Abstract
Tenascin-C is a large, multimodular, extracellular matrix glycoprotein that exhibits a very restricted pattern of expression but an enormously diverse range of functions. Here, we discuss the importance of deciphering the expression pattern of, and effects mediated by, different forms of this molecule in order to fully understand tenascin-C biology. We focus on both post transcriptional and post translational events such as splicing, glycosylation, assembly into a 3D matrix and proteolytic cleavage, highlighting how these modifications are key to defining tenascin-C function.
Keywords: biosynthesis, cancer, development, glycosylation, matrix assembly, proteolytic cleavage, splicing, tenascin-C, therapeutics, transcription
Abbreviations
- AD1/AD2
additional domain 1/ additional domain 2
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- ASMCs
aortic smooth muscle cells
- BDNF
brain derived neurotrophic factor
- bFGF
basic fibroblast growth factor
- BHKs
baby hamster kidney cells
- BMP
bone morphogenetic protein
- c
charged
- CA19–9
carbohydrate antigen 19–9
- CALEB
chicken acidic leucine-rich EGF-like domain containing brain protein
- ccRCC
clear cell renal cell carcinoma
- CEA
carcinoembryonic antigen
- chRCC
chromophobe-primary renal cell carcinoma
- CNS
central nervous system
- CRC
colorectal carcinomas
- CTGF
connective tissue growth factor
- DCIS
ductal carcinoma in-situ
- ECM
extracellular matrix
- EDA-FN
extra domain A containing fibronectin
- EDB-FN
extra domain B containing fibronectin
- EGF-L
epidermal growth factor-like
- EGF-R
epidermal growth factor receptor
- ELISPOT
enzyme-linked immunospot assay
- FBG
fibrinogen-like globe
- FGF2
fibroblast growth factor 2
- FGF4
fibroblast growth factor 4
- FN
fibronectin
- FNIII
fibronectin type III-like repeat
- GMEM
glioma-mesenchymal extracellular matrix antigen
- GPI
glycosylphosphatidylinositol
- HB-EGF
heparin-binding EGF-like growth factor
- HCEs
immortalized human corneal epithelial cell line
- HGF
hepatocyte growth factor
- HNK-1
human natural killer-1
- HSPGs
heparan sulfate proteoglycans
- HUVECs
human umbilical vein endothelial cells
- ICC
immunocytochemistry
- IF
immunofluorescence
- IFNγ
interferon gamma
- IGF
insulin-like growth factor
- IGF-BP
insulin-like growth factor-binding protein
- IHC
immunohistochemistry
- IL
interleukin
- ISH
in situ hybridization
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- mitogen-activated protein kinase
MAPK
- MMP
matrix metalloproteinase
- MPNSTs
malignant peripheral nerve sheath tumors
- Mr
molecular mass
- NB
northern blot
- NF-kB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
natural killer cells
- NSCLC
non-small cell lung carcinoma
- NSCs
neural stem cells
- NT
neurotrophin
- PAMPs
pathogen-associated molecular patterns
- PDGF
platelet derived growth factor
- PDGF-Rβ
platelet derived growth factor receptor β
- pHo
extracellular pH
- PIGF
phosphatidylinositol-glycan biosynthesis class F protein
- PLCγ
phospholipase-C gamma
- PNS
peripheral nervous system
- pRCC
papillary renal cell carcinoma
- PTPRζ1
receptor-type tyrosine-protein phosphatase zeta
- RA
rheumatoid arthritis
- RCC
renal cell carcinoma
- RD
rhabdomyosarcoma
- RGD
arginylglycylaspartic acid
- RT-PCR
real-time polymerase chain reaction
- SB
Southern blot
- SCC
squamous cell carcinoma
- siRNA
small interfering RNA
- SMCs
smooth muscle cells
- SVZ
sub-ventricular zone
- TA
tenascin assembly domain
- TGFβ
transforming growth factor β
- TIMP
tissue inhibitor of metalloproteinases
- TLR4
toll-like receptor 4
- TNFα
tumor necrosis factor α
- TSS
transcription start site
- UBC
urothelial bladder cancer
- UCC
urothelial cell carcinoma
- VEGF
vascular endothelial growth factor
- VSMCs
vascular smooth muscle cells
- VZ
ventricular zone
- WB
immunoblot/ western blot
The Multifunctional Nature of Tenascin-C
