Abstract
Obese subjects with a similar body mass index (BMI) exhibit substantial heterogeneity in gluco- and cardio-metabolic heath phenotypes. However, defining genes that underlie the heterogeneity of metabolic features among obese individuals and determining metabolically healthy and unhealthy phenotypes remain challenging. We conducted unsupervised hierarchical clustering analysis of subcutaneous adipose tissue transcripts from 30 obese men and women ≥40 years old. Despite similar BMIs in all subjects, we found two distinct subgroups, one metabolically healthy (Group 1) and one metabolically unhealthy (Group 2). Subjects in Group 2 showed significantly higher total cholesterol (p=0.005), LDL cholesterol (p=0.006), 2h-Insulin during OGTT (p=0.015) and lower insulin sensitivity (SI, p=0.029) compared to Group 1. We identified significant up-regulation of 141 genes (e.g. MMP9 and SPP1) and down-regulation of 17 genes (e.g. NDRG4 and GINS3) in group 2 subjects. Intriguingly, these differentially expressed transcripts were enriched for genes involved in cardiovascular disease-related processes (p=2.81×10−11–3.74×10−02) and pathways involved in immune and inflammatory response (p=8.32×10−5–0.04). Two down-regulated genes, NDRG4 and GINS3, have been located in a genomic interval associated with cardiac repolarization in published GWASs and zebra fish knockout models. Our study provides evidence that perturbations in the adipose tissue gene expression network are important in defining metabolic health in obese subjects.
Keywords: Adipose tissue, obesity, transcript, genomics, metabolic health
Introduction
Many diseases of high public health importance, such as type 2 diabetes and metabolic syndrome, are profoundly influenced by obesity status (1). However, individuals with a similar body mass index (BMI) can exhibit substantially different metabolic profiles for glucose tolerance, lipids, and blood pressure. In the NHANES study, an estimated 31.7% of all obese individuals (~10% of the U.S. adult population) exhibit a better metabolic profile than age-adjusted BMI expectations and are categorized as “metabolically healthy obese” (MHO) (2;3).
An increase in white adipose tissue (WAT) mass in response to chronic positive energy imbalance is a defining feature of obesity. However, several structural/histological characteristics and depot-specific (subcutaneous and visceral) distributions of adipose tissue differ between metabolically healthy and unhealthy obese individuals (4). The location and cellular mechanisms of tissue expansion (hyperplasia and hypertrophy) greatly affect the normal physiological function of adipose tissue (5). This process may be mechanistically linked to the development of obesity-associated metabolic disorders (6;7). We and others have shown significant dysregulation in the adipose tissue transcriptome with obesity (8–11). Many genes are correlated with obesity (BMI) and adiposity (% fat mass). However, pinpointing which defining genes underlie the heterogeneity of metabolic features among obese individuals and determine metabolically healthy and unhealthy phenotypes remains challenging.
The purpose of this study is to identify an adipose tissue transcriptomic signature that may determine metabolic health in obese subjects. Defining metabolically healthy and unhealthy obese subjects based on clinical and biochemical characteristics is difficult (2;3). Thus, we performed unsupervised hierarchical clustering analysis using data from obesity-associated adipose tissue transcripts to first identify comparatively homogenous sub-groups among obese subjects. We hypothesized that these transcriptionally homogenous sub-groups could differ in metabolic characteristics, and that a subset of obesity-associated transcripts in subcutaneous adipose tissue are involved in defining metabolic health in obese subjects. We then performed additional statistical and bioinformatics analyses to compare sub-groups of obese subjects.
