Abstract
DNA methylation is an epigenetic mechanism that plays a key role in regulating gene expression and other functions. Although this modification is seen in different sequence contexts, the most frequently detected DNA methylation in mammals involves cytosine-guanine dinucleotides. Pathological alterations in DNA methylation patterns are described in a variety of human diseases, including cancer. Unlike genetic changes, DNA methylation is heavily influenced by subtle modifications in the cellular microenvironment. In all cancers, aberrant DNA methylation is involved in the alteration of a large number of oncological pathways with relevant theranostic utility. Several technologies for DNA methylation mapping were recently developed and successfully applied in cancer studies. The scope of these technologies varies from assessing a single cytosine-guanine locus to genome-wide distribution of DNA methylation. Here, we review the strengths and weaknesses of these approaches in the context of clinical utility for the molecular diagnosis of human cancers.
Keywords: DNA methylation, cancer diagnosis, epigenomics, sequencing, microarray
DNA methylation and gene expression: A complex relationship that depends on the context
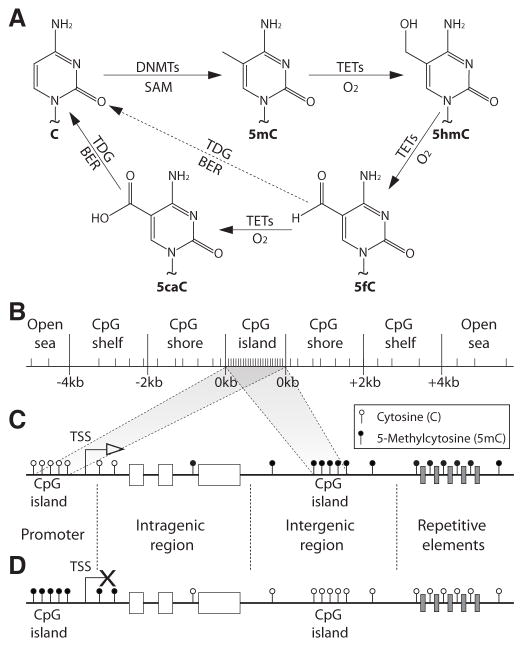
DNA methylation commonly refers to the covalent addition of a methyl (−CH3) group from the s-adenosylmethionine (SAM) to the fifth carbon of the cytosine base (5mC) which is catalyzed by DNA methyltransferases (DNMTs) enzymes (Figure 1A). It is extensively demonstrated that DNA methylation plays a key role in chromosomal stability, gene expression, genome imprinting, and transcriptional silencing of foreign DNA fragments [1–3]. The most abundant DNA methylation in mammalian genomes occurs in cytosines that belong to CpG dinucleotides [4]. Nevertheless, non-CpG DNA methylation mainly occurring in CpHpG trinucleotides (where H can be adenine [A], thymine [T], or C) was also reported in healthy and pathological processes [5]. Interestingly, the human genome is particularly poor in CpG dinucleotides. While at least 132 million CpG dinucleotides are expected (the human genome is composed of ~21% C and ~21% G nucleotides; therefore, the probability of a CpG dinucleotide to occurs is 4.41%), only ~28 million (0.93%) are actually observed, of which 60–80% are generally methylated [4,6]. In fact, DNA methylation is considered a mutagenic event. Deamination of 5mC generates T, which in some cases are identified and removed by mismatch repair mechanisms. But, in other cases, these are considered wild type information and maintained in the mammalian genome. In consequence, the content of TpG dinucleotides in the human genome is higher than expected and presents a negative correlation with CpG abundance [7]. In addition to 5mC, other chemical modifications of C residues were recently identified. These modifications include 5-hydroxymethyl-cytocine (5hmC), 5-formyl-cytosine (5fC), and 5-carboxyl-cytosine (5caC) [8]. 5hmC, 5fC, and 5caC are generated by the successive oxidation of 5mC catalyzed by the ten-eleven translocation methylcytosine dioxygenases (TETs) (Figure 1A) [8]. Finally, 5fC and 5caC can be excised and repaired by thymine-DNA glycosylase (TDG) and base excision repair (BER) systems to generate unmethylated Cs (Figure 1A) [8].
Figure 1. Cytosine modifications and distribution of 5-methylcytosine (5mC in the human genome.
A- Chemical structures of the unmodified cytosine (C), 5mC and its oxidation products 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). First, a methyl group is transferred from the s-adenosylmethionine (SAM) to the fifth carbon of cytosine by DNA methyltransferase enzymes (DNMTs). Second, the oxidation of 5mC is carried out by the ten-eleven translocation methylcytosine dioxygenases (TETs) to 5hmC, 5fC, and 5caC. Finally, 5fC and 5caC can be excised and repaired by thymine-DNA glycosylase (TDG) and base excision repair (BER) systems to generate unmethylated Cs. B- Representation of the CpG context. C–D. Distribution of 5mC in normal (C) and cancer (D) cells. CpG: cytosine – guanine
The levels of gene expression are influenced by a multitude of factors, including DNA methylation. Initially, 5mC was proposed to interfere with transcription factor binding; however, its influence appears to involve more complex and indirect mechanisms in which methyl-cytosine binding domain (MBD) proteins recruit multi-protein complexes that ultimately induce chromatin conformation changes [9]. In this model, the strength of the repression would directly depend on the CpG concentration. Far from having a definitive answer, the relationship between DNA methylation and gene expression remains a topic of intense debate. Recent advancements in high-throughput DNA methylation mapping provided new insights about the complexity of this regulatory mechanism.
The CpG context
The distribution of CpG dinucleotides defines regions of importance for the epigenetic regulation of gene transcription. Genomic regions of at least 200 base pairs (bp), in which the G and C content accounts for at least 50% of the nucleotides with an observed/expected CpG ratio>0.6, are denominated ‘CpG islands’ [10]. Because of the previously mentioned spontaneous or enzymatically-mediated deamination of 5mCs to Ts, from an evolutionary point of view, it has been hypothesized that the conservation of CpG islands is due to absent or infrequent DNA methylation on CpG sites located at these regions [10]. In fact, CpG islands are usually unmethylated in normal cells, and its methylation only occurs in specific and well-controlled physiological situations such as development, differentiation, memory acquisition, genomic imprinting, and X chromosome inactivation [3,4,11–13]. DNA methylation at CpG islands, especially when overlapping gene promoters, was traditionally considered synonymous with gene expression silencing. However, in light of current studies, this statement may need to be reevaluated. In addition to CpG islands, the CpG context defines regions of lower CpG density located around the islands called ‘CpG shores’ (up to 2 kb away from the CpG island) and ‘CpG shelves’ (2 to 4 kb away from the CpG island), and regions distant from CpG islands (>4 kb away from a CpG island), named by some authors as ‘open sea’ (Figure 1B) [14,15]. In spite of a lowered CpG density, these regions have demonstrated to be functionally important for gene expression regulation. For example, CpG shores were found to be associated with cancer-, tissue, and reprogramming-specific differentially methylated regions (C-DMRs, T-DMRs, and R-DMRs) with mechanistic relevance for the expression of associated genes [15,16]. Furthermore, in cell differentiation, the methylation of CpG shores is more dynamic than the methylation of CpG islands, indicating a key role of CpG shore methylation on cell fate determination [16]. In addition, differential methylation of CpG shelves as well as open sea regions have been associated with hepatocellular carcinoma [17], and metastatic melanomas [18].
Gene structure and regulatory elements
The influence on gene expression levels also depends on the location of DNA methylation with regard to gene structures, specifically at promoter, intragenic, and intergenic CpG sites [19].
More than half of human genes contain CpG islands located upstream of their transcription start sites (TSSs). These regions, denominated promoter elements, were traditionally considered the regions where DNA methylation has the strongest influence on gene expression [20]. However, recent discoveries describe a more complex picture. Based on CpG ratio, CpG content, and length of CpG-enriched region, three classes of gene promoters have been recognized [21]. High-, intermediate- and low-CpG density promoters (HCPs, ICPs, and LCPs) represent strong, weak, and non-CpG island promoters, respectively. HCPs are usually unmethylated, even when the transcription of the associated gene is inactive. LCPs are usually methylated without having a blocking effect on the promoter activity. Yet, substantial fluctuation occurs in the DNA methylation level on LCPs according to the developmental stage of the cells. Conversely, ICPs are frequently methylated with a repressive effect on gene expression [21]. Interestingly, these ICP promoters were suggested to be targets of de-novo DNA methylation by DNMT3A and DNMT3B with a role in the microenvironment-induced fine-tuning of gene expression variations [22].
Novel evidence indicates that intragenic DNA methylation also influences gene transcription. For instance, methylation of CpG sites located in the 5′ untranslated region (5′UTR) inhibits gene transcription initiation [23]. Conversely, methylation of non-CpG sites located in the gene body can stabilize transcription elongation and influence the selection of alternative splicing sites [5,19]. In addition, methylation of 3′UTR regions is significantly associated with an increased gene expression rate [24]. Furthermore, 5hmC found in gene bodies is positively correlated with gene expression levels, and in contrast to 5mC, is also enriched in active enhancers and CpG-rich promoter regions but it is largely absent from intergenic regions [25].
DNA methylation of intergenic genomic intervals can affect the accessibility to cis- and trans-regulatory elements, such as enhancers and silencers, greatly influencing the cell-type specific gene expression programs [26]. For example, differences in DNA methylation levels between neuronal and glial cells are concentrated in enhancer elements which contribute to cell type-specific gene expression programs [27]. Additionally, methylation of repetitive sequences such as LINE-1 and Alu plays an important role in chromosomal stability, mitotic segregation, and suppression of transposable element expression [28,29]. As a general finding, in normal cells, promoter CpG islands present low or null DNA methylation levels, while intragenic and intergenic CpG sites are usually methylated (Figure 1C).
In summary, the relationship between DNA methylation and gene expression is highly complex, and in light of current information, the traditional concept of DNA methylation silencing gene expression by affecting CpG island promoters needs to be carefully reevaluated and expanded.