Tenascin-C was independently and concurrently characterized in the 1980s by several research groups with interests in the fields of cancer, matrix biology and embryonic/neural development. Tenascin-C is highly expressed in the developing embryo in a strictly regulated spatio-temporal pattern, but most healthy adult tissues exhibit negligible tenascin-C levels. Here, expression is constrained to sites where high cell turnover, plasticity and tissue-remodeling are obligatory; such as stem cell niches and the central nervous system (CNS), in addition to regions which undergo significant tensile stress; such as tendons, ligaments and smooth muscle fibers . However, transient expression of tenascin-C is observed at sites of active tissue remodeling in the adult, such as during the healing of wounds, and persistent tenascin-C expression is detected in pathological states such as cancer or rheumatoid arthritis (RA) (reviewed in1-3)
Tenascin-C consists of 4 distinct domains, which can interact with pathogenic components, matrix constituents, soluble factors and cell surface proteins; conferring upon tenascin-C the ability to bind to more than 25 different molecules identified thus far.3 For example, the tenascin assembly (TA) domain forms inter-molecular hydrophobic interactions and disulphide bridges, the epidermal growth factor-like (EGF-L) repeats act as a low affinity ligand for the EGF-receptor (EGF-R), inducing mitogen-activated protein kinase (MAPK), and phospholipase-C (PLC) -γ signaling, the fibronectin (FN) type III-like repeats (FNII) interact with proteins as diverse as integrins, aggrecan, and perlecan, as well as binding to growth factors including members of the platelet-derived growth factor (PDGF), fibroblast growth factor (FGF) and transforming growth factor-β (TGFβ) families (Table 1), and the fibrinogen-like globe (FBG) binds to integrins, receptor-type tyrosine-protein phosphatase zeta (PTPRζ1) and activates Toll-like receptor-4 (TLR4). This enables tenascin-C to drive a range of processes from oligomerization to induction of mitogenic responses, cell migration, cell attachment, cell spreading, focal adhesion, cell survival, matrix assembly, neurite outgrowth and potentiation and protease and pro-inflammatory cytokine synthesis (reviewed in 3).
Table 1.
Interactions of tenascin-C with growth factors and growth factor receptors. For studies on EGF-L repeats,198-200 FNIII 4–5,201 and FBG.202 Vascular endothelial growth factor (VEGF), phosphatidylinositol-glycan biosynthesis class F protein (PIGF), bone morphogenetic protein (BMP), neurotrophin (NT), brain-derived neurotrophic factor (BDNF), nerve growth factor (NGF), insulin-like growth factor (IGF), insulin-like growth factor –binding protein (IGF-BP), connective tissue growth factor (CTGF), heparin-binding EGF-like growth factor (HB-EGF), hepatocyte growth factor (HGF), chicken acidic leucine-rich EGF-like domain containing brain protein (CALEB)
| Tenascin-C Domain | Growth Factor/ Receptor Family | Growth Factor/ Receptor | Interaction Binding Affinity (+) No Binding Affiniy (−) |
|---|---|---|---|
| EGF-L Repeats | EGF-R | EGF-R | + |
| FNIII 4–5 | FGF | FGF-1/ -9/ -16/ -19/ -20/ -21 | - |
| FGF-2/ -4/ -6/ -7/ -8/ -10/ -17/ -18 | + | ||
| PDGF/VEGF | PDGF-AA/ -AB/ -BB/ -DD | + | |
| VEGF-A165 | + | ||
| VEGF-A121 | - | ||
| VEGF-B/ -C | + | ||
| PIGF | PIGF-1 | - | |
| PIGF-2/ -3 | + | ||
| TGFβ | BMP-2 | + | |
| BMP-4/ -6/ -7 | - | ||
| TGFβ-1/ -2 | + | ||
| TGFβ-3 | - | ||
| Neurotrophic Factors | NT-3 | + | |
| BDNF | + | ||
| NGF | - | ||
| IGF | IGF-1/ -2 | - | |
| IGF-BP3/ -BP5 | + | ||
| CCN | CTGF | - | |
| EGF | EGF | - | |
| HB-EGF | - | ||
| S1 Plasminogen | HGF | + | |
| FBG | EGF | CALEB-80 | + |
| CALEB-140 | - |
This collection of functions is reflected in the wide ranging consequences of genetic deletion of tenascin-C in mice. Reported abnormalities include reduced FN during dermal wound healing,4 hyperactivity,5 reduced kidney regeneration,6 reduced haematopoiesis,7 increased tumor monocyte population,8 abnormal tumor organization and angiogenesis,9,10 and aberrant immune responses,11-13 among many others (reviewed in14,15). Genetic variation at the human tenascin-C gene locus is associated with 6-fold increase in risk of Achilles tendon injury,16 non-syndromic hearing loss,17 and increased risk of developing adult asthma.18
Together these data point toward very diverse biological and pathological roles for tenascin-C. Here, we discuss some of the mechanisms that exist in order to dictate how this extracellular matrix (ECM) glycoprotein can exert so many different effects.
