/3-mannosidase (D) in order to reveal whether GlcNAc residues were present on the α1,3-arm (i.e., the GlcNAc transferred by GlcNAc-TI). All glycans in these fractions were subject to MS/MS and examples for two isomers of Man3GlcNAc3 and the single isomer of Man3GlcNAc5 are shown (E-G). The residues cleaved enzymatically or the bonds putatively cleaved during MS/MS fragmentation are indicated with dashed lines; losses of N-acetylglucosamine or α1,3-mannose are indicated by the Δ values.
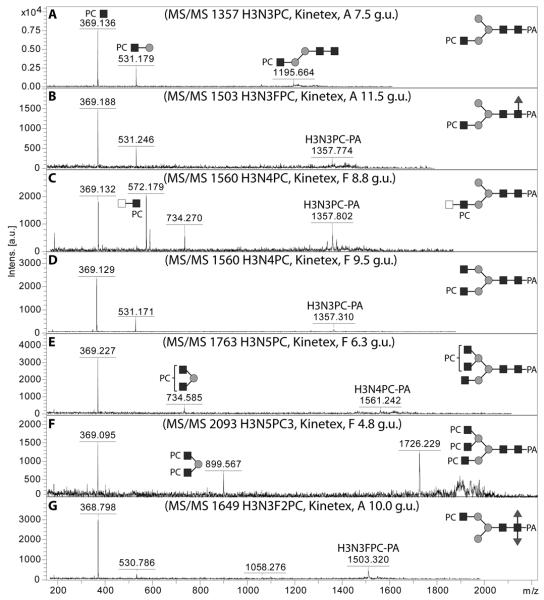
Phosphorylcholine-modified glycans
The modification of glycans by phosphorylcholine is a feature of a number of non-vertebrate species and has been previously reported on N-glycans from nematodes (47,48). In terms of MS/MS, a signature for a phosphorylcholine linked to GlcNAc on glycans is an intense MS/MS fragment of m/z 369 (Figure 8 A and B) (48); in the case of Hex3HexNAc4PC1 (m/z 1560) there were two elution positions and MS/MS of one of these yielded a fragment of m/z 572, indicative of a phosphorylcholine modification of a diHexNAc motif (compare Figure 8 C and D); without further specific data, the presence of a LacdiNAc (GalNAcβ1,4GlcNAc) in this context can only be assumed. MS/MS of glycans with the compositions Hex3HexNAc5PC1 (m/z 1763) and Hex3HexNAc5PC3 (m/z 2094) yielded fragments of respectively m/z 369 and 734 or m/z 369, 899 and 1726, which are compatible with phosphorylcholine modifications of triantennary structures (Figure 8E and F; see also Supplementary Figure 4 for MS/MS spectra of related structures). Hydrofluoric acid effectively removes phosphorylcholine (47) and a number of fractions containing putatively phosphorylcholine-modified N-glycans were treated with this reagent (Figure 9).
Figure 8. MALDI-TOF MS/MS of pyridylamino-labelled N-glycans modified with phosphorylcholine.
The fragmentation patterns for selected glycans predicted to be modified with phosphorylcholine and present in various fractions of PNGase F or PNGase A released pools chromatographed with the Kinetex XB-C18 column are shown: (A,B) one isomer each of glycans with m/z 1357 and m/z 1503, (C,D) two differently-eluting isomers with m/z 1560, (E) one isomer with m/z 1763, (F) the single isomer with m/z 2093 and (G) the single isomer with m/z 1649. The typical fragments of m/z 369, 531, 572 and 734 are diagnostic for Hex0-1HexNAc1-2PC1.