Role of DNA methylation in cancer: A misbalance between methylation and demethylation
As previously reviewed [30–33], aberrant DNA methylation interacts with genomic and other epigenomic alterations, such as histone modification and nucleosome positioning during tumor initiation and progression. The DNA methylation landscape of normal cells is a well-controlled balance between methylation and demethylation [33]. In this regard, we consider aberrant DNA methylation as deficient, excessive or ectopic methylation/demethylation. Aberrant DNA methylation is observed in both sporadic and inherited cancers; however, specific patterns can be identified in each case [34]. Even though several tumor type-specific DNA methylation alterations have been reported, an epigenetic hallmark for all human cancers appears to be global DNA hypomethylation and a loci-specific DNA hypermethylation [31]. Yet, the timing and the pattern of aberrantly methylated regions vary in different stages and cancer types [35]. Unlike the distribution of DNA methylation in normal cells, cancer cells present increased DNA methylation in CpG-rich regions and decreased DNA methylation in CpG-poor regions (Figure 1D). Traditionally, gene promoter CpG islands hypermethylation has been largely studied in cancer, however, new studies show that aberrant methylation in regions other than gene promoter CpG islands may potentially be novel theranostic markers [36].
In addition to gene-specific aberrant methylation, the characterization of tumor type- and stage-specific genome-wide DNA methylation patterns offers an opportunity to identify unexplored molecular biomarkers. Currently, the multi institutional cooperation effort with The Cancer Genome Atlas (TCGA) aims to unravel novel and functional DNA methylation alterations in a wide variety of human cancers [37]. TCGA has already analyzed the genome-wide DNA methylation patterns of >10,000 human specimens from 33 different tumor types and paired normal controls (https://tcga-data.nci.nih.gov/tcga/). This unprecedented large scale genomic, transcriptomic, epigenomic, and proteomic data, in addition to other independent laboratory efforts, represents a milestone in cancer research and a revolution in the translation of the ‘-omic’ era into clinical practices.
Clinical translation of DNA methylation research
The list of clinically-relevant DNA methylation alterations in human tumors is extensive. Examples about the role of DNA methylation markers associated with diagnostic, prognostic, and drug response for different cancer-types have been reviewed [31,32,38,39]. Here, we discuss the most relevant advancements on the clinical utility of DNA methylation mapping of specific human cancers.
Glioblastoma
Perhaps one of the most significant clinical translations of DNA methylation research in solid tumors has been studied in patients with glioblastoma (GBM). These malignant gliomas are the most common and deadly type of primary brain tumors. Nevertheless, survival of these patients has improved from 10 to 14 months after diagnosis in the last 5 years due in part to the identification of patients who benefit from specific treatment strategies [40]. For example, it is now well recognized that O-6-methylguanine-DNA methyltransferase (MGMT) silencing by DNA methylation predicts a better tumor response to the combination treatment of temozolomide with radiation [41]. In addition to its predictive value, MGMT methylation is associated with better overall survival, even in absence of temozolomide treatment [42]. This observation was recently confirmed by a randomized phase III clinical trial in which GBMs with MGMT methylation improved overall and progression free survival [43]. GBMs were one of the first tumor types explored by TCGA. With >500 surgically-resected untreated primary tumors, integrative analysis of TCGA data identified four major molecular subtypes of GBM [44]. One of those subtypes (pro-neural) contains GBMs with frequent mutations in Isocitrate Dehydrogenase 1 (NADP+), Soluble (IDH1) gene [44]. IDH1 mutations are associated with a Glioma CpG Island Methylator Phenotype (G-CIMP), in which the global methylation level is significantly higher than that observed in non-G-CIMP GBMs [45,46]. Both G-CIMP and IDH1 gene mutations are favorable prognostic markers for GBM [47]. More recently, a similar molecular classification scheme was developed using the same DNA methylation mapping approach [48]. This study incorporated two hotspot mutations on the H3 Histone, Family A3 (H3F3A) gene to the molecular subtypes [48]. Interestingly, the authors identified a significant mutual exclusivity between mutations in H3F3A and IDH1 genes, defining a new GBM epigenetic subtype characterized by a global hypomethylation, which has been denominated Glioma CpG Hypomethylator Phenotype (G-CHOP) [48]. G-CHOP tumors present frequent H3F3AmutG34 mutations, specific tumor development location in the brain, and a higher incidence of pediatric GBM [48]. Although the prognostic and predictive significance of these new findings have yet to be fully understood and validated, the identification of these novel and well-characterized molecular subtypes may translate into the development of new targeted therapies. The discovery of G-CIMP and G-CHOP is an example of the importance of performing on a genome-wide scale, as opposed to gene-specific DNA methylation analyses.
Colorectal cancer
Colorectal cancer (CRC) is the third most commonly diagnosed and fatal type of cancer in men and women worldwide [49]. CRC progresses slowly through histologically well-defined stages [50]. The progression to invasive and metastatic stages is prompted by the successive accumulation of mutations in genes that greatly contribute to genome instability reflected by microsatellite instability (MSI) [51]. In parallel, this process is accompanied by the accumulation of a multitude of aberrantly methylated gene promoters [52]. In sporadic CRC, ~15% of the cases display MSI caused by MutL Homolog 1 (MLH1) gene aberrant methylation [50]. Interestingly, MLH1 methylation presents a significant mutual exclusivity with MGMT methylation [53]. Similar to GBM, ~40% of CRCs present transcriptional silencing of MGMT throughout aberrant DNA methylation [54]. However, unlike GBM, CRC patients with MGMT methylation have a lower response rate to temozolomide [55]. In a phase II clinical trial, patients with metastatic CRC and MGMT methylation showed favorable clinical response to dacarbizine [54]. Additionally, MGMT methylation is associated with G>A transitions, low MSI (L-MSI), and higher frequency of Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations [53,56]. CRCs can be also classified as high-CIMP (H-CIMP), low-CIMP (L-CIMP), and CIMP-negative [57,58]. Patients with H-CIMP present tumors with frequent proximal colon location, high-MSI (H-MSI), frequent v-raf murine sarcoma viral oncogene homolog B (BRAF) V600E mutation, and low frequency of MGMT methylation [59,60]. A recent study reveals that BRAF (V600E) mutation is responsible for H-CIMP and MLH1 aberrant methylation through the activation of the transcriptional repressor v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G (MAFG) [61]. This study represents an example of the link between ‘driver’ genetic alterations and genome-wide DNA methylation patterns. Additionally, our studies indicate that global hypomethylation, assessed by the methylation status of LINE-1, is an early event in the adenomatous change of colorectal cells and the level of LINE-1 hypomethylation increases with the stage of disease [62,63].
During the past decade, a large number of locus-specific DNA methylation alterations were associated with clinical features of CRC in tumor tissue as well as in blood and stool cell-free DNA [62,64–74]. More recently, a new set of genome-wide DNA methylation signatures were identified with potential prognostic value in CRC [75–78]. Integrative TCGA analyses also revealed new DNA methylation markers with clinical utility for aggressive CRC [79]. Advancements in DNA methylation mapping and analysis techniques will continue to expand discoveries for colorectal-specific biomarkers, and hopefully theranostic markers in order to improve disease management.
Breast cancer
Breast cancer (BC), the most frequent tumor diagnosed in women [49], is often considered a single disease, but it is in fact a compilation of different types of tumors affecting the mammary gland. Molecular stratification based on gene expression profiles has improved the treatment strategy of BC patients [80]. However, large clinical heterogeneity is still observed among patients with the same BC subtype. In this regard, DNA methylation analysis appears to have the potential to improve molecular classification. DNA methylation research in BC pathogenesis has grown exponentially since the early 90’s with the seminal description of differential DNA methylation of the estrogen receptor (ER) gene between BCs with and without ER expression [81]. For example, a recent study found that the methylation level of matrix metallopeptidase 7 (MMP7) gene can effectively distinguish basal-like breast cancers (BLBCs) from other types of triple negative breast cancers (TNBCs) [82]. Another recent study suggests that aberrant methylation of breast cancer 1, early onset (BRCA1) gene has prognostic utility in early-stage TNBCs [83]. Our laboratory and others have described a multitude of aberrantly methylated genes that contribute to BC progression [84–96]. In fact, using independent cohorts, our studies demonstrated a strong association of retinoic acid receptor, beta (RARB) gene methylation and macroscopic lymph node metastasis [86,96]. New studies have identified functional DNA methylation alterations outside of the CpG islands. For example, increased DNA methylation of the 5′CpG shore located on caveolin 1, caveolae protein, 22kDa (CAV1) gene correlates with increased caveolin-1 expression levels [97]. Interestingly, genome-wide DNA methylation landscapes of in-situ and invasive BC has been recently investigated [98–104]. To date, TCGA has profiled genome-wide methylomes for a large number of BC tumors and normal tissues (n=1,044). The partial analysis of these data (n=802) revealed that the methylation signature using 574 CpG sites identified five DNA methylation BC subgroups [105]. One subgroup presented a CIMP-like phenotype and contained a higher proportion of luminal B subtype and lobular histology tumors, while another subgroup presented a CHOP-like phenotype highly enriched in BLBCs [105,106]. Further classification of BCs through DNA methylation is needed because it may enable the development of more effective therapeutic regimens for better clinical outcomes.
Melanoma
Melanoma is by far considered the most deadly type of human skin cancer. With a steadily rising incidence during the last three decades, this tumor has become a major challenge for clinic management [49]. Melanoma pathogenesis is greatly influenced by genomic mutations induced by ultraviolet light [107]. A consequence of this mutational landscape is the frequent presence of driver mutations in genes such as BRAF, NRAS, and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT), among others [107]. However, once the primary melanoma is established, a rapid progression to regional and distant organ metastasis is frequently observed [108]. This process requires a highly adaptable gene expression program which, in addition to somatic mutations, reflects the influence of epigenomic alterations. During the past decade, the identification of DNA methylation alterations by our laboratory and others has evolved from targeted single loci to genome-wide approaches [109–120]. Such studies have produced a number of complementary prognostic biomarkers, and identified unexplored therapeutic targets [121]. This new approach identified that alterations in DNA methylation patterns appear early in the progression from benign nevus to primary melanoma [122], and are associated with the prognosis of AJCC Stage IIIC patients [123]. We identified that aberrant methylation of absent in melanoma 1 (AIM1) gene and LINE-1 in tumor or serum are related to melanoma progression and outcome [117]. Our integrative genome-wide DNA methylation, copy number variation, and gene expression analysis identified functional DNA methylation alterations in the progression from primary melanoma to brain metastasis [18,124–126]. In these studies, aberrant methylation of previously unexplored genes in melanoma, such as homeobox (HOX) and spliceosome factor genes were identified to be associated with progression to brain metastasis. Our laboratory has also identified a melanoma CIMP associated with the development of malignancy through the coordinated inactivation of tumor-related genes and methylation of multiple noncoding, methylated-in-tumor (MINT) loci [127]. In addition, similar to CRC, BRAF (V600E) mutation, a frequent alteration of cutaneous melanomas, also causes genome-wide DNA methylation changes in the melanoma epigenome [128].