Transcriptional Regulation of Tenascin-C
One of the foremost means of regulating tenascin-C function is via control of its expression at the transcriptional level; and the cloning, and subsequent characterization, of the tenascin-C gene has begun to shed some light on the molecular mechanisms that underpin this tightly regulated control.
Cloning the TNC gene
The tenascin-C gene (TNC) is a large intron rich gene, which in humans spans 97.68 kb of DNA on the antisense strand of chromosome 9, at locus 9q32–34/9q33.119,20 and of which only ∼7.9% is protein coding (reviewed in 21). The first partial gene sequence derived for chicken tenascin-C identified several transcripts of various lengths prompting the speculation that the transcript is alternatively spliced.22 The first human tenascin-C exon sequence was published using cDNA clones isolated from U-373MG glioblastoma cells, identifying a clone with 8 consecutive FNIII like repeats, and another clone containing the same repeats but with a 1.9 kb insert between FNIII 5 and 6; providing clear genetic evidence of alternative splicing within the tenascin-C transcript.23
Transcriptional regulators of tenascin-C expression
Expression of tenascin-C is regulated in a stimulus specific manner in humans, mice, rats and chickens; the promoter elements of which are well conserved up to around 250 bp upstream of the transcription start site (TSS), including a TATA box located 21 nucleotides downstream of the TSS.24 Regulation of the tenascin-C promoter is influenced by many transcription factors (Table 2)(reviewed in21). These data illustrate how tenascin-C transcription may be induced or repressed in response to different subsets of stimuli including pathogen-associated molecular patterns (PAMPs), cytokines, reactive oxygen species, growth factors and mechanical stress. More about the transcriptional regulation of all tenascin family genes is described in detail by Chiovaro et al in this issue. However, it is likely that not all of the transcription factors regulating tenascin-C expression are known; and our understanding of the details of this large TNC locus is only in its infancy. Moreover, in addition to simply turning TNC on and off, some stimuli specifically induce particular forms of tenascin-C, and this is discussed in more detail below.
Table 2.
Transcriptional regulators of tenascin-C expression
| Stimulus | Transcription Factor/ Promoter | +ve/−ve regulation | Features of Study | Reference |
|---|---|---|---|---|
| PAMPs | ||||
| Lipopolysaccharide (LPS) | NF-kB | +ve | Identified 33 binding sites in TNC locus. In primary human monocyte derived dendritic cells tenascin-C expression was PI3K/Akt dependent | Goh et al.203 |
| Cytokines | ||||
| TNFα/ Sphingomyelinase | Postulated ATF-2/c-jun | +ve | JNK/SAPK-1 pathway activation increased tenascin-C expression in human epidermal keratinocytes. | Latijnhouwers et al.204 |
| IL-4/ IFN-y | Postulated STATs | +ve | Activate JAK-STAT pathway | Latijnhouwers et al.204 |
| IL-13 | unknown | +ve | Cultured human dermal fibroblasts, regulation was PI3K/Akt and/or PKC pathway dependent | Jinnin et al.205 |
| TNFα | NF-kB | +ve | TNFα driven tenascin-C expression was NF-kB p65 subunit (RelA) dependent in cultured human articular chondrocytes | Nakoshi et al.206 |
| Growth Factors | ||||
| TGFβ | Smad3/4, Ets1 Sp1, p300/ CREB-binding protein | +ve | Utilized WS-1 human dermal fibroblasts. | Jinnin et al.207 |
| PDGF | Sp1 Ets1/Ets2 | +ve | Cultured human dermal fibroblasts, PI3K/Akt dependent | Jinnin et al.208 |
| Fli1 | −ve | Overexpression in human dermal fibroblasts inhibits effects of PDGF on tenascin-C expression | ||
| Mechanical Strain | ||||