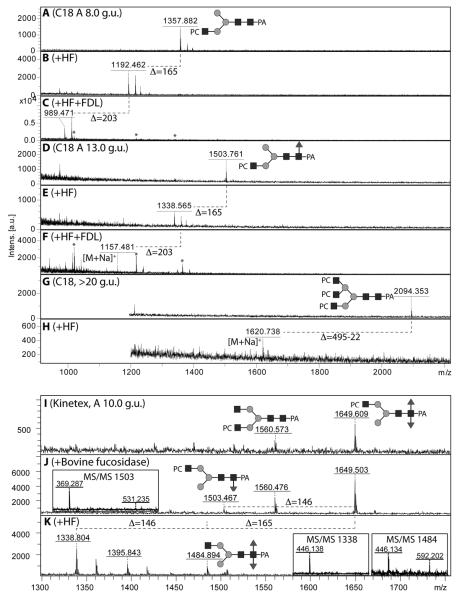
Figure 9. Chemical and enzymatic treatments of pyridylamino-labelled phosphorycholine-modified N-glycans.
Selected C18 fractions (A, D, G) were subject to hydrofluoric acid treatment in order to demonstrate the presence of phosphodiesters (loss of one phosphorylcholine is indicated by a loss of 165 Da; B, E, H); in order to determine on which arm the HexNAc1PC1 motif was present, two of these fractions were subsequently subject to treatment with the specific FDL hexosaminidase (C,F) to determine whether a GlcNAc was released from the α1,3-arm. Furthermore, a specific Kinetex-fractionated glycan containing a putative phosphorylcholine-modified difucosylated N-glycan (I, m/z 1649, for MS/MS see Figure 8G) was subject to fucosidase and hydrofluoric acid treatments (J,K); the latter resulted in full removal of the phosphorylcholine moiety and incomplete removal of an α1,3-fucose (see insets for relevant MS/MS of the digestion products); the co-eluting m/z 1560 glycan (Hex3HexNAc4PC1) is also sensitive to this treatment (product of m/z 1395 in panel K).
In the case of Hex3HexNAc3Fuc0-1PC1 structures, isomers were apparent. The later eluting structures were hypothesised to have the HexNAc1PC1 attached to the α1,6-mannose, the earlier on the α1,3-mannose. In order to determine this more specifically, the Apis mellifera FDL hexosaminidase was employed after hydrofluoric acid treatment. As predicted, the Hex3HexNAc3Fuc0-1PC1 (m/z 1357 and 1503) eluting at 8.0 and 13.0 g.u. each lost one HexNAc after consecutive hydrofluoric acid and FDL hexosaminidase treatment (Figure 9 A-C and D-F); on the other hand, the later-eluting isomer with m/z 1357 lost one hexose upon α1,2/3-mannosidase digestion (compare Figure 6C and D). An unusual structure (m/z 1649) was predicted to possess two fucoses in addition to one phosphorylcholine; hydrofluoric acid treatment overnight resulted in a serial loss of phosphorylcholine (m/z 1484) and then of fucose (m/z 1338). Whereas the MS/MS spectrum of the untreated glycan was dominated by the m/z 369 fragment indicative for phosphorylcholine (Figure 8G), the intermediate product (m/z 1484) displayed a fragment of m/z 592 indicative of difucosylation of the core, while the lowest mass product carried only one core fucose as shown by the m/z 446 fragment (Figure 9I-K). This is the first time we have detected a nematode glycan carrying both core α1,3-fucose and antennal phosphorylcholine, whereby the non-reducing terminal GlcNAc is predicted to be on the α1,6-arm, compatible with the substrate specificity of the nematode core α1,3-fucosyltransferase (FUT-1), which is unable to modify glycans with a GlcNAc on the α1,3-arm (27).
Glycans with two or three phosphorylcholine moieties (Hex3HexNAc4PC2 and Hex3HexNAc5Fuc0-1PC3; m/z 1726, 2094 and 2240) lost as expected 330 or 495 Da upon hydrofluoric acid treatment (see Figure 9H and Supplementary Figures 5 and 6), but the treated glycans were then relatively poorly detected as it appears that phosphorylcholine increases the ability of glycans to ionise. This effect is also obvious upon MS/MS as phosphorylcholine-containing fragments dominate these spectra, an effect only lost after hydrofluoric acid treatment (Supplementary Figure 6). Multiple modifications with phosphorylcholine led to rather different elution properties on the three columns used (Figure 2): whereas the presence of three phosphorylcholine moieties caused very high retention on the classical C18 material (>20 g.u.), rather low retention was observed (in the range 4-7 g.u.) for such glycans on the two other columns (RP-amide and Kinetex XB-C18) based on fused cores.