TCGA network has analyzed >450 melanoma specimens, however, this cohort presents an important limitation. Unlike other solid tumors included in TCGA which are predominantly primary tumors, the melanoma cohort is mainly (~80%) composed of metastatic specimens. This is a consequence of the usually limited size and availability of primary melanomas to perform multiplatform assays. To date, there are no reports summarizing the findings in this TCGA melanoma cohort that can provide an integrative view of melanoma alterations.
Non-small cell lung cancer
Recent advancements in non-small cell lung cancer (NSCLC) treatment involve the inhibition of genes harboring driver mutations in epidermal growth factor receptor (EGFR), v-akt murine thymoma viral oncogene homolog 1 (AKT1), phosphoinositide 3-kinase (PI3K) and erb-b2 receptor tyrosine kinase 2 (ERBB2), and fusion of the echinoderm microtubule-associated protein-like 4 (EML4) and the anaplastic lymphoma receptor tyrosine kinase (ALK) genes (EML4-ALK) [129]. However, patients without these targetable mutations (>50%) are generally treated by non-specific systemic chemotherapy [130] and have a median survival <14 months [131]. The characterization of DNA methylation alterations that can serve as theranostic markers appears to be an alternative for these patients. A recent study identified a NSCLC DNA methylation signature associated with shorter disease-free survival [132]. Importantly, this signature, consisting of five genes (ALX1, HOXA9, HIST1H4F, PCDHGB6, and NPBWR1) identifies NSCLC stage I patients who have a high risk of recurrence and may benefit from adjuvant chemotherapy. Two studies have reported TCGA integrative genomic and epigenomic data analysis of lung cancers [133,134]. One study, focused on lung adenocarcinomas [133], identified a higher frequency of methylation of the gene CDKN2A on proximal inflammatory tumors and classified the tumors into H-CIMP, L-CIMP and intermediate CIMP [133]. Tumors with CIMP-H phenotype presented aberrant methylation of CDKN2A, GATA2, GATA4, GATA5, HIC1, HOXA9, HOXD13, RASSF1, SFRP1, SOX17 and WIF1 [133]. The other study, focused on lung squamous cell carcinoma (SQCC) [134], identified a higher overall methylation level in the classical subtype. This study identified that 72% of SQCCs harbor CDKN2A gene alterations, of which 21% are due to aberrant DNA methylation [134]. These recent integrative analyses support the important role of aberrant DNA methylation on the etiology of lung cancers, in addition to the mutations on driver genes.
Prostate cancer
Prostate cancer (PCa) is the most commonly detected cancer in men and the second highest cause of cancer deaths in this gender [49]. Patients with high prostate-specific antigen (PSA) levels are regularly subject to invasive needle biopsies. The low specificity of the PSA test leads to an overdiagnosis of clinically insignificant tumors, thus resulting in potentially unnecessary clinical interventions [135]. Identifying tumor biomarkers that provide earlier, more sensitive detection and that can assess the potential biological and clinical aggressiveness is urgently needed for PCa. Aberrant DNA methylation of glutathione s-transferase pi 1 (GSPT1) gene is the most common (>81%) molecular alteration reported in PCa [136,137]. The evaluation of GSTP1 promoter methylation in body fluids, including plasma, urine, ejaculate, prostatic secretion, and biopsy washes [138–140] complements PSA testing and increases the specificity compared with PSA testing alone [141]. This exemplifies the importance of combining current diagnostics tests with DNA methylation-based biomarkers in clinical practice.
Emerging technologies for DNA methylation mapping: New insights on cancer diagnosis
Technologies for DNA methylation profiling utilize a great variety of strategies. The diverse approaches employed vary in the size and the scope of data generated, and in the costs. Consideration of these aspects is critical for making the right decision when designing a cancer diagnostic and prognostic algorithm. For didactical purposes, methods to study DNA methylation can be classified in three categories according to the number and the distribution of CpG sites being evaluated.
Global 5meC quantification approaches
Methods within this category focus on determining the total 5mC content of a genome. Initially, 5mC quantification strategies were based on high profile liquid chromatography (HPLC) [142] and the incorporation of radioactive methyl groups [143]. Most recently, these approaches have been updated to capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry [144], and 5mC antibodies for immunohistochemistry [145], and ELISA (methDNA-ELISA) [146] analyses. Overall, these approaches allow evaluating major changes in the global DNA methylation level in a relatively inexpensive way. However, their main limitation is that region-specific DNA methylation cannot be analyzed.
Locus-specific DNA methylation approaches
Methods included in this category involve determination of DNA methylation status of preselected genomic regions. The surveyed regions contain a variable number of CpG sites, but this number is always limited to the extent of the targeted segment. Generally, locus-specific approaches have a relatively low cost and generate data that are easy to interpret. This makes it a cost-effective strategy and is especially useful when the targeted region contains well-validated biomarkers. Two major strategies are employed to investigate locus-specific DNA methylation:
Sodium bisulfite modification
Sodium bisulfite modification (SBM) converts C residues to uracil, but leaves 5mC residues unaffected. The seminal sodium bisulfite-based methylation-specific PCR (MSP) [147] was improved with real-time PCR-based approaches such as quantitative-MSP (qMSP) [148] and MethyLight [149], and sequencing-based technologies such as Pyrosequencing [150] and direct bisulfite-sequencing [151]. Our laboratory enhanced MSP capabilities to achieve an Absolute Quantitative Assessment of Methylated Alleles (AQAMA) [68]. This strategy allowed the identification of clinically relevant DNA methylation aberrations in breast, colorectal, and squamous-cell esophageal carcinomas [62,68,92–94,152]. Limitations of SBM are discussed later in the section ‘Genome-wide DNA methylation approaches.
Methylation-sensitive restriction enzymes
Methylation-sensitive restriction enzymes (MSREs) recognize and cut genomic sequences containing unmethylated CpG dinucleotides. Thus, the initial MSRE coupled with PCR or Southern Blot techniques [153] were improved into genome-wide DNA methylation techniques such as the HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) [154] and the Microarray-based Integrated Analysis of Methylation by Isoschizomers (MIAMI) [155]. The combination of SBM and MSRE denominated COmbined Bisulfite Restriction Analysis (COBRA) has also been largely used in cancer research [156]. Limitations of the use of MSREs are discussed later in the section ‘Genome-wide DNA methylation approaches’.
The main disadvantages of locus-specific techniques involve the requirement of knowing the region(s) of interest beforehand, the limited number of interrogated CpG sites, and the challenge of targeting specific CpG sites located in CpG-rich regions.
Genome-wide DNA methylation approaches
Next-generation high-throughput technologies have had a great impact in improving genome-wide DNA methylation mapping. The approaches used for genome-wide DNA methylation analyses can be classified according to the strategy employed to expose the methylation status: 5mC-specific immunoprecipitation (IP), MSRE digestion, SBM, and third-generation sequencing (TGS) technologies. With the exception of TGS technologies, in spite of the initial strategy, each approach can be evaluated by either microarray or next-generation sequencing (NGS) technologies (Figure 2). Here, we summarize the design, advantages/disadvantages, and cost/benefit ratio for the most relevant technologies that are revolutionizing DNA methylation mapping on human cancer.
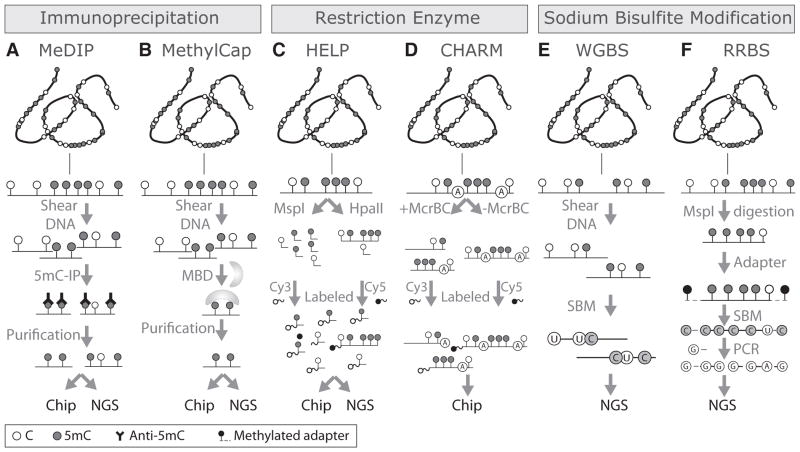
Figure 2. Genome-wide DNA methylation mapping technologies.
Technologies based on 5mC immunoprecipitation (IP). A- Methylated DNA IP (MeDIP) is based on the use of 5mC-specific antibodies. The product is a fraction of genomic DNA enriched in methylated DNA that can be analyzed by either DNA microarrays (chip; MeDIP-chip) or next-generation sequencing (NSG; MeDIP-Seq). B- MethylCap is based on the capture of methylated DNA by methyl-CpG binding domain (MBD) proteins. The fraction of methylated DNA enriched by this approach can also be analyzed by either chip (MethylCap-chip) or NGS (MethylCap-Seq). C- HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) is based on the differential labeling of the products of two restriction enzymes, one methylation-resistant (MspI) and other methylation-sensitive (HpaII). D- Comprehensive High-throughput Arrays for Relative Methylation (CHARM) uses the restriction enzyme McrBC which cuts 5mC that are preceded by a purine (A or G) base. The restricted (+McrBC) DNA is differentially labeled and compared with the non- McrBC restricted (-McrBC) DNA fragments in a DNA microarray. E- Whole genome bisulfite sequencing (WGBS) involves the fragmentation and sodium bisulfite modification (SBM) of genomic DNA followed by NGS. F- Reduced Representation Bisulfite Sequencing (RRBS) involves the digestion of genomic DNA by the methylation-resistant restriction enzyme MspI. Each fragment is coupled to a methylated adapter and SBM. Finally, only the converted fragments are amplified by PCR using primers that are complementary to the adaptor, and the products are analyzed by NGS.