| Mechanical strain | NF-kB | +ve | Induced during mechanical strain via ROS in rat neonatal cardiac myocytes. Inhibited by anti-oxidants | Yamamoto et al.209 |
| Cyclic strain | MKL1 | +ve | Binds CArG box (c-fos promoter) | Asparuhova et al.210 |
| Biomechanical stretch | NFAT5 | +ve | Activates TNC expression following mechanical stress in vascular smooth muscle cells (VSMCs). May improve migratory activity in VSMCs | Scherer et al.211 |
| Other/ Unknown | ||||
| Evx-1 | +ve | Dependent on 89 bp region containing TRE/AP-1 site in chicken tenascin-c promoter | Jones et al.212 | |
| Strain responsive element | Chicken tenascin-C promoter | Chiquet-Ehrismann et al.213 | ||
| TATA box, Sp1, NF-1, C/EBP, AP-1, AP-2, Krox-20, Pax | +ve/ unknown | Sequence analysis of the promoter region identified multiple putative transcription factor binding sites | Gherzi et al.214 | |
| NF-1, TN control-element | +ve | +ve regulation in mouse NIH-3T3 fibroblasts, C6 rat glioma and N2A mouse neuroblastoma cells | Copertino et al.215 | |
| OCT | +ve | Required for +ve regulation by Brn2 in mouse N2A, inactive in C6 glioma | Copertino et al.215 | |
| Krox24/EGR-1 element | +ve/−ve | +ve regulation in mouse C6 glioma −ve regulation in mouse N2A neuroblastoma No effect in mouse NIH-3T3 fibroblasts | Copertino et al.215 | |
| OTX2 | −ve | Homeodomain protein involved in anterior head formation. Represses tenascin-C expression in OTX2 transfected cells; U87-MG glioma cells, C6 rat glial tumor cells, O1 human primary glioblastoma, MRC-5 human fibroblasts, NIH-3T3 mouse fibroblasts and SEK-MEL-28 human melanoma cells | Gherzi et al.216 | |
| NF-kB/c-Jun | NF-kB and c-Jun synergistically trans-activate the tenascin-C promoter with c-Jun binding at a GCN4/AP-1 site in rat REF and RF3T3 fibroblast cell lines | Mettouchi et al.217 | ||
| Denatured type-I collagen | Unknown | +ve | Rat aortic A10 VSMCs cultured on denatured type-I collagen express tenascin-C in ERK1/2 and β3 dependent. Promoter for transcription factor is in -43 to -165 bp 5’ of TSS | Jones et al.218 |
| Fli1 | +ve | Overexpression in human dermal fibroblasts results in +ve regulation. Modest activation observed with Ets1 and Ets2 | Shirasaki et al.219 | |
| Denatured type-I collagen | Prx1 | +ve | Prx1/2 expression increases when rat aortic A10 VSMCs are cultured on denatured collagen substrate. Prx1 expression enhances TNC promoter 20-fold | Jones et al.220 |
| Focal Adhesion Kinase | Prx1 | +ve | Mouse fibroblast cell-lines. FAK induces Prx1, promoting tenascin-C dependent migration | McKean et al.221 |
| GATA-6 | −ve | Overexpression inhibited basal and decreased IL-4 /TGFβ induced tenascin-C mRNA/protein levels in human foreskin fibroblasts | Ghatnekar and Trojanowska.222 | |
| Notch2 | RBPJk | +ve | Required for Notch-2 dependent transactivation of TNC promoter | Sivasankaran et al.223 |
Post Transcriptional Regulation of Tenascin-C
Alternative splicing allows a single gene to encode multiple proteins by the inclusion or exclusion of selected exons into the mature mRNA, dramatically increasing the size and diversity of the proteome. Up to 95% of the ∼21,000 protein coding genes in humans are alternatively spliced,25 and 85% of these have a minor splice isoform with an expression frequency exceeding 15%. As a result this moderate number of genes is able to produce >290,000 non-redundant peptide combinations.26 Following on from the first observations by Jones et al.22 and Gulcher et al.23 that tenascin-C is subject to alternative splicing, many studies have expanded on this theme revealing that post transcriptional modification of tenascin-C has a profound effect on tenascin biology.