Discussion
Comparisons of nematode N-glycomes
In terms of nematodes, the range of N-linked oligosaccharides in Pristionchus pacificus is of middling complexity with about seventy detectable structures (Table 1), whereas the wild-type strain of the more familiar Caenorhabditis elegans probably expresses twice this number. The major types of N-glycans are the same in both organisms, but highly-fucosylated as well as methylated structures are rather absent from P. pacificus and only trace amounts of trifucosylated N-glycans or glycans with galactosylated core fucose were detected; indeed, as compared to analysing the C. elegans N-glycome with RP-amide-HPLC and MALDI-TOF (unpublished data), only two structures with galactosylated core α1,6-fucose and three with three fucose residues were found in P. pacificus as opposed to at least ten and eight, respectively, in C. elegans. Nevertheless, there are a few structures in P. pacificus previously not reported from any other species; a particularly unusual example is (Manβ1,4GlcNAcβ1,4(Fucα1,6)(MeFucα1,3)GlcNAc) which carries a methyl group on a core fucose residue.
Considering that there are phosphorylcholine-modified as well as trifucosylated N-glycans, the N-glycome of P. pacificus also contains features present in some parasitic nematodes such as Haemonchus contortus, Oesophagostomum dentatum and Ascaris suum (17,18,49). LacdiNAc motifs with or without phosphorylcholine are known from Trichinella spiralis and Dirofilaria immitis (50,51) and multiantennary glycans have been described for Acanthocheilonema viteae and Onchocerca volvulus and T. spiralis (47,50). Missing from C. elegans and P. pacificus are apparently the Lewis-type modifications with α1,3-fucose on the antennae, which are a feature of T. spiralis and Dictyocaulus viviparus (52,53). These similarities and differences are not necessarily due to the evolutionary distance (C. elegans, P. pacificus, H. contortus, O. dendatum and D. viviparus are members of the Rhabditina clade, D. immitis and A. suum are members of the Spirurina and T. spiralis belongs to the Dorylaimia), but may be due to the distinctive lifestyles (parasitic/non-parasitic). Indeed, ‘copying’ host-type or host-like glycomotifs may be a factor contributing to parasite survival and immunomodulation of host immune systems (54).
Variations in the glycosylation between different nematode species have presumably a genetic origin in terms of either the presence or absence of certain genes or different expression levels (whether temporal, cell-specific or total amount of transcript). Although the P. pacificus genome has been sequenced, its annotation is incomplete; thus, the total number of transcriptionally-active glycosyltransferase genes cannot be easily surmised. Nevertheless, it can be stated that there are a number of potential α1,3-fucosyltransferase homologues, including an obvious FUT-1 orthologue, and that there are some predicted reading frames of relatively low homology to the fucose-modifying galactosyltransferase GALT-1. None of the encoded proteins have been tested regarding their enzymatic activity. However, although P. pacificus is the second non-parasitic nematode to have its N-glycome analysed and it is a member of the same clade, it is not a particularly close relative of C. elegans; it is estimated that they separated as species some 280–430 million years ago (13).
RP-HPLC of unusual N-glycans from nematodes
The purpose of our study was primarily to examine the N-glycome of P. pacificus, which is a second model nematode, although it not so extensively used in biology as C. elegans. However, a secondary purpose was to compare different RP-HPLC columns for fractionation of nematode pyridylaminated N-glycans. Therefore, a direct comparison was made between the alkyl amide (Ascentis Express) and two different octadecyl RP columns (Agilent Hypersil C18 and Kinetex XB-C18) while using the same buffers for the mobile phase; C18 silica is a well-established material for N-glycan analysis with first reports on its use with pyridylaminated N-glycans some thirty years ago (55), whereas Ascentis Express RP-amide and Kinetex XB-C18 columns were first on the market in 2008 and 2010 respectively. Both the latter can either be run at a lower flow rates, as in the present study, or at higher pressures, on a UPLC system, as compared to standard C18 columns.