5mC: 5-Methylcytosine; CpG: Cytosine – guanine.
Methods based on 5mC immunoprecipitation
These methods consist of the enrichment and isolation of fragments of genomic methylated DNA. 5mC, as well as the proteins that specifically bind to 5mC such as MBD proteins, can be targeted using specific antibodies. The advantage of these approaches is that they are not affected by either incomplete digestion or bisulfite conversion. Furthermore, since there is no SBM, data processing does not require alignment to a reference sodium bisulfite-converted human genome. However, depending on the quality and potential cross-reactivity of 5mC antibodies, incomplete or non-specific IP can be detected. In addition, 5mC IP-based methods tend to overrepresent high-density CpG regions.
Methylated DNA immunoprecipitation
Methylated DNA immunoprecipitation (MeDIP) provides regional DNA methylation levels in a genome-wide manner [157]. This technique is based on the IP of methylated DNA fragments by using 5mC-specific antibodies. Briefly, genomic DNA is either sonicated or fragmented by restriction enzymes to generate fragments between 100 and 400 bp. Then, the fragmented DNA is heat-denatured to single-stranded DNA and immunocaptured by a 5mC-specific antibody. The enriched methylated DNA can be analyzed by DNA microarrays (MeDIP-Chip) or NGS (MeDIP-Seq). Genomic regions significantly overrepresented in the microarray or in the sequencing data are identified as methylated regions (Figure 2A). Thus, MeDIP-Chip and MeDIP-Seq generate a large amount of information and have been used to identify differentially methylated coding, non-coding, and regulatory genomic regions in breast [158,159], cervical [160], endometrial [161], leukemia [162], ovarian [163], renal [164], and testicular [165] cancers.
Methyl-CpG binding domain (MBD) protein capture
Methyl-CpG binding domain (MBD) protein capture (MethylCap) assay is based on the high affinity purification of methylated genomic DNA using a MBD-specific antibody [166,167]. The procedure and generated data are similar to MeDIP. However, MethylCap has demonstrated to be more efficient at testing CpG islands than MeDIP [168]. Methylated DNA enriched by MethylCap can also be analyzed by both microarrays (MethylCap-Chip) and NGS (MethylCap-Seq) (Figure 2B). MethylCap was recently applied to identify genome-wide DNA methylation biomarkers for bladder [169], colorectal [76], leukemia [170], lung [171,172], ovarian [173,174], and renal [175] cancer diagnoses.
However, both MeDIP and MethylCap have important limitations for routine application in clinical diagnoses. The quality of formalin-fixed paraffin embedded (FFPE) DNA is not appropriate for MeDIP or MethylCap. Moreover, the data generated have a low resolution; there is no information about the methylation status of single CpG sites within the ~150 bp size fragments. Additionally, since it has been demonstrated that some isoforms of MBD1 can bind to both, methylated and unmethylated CpGs [176] via different domains [177], selecting the right MBD domain is critical to exclude false positive results on this technique.
Methods based on methylation-sensitive restriction enzymes
These methods are based on the selective restriction of methylated and unmethylated DNA sequences. Different combinations of MSREs have been employed. Like IP-based techniques, these methods do not require alignment to a reference sodium bisulfite-converted human genome.
HpaII tiny fragment enrichment by ligation-mediated PCR
In HpaII tiny fragment enrichment by ligation-mediated PCR (HELP), the genomic DNA is separately digested with MspI (resistant to 5mC) and HpaII (sensitive to 5mC). The restriction fragments are then amplified by ligation-mediated PCR labeled with two specific fluorochromes (one for MspI and the other for HpaII fragment products) and hybridized to a microarray (HELP-Chip) or directly sequenced (HELP-Seq) (Figure 2C). The advantage of this method is that it can test methylation levels of genomic regions in spite of their CpG density. However, it is limited to regions that present the HpaII/MspI restriction site (5′-C*CGG-3′), which comprises ~7% of all CpG sites. A recent study using the HELP-Chip assay identified a methylation signature involving 15 loci predictive of survival on acute myeloid leukemia patients [178].
Comprehensive high-throughput arrays for relative methylation
This method uses the enzyme McrBC coupled with a customized microarray hybridization [179]. McrBC is a type IV deoxyribonuclease that, unlike methylation-sensitive enzymes, specifically cuts methylated DNA without affecting unmethylated DNA. Its recognition site contains a purine (A or G) base before the methylated cytosine (5′-A/G5mC-3′). The probes contained in the Comprehensive High-throughput Arrays for Relative Methylation (CHARM) microarray are designed based on the CpG location and density [179] (Figure 2D). Compared with MeDIP-Chip and HELP-Chip, CHARM has better resolution, sensitivity, and specificity [179]. Using this approach, novel DMRs were identified in colon cancer and the importance of CpG shore methylation was evidenced [179].
Unlike IP-based approaches, MSREs-based technologies can be applied to FFPE clinical specimens. However, three important aspects should be considered. First, FFPE-derived DNA has a multitude of fragmentation sites which increases with age or time before fixation. Second, incomplete digestion represents an important issue. Third, these analyses are limited to the number and location of restriction sites in the genome. In addition, these approaches lack single CpG resolution.
Methods based on sodium bisulfite conversion
These methods rely on the mutagenic potential of sodium bisulfite. Importantly, this modification enables the generation of genome-wide maps of 5mC at a single-base resolution. However, the conversion leads to a reduced genome complexity (global decrease of C abundance), generating greater sequence redundancy, increasing the complexity of the sequence alignment (for sequencing-based methods), and decreasing the hybridization specificity (for microarray-based methods). Another disadvantage of SBM-based approaches is that it cannot discriminate between 5mC and 5hmC.
Human Methylation 450k BeadChip
Human Methylation 450k BeadChip (HM450K) is an expansion of the previously clinically-validated GoldenGate and Human Methylation 27k BeadChips [14]. This method quantifies methylation levels at single-base resolution in a genome-wide manner. Genomic DNA is sodium bisulfite modified and whole-genome amplified. The products are then enzymatically fragmented, purified, and hybridized to the BeadChip [14]. The oligonucleotide probes contained in this array survey the methylation level of over 485,000 cytosines (482,169 CpG dinucleotides and 3,343 CpNpG trinucleotides). Even though these CpG sites only represent 1.73% of all the CpGs, the design covers 21,231 (99%) RefSeq annotated genes with an average of 17.2 probes per gene distributed across the promoter, 5′UTR, first exon, gene body, and 3′UTR regions and 26,658 (96%) annotated CpG islands and their CpG context of shores, shelves, and distant regions identified as ‘open sea’ [14]. We observed that in this array, CpG island and CpG shore probes are predominantly located at gene promoter regions, while CpG shelf and ‘open sea’ probes are predominantly located at gene body and intergenic regions [18]. HM450K has demonstrated to be one of the most cost-effective practices for surveying genome-wide DNA methylation variations in human cancers [18,100,106,119,125,126,180–184]. Importantly, this is the method of choice by TCGA, where more than 10,000 specimens have been analyzed. However, some technical limitations with the probe design and analysis pipeline were recently described. First, several probes (~30,000) have a high likelihood of cross-hybridizing to non-targeted genomic regions [185]. Second, the CpG recognition site of many probes (~70,000) contains known single nucleotide polymorphisms (SNPs) [185]. As we recently reported, probes with affinity for non-targeted regions, as well as probes overlapping SNPs should be excluded from the analysis [18]. A general disadvantage of microarray-based approaches is that they only interrogate genomic regions previously determined in which specific synthetic probes can be designed.
Whole-genome bisulfite sequencing
Bisulfite conversion of genomic DNA combined with NGS is the most effective approach to evaluate at a single base resolution the methylation status of almost every C on the genome (~28M) (Figure 2E). This technology has provided evidence about non-CpG methylation [5]. More recently, it has been successfully applied to generate entire methylomes of different normal and respective cancer cell types [186–188]. This approach is generating the most comprehensive DNA methylation maps and promise to provide information about unexplored cancer genomic regions to design most informative diagnostic algorithms and potential therapeutic targets. However, despite of whole-genome bisulfite sequencing (WGBS) advantages, it is still extremely expensive and the data generated represent a challenge for bioinformatics approaches. Also, the large amount of DNA required for WGBS is prohibitive for cancer studies. For all these, WGBS is not yet suitable for routine cancer diagnostics. To maintain the advantages of bisulfite sequencing, while dramatically reducing the costs and the amount of input DNA, different adaptations of WGBS have been developed. Targeted bisulfite sequencing methods, such as reduced representation bisulfite sequencing (RRBS) [189] and bisulfite sequencing of padlock probes (BSPP) [190], which dramatically decrease the costs of WGBS and the complexity of the data analysis. By the other hand, the transposase-based in vitro shotgun library construction (‘tagmentation’) enables a >100-fold reduction in starting material (~20ng) relative to conventional WGBS protocols [191].
Reduced representation bisulfite sequencing
This is the most widely used adaptation of WGBS, in which a smaller fraction of genomic DNA is bisulfite sequenced [189]. Genomic DNA is first digested with a methylation-insensitive restriction enzyme, such as MspI. Then, methylated adaptors are linked to the restriction fragments and the fraction of 40–220 bp is purified and sodium bisulfite modified. The converted fragments are amplified by PCR using primers that are complementary to the adaptor, and then the PCR products are finally sequenced (Figure 2F). Reduced representation bisulfite sequencing (RRBS) generates information about the methylation statuses of 4–6 million of CpG sites at single base resolution. Recently, this protocol was optimized to analyze nanograms of genomic DNA. RRBS has been used to generate genome-wide DNA methylation maps of gliomas [192], leukemia [193], and colon cancer [194].
5mC and 5hmC-specific approaches for sodium bisulfite modification
A significant limitation of current bisulfite-based approaches is the inability to distinguish 5mC from 5hmC. In order to solve this issue, two strategies were recently developed. First is the oxidative bisulfite (ox-BS) modification, which is based on the differential oxidation of 5mC and 5hmC by potassium perruthenate (KruO4). Under this condition, 5mC is not affected, while 5hmC is oxidized to 5fC, which is eventually converted to uracil residues during the SBM step. This approach is coupled with NGS [195] and HM450K BeadChips [196]. The other approach is the TET-assisted bisulfite (TAB) modification, where 5hmCs are protected by glucosylation, and then the 5mCs are oxidized to 5caC by the TET enzymes and then finally converted to uracil residues by SBM. This approach is coupled with NGS (TAB-Seq) [197].