Tenascin-c exon structure
The first TNC human exon sequences published showed the gene to comprise 28 exons (accession number NM_002610). However, 2 further alternatively spliced FNIII domains (additional domains 1 and 2: FNIII AD127 and AD228 (accession numbers U88892.1 and EU295718.1 respectively) were later discovered. FNIII AD1 was identified within human U251 glioma cDNA clones as a single exon between FNIII repeats B and C. This study also showed that while AD1 was present in human glioblastoma, neuroblastoma and osteosarcoma tumor cells, it was absent in healthy human lung fibroblasts and umbilical vein endothelial cells (HUVECs) providing the first indication that tenascin-C splicing may play a role in tumorigenesis.27 Human FNIII AD2 was discovered between FNIII B and AD1 in oral mucosa carcinoma samples and was named so on account of being in the same respective location as the avian FNIII AD2; situated between FNII B and AD1, in addition to it sharing 70% amino acid and 55% nucleotide sequence homology with the avian FNIII AD2.28,29 Thus human TNC contains 30 exons (Fig. 1).
Figure 1.
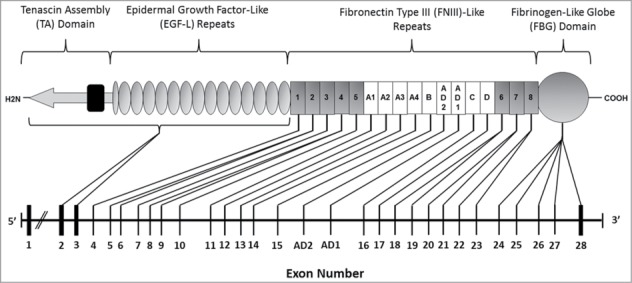
The exon structure of TNC with corresponding protein domains in tenascin-C. The human tenascin-C protein comprises 4 domains: a TA domain, 14.5 EGF-L repeats, up to 17 FNIII like repeats and an FBG domain. Eight of the FNIII repeats are constitutively expressed (FNIII 1–8 (gray), and 9 can be alternatively spliced (FNIIIA1-D (white). The TNC gene comprises 30 exons (1–28, plus AD1 and AD2). All exons are translated excluding the first. Exon 2 encodes the start sequence for translation of mRNA, and together exons 2 and 3 code for the signal peptide, the TA domain and all the EGF-L repeats. The 8 constitutively expressed FNIII repeats are coded for between exons 4–10 and 18–23, and the 9 alternatively spliced FNIII from exons 11–17. Each alternatively spliced FNIII repeat is encoded by its own exon, In contrast only the constitutive FNIII repeats 1 and 3 are encoded by a single exon; the remainder of the modules FNIII 2 and 4–8 are encoded by 2 exons each. Alternative splicing of FNIII domains within the tenascin-C pre-mRNA transcript means that the human TNC exon sequence varies in size from a maximum of 9154 bp to a minimum of 6251 bp. The FBG domain is coded for by exons 24–28.23,27,28,248
Tenascin-C in other vertebrate species including chickens, rats, mice and pigs, exhibits similar domain organizations to the human protein (Fig. 2). Each contains a TA domain, a series of EGF-L and FNIII like domains and an FBG domain. However, a number of differences exist between orthologs. For example, all vertebrate tenascin-C contains 13.5 EGF-L repeats, with the exception of mammals which possess 14.5 EGF-L repeats.30 Furthermore, tenascin-C from humans, rats, mice and chickens each contain 8 constitutive FNIII repeats, with an additional cassette of FNIII which may be alternatively spliced between FNIII domains 5 and 6. However, the number of alternatively spliced FNIII repeats varies by species; with chickens and mice, rats, and humans having 6, 7 and 9 respectively.28,31,32
Figure 2.