Subjects and Methods
The sample included a subset (30 obese men and women) from the 170 subjects recruited for the University of Arkansas for Medical Sciences (UAMS) gene expression study (8;12). These 30 subjects were all obese (BMI ≥ 30 kg/m2), non-diabetic, and ≥ 40 years old. They had been characterized by frequently sampled intravenous glucose tolerance tests (FSIVGT), and genome-wide gene expression data were available from subcutaneous adipose tissue biopsy samples. Three additional subjects met the selection criteria mentioned above, but genome wide expression data of acceptable quality were unobtainable for these subjects. Methods for recruitment, physical examination, physiological experiments, and obtaining biopsies have been published. In brief, European-American or African-American men and women in general good health, between 19 and 60 years of age, and had a BMI between 19 and 45 kg/m2 were recruited. All participants had a screening visit, at which time height, weight, and waist and hip circumference were measured; body fat was determined by dual x-ray absorptiometry (DXA) scan (Hologic QDR-4500); fasting blood samples for lipid measurements were taken; and a standard 75-g oral glucose tolerance test (OGTT) was done. At the second visit, adipose biopsies were obtained from fasting subjects. Insulin modified (0.03 U/kg) frequently sampled intravenous glucose tolerance tests (FSIGT) were performed to determine insulin sensitivity (SI) and acute insulin response (AIRG). Biopsies were obtained from abdominal subcutaneous fat near the umbilicus under local anesthesia (Lidocaine) using a Bergstrom needle. Each biopsy is made by opening the Bergstrom needle and applying suction using a 120 ml syringe. Samples were rinsed immediately in sterile normal saline, quick-frozen in liquid nitrogen, and stored at −80°C for further use. All participants provided written informed consent under protocols originally approved by the UAMS. Our current study was approved by the Institutional Review Board at Wake Forest School of Medicine.
Insulin was measured by the UAMS Clinical Research Center core laboratory using an immuno-chemiluminometric assay (Invitron Limited, Monmouth, UK). Plasma glucose was measured by glucose oxidase methods at LabCorp, Inc. (Burlington, NC). Plasma triglyceride, total cholesterol, and HDL cholesterol concentrations were measured directly (at LabCorp) by enzymatic colorimetric methods. LDL cholesterol and VLDL cholesterol concentrations were calculated indirectly by the Friedewald equation.
Total RNA was isolated from whole adipose tissue using the RNAeasy Lipid Tissue Mini kit (QIAGEN Inc-USA, Valencia, CA). The quantity and quality of the isolated total RNA samples were determined by ultraviolet spectrophotometry (Nanodrop, Thermo Scientific, Pittsburgh, PA) and electrophoresis (Experion nucleic acid analyzer, BioRad Laboratories, Inc., Hercules, CA), respectively. High-quality RNA with RIN (RNA integrity number) >8 was used for genome-wide transcriptome analysis.
Genome-wide transcriptome analysis and initial array processing were done at the Center for Human Genomics Core Laboratory (Wake Forest School of Medicine) using HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA) whole genome gene expression arrays according to the vendor-recommended standard protocol (8). Raw expression intensity was background subtracted and normalized by the average normalization algorithm as implemented in GenomeStudio Gene Expression Module v1.0 application software (Illumina). Normalized data were used for further analysis.
Data Analysis
We performed unsupervised hierarchical clustering to identify homogeneous groups among obese subjects based on adipose tissue transcript expression. A dissimilarity matrix based on Pearson’s correlation coefficient between each individual was generated using Z-score-normalized values for each probe and seriation under multiple fragment heuristics. The hierarchical clustering-based tree and heat map was generated using McQuitty’s criteria implemented in PermutMatrix version 1.9.3EN software (13). For the current analysis, we chose a set of 1595 obesity-associated probes (associated with % fat mass and BMI independent of age, gender and race) that may define the heterogeneity of metabolic features among obese individuals, based on our previous study (8). To estimate the differences in gluco- and cardio-metabolic phenotypes between the two groups (two major groups/clusters assigned by unsupervised hierarchical clustering analysis), we used nonparametric Mann-Whitney U-test and univariate analysis of variance, with age, gender, and race as covariates. To identify the transcripts with significantly different expression between the two groups, we utilized nonparametric Wilcoxon statistics on normalized data as implemented in Statistical Analysis for Microarray (SAM) software (14). We considered results significant for a false discovery rate (q value) ≤ 5%, and fold difference between groups of >1.5, based on 1000 permutations. Additional validation of SAM analysis was performed by t-test (p ≤0.05) on log2 transformed expression data. Bioinformatic analysis was performed to investigate the enrichment of differentially expressed genes in known biological pathways or Gene Ontology (GO) categories. We performed canonical pathway analysis and interaction network analysis of differentially expressed genes using Ingenuity Pathway analysis (IPA; https://analysis.ingenuity.com, Build version: 313398M; Content version: 18841524). Pathways with a corrected p-value (Benjamini and Hochberg p-value) ≤0.05 were considered significantly enriched in our gene lists. Additional annotations of differentially expressed genes were performed by singular enrichment analysis (SEA) using the DAVID v6.7 functional annotation tool (http://david.abcc.ncifcrf.gov/)(15). Detailed analysis parameters for SEA using the functional annotation chart module of DAVID are described elsewhere (9). In the SEA analysis, a category with a gene count >5, an EASE score ≤0.05 (modified Fisher’s exact p-value), and FDR ≤5% was considered as a significant threshold.