Methods based on third-generation sequencing (TGS) technologies
SBM-based approaches cannot distinguish 5mC from 5hmC, which means that previous studies relying on this approach have pooled both modifications as 5mC. Today, we know that due to their contradictory effect on gene expression, grouping them as 5mC is a mistake that needs to be solved. Single-molecule real-time (SMRT) DNA sequencing method is a TGS platform that allows sequencing of the 5mC without SBM. This is based on the fluorescently labeled nucleotide incorporation kinetics [198]. The covalent modifications of C, such as 5mC and 5hmC, differentially affect the kinetics of the DNA polymerase generating a kinetics signature that allows the discrimination between C, 5mC, and 5hmC [198]. This alternative appears to be a solution for the limitation of SBM-based sequencing approaches. However, it still needs to be properly validated in cancer methylome studies and the kinetic detection systems need higher sensitivity.
Genome-wide DNA methylation pattern as a cancer biomarker: strengths, and weaknesses
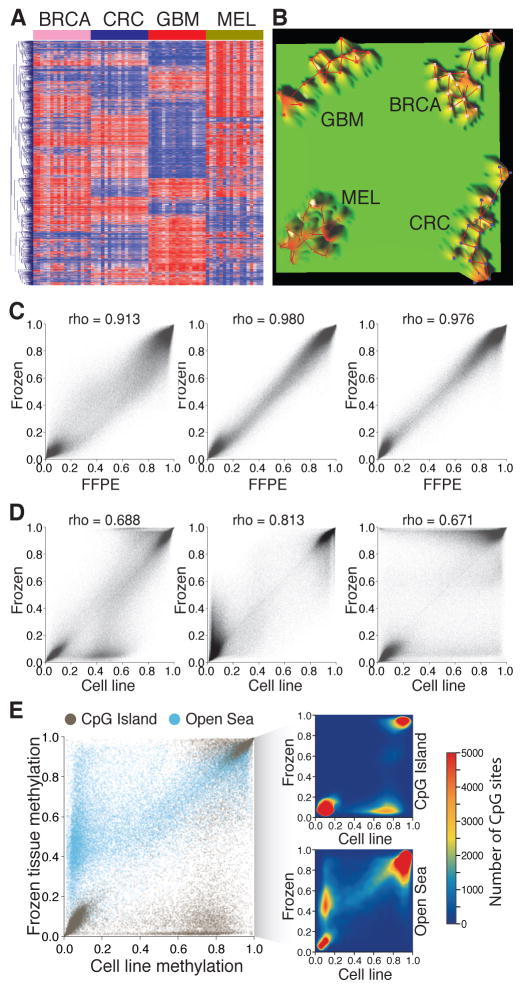
Compared with other cancer-specific biochemical modifications, 5mC is a cancer-specific stable modification that can be detected in DNA samples obtained from diverse body fluids, including whole blood [199], plasma [200], circulating tumor cells [201], serum [112], saliva [202], urine [203], bronchoalveolar lavage [204], sputum [205], stool [206], and fine needle aspirate [207,208], as reviewed elsewhere [209]. These features make the 5mC modification a suitable target for alternative non-invasive diagnostic strategies. Additionally, compared to genetic-based approaches, DNA methylation-based diagnostics present important advantages. First, the number of aberrantly methylated CpG sites is significantly larger than the number of genetic mutations [210]. This allows for the design of more flexible diagnostic strategies. Furthermore, this is particularly important for cancer types with low prevalence of somatic mutations [211–213]. Second, DNA methylation-based diagnostic approaches have the potential to identify tumors of unknown origin. In the example presented in figure 3, using TCGA data, we analyzed the top 4,000 most variable CpG sites across four cancer types (breast, colorectal, glioblastoma, and melanoma). As a representative sample, we included 17 primary tumors for each cancer type. Based on the DNA methylation signatures, the samples clustered into four independent clades corresponding to the tumor type (Figure 3A–B).
Figure 3. Strengths and weaknesses of DNA methylation as a cancer biomarker.
A–B: DNA methylation profiling can identify the tumor types. A- Heat map showing the unsupervised hierarchical cluster analysis using the DNA methylation level of the top 2,000 most variable CpG sites in four different cancer types: breast cancer (BRCA), colorectal cancer (CRC), glioblastoma (GBM), and melanoma (MEL). B- Terrain maps generated with the same DNA methylation signatures showing four separate conglomerates of samples representing the four different tumor types. C–E: Culture conditions induce genome-wide variations in DNA methylation level. C- Correlations between DNA methylation levels of three pairs of melanoma brain metastases stored as frozen and as formalin-fixed paraffin embedded tissues. D- Correlations between DNA methylation levels of the same three pairs of melanoma brain metastases stored frozen and established as cell cultures. E- Comparison of DNA methylation levels according to the CpG context of the assessed CpG sites. CpG sites located in CpG islands and in open sea regions are represented. The density plots represent the variations observed in CpG islands (upper) and open sea (lower).
CpG: Cytosine – guanine.
On the other hand, three important caveats should be considered with regard to the clinical utility of DNA methylation biomarkers:
First, there is a major interference of intratumor DNA methylation heterogeneity. The cell population under study usually comprises a mixture of phenotypically and epigenetically diverse cell types, and therefore the DNA methylation level is an average value determined by the methylation status of the specific locus and the proportion of cell type under study. This issue has been recently addressed in PCa [214]. In consequence, tissue heterogeneity is a significant issue in DNA methylation studies of cancer. This limitation may be reduced by improving the tissue microdissection techniques to guarantee a high percentage of tumor cell populations. Certainly, this drawback is not limited to DNA methylation since it has also been reported in genomic mutations [215].
Second, culture conditions have the potential to induce variation on DNA methylation patterns that are not present in the original tumor. To evaluate this phenomenon, we generated genome-wide DNA methylation landscapes of DNA samples obtained from brain metastases from melanoma patients processed in three different storage conditions: frozen tissues, FFPE sections, and established cultures. We identified a high correlation between FFPE and frozen tissues (Spearman’s rho = 0.913, 0.981, and 0.976; Figure 3C) in the three cases. However, the correlation between DNA methylation profiles obtained from established cultures and either frozen or FFPE tissues was significantly lower (Spearman’s rho 0.671, 0.688, and 0.813; Figure 3D). We separately evaluated variations on DNA methylation levels of CpG sites located at CpG islands and CpG sites located at ‘open sea’ regions. We observe an increase in DNA methylation within a subset of CpG sites located at CpG islands and a decrease in methylation with a subset of CpG sites located at ‘open sea’ regions (Figure 3E). In summary, due to the DNA methylation variations induced by cell culture conditions, DNA methylation biomarkers identified on in-vitro models have to be verified and validated in larger clinical specimen cohorts.
Third, as in the detection of genetic variations, the characterization of aberrant DNA methylation patterns depends on having a normal reference DNA methylome. However, since each cell type presents a specific DNA methylation pattern, defining a reference DNA methylome is a major challenge. In consequence, cancer methylome studies require the evaluation of a large number of normal tissues. Today, the Human Epigenomic Project (HEP) consortium, the International Human Epigenomic Consortium (IHEC), the U.S. National Health Institute Roadmap Epigenomics Program, and the ENCyclopedia of DNA Elements (ENCODE) project are delineating reference maps for DNA methylation, and other epigenomic modifications, for a large variety of normal adult, fetal, and embryonic human cell types. Data generated by these projects are currently used as normal references for different cancer epigenomic studies. We expect that upon competition of these key endeavors, the identification of cancer-related methylation alterations will be significantly enhanced.
Summary.
The understanding of epigenomic alterations in cancers has rapidly increased due to the emergence of more sensitive high-throughput technologies. DNA methylation, one of the most widely studied epigenetic mechanisms, represents an attractive target for cancer diagnosis, prognosis, and treatment. Seminal studies in cancer epigenetics focused on candidate genes mainly centered on the gene promoter. Today, with the advent of recently improved technologies, the focus shifted to genome-wide and whole-genome mapping of DNA methylation distribution. These new approaches revolutionized our understanding of epigenomic regulation and began to modify our preconceptions about the role and clinical utility of DNA methylation in cancer patients.
This Review explores emerging technologies for the study of DNA methylation as a cancer biomarker for diagnosis and prognosis. First, it provides new insight into the role of DNA methylation in gene expression, with particular focus on the importance of the cytosine-guanine (CpG) dinucleotide context. Second, it summarizes recent clinical applications of DNA methylation as a cancer theranostic biomarker. Finally, emerging technologies for studying DNA methylation are discussed, including considerations about strengths and weaknesses for each situation.
Expert commentary.
DNA methylation is an epigenetic modification with high value for cancer diagnostics. There is a large list of clinically-relevant DNA methylation alterations, with some showing utility as diagnostic, prognostic and/or therapeutic markers. During the past few years, the introduction of NGS and high-throughput microarrays to cancer research led to a paradigm shift in the approaches of characterizing aberrant DNA methylomes. Genome-wide mapping as opposed to single CpG assessment is enhancing the potential use of DNA methylation as a cancer biomarker. In addition, our understanding of the role of DNA methylation marks in the genome has increased. The concept that DNA methylation was only restricted to CpG dinucleotides and that its influence on gene expression regulation was centered on gene promoter CpG islands have been recently challenged by improvements in DNA methylation mapping technologies. With the discovery of additional functions of DNA methylation in transcription regulation, another paradigm that has been modified is the concept that DNA methylation always induces gene silencing. It is now clear that under different CpG and gene structure contexts, DNA methylation can also enhance, stabilize, or define alternative splicing sites, among other functions.
Five-year view.