Subtle variations were observed with the similar trends in terms of elution of fucosylated and oligomannosidic glycans (order of retention and relative retention in terms of glucose units), although the fucosylated glycans do not elute as late on the RP-amide column The Kinetex XB-C18 column can be concluded to have the finest resolution as expected for a fused core column with, e.g., Man3GlcNAc2 and Man5GlcNAc2 being resolved into two different fractions; also, the relative order of retention times were similar but still different as compared to the classical C18 column (with the core α1,6-fucosylated glycans eluting rather late in terms of g.u.) and the difucosylated structure Man3GlcNAc2Fuc2 co-eluted with the Man5GlcNAc2 glycan on the Kinetex column, while eluting either slightly earlier or slightly later on the other two. Under the conditions used, the longest run times in terms of actual minutes and of glucose units were for the Kinetex column; however, its use resulted in the detection of the highest number of glycans (Table 1). Whereas we have previously used the RP-amide column for N-glycan analysis (31), we are not aware of any glycomic applications to date for the Kinetex column.
Based on studies from our and other laboratories over the years (37,41,43,45,56-58), it is clear that RP-HPLC, as necessary also combined with size-based fractionation for 2D-HPLC, is an important tool in glycomic analyses due to the ability to distinguish certain types of isomers or other structural features. Regardless of the exact nature of the column used, factors causing earlier elution of pyridylamino-labelled N-glycans include a longer A branch (α1,3-antenna) and proximal core α1,3-fucosylation; later elution is associated with elongation of the α1,6-mannose (including the retention of an α1,2-mannose on the B branch) or with core α1,6-fucosylation. Thus, one can contrast the behaviour of isomer pairs such as 0M with M0, MGn with GnM, Man8B with Man8A or MMF3 with MMF6 (see Table 1 for structural nomenclature). Furthermore, the addition to a residue can result in a predictable shift as can be observed when comparing MM with MMF6 or MGn with GnGn (in both cases, the increased retention follows the expectation). These general rules, which have been verified in many studies, can now be supplemented by other effects, such as distal core α1,3-fucosylation, galactosylation of the core α1,6-fucose or methylation increasing retention. Often similar effects are observed with porous graphitised carbon, but this is generally used in combination with ESI-MS (59).
The most obvious difference between the three columns was in terms of the elution of phosphorylcholine-modified glycans: these displayed late retention times on a classical C18 column, but eluted earlier on the ‘fused core’ RP-amide and Kinetex XB-C18 materials. This is advantageous in that the gradients do not have to be extended to high methanol concentrations; thus, loss of such glycans due to premature termination of the gradient is avoided. Indeed that the presence of more phosphorylcholine residues results in even earlier retention on the RP amide column (the triply-modified glycans elute between 4-5 g.u., whereas the singly-modified Hex3HexNAc3PC1 elutes around 6 g.u.). This property is particularly important as many of these glycans are also core α1,6-fucosylated which leads to even later elution on standard C18. The reason for this behaviour may be the properties of the particles used in the different materials: possibly phosphorylcholine-modified glycans are trapped within the typical C18 beads and so elute slowly, but these structures cannot penetrate the fused core beads.
Conclusion
By the use of HPLC in conjunction with MALDI-TOF MS, we can propose the structure of some seventy N-glycans from Pristionchus pacificus; it is certain that definition of all these structures would not have been possible if MS/MS had been performed from a non-fractionated ‘whole glycome’. Such a typical glycomic analysis, even if based on permethylation or perdeuteromethylation, would almost certainly hide the presence of isomers, especially those present in low quantities. Thereby, adequate HPLC fractionation is a key part of our glycomic analytical strategy. Certainly, the use of ‘new generation’ RP-HPLC columns with fused core technologies (60) offers further possibilities to resolve complex mixtures of glycans from whole organisms and so aid the discovery of further unusual oligosaccharide modifications in non-mammalian systems.