Results and Discussion
White adipose tissue modulates both glucose and lipid homeostasis in humans via its master regulatory role in controlling whole-body lipid flux (16). Under conditions of chronic energy overload, adipose tissue sequesters excess fuel via expansion of adipocytes and leads to obesity. However, decompensation of this adaptive homeostatic mechanism (largely regulated by the interaction of genetic and epigenetic factors with dietary components) may lead to adipose tissue dysfunction and is likely to be the causal link for metabolic abnormalities in a subset of obese subjects.
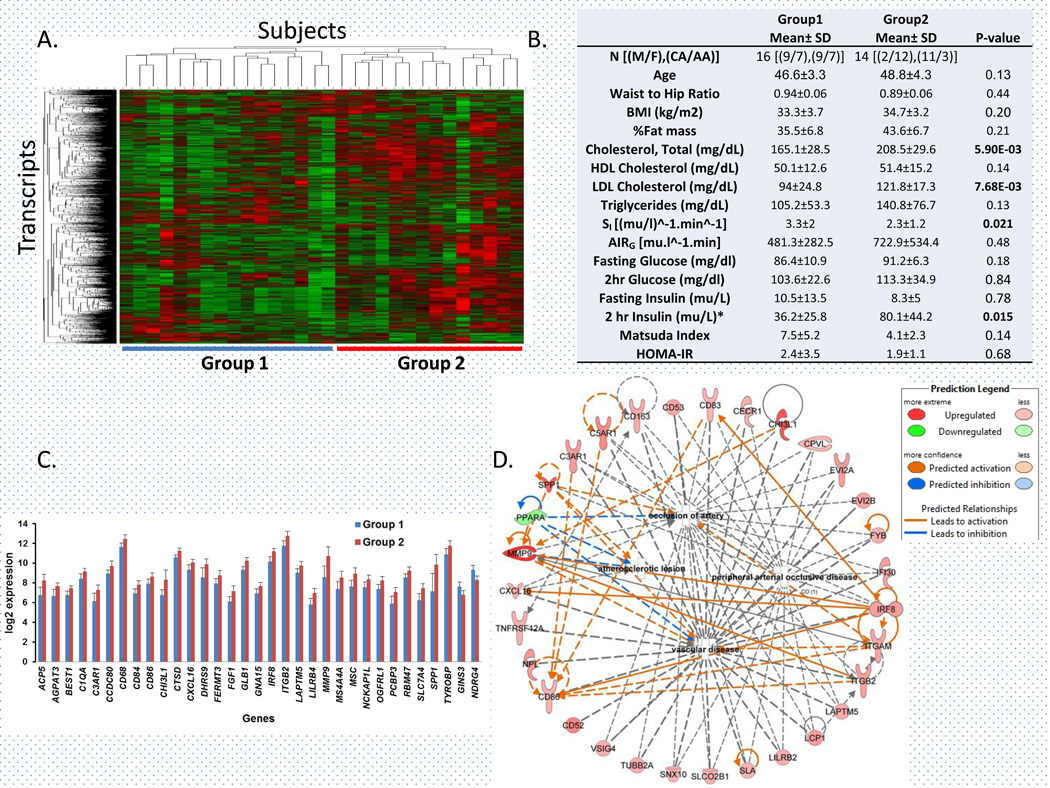
After unsupervised hierarchical clustering of 1595 obesity-associated (independent of age, gender and race) transcripts, we classified the 30 obese subjects into two groups of 16 and 14 subjects, respectively (Figure 1A). These two groups were not significantly different in BMI. However, group 2 subjects had significantly higher total cholesterol (p= 0.005), LDL-cholesterol (p= 0.006), 2h-Insulin during OGTT (p=0.015) and lower insulin sensitivity (SI, p= 0.029) compared to group 1 (Figure 1B). Despite similar BMI, obese subjects in group 2 showed poor gluco-and cardio-metabolic phenotype. Thus, our study revealed a distinct signature of adipose tissue transcripts that can differentiate obese subjects with comparatively better metabolic health (Group 1) from obese subjects with poor metabolic heath (Group 2). Group 2 subjects showed higher fat mass (43.6 ± 6.7%) compared to Group 1 subjects (35.5 ± 6.8%), but this difference was not statistically significant after adjustment for age, gender, and race. Similarly, differences in waist-hip ratio were also not significant. Thus, the observed difference in metabolic characteristics between the two groups cannot be attributed to the differences in total adiposity. However, we have not analyzed depot-specific distribution or histological characteristics of white adipose tissue in these subjects.