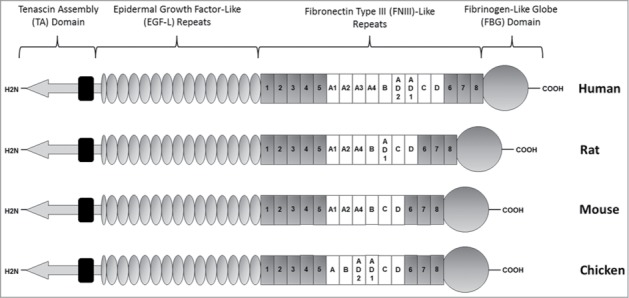
Schematic representation of human, rat, mouse and chicken tenascin-C. While each species contains 8 constitutively expressed FNIII repeats, the number and content of alternatively spliced FNIII repeats varies. Human tenascin-C contains 9 alternatively spliced FNIII repeats, rat 7, and mouse and chicken 6 each. Alternatively spliced repeats are typically more homologous than constitutive repeats. For example, constitutive mouse FNIII repeats share on average 44% nucleotide sequence identity to each other, in contrast to the alternatively spliced FNIII which share 52% identity. Of the mouse alternatively spliced FNIII, A2 and D share the lowest nucleotide identity at 41%, while A1 and A4 share 80%. Analysis of human tenascin-C also noted 80% amino acid sequence homology between the first 4 alternatively spliced modules (A1, A2, A3 and A4), in contrast to the other alternatively spliced FNIII repeats raising the possibility that these domains are the result of gene duplication of an ancestral FNIII module. The absence of any comparable homology between avian alternatively spliced FNIII repeats, allows for speculation that any such duplication occurred after the divergence of avian and mammalian lineages.19,33
Theoretically there are 511 possible human tenascin-C splice isoforms; this figure was calculated via the equation as follows, where 9 represents the total number of alternatively spliced FNIII, “r” represents the total number of alternatively spliced FNIII included in a single variant, and “!” denotes a factorial e.g. 4! = 4 × 3 × 2 × 1. Despite this large theoretical variation, the actual number of recorded splices does not exceed 100. This may partly be explained by the finding that some alternatively spliced FNIII domains are preferentially expressed together. Human FNIII AD2 is observed to be included in the transcript only when linked to AD1, FNIII C only when alongside D and interestingly FNIII A4 and C have never been observed to be linked together in mice or humans. 28,31,33This partnering of multiple alternatively spliced FNIII repeats significantly reduces the possible number of tenascin-C splice variants which are observed.
Alternatively spliced isoforms of tenascin-C have been shown to play integral roles in many different processes. Here we focus on how splicing controls tenascin-C action during development and in cancer.
Tenascin-C splicing - development
Tenascin-C is abundantly expressed during embryogenesis in neuroectodermal tissues, and subsequently in several non-neural sites where high cell-turnover, tissue remodeling and epithelial-mesenchymal interactions occur (reviewed in24). Typically, the overall expression of tenascin-C decreases with increasing age, generally peaking during embryonic development and shortly after birth before decreasing into adulthood when expression is restricted to a few sites at relatively low levels.34,35 Prior to the development of tools permitting the detection of specific alternatively spliced FNIII repeats, many studies reported the presence of high and low molecular mass (Mr) isoforms of tenascin-C, differentiated by their apparent Mr upon PAGE (PAGE) under reducing conditions, at various stages of embryogenesis in chickens, mice and rats. Further analysis utilizing in situ hybridization (ISH) and immunostaining/protein gel blotting (WB) with cDNA probes and monoclonal antibodies (mAbs) respectively, could distinguish between ‘long’ and ‘short’ tenascin-C forms; revealed that differently size splice variants are expressed in cell and tissue specific patterns that change over the course of development. These studies are described below and summarized in Table 3.
Table 3.