Supplementary Material
Acknowledgements
This work was funded in part by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF) [grants P21946 to K.P. and P23922 to I.B.H.W.]. The Pristionchus and E. coli strains used were provided by the Caenorhabditis Genetics Centre (CGC), which is funded by the NIH Office of Research Infrastructure Programmes (P40 OD010440). We also thank Dr. Markus Künzler (ETH Zürich) for the independent sample of Pristionchus and Dr. Martin Dragosits for preparation of the recombinant Aspergillus β-galactosidase and Apis FDL β-hexosaminidase.
References
- 1.Schiller B, Hykollari A, Yan S, Paschinger K, Wilson IBH. Complicated N-linked glycans in simple organisms. Biol. Chem. (Hoppe Seyler) 2012;393:661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, Yang L, Zeng N, Xu X, Xiong Z, Chen F, Wu X, Zhang G, Fang X, Kang Y, Anderson GA, Harris TW, Campbell BE, Vlaminck J, Wang T, Cantacessi C, Schwarz EM, Ranganathan S, Geldhof P, Nejsum P, Sternberg PW, Yang H, Wang J, Wang J, Gasser RB. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- 3.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CK, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrin-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, Holroyd N, Bartley DJ, Beasley H, Britton C, Curran D, Devaney E, Gilabert A, Hunt M, Jackson F, Johnston SL, Kryukov I, Li K, Morrison AA, Reid AJ, Sargison N, Saunders GI, Wasmuth JD, Wolstenholme A, Berriman M, Gilleard JS, Cotton JA. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome biology. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, Tyagi R, Heizer E, Zhang X, Bhonagiri-Palsikar V, Minx P, Warren WC, Wang Q, Zhan B, Hotez PJ, Sternberg PW, Dougall A, Gaze ST, Mulvenna J, Sotillo J, Ranganathan S, Rabelo EM, Wilson RK, Felgner PL, Bethony J, Hawdon JM, Gasser RB, Loukas A, Mitreva M. Genome of the human hookworm Necator americanus. Nature genetics. 2014;46:261–269. doi: 10.1038/ng.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, Yang SP, Warren WC, Fulton RS, Bhonagiri V, Zhang X, Hallsworth-Pepin K, Clifton SW, McCarter JP, Appleton J, Mardis ER, Wilson RK. The draft genome of the parasitic nematode Trichinella spiralis. Nature genetics. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jex AR, Nejsum P, Schwarz EM, Hu L, Young ND, Hall RS, Korhonen PK, Liao S, Thamsborg S, Xia J, Xu P, Wang S, Scheerlinck JP, Hofmann A, Sternberg PW, Wang J, Gasser RB. Genome and transcriptome of the porcine whipworm Trichuris suis. Nature genetics. 2014;46:701–706. doi: 10.1038/ng.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, Takanashi T, Tsai IJ, Assefa SA, Cock PJ, Otto TD, Hunt M, Reid AJ, Sanchez-Flores A, Tsuchihara K, Yokoi T, Larsson MC, Miwa J, Maule AG, Sahashi N, Jones JT, Berriman M. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS pathogens. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- 10.The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 11.Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, Wold BJ, Sternberg PW. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome research. 2010;20:1740–1747. doi: 10.1101/gr.111021.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan J, Dillman AR, Macchietto MG, Heikkinen L, Lakso M, Fracchia KM, Antoshechkin I, Mortazavi A, Wong G, Sternberg PW. The draft genome and transcriptome of Panagrellus redivivus are shaped by the harsh demands of a free-living lifestyle. Genetics. 2013;193:1279–1295. doi: 10.1534/genetics.112.148809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, Mitreva M, Roeseler W, Tian H, Witte H, Yang SP, Wilson RK, Sommer RJ. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nature genetics. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haslam SM, Dell A. Hallmarks of Caenorhabditis elegans N-glycosylation: complexity and controversy. Biochimie. 2003;85:25–32. doi: 10.1016/s0300-9084(03)00041-5. [DOI] [PubMed] [Google Scholar]