As the cost of NGS-based technologies continues to decrease, the clinical utility of DNA methylation patterns on cancer diagnostics will rapidly increase. The introduction of TGS technologies to cancer studies promises to bring a solution to the multiple problems of DNA methylation mapping using bisulfite-based approaches. Defining the reference DNA methylomes will significantly improve the identification of cancer-specific DNA methylation patterns. This will result in a faster and easier process to identify novel and cancer-specific DNA methylation alterations. However, some improvements should be made before there can be a definite translation of DNA methylation to the clinical practice. First, methylation assays should be improved for low quality/amount of DNA samples, especially those obtained from microdissected FFPE tissues. Second, for current and upcoming cancer diagnostic DNA methylation markers, there is an urgent need for validation studies in large multicenter validation cohorts. Third, comparisons between DNA methylation markers with gene expression and mutation markers should be carried out. This comparison should address the question of whether DNA methylation markers can be used as surrogated for gene expression or mutation markers and identify novel diagnostic strategies in which DNA methylation can be integrated to existing gene expression or mutation markers.
Key issues.
Cancer DNA methylation mapping has rapidly increased with the emergence of high-throughput technologies.
The traditional view that DNA methylation occurs predominantly at CpG dinucleotides has been challenged with new findings based on NGS approaches.
The preconception that gene expression is mainly regulated by DNA methylation at promoter CpG islands has been expanded to other functional intra- and intergenic regions.
Non-CpG methylation in gene bodies is positively correlated with gene expression levels.
DNA methylation of CpGs outside of a CpG island context, such as CpG shores, shelves, and the so called ‘open sea’, demonstrate functionality for gene expression in normal and tumor cells.
DNA methylation maps are starting to gain importance as current theranostic markers for cancer management.
Genome-wide and whole-genome DNA methylation mapping approaches are replacing the traditional locus-specific approaches for the discovery of novel cancer biomarkers.
DNA methylomes generated from FFPE tissues maintain a high correlation with those generated from frozen tissues. However, DNA methylomes generated from cultured cells present significant differences with FFPE and frozen tissues.
In contrast to genetic profiling, DNA methylation mapping can identify the cancer-type.
The human reference methylome of normal tissues is needed to accelerate the discovery of cancer-related DNA methylation variations.
Acknowledgments
Financial support: This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (D.H.), the Ruth and Martin H. Weil Foundation (D.H.), the Margie and Robert E. Petersen Foundation (D.H., D.M.), Gonda Research Foundation, the National Institute of Health, National Cancer Institute, USA [1R01CA167967-01A1 (D.H.), P01CA029605 Project II/Core C (D.H.)], and the Associates for Breast and Prostate Cancer Studies (ABCs) Grant Award (D.M.).
We want to thank Jamie Huynh, Michelle Liu, and Nellie Nelson for their critical revision of this manuscript.
Footnotes
Conflicts of interest: The authors have declared that no competing interest exists.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute or the National Institutes of Health.
References
- 1.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12(9):615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33 (Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15(8):517–530. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- 4.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 5**.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. This study was the first whole-genome DNA methylation mapping of a human genome and describes the presence of non-CpG dinucleotide methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens M, Cheng JB, Li D, et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Research. 2013;23(9):1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmen MW. Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals. Genomics. 2008;92(1):33–40. doi: 10.1016/j.ygeno.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 8*.Shen L, Wu H, Diep D, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153(3):692–706. doi: 10.1016/j.cell.2013.04.002. This article summarizes the newly identified functional modifications of cytosine bases in the mammalian genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 10.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. Journal of molecular biology. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 11.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day JJ, Sweatt JD. DNA methylation and memory formation. Nature neuroscience. 2010;13(11):1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15(6):367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. This article presents a comprehensive analysis of the more than 450,000 genomic regions covered by the Human Methylation 450K BeadChip and introduces the denomination of open sea for CpG sites located more than 4 kb away of a CpG island. [DOI] [PubMed] [Google Scholar]
- 15**.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature genetics. 2009;41(2):178–186. doi: 10.1038/ng.298. This study identified tissue- and cancer-specific differentially methylated regions (DMRs) located outside of the traditional CpG islands, in regions denominated CpG Shores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen J, Wang S, Zhang YJ, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8(1):34–43. doi: 10.4161/epi.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzese DM, Scolyer RA, Huynh JL, et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum Mol Genet. 2014;23(1):226–238. doi: 10.1093/hmg/ddt420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. This article reviews the function of DNA methylation in regards to the CpG density and gene structure. [DOI] [PubMed] [Google Scholar]
- 20.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 21*.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics. 2007;39(4):457–466. doi: 10.1038/ng1990. This study classified gene promoters according to their CpG density and functionality. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Mav D, Grimm SA, et al. Fine-tuning of epigenetic regulation with respect to promoter CpG content in a cell type-specific manner. Epigenetics. 2014;9(5):747–759. doi: 10.4161/epi.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature structural & molecular biology. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 24.Smith JF, Mahmood S, Song F, et al. Identification of DNA methylation in 3′ genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2(3):161–172. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 25.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome biology. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Schmidl C, Klug M, Boeld TJ, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19(7):1165–1174. doi: 10.1101/gr.091470.109. This study identified a key role of DNA methylation at enhancer regions in determining cellular fate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozlenkov A, Roussos P, Timashpolsky A, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42(1):109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21(6):1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annual review of genomics and human genetics. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 31**.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. This article reviews the DNA methylation misbalanced in cancer cells. [DOI] [PubMed] [Google Scholar]
- 32.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 34.Esteller M, Fraga MF, Guo M, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10(26):3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 35.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature genetics. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Han H, De Carvalho DD, et al. Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell. 2014 doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 39.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60(6):376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 40.Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 42.Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. This study identified a new molecular classification of glioblastoma. [DOI] [PubMed] [Google Scholar]
- 49.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 50.Deschoolmeester V, Baay M, Specenier P, Lardon F, Vermorken JB. A review of the most promising biomarkers in colorectal cancer: one step closer to targeted therapy. Oncologist. 2010;15(7):699–731. doi: 10.1634/theoncologist.2010-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goel A, Arnold CN, Niedzwiecki D, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer research. 2003;63(7):1608–1614. [PubMed] [Google Scholar]
- 52.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nature reviews. Gastroenterology & hepatology. 2011;8(12):686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fox EJ, Leahy DT, Geraghty R, et al. Mutually exclusive promoter hypermethylation patterns of hMLH1 and O6-methylguanine DNA methyltransferase in colorectal cancer. J Mol Diagn. 2006;8(1):68–75. doi: 10.2353/jmoldx.2006.050084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amatu A, Sartore-Bianchi A, Moutinho C, et al. Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer. Clin Cancer Res. 2013;19(8):2265–2272. doi: 10.1158/1078-0432.CCR-12-3518. [DOI] [PubMed] [Google Scholar]
- 55.Hochhauser D, Glynne-Jones R, Potter V, et al. A phase II study of temozolomide in patients with advanced aerodigestive tract and colorectal cancers and methylation of the O6-methylguanine-DNA methyltransferase promoter. Molecular cancer therapeutics. 2013;12(5):809–818. doi: 10.1158/1535-7163.MCT-12-0710. [DOI] [PubMed] [Google Scholar]
- 56.Qi J, Zhu YQ, Huang MF, Yang D. Hypermethylation of CpG island in O6-methylguanine-DNA methyltransferase gene was associated with K-ras G to A mutation in colorectal tumor. World journal of gastroenterology : WJG. 2005;11(13):2022–2025. doi: 10.3748/wjg.v11.i13.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Yagi K, Akagi K, Hayashi H, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. This is the most update study in the classification of colorectal cancer based on DNA methylation signatures. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Cho NY, Choi M, et al. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathology international. 2008;58(2):104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 60.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. The American journal of pathology. 2001;159(3):1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang M, Ou J, Hutchinson L, Green MR. The BRAF Oncoprotein Functions through the Transcriptional Repressor MAFG to Mediate the CpG Island Methylator Phenotype. Mol Cell. 2014;55(6):904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6(4):e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benard A, van de Velde CJ, Lessard L, et al. Epigenetic status of LINE-1 predicts clinical outcome in early-stage rectal cancer. British journal of cancer. 2013;109(12):3073–3083. doi: 10.1038/bjc.2013.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umetani N, Takeuchi H, Fujimoto A, et al. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10(22):7475–7483. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 65.Umetani N, Fujimoto A, Takeuchi H, et al. Allelic imbalance of APAF-1 locus at 12q23 is related to progression of colorectal carcinoma. Oncogene. 2004;23(50):8292–8300. doi: 10.1038/sj.onc.1208022. [DOI] [PubMed] [Google Scholar]
- 66.Umetani N, de Maat MF, Sunami E, et al. Methylation of p16 and Ras association domain family protein 1a during colorectal malignant transformation. Molecular cancer research : MCR. 2006;4(5):303–309. doi: 10.1158/1541-7786.MCR-05-0199. [DOI] [PubMed] [Google Scholar]
- 67.Taback B, Saha S, Hoon DS. Comparative analysis of mesenteric and peripheral blood circulating tumor DNA in colorectal cancer patients. Annals of the New York Academy of Sciences. 2006;1075:197–203. doi: 10.1196/annals.1368.027. [DOI] [PubMed] [Google Scholar]
- 68.de Maat MF, Umetani N, Sunami E, Turner RR, Hoon DS. Assessment of methylation events during colorectal tumor progression by absolute quantitative analysis of methylated alleles. Molecular cancer research : MCR. 2007;5(5):461–471. doi: 10.1158/1541-7786.MCR-06-0358. [DOI] [PubMed] [Google Scholar]