Association of ‘long’ and ‘short’ tenascin-C splice variants with stages of embryonic development
| Species | Size of splice variants detected (kb, kDa, if known) | Tissue or cell type | Features of study | Reference |
|---|---|---|---|---|
| Chicken | Small (150 kDa, 170 kDa, 190 kDa, 200 kDa) and large (220 kDa) | Embryonic skin fibroblasts, breast muscle | E11 fibroblasts predominantly express Tn220 in ratio 4:1:1 with Tn200/190 respectively, and express 7x more tenascin-C than muscle cells. E11 myoblasts express Tn220, 200 and 190 in ratio 2:1:1 respectively | Chiquet and Fambrough.42 |
| Small and large | Embryonic brain, gizzard, wing and skin fibroblasts | E10 skin fibroblasts predominantly express Tn220, but also Tn200 isoform. Doublet at ∼190 kDa predominant in E11 brain, gizzard and wing. Bands also detected include 210, 220 kDa and ∼400 kDa in brain | Erickson and Taylor.224 | |
| Small (170 kDa) and large (195 kDa, 205 kDa and 220 kDa) | Sterna | Identified 6-armed oligomer from E17 sterna. Reducing-PAGE gave prominent major bands at 195/205 kDa, and minor bands at 220/170 kDa | Vaughan et al.176 | |
| Small (190/ 200 kDa) | Embryonic chick retina | In E8 retina Tn 190/200 abundant along with ligand contactin/F11 in inner and outer plexiform layers. Identified possible binding site for HSPGs within FNIII-5 | Vaughan et al.176 | |
| Small (180 kDa, 160 kDa) and large (200 kDa, 220 kDa, 250 kDa). | Brain | Identified novel 250 kDa chondroitin sulfate containing isoform. Larger isoforms are expressed extensively at E6 to E15, but prevalence of smaller isoforms increases over this time | Hoffman et al.43 | |
| Small (220 kDa, 200 kDa/ 7.2 kDa) and large (240 kDa, 220 kDa/ 8 kb) | Embryonic gizzard, brain, liver | 7.2 kb and 8 kb mRNA isoforms increased in expression in E9/E15 gizzard and brain respectively. Corresponding peptides of 220/240 kDa and 200/220 kDa were identified in gizzard and brain respectively | Jones et al.22 | |
| Small (190 kDa) and large (230 kDa/ 200 kDa) | Embryonic chick fibroblasts | Identified 3 cDNA clones generated from E11 skin fibroblasts with open reading frames of 1808, 1626 and 1535 amino acids, which correspond with in vitro translated tenascin identified at 200/ 180 and 170 kDa | Spring et al.38 | |
| Small (190 kDa) and large (230 kDa/ 200 kDa) | Primary chick fibroblasts | Large isoform associated with gizzard smooth muscle layer and connective tissue below villi epithelium. Shorter isoform predominant in tendons and intramuscular connective tissue. Transfection with middle-T polyomavirus antigen induces preferential secretion of large but not small isoforms | Matsuoka et al.225 | |
| Small (6.6 kb, 6.4 kb) and large (220 kDa/ 7.2 kb) | Cerebellum | Total tenascin-C increased E8 to E15, then decreased until barely detectable at P3. Seven.2 kb mRNA prominent in E6–15 cerebellum while 6.4 kb decreases over this time. ISH probe for FNIII-B,C hybridized only to 7.2 kb message in CNS, and was absent in non-neural tissues (chondroblasts, tendons and lung mesenchyme) | Prieto et al.44 | |
| Large | Embryonic skin fibroblasts | Proteolytic cleavage of the 230 kDa variant isolated from E11 skin fibroblasts by pronase, and detected by mAb Tn68, produced C-terminal heparin binding peptide fragment specific only to cleavage of large isoform | Chiquet et al.37 | |
| Small (190 kDa) and large (200 kDa and 220 kDa) | Embryonic cornea | Close association between 220 Kda isoform expression and embryonic corneal cell migration in E3–19 | Kaplony et al.50 | |
| Small (190 kDa) and large (230 kDa) | Embryonic lung bud/ bronchiole tube epithelium | Used ISH to detect mRNAs corresponding to Tn190 in tips of budding bronchioles but not older epithelia or dense mesenchyme. Tn230 probes had identical association to Tn190 | Koch et al.226 | |
| Small (200 kDa, 190 kDa) and large (220 kDa) | Neural crest | Identified Tn230, 200, 190 in E3 neural crest cell conditioned media. Identified neural crest as major expresser of tenascin-C in developing spinal cord until E18 after which all splices not expressed | Tucker and McKay.48 | |
| Small (190 kDa, 200 