- 15.Paschinger K, Gutternigg M, Rendić D, Wilson IBH. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydrate research. 2008;343:2041–2049. doi: 10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Hanneman AJ, Rosa JC, Ashline D, Reinhold V. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- 17.Yan S, Bleuler-Martinez S, Plaza Gutierrez DF, Künzler M, Aebi M, Joachim A, Razzazi-Fazeli E, Jantsch V, Geyer R, Wilson IBH, Paschinger K. Galactosylated fucose epitopes in nematodes: increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J Biol Chem. 2012;287:28276–28290. doi: 10.1074/jbc.M112.353128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haslam SM, Coles GC, Munn EA, Smith TS, Smith HF, Morris HR, Dell A. Haemonchus contortus glycoproteins contain N-linked oligosaccharides with novel highly fucosylated core structures. J. Biol. Chem. 1996;271:30561–30570. doi: 10.1074/jbc.271.48.30561. [DOI] [PubMed] [Google Scholar]
- 19.Yan S, Serna S, Reichardt NC, Paschinger K, Wilson IBH. Array-assisted Characterization of a Fucosyltransferase Required for the Biosynthesis of Complex Core Modifications of Nematode N-Glycans. J Biol Chem. 2013;288:21015–21028. doi: 10.1074/jbc.M113.479147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang YT, Shien JH, Tan DH, Shieh MK, Liu CC, Chen YS, Chang PC. Identification of the lic1ABCD operon that controls the phase-variable expression of phosphorylcholine on lipopolysaccharide from Avibacterium paragallinarum. Avian pathology: journal of the W.V.P.A. 2013;42:72–78. doi: 10.1080/03079457.2012.760840. [DOI] [PubMed] [Google Scholar]
- 21.Unkefer CJ, Jackson C, Gander JE. The 5-O-β-D-galactofuranosyl-containing glycopeptide from Penicillium charlesii. Identification of phosphocholine attached to C-6 of mannopyranosyl residues of the mannan region. J Biol Chem. 1982;257:2491–2497. [PubMed] [Google Scholar]
- 22.Paschinger K, Gonzalez-Sapienza GG, Wilson IBH. Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int J Parasitol. 2012;42:279–285. doi: 10.1016/j.ijpara.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harnett W, Harnett MM. Modulation of the host immune system by phosphorylcholine-containing glycoproteins secreted by parasitic filarial nematodes. Biochimica et biophysica acta. 2001;1539:7–15. doi: 10.1016/s0167-4889(01)00101-x. [DOI] [PubMed] [Google Scholar]
- 24.Volanakis JE. Human C-reactive protein: expression, structure, and function. Molecular immunology. 2001;38:189–197. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 25.Butschi A, Titz A, Wälti M, Olieric V, Paschinger K, Nöbauer K, Guo X, Seeberger PH, Wilson IBH, Aebi M, Hengartner M, Künzler M. Caenorhabditis elegans N-glycan core β-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS pathogens. 2010;6:e1000717. doi: 10.1371/journal.ppat.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert M, Bleuler-Martinez S, Butschi A, Walti MA, Egloff P, Stutz K, Yan S, Wilson IBH, Hengartner MO, Aebi M, Allain FH, Kunzler M. Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System. PLoS pathogens. 2012;8:e1002706. doi: 10.1371/journal.ppat.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paschinger K, Rendić D, Lochnit G, Jantsch V, Wilson IBH. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J. Biol. Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 28.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IBH. Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology. 2005;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 29.Titz A, Butschi A, Henrissat B, Fan YY, Hennet T, Razzazi-Fazeli E, Hengartner MO, Wilson IBH, Künzler M, Aebi M. Molecular basis for galactosylation of core fucose residues in invertebrates: Identification of Caenorhabditis elegans N-glycan core a1,6-fucoside b1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J Biol Chem. 2009;284:36223–36233. doi: 10.1074/jbc.M109.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Tan J, Reinhold VN, Spence AM, Schachter H. UDP-N-acetylglucosamine:α-3-D-mannoside β-1,2-N-acetylglucosaminyltransferase I and UDP-N-acetylglucosamine:α-6-D-mannoside β-1,2-N-acetylglucosaminyltransferase II in Caenorhabditis elegans. Biochimica et biophysica acta. 2002;1573:271–279. doi: 10.1016/s0304-4165(02)00393-8. [DOI] [PubMed] [Google Scholar]