Figure 1. Adipose tissue transcripts play a role in defining gluco- and cardio-metabolic phenotypes in obese subjects.
A) Unsupervised hierarchical clustering of 1595 obesity-associated transcripts in subcutaneous adipose tissue classified 30 obese subjects into two major groups. B) Metabolic characteristics of two groups of obese subjects identified by clustering analysis. M, Male; F, Female; CA, Caucasian; AA, African-American; P-value, statistical significance based on univariate analysis of variance. C) Expression (log2) of transcripts (mean ± SD) most significantly (p ≤ 0.0001, fold change ≥ 1.5 and FDR ≤ 5%) differentially expressed between two groups of obese subjects. D) Network and predicted molecular activity of 31 differentially expressed genes involved in different cardiovascular disease-related processes.
We identified 158 transcripts with ≥1.5 fold difference in expression between groups 1 and 2 (Supplementary table 1). Group 2 had significant up-regulation of 141 genes and down-regulation of 17 genes. Highly up-regulated genes included MMP9 (Matrix metallopeptidase 9; 4.1 fold, p= 5.68×10−6) and SPP1 (Secreted phosphoprotein 1 or osteopontin; 3.2 fold, p= 3.54×10−5); while the top down-regulated genes included NDRG4 (N-myc downstream-regulated gene family member 4, −2.1 fold, p= 7.01×10−7) and GINS3 (GINS complex subunit 3; −1.8 fold, p= 1.58×10−5) (Figure 1C). Two large genome-wide association studies (GWAS) identified significant association of ECG QT interval (a quantitative measure of cardiac repolarization) in human chromosome 16q21, a genomic interval which includes several candidate genes including NDRG4 and GINS3 (17;18). NDRG4 knockdown in zebra fish embryos is associated with defective cardiac morphogenesis and function, including weak contractility due to marked reduction in proliferative myocytes (19). Similarly GINS3 −/− zebra fish embryos show a significant defect in cardiac repolarization (20).
In our study, the distribution of age and race was not significantly different between groups, but distribution of gender was significant. Thus, we compared only women from Group 2 (N= 12) and Group 1 (N=7) to identify differentially expressed genes. Despite the lower statistical power, results remain consistent. Of 158 differently expressed genes, 126 were significantly differentially expressed (p≤ 0.05) in the women-only analysis. The fold changes of these genes were strongly correlated between the overall cohort and the women only (overall vs women-only analysis, r=0.83, p= 8.76×10−42). Thus, the differential expression of genes between groups as identified by unsupervised hierarchical clustering analysis can mainly be attributed to differences in metabolic characteristics of the subjects.
Transcripts differentially expressed between the two groups were significantly enriched for the functional categories of genes (Table 1). These genes were primarily associated with cardiovascular disease, including peripheral arterial occlusive disease due to atherosclerotic lesions (enrichment p = 2.81×10−11). The Ingenuity knowledge base shows an interaction network of 31 differentially expressed genes involved in different cardiovascular disease-related processes (Figure 1D). These differentially expressed genes were also enriched in several canonical pathways involved in immune and inflammatory response, including the complement system (p= 8.32×10−5), TREM1 signaling (p= 3.09×10−3) and IL-8 signaling (p= 3.09×10−3) (Table 1). Similarly DAVID analysis identified signaling in the immune system (p= 3.94×10−10, 21 genes, Reactome pathway database) as the most strongly enriched pathway. Stronger upregulation of genes in pathways involved in immune and inflammatory responses, in metabolically unhealthy obese subjects, may be the link between obesity, cardiovascular disease, and type 2 diabetes (7). Genes in immune and inflammatory response pathway in adipose tissue were associated with insulin resistance (9), which could explain the significantly lower insulin sensitivity among Group 2 subjects in our study. Genes in these pathways may become novel therapeutic targets in preventing obesity-associated diseases. In conclusion, our study provides evidence that the perturbation in the adipose tissue gene expression network may be critical in defining metabolic health, including cardiometabolic phenotypes in obese subjects. Further functional studies will be required to define the causal relationship of these genes with cardio- and gluco-metabolic phenotypes in obesity.