- 69.Melotte V, Lentjes MH, van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. Journal of the National Cancer Institute. 2009;101(13):916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 70.de Maat MF, Narita N, Benard A, et al. Development of sporadic microsatellite instability in colorectal tumors involves hypermethylation at methylated-in-tumor loci in adenoma. The American journal of pathology. 2010;177(5):2347–2356. doi: 10.2353/ajpath.2010.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer metastasis reviews. 2010;29(1):181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 72.Carmona FJ, Azuara D, Berenguer-Llergo A, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer prevention research. 2013;6(7):656–665. doi: 10.1158/1940-6207.CAPR-12-0501. [DOI] [PubMed] [Google Scholar]
- 73.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 74.Benard A, Zeestraten EC, Goossens-Beumer IJ, et al. DNA methylation of apoptosis genes in rectal cancer predicts patient survival and tumor recurrence. Apoptosis : an international journal on programmed cell death. 2014;19(11):1581–1593. doi: 10.1007/s10495-014-1022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi JM, Dhir M, Van Neste L, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17(6):1535–1545. doi: 10.1158/1078-0432.CCR-10-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simmer F, Brinkman AB, Assenov Y, et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7(12):1355–1367. doi: 10.4161/epi.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22(2):271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sipos F, Muzes G, Patai AV, et al. Genome-wide screening for understanding the role of DNA methylation in colorectal cancer. Epigenomics. 2013;5(5):569–581. doi: 10.2217/epi.13.52. [DOI] [PubMed] [Google Scholar]
- 79.TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 81.Piva R, Rimondi AP, Hanau S, et al. Different methylation of oestrogen receptor DNA in human breast carcinomas with and without oestrogen receptor. British journal of cancer. 1990;61(2):270–275. doi: 10.1038/bjc.1990.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sizemore ST, Sizemore GM, Booth CN, et al. Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast cancer research and treatment. 2014;146(1):25–40. doi: 10.1007/s10549-014-2989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma P, Stecklein SR, Kimler BF, et al. The prognostic value of promoter methylation in early stage triple negative breast cancer. Journal of cancer therapeutics & research. 2014;3(2):1–11. doi: 10.7243/2049-7962-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kagara N, Huynh KT, Kuo C, et al. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. The American journal of pathology. 2012;181(1):257–267. doi: 10.1016/j.ajpath.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 85.Lee H, Zhang P, Herrmann A, et al. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109(20):7765–7769. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shinozaki M, Hoon DS, Giuliano AE, et al. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11(6):2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 87.Sunami E, Shinozaki M, Higano CS, et al. Multimarker circulating DNA assay for assessing blood of prostate cancer patients. Clinical chemistry. 2009;55(3):559–567. doi: 10.1373/clinchem.2008.108498. [DOI] [PubMed] [Google Scholar]
- 88.Sunami E, Shinozaki M, Sim MS, et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast cancer research : BCR. 2008;10(3):R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sunami E, Vu AT, Nguyen SL, Hoon DS. Analysis of methylated circulating DNA in cancer patients’ blood. Methods in molecular biology. 2009;507:349–356. doi: 10.1007/978-1-59745-522-0_25. [DOI] [PubMed] [Google Scholar]
- 90.Taback B, Giuliano AE, Lai R, et al. Epigenetic analysis of body fluids and tumor tissues: application of a comprehensive molecular assessment for early-stage breast cancer patients. Annals of the New York Academy of Sciences. 2006;1075:211–221. doi: 10.1196/annals.1368.029. [DOI] [PubMed] [Google Scholar]
- 91.Umetani N, Mori T, Koyanagi K, et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24(29):4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 92.van Hoesel AQ, Sato Y, Elashoff DA, et al. Assessment of DNA methylation status in early stages of breast cancer development. British journal of cancer. 2013;108(10):2033–2038. doi: 10.1038/bjc.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Hoesel AQ, van de Velde CJ, Kuppen PJ, et al. Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast cancer research and treatment. 2012;134(3):1103–1114. doi: 10.1007/s10549-012-2038-0. [DOI] [PubMed] [Google Scholar]
- 94.van Hoesel AQ, van de Velde CJ, Kuppen PJ, et al. Primary tumor classification according to methylation pattern is prognostic in patients with early stage ER-negative breast cancer. Breast cancer research and treatment. 2012;131(3):859–869. doi: 10.1007/s10549-011-1485-3. [DOI] [PubMed] [Google Scholar]
- 95.Marzese DM, Gago FE, Vargas-Roig LM, Roque M. Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes. 2010;24(5):271–280. doi: 10.1016/j.mcp.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 96.Marzese DM, Hoon DS, Chong KK, et al. DNA methylation index and methylation profile of invasive ductal breast tumors. J Mol Diagn. 2012;14(6):613–622. doi: 10.1016/j.jmoldx.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Rao X, Evans J, Chae H, et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene. 2013;32(38):4519–4528. doi: 10.1038/onc.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fleischer T, Frigessi A, Johnson KC, et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome biology. 2014;15(8):435. doi: 10.1186/s13059-014-0435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rhee JK, Kim K, Chae H, et al. Integrated analysis of genome-wide DNA methylation and gene expression profiles in molecular subtypes of breast cancer. Nucleic Acids Res. 2013;41(18):8464–8474. doi: 10.1093/nar/gkt643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ambrosone CB, Young AC, Sucheston LE, et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget. 2014;5(1):237–248. doi: 10.18632/oncotarget.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghosh S, Gu F, Wang CM, et al. Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast cancer research and treatment. 2014;147(3):653–659. doi: 10.1007/s10549-014-3132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson KC, Koestler DC, Cheng C, Christensen BC. Age-related DNA methylation in normal breast tissue and its relationship with invasive breast tumor methylation. Epigenetics. 2014;9(2):268–275. doi: 10.4161/epi.27015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lindqvist BM, Wingren S, Motlagh PB, Nilsson TK. Whole genome DNA methylation signature of HER2-positive breast cancer. Epigenetics. 2014;9(8):1149–1162. doi: 10.4161/epi.29632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang F, Yang Y, Fu Z, et al. Differential DNA methylation status between breast carcinomatous and normal tissues. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2014;68(6):699–707. doi: 10.1016/j.biopha.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 105**.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. This TCGA study identified five breast cancer DNA methylation subtypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roessler J, Ammerpohl O, Gutwein J, et al. The CpG island methylator phenotype in breast cancer is associated with the lobular subtype. Epigenomics. 2014:1–13. doi: 10.2217/epi.14.74. [DOI] [PubMed] [Google Scholar]
- 107.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 109.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer research. 2003;63(7):1639–1643. [PubMed] [Google Scholar]
- 110.Hoon DS, Spugnardi M, Kuo C, et al. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23(22):4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mori T, Kim J, Yamano T, et al. Epigenetic up-regulation of C-C chemokine receptor 7 and C-X-C chemokine receptor 4 expression in melanoma cells. Cancer research. 2005;65(5):1800–1807. doi: 10.1158/0008-5472.CAN-04-3531. [DOI] [PubMed] [Google Scholar]
- 112.Mori T, O’Day SJ, Umetani N, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005;23(36):9351–9358. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koyanagi K, Mori T, O’Day SJ, et al. Association of circulating tumor cells with serum tumor-related methylated DNA in peripheral blood of melanoma patients. Cancer research. 2006;66(12):6111–6117. doi: 10.1158/0008-5472.CAN-05-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mori T, Martinez SR, O’Day SJ, et al. Estrogen receptor-alpha methylation predicts melanoma progression. Cancer research. 2006;66(13):6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kitago M, Martinez SR, Nakamura T, Sim MS, Hoon DS. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15(9):2988–2994. doi: 10.1158/1078-0432.CCR-08-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nguyen T, Kuo C, Nicholl MB, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6(3):388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoshimoto S, Kuo CT, Chong KK, et al. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. The Journal of investigative dermatology. 2012;132(6):1689–1697. doi: 10.1038/jid.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huynh KT, Takei Y, Kuo C, et al. Aberrant hypermethylation in primary tumours and sentinel lymph node metastases in paediatric patients with cutaneous melanoma. The British journal of dermatology. 2012;166(6):1319–1326. doi: 10.1111/j.1365-2133.2012.10867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sato Y, Marzese DM, Ohta K, et al. Epigenetic regulation of REG1A and chemosensitivity of cutaneous melanoma. Epigenetics. 2013;8(10):1043–1052. doi: 10.4161/epi.25810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fratta E, Sigalotti L, Covre A, et al. Epigenetics of melanoma: implications for immune-based therapies. Immunotherapy. 2013;5(10):1103–1116. doi: 10.2217/imt.13.108. [DOI] [PubMed] [Google Scholar]
- 121.Greenberg ES, Chong KK, Huynh KT, Tanaka R, Hoon DS. Epigenetic biomarkers in skin cancer. Cancer letters. 2014;342(2):170–177. doi: 10.1016/j.canlet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Conway K, Edmiston SN, Khondker ZS, et al. DNA-methylation profiling distinguishes malignant melanomas from benign nevi. Pigment cell & melanoma research. 2011;24(2):352–360. doi: 10.1111/j.1755-148X.2011.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sigalotti L, Covre A, Fratta E, et al. Whole genome methylation profiles as independent markers of survival in stage IIIC melanoma patients. Journal of translational medicine. 2012;10:185. doi: 10.1186/1479-5876-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marzese DM, Huynh JL, Kawas NP, Hoon DSB. Multi-platform genome-wide analysis of melanoma progression to brain metastasis. Genomics Data. 2014;2(0):150–152. doi: 10.1016/j.gdata.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marzese DM, Scolyer RA, Roque M, et al. DNA methylation and gene deletion analysis of brain metastases in melanoma patients identifies mutually exclusive molecular alterations. Neuro Oncol. 2014 doi: 10.1093/neuonc/nou107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marzese DM, Liu M, Huynh JL, et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment cell & melanoma research. 2014 doi: 10.1111/pcmr.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tanemura A, Terando AM, Sim MS, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15(5):1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell cycle. 2012;11(2):286–295. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rooney M, Devarakonda S, Govindan R. Genomics of squamous cell lung cancer. Oncologist. 2013;18(6):707–716. doi: 10.1634/theoncologist.2013-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paz-Ares LG, de Marinis F, Dediu M, et al. PARAMOUNT: Final overall survival results of the phase III study of maintenance pemetrexed versus placebo immediately after induction treatment with pemetrexed plus cisplatin for advanced nonsquamous non-small-cell lung cancer. J Clin Oncol. 2013;31(23):2895–2902. doi: 10.1200/JCO.2012.47.1102. [DOI] [PubMed] [Google Scholar]
- 132.Sandoval J, Mendez-Gonzalez J, Nadal E, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31(32):4140–4147. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 133.TCGA. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.TCGA. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Draisma G, Boer R, Otto SJ, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. Journal of the National Cancer Institute. 2003;95(12):868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 136.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91(24):11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bastian PJ, Yegnasubramanian S, Palapattu GS, et al. Molecular biomarker in prostate cancer: the role of CpG island hypermethylation. European urology. 2004;46(6):698–708. doi: 10.1016/j.eururo.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 138.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9(7):2673–2677. [PubMed] [Google Scholar]