- 31.Kurz S, Jin C, Hykollari A, Gregorich D, Giomarelli B, Vasta GR, Wilson IBH, Paschinger K. Haemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulphated and blood group A-modified N-glycans. J Biol Chem. 2013;288:24410–24428. doi: 10.1074/jbc.M113.478933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hykollari A, Balog CI, Rendic D, Braulke T, Wilson IBH, Paschinger K. Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. Journal of proteome research. 2013;12:1173–1187. doi: 10.1021/pr300806b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong RL, Sommer RJ. Pristionchus pacificus: a well-rounded nematode. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:651–659. doi: 10.1002/bies.20404. [DOI] [PubMed] [Google Scholar]
- 34.Sommer RJ, McGaughran A. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Molecular ecology. 2013;22:2380–2393. doi: 10.1111/mec.12286. [DOI] [PubMed] [Google Scholar]
- 35.Herrmann M, Mayer WE, Hong RL, Kienle S, Minasaki R, Sommer RJ. The nematode Pristionchus pacificus (Nematoda: Diplogastridae) is associated with the oriental beetle Exomala orientalis (Coleoptera: Scarabaeidae) in Japan. Zoological science. 2007;24:883–889. doi: 10.2108/zsj.24.883. [DOI] [PubMed] [Google Scholar]
- 36.Hase S, Ibuki T, Ikenaka T. Reexamination of the pyridylamination used for fluorescence labelling of oligosaccharides and its application to glycoproteins. J Biochem (Tokyo) 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- 37.Paschinger K, Hykollari A, Razzazi-Fazeli E, Greenwell P, Leitsch D, Walochnik J, Wilson IBH. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 2012;22:300–313. doi: 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dragosits M, Pflugl S, Kurz S, Razzazi-Fazeli E, Wilson IB, Rendic D. Recombinant Aspergillus beta-galactosidases as a robust glycomic and biotechnological tool. Applied microbiology and biotechnology. 2014;98:3553–3567. doi: 10.1007/s00253-013-5192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dragosits M, Yan S, Razzazi-Fazeli E, Wilson IBH, Rendić D. Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology. 2015 doi: 10.1093/glycob/cwu132. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tretter V, Altmann F, März L. Peptide-N4-(N-acetyl-b-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached a1®3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 41.Tomiya N, Lee YC, Yoshida T, Wada Y, Awaya J, Kurono M, Takahashi N. Calculated two-dimensional sugar map of pyridylaminated oligosaccharides: Elucidation of the jack bean α-mannosidase digestion pathway of Man9GlcNAc2. Analytical biochemistry. 1991;193:90–100. doi: 10.1016/0003-2697(91)90047-w. [DOI] [PubMed] [Google Scholar]
- 42.Schachter H. Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem Cell Biol. 1986;64:163–181. doi: 10.1139/o86-026. [DOI] [PubMed] [Google Scholar]
- 43.Tomiya N, Awaya J, Kurono M, Endo S, Arata Y, Takahashi N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Analytical biochemistry. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- 44.Wuhrer M, Deelder AM, van der Burgt YE. Mass spectrometric glycan rearrangements. Mass spectrometry reviews. 2011;30:664–680. doi: 10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- 45.Fabini G, Freilinger A, Altmann F, Wilson IBH. Identification of core α1,3-fucosylated glycans and the requisite fucosyltransferase in Drosophila melanogaster. Potential basis of the neural anti-horseradish peroxidase epitope. J. Biol. Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- 46.Warren CE, Krizius A, Roy PJ, Culotti JG, Dennis JW. The C. elegans gene, gly-2, can rescue the N-acetylglucosaminyltransferase V mutation of Lec4 cells. J. Biol. Chem. 2002;277:22829–22838. doi: 10.1074/jbc.M201390200. [DOI] [PubMed] [Google Scholar]