Table 1. Enrichment of important biological pathways among genes differentially expressed between metabolically healthy and unhealthy obese subjects.
Differentially expressed genes and numbers of differentially up- or down-regulated transcripts in each enriched pathway/functional category within metabolically unhealthy obese subjects are shown (significant differences at Benjamini and Hochberg corrected p≤0.05). All significantly differentially expressed genes are shown in Supplementary Table 1.
| Diseases or Function Annotation | # Up-regulated |
# Down-regulated |
p-Value | Genes |
|---|---|---|---|---|
| Proliferation of blood cells (Cellular Development, Cellular Growth and Proliferation) | 28 | 0 | 2.81E-11 | BTK, CCNA2, CD163, CD209, CD33, CD37, CD4, CD83, CD84, CD86, CTSD, CTSZ, FCGR2B, IRF8, ITGB2, LGALS9, LILRB2, LILRB4, MMP9, MNDA, NCKAP1L, PTPRO, RAC2, SPI1, SPP1, TNFRSF12A, VSIG4, WAS |
| Peripheral arterial occlusive disease (Cardiovascular Disease) | 20 | 0 | 2.81E-11 | C3AR1, C5AR1, CD163, CD53, CD83, CECR1, CPVL, EVI2A, EVI2B, FYB, IFI30, IRF8, LAPTM5, LCP1, LILRB2, SLA, SLCO2B1, SNX10, SPP1, VSIG4 |
| Occlusion of artery (Cardiovascular Disease) | 26 | 1 | 1.91E-10 | C3AR1, C5AR1 , CD163, CD53, CD83, CECR1, CHI3L1, CPVL, CXCL16, EVI2A, EVI2B, FYB, IFI30, IRF8, ITGAM, ITGB2, LAPTM5, LCP1, LILRB2, MMP9, PPARA, SLA, SLCO2B1, SNX10, SPP1, TUBB2A, VSIG4 |
| Vascular disease (Cardiovascular Disease) | 30 | 1 | 1.29E-09 | C3AR1, C5AR1, CD163, CD52, CD53, CD83, CD86, CECR1, CHI3L1, CPVL, CXCL16, EVI2A, EVI2B, FYB, IFI30, IRF8, ITGAM, ITGB2, LAPTM5, LCP1, LILRB2, MMP9, NPL, PPARA, SLA, SLCO2B1, SNX10, SPP1, TNFRSF12A, TUBB2A, VSIG4 |
| Proliferation of immune cells (Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function) | 24 | 0 | 2.04E-09 | BTK, CCNA2, CD209, CD33, CD37, CD4, CD83, CD84, CD86, CTSD, CTSZ, FCGR2B, IRF8, ITGB2, LGALS9, LILRB2, LILRB4, MMP9, MNDA, NCKAP1L, SPI1, SPP1, VSIG4, WAS |
| Ingenuity Canonical Pathways | ||||
| Complement System | 5 | 1 | 8.32E-05 | C1QC, C5AR1, C1QB, C6, C3AR1, C1QA |
| TREM1 Signaling | 6 | 0 | 3.09E-03 | TLR8, CD83, FCGR2B, TYROBP, ITGAX, CD86 |
| IL-8 Signaling | 9 | 0 | 3.09E-03 | ITGAM, RRAS2, GPLD1, MMP9, ITGAX, ITGB2, RAC2, CYBB, NCF2 |
| Leukocyte Extravasation Signaling | 8 | 0 | 0.020 | ITGAM, WAS, MMP9, BTK, ITGB2, RAC2, CYBB, NCF2 |
| Actin Cytoskeleton Signaling | 8 | 0 | 0.025 | WAS, FGF1, RRAS2, NCKAP1L, IQGAP2, LBP, MATK, RAC2 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 6 | 0 | 0.025 | TLR8, C1QC, C5AR1, C1QB, C3AR1, C1QA |
| Role of NFAT in Regulation of the Immune Response | 7 | 0 | 0.025 | FCGR2B, RRAS2, BTK, CD4, FCER1G, CD86, GNA15 |
| Communication between Innate and Adaptive Immune Cells | 5 | 0 | 0.026 | TLR8, CD83, CD4, FCER1G, CD86 |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 5 | 0 | 0.041 | WAS, GPLD1, FGR, FYB, RAC2 |
Supplementary Material
Acknowledgements
This work was supported by grant R01 DK039311 and R01 DK090111 from the National Institutes of Health/NIDDK. We thank the Clinical Research Center staff of University of Arkansas for Medical Sciences for their outstanding support in the physiologic studies and assistance with data management. We thank Prof. Siqun Zheng, Director, and the technical staff of the Center for Human Genomics, Wake Forest School of Medicine, especially Ms. Shelly Smith and Dr. Ge Li for their extensive support in gene expression analysis. We also thank Ethel Kouba (Internal Medicine, Endocrinology) and Karen Klein (Biomedical Research Services and Administration) for critical reading and editing of our manuscript.