- 139.Goessl C, Muller M, Heicappell R, Krause H, Miller K. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Annals of the New York Academy of Sciences. 2001;945:51–58. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 140.Jeronimo C, Usadel H, Henrique R, et al. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60(6):1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 141.Wu T, Giovannucci E, Welge J, et al. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a meta-analysis. British journal of cancer. 2011;105(1):65–73. doi: 10.1038/bjc.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wagner I, Capesius I. Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochimica et biophysica acta. 1981;654(1):52–56. doi: 10.1016/0005-2787(81)90135-0. [DOI] [PubMed] [Google Scholar]
- 143.Soares J, Pinto AE, Cunha CV, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85(1):112–118. [PubMed] [Google Scholar]
- 144.Chen ML, Shen F, Huang W, et al. Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clinical chemistry. 2013;59(5):824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Poloni A, Goteri G, Zizzi A, et al. Prognostic role of immunohistochemical analysis of 5 mc in myelodysplastic syndromes. Eur J Haematol. 2013;91(3):219–227. doi: 10.1111/ejh.12145. [DOI] [PubMed] [Google Scholar]
- 146.Riganti C, Salaroglio IC, Caldera V, et al. Temozolomide downregulates P-glycoprotein expression in glioblastoma stem cells by interfering with the Wnt3a/glycogen synthase-3 kinase/beta-catenin pathway. Neuro Oncol. 2013;15(11):1502–1517. doi: 10.1093/neuonc/not104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lo YM, Wong IH, Zhang J, et al. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer research. 1999;59(16):3899–3903. [PubMed] [Google Scholar]
- 149.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. BioTechniques. 2003;35(1):146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 151.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nature protocols. 2006;1(5):2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 152.Hoshimoto S, Takeuchi H, Ono S, et al. Genome-wide Hypomethylation and Specific Tumor-Related Gene Hypermethylation are Associated with Esophageal Squamous Cell Carcinoma Outcome. J Thorac Oncol. 2014 doi: 10.1097/JTO.0000000000000441. [DOI] [PubMed] [Google Scholar]
- 153.Hu WS, Fanning TG, Cardiff RD. Mouse mammary tumor virus: specific methylation patterns of proviral DNA in normal mouse tissues. Journal of virology. 1984;49(1):66–71. doi: 10.1128/jvi.49.1.66-71.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hatada I, Fukasawa M, Kimura M, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25(21):3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- 156.Lee EJ, Luo J, Wilson JM, Shi H. Analyzing the cancer methylome through targeted bisulfite sequencing. Cancer letters. 2013;340(2):171–178. doi: 10.1016/j.canlet.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157**.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37(8):853–862. doi: 10.1038/ng1598. This article introduced the methylated DNA immunoprecipitation technique. In this first publication, MeDIP was coupled with a microarray. [DOI] [PubMed] [Google Scholar]
- 158.Flanagan JM, Cocciardi S, Waddell N, et al. DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. American journal of human genetics. 2010;86(3):420–433. doi: 10.1016/j.ajhg.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lendvai A, Johannes F, Grimm C, et al. Genome-wide methylation profiling identifies hypermethylated biomarkers in high-grade cervical intraepithelial neoplasia. Epigenetics : official journal of the DNA Methylation Society. 2012;7(11):1268–1278. doi: 10.4161/epi.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang B, Xing X, Li J, et al. Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC genomics. 2014;15(1):868. doi: 10.1186/1471-2164-15-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yalcin A, Kreutz C, Pfeifer D, et al. MeDIP coupled with a promoter tiling array as a platform to investigate global DNA methylation patterns in AML cells. Leukemia research. 2013;37(1):102–111. doi: 10.1016/j.leukres.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 163.Gloss BS, Patterson KI, Barton CA, et al. Integrative genome-wide expression and promoter DNA methylation profiling identifies a potential novel panel of ovarian cancer epigenetic biomarkers. Cancer letters. 2012;318(1):76–85. doi: 10.1016/j.canlet.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 164.Morris MR, Ricketts CJ, Gentle D, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30(12):1390–1401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- 165.Cheung HH, Lee TL, Davis AJ, et al. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. British journal of cancer. 2010;102(2):419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brinkman AB, Simmer F, Ma K, et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 167.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28(10):1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhao Y, Guo S, Sun J, et al. Methylcap-seq reveals novel DNA methylation markers for the diagnosis and recurrence prediction of bladder cancer in a Chinese population. PLoS One. 2012;7(4):e35175. doi: 10.1371/journal.pone.0035175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Niederwieser C, Kohlschmidt J, Volinia S, et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia. 2014 doi: 10.1038/leu.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Carvalho RH, Haberle V, Hou J, et al. Genome-wide DNA methylation profiling of non-small cell lung carcinomas. Epigenetics Chromatin. 2012;5(1):9. doi: 10.1186/1756-8935-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Carvalho RH, Hou J, Haberle V, et al. Genomewide DNA methylation analysis identifies novel methylated genes in non-small-cell lung carcinomas. J Thorac Oncol. 2013;8(5):562–573. doi: 10.1097/JTO.0b013e3182863ed2. [DOI] [PubMed] [Google Scholar]
- 173.Yu W, Jin C, Lou X, et al. Global analysis of DNA methylation by Methyl-Capture sequencing reveals epigenetic control of cisplatin resistance in ovarian cancer cell. PLoS One. 2011;6(12):e29450. doi: 10.1371/journal.pone.0029450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang RL, Gu F, Kirma NB, et al. Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics. 2013;8(6):624–634. doi: 10.4161/epi.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sharpe K, Stewart GD, Mackay A, et al. The effect of VEGF-targeted therapy on biomarker expression in sequential tissue from patients with metastatic clear cell renal cancer. Clin Cancer Res. 2013;19(24):6924–6934. doi: 10.1158/1078-0432.CCR-13-1631. [DOI] [PubMed] [Google Scholar]
- 176.Jorgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Molecular and cellular biology. 2004;24(8):3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bianchi C, Zangi R. Molecular Dynamics Study of the Recognition of dimethylated CpG sites by MBD1 Protein. Journal of chemical information and modeling. 2015 doi: 10.1021/ci500657d. [DOI] [PubMed] [Google Scholar]
- 178.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Busche S, Ge B, Vidal R, et al. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer research. 2013;73(14):4323–4336. doi: 10.1158/0008-5472.CAN-12-4367. [DOI] [PubMed] [Google Scholar]
- 181.Naumov VA, Generozov EV, Zaharjevskaya NB, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8(9):921–934. doi: 10.4161/epi.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cheng Y, Yan Z, Liu Y, et al. Analysis of DNA methylation patterns associated with the gastric cancer genome. Oncology letters. 2014;7(4):1021–1026. doi: 10.3892/ol.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gasperov NM, Farkas SA, Nilsson TK, Grce M. Epigenetic activation of immune genes in cervical cancer. Immunology letters. 2014 doi: 10.1016/j.imlet.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 184.Karlsson A, Jonsson M, Lauss M, et al. Genome-wide DNA methylation analysis of lung carcinoma reveals one neuorendocrine and four adenocarcinoma epitypes associated with patient outcome. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-14-1087. [DOI] [PubMed] [Google Scholar]
- 185.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109(26):10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature genetics. 2012;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189**.Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33(18):5868–5877. doi: 10.1093/nar/gki901. This article introduced the adaptation of whole-genome bisulfite sequencing named reduced representation bisulfite sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Diep D, Plongthongkum N, Gore A, et al. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9(3):270–272. doi: 10.1038/nmeth.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang Q, Gu L, Adey A, et al. Tagmentation-based whole-genome bisulfite sequencing. Nature protocols. 2013;8(10):2022–2032. doi: 10.1038/nprot.2013.118. [DOI] [PubMed] [Google Scholar]
- 192.Li S, Chowdhury R, Liu F, et al. Tumor-Suppressive miR148a Is Silenced by CpG Island Hypermethylation in IDH1-Mutant Gliomas. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-14-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pei L, Choi JH, Liu J, et al. Genome-wide DNA methylation analysis reveals novel epigenetic changes in chronic lymphocytic leukemia. Epigenetics. 2012;7(6):567–578. doi: 10.4161/epi.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ashktorab H, Daremipouran M, Goel A, et al. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9(4):503–512. doi: 10.4161/epi.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195*.Booth MJ, Ost TW, Beraldi D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nature protocols. 2013;8(10):1841–1851. doi: 10.1038/nprot.2013.115. Describes an adaptation of bisulfite sequencing to distinguish 5mC from 5hmC by inducing the oxidation of 5hmC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stewart SK, Morris TJ, Guilhamon P, et al. oxBS-450K: A method for analysing hydroxymethylation using 450K BeadChips. Methods. 2014 doi: 10.1016/j.ymeth.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198*.Flusberg BA, Webster DR, Lee JH, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. Introduces the third-generation of sequencing technologies to study DNA methylation maps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16(1):108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 200.Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clinical chemistry. 2008;54(2):414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 201.Chimonidou M, Strati A, Tzitzira A, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clinical chemistry. 2011;57(8):1169–1177. doi: 10.1373/clinchem.2011.165902. [DOI] [PubMed] [Google Scholar]
- 202.Righini CA, de Fraipont F, Timsit JF, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13(4):1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 203.Yu J, Zhu T, Wang Z, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13(24):7296–7304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 204.Ahrendt SA, Chow JT, Xu LH, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. Journal of the National Cancer Institute. 1999;91(4):332–339. doi: 10.1093/jnci/91.4.332. [DOI] [PubMed] [Google Scholar]
- 205.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95(20):11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Glockner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer research. 2009;69(11):4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jeronimo C, Costa I, Martins MC, et al. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin Cancer Res. 2003;9(9):3413–3417. [PubMed] [Google Scholar]
- 208.Pellise M, Castells A, Gines A, et al. Detection of lymph node micrometastases by gene promoter hypermethylation in samples obtained by endosonography- guided fine-needle aspiration biopsy. Clin Cancer Res. 2004;10(13):4444–4449. doi: 10.1158/1078-0432.CCR-03-0600. [DOI] [PubMed] [Google Scholar]
- 209**.Ma Y, Wang X, Jin H. Methylated DNA and microRNA in Body Fluids as Biomarkers for Cancer Detection. International journal of molecular sciences. 2013;14(5):10307–10331. doi: 10.3390/ijms140510307. Reviews the utility of DNA methylation as a biomarker in body fluids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer research. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 211.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Brocks D, Assenov Y, Minner S, et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8(3):798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 215.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]