- 47.Haslam SM, Houston KM, Harnett W, Reason AJ, Morris HR, Dell A. Structural studies of N-glycans of filarial parasites. Conservation of phosphorylcholine-substituted glycans among species and discovery of novel chito-oligomers. J Biol Chem. 1999;274:20953–20960. doi: 10.1074/jbc.274.30.20953. [DOI] [PubMed] [Google Scholar]
- 48.Paschinger K, Hackl M, Gutternigg M, Kretschmer-Lubich D, Stemmer U, Jantsch V, Lochnit G, Wilson IBH. A deletion in the Golgi α-mannosidase II gene of Caenorhabditis elegans results in unexpected non-wild type N-glycan structures. J. Biol. Chem. 2006;281:28265–28277. doi: 10.1074/jbc.M602878200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pöltl G, Ahrazem O, Paschinger K, Ibañez MD, Salcedo G, Wilson IBH. Molecular and immunological characterization of the glycosylated orange allergen Cit s 1. Glycobiology. 2007;17:220–230. doi: 10.1093/glycob/cwl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morelle W, Haslam SM, Olivier V, Appleton JA, Morris HR, Dell A. Phosphorylcholine-containing N-glycans of Trichinella spiralis: identification of multiantennary lacdiNAc structures. Glycobiology. 2000;10:941–950. doi: 10.1093/glycob/10.9.941. [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Cummings RD, McCall JW. Characterization of the N-linked oligosaccharides in glycoproteins synthesized by microfilariae of Dirofilaria immitis. The Journal of parasitology. 1993;79:815–828. [PubMed] [Google Scholar]
- 52.Haslam SM, Coles GC, Morris HR, Dell A. Structural characterisation of the N-glycans of Dictyocaulus viviparus: discovery of the Lewisx structure in a nematode. Glycobiology. 2000;10:223–229. doi: 10.1093/glycob/10.2.223. [DOI] [PubMed] [Google Scholar]
- 53.Reason AJ, Ellis LA, Appleton JA, Wisnewski N, Grieve RB, McNeil M, Wassom DL, Morris HR, Dell A. Novel tyvelose-containing tri- and tetra-antennary N-glycans in the immunodominant antigens of the intracellular parasite Trichinella spiralis. Glycobiology. 1994;4:593–604. doi: 10.1093/glycob/4.5.593. [DOI] [PubMed] [Google Scholar]
- 54.van Die I, Cummings RD. Glycan gimmickry by parasitic helminths: a strategy for modulating the host immune response? Glycobiology. 2010;20:2–12. doi: 10.1093/glycob/cwp140. [DOI] [PubMed] [Google Scholar]
- 55.Hase S, Kikuchi N, Ikenaka T, Inoue K. Structures of sugar chains of the third component of human complement. Journal of biochemistry. 1985;98:863–874. doi: 10.1093/oxfordjournals.jbchem.a135366. [DOI] [PubMed] [Google Scholar]
- 56.Gutternigg M, Kretschmer-Lubich D, Paschinger K, Rendić D, Hader J, Geier P, Ranftl R, Jantsch V, Lochnit G, Wilson IBH. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants and insects. J. Biol. Chem. 2007;282:27825–27840. doi: 10.1074/jbc.M704235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomiya N, Kurono M, Ishihara H, Tejima S, Endo S, Arata Y, Takahashi N. Structural analysis of N-linked oligosaccharides by a combination of glycopeptidase, exoglycosidases, and high-performance liquid chromatography. Analytical biochemistry. 1987;163:489–499. doi: 10.1016/0003-2697(87)90253-3. [DOI] [PubMed] [Google Scholar]
- 58.Hase S. Pyridylamination as a means of analyzing complex sugar chains. Proceedings of the Japan Academy. Series B, Physical and biological sciences. 2010;86:378–390. doi: 10.2183/pjab.86.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruhaak LR, Deelder AM, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Analytical and bioanalytical chemistry. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 60.Ali I, Al-Othman ZA, Nagae N, Gaitonde VD, Dutta KK. Recent trends in ultra-fast HPLC: new generation superficially porous silica columns. Journal of separation science. 2012;35:3235–3249. doi: 10.1002/jssc.201200454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.