Footnotes
Conflict of interest:
The authors declare no conflict of interest
Supplementary information is available at the International Journal of Obesity's website.
References
- 1.Ahima RS, Lazar MA. Physiology. The health risk of obesity--better metrics imperative. Science. 2013;341(6148):856–858. doi: 10.1126/science.1241244. [DOI] [PubMed] [Google Scholar]
- 2.Bluher M. The distinction of metabolically 'healthy' from 'unhealthy' obese individuals. Curr Opin Lipidol. 2010;21(1):38–43. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- 3.Phillips CM. Metabolically healthy obesity: definitions, determinants and clinical implications. Rev Endocr Metab Disord. 2013;14(3):219–227. doi: 10.1007/s11154-013-9252-x. [DOI] [PubMed] [Google Scholar]
- 4.Tchkonia T, Thomou T, Zhu Y, Karagiannides I, Pothoulakis C, Jensen MD, et al. Mechanisms and Metabolic Implications of Regional Differences among Fat Depots. Cell Metab. 2013;17(5):644–656. doi: 10.1016/j.cmet.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jo J, Gavrilova O, Pack S, Jou W, Mullen S, Sumner AE, et al. Hypertrophy and/or Hyperplasia: Dynamics of Adipose Tissue Growth. PLoS Comput Biol. 2009;5(3):e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Despres JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 7.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444(7121):875–880. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- 8.Das SK, Sharma NK, Hasstedt SJ, Mondal AK, Ma L, Langberg KA, et al. An integrative genomics approach identifies activation of thioredoxin/thioredoxin reductase-1-mediated oxidative stress defense pathway and inhibition of angiogenesis in obese nondiabetic human subjects. J Clin Endocrinol Metab. 2011;96(8):E1308–E1313. doi: 10.1210/jc.2011-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes. 2011;60(3):1019–1029. doi: 10.2337/db10-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller MP, Attie AD. Physiological insights gained from gene expression analysis in obesity and diabetes. Annu Rev Nutr. 2010;30:341–364. doi: 10.1146/annurev.nutr.012809.104747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qatanani M, Tan Y, Dobrin R, Greenawalt DM, Hu G, Zhao W, et al. Inverse regulation of inflammation and mitochondrial function in adipose tissue defines extreme insulin sensitivity in morbidly obese patients. Diabetes. 2013;62(3):855–863. doi: 10.2337/db12-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma NK, Langberg KA, Mondal AK, Das SK. Phospholipid biosynthesis genes and susceptibility to obesity: analysis of expression and polymorphisms. PLoS One. 2013;8(5):e65303. doi: 10.1371/journal.pone.0065303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21(7):1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(5):367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41(4):407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu X, Jia H, Garrity DM, Tompkins K, Batts L, Appel B, et al. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev Biol. 2008;317(2):486–496. doi: 10.1016/j.ydbio.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120(7):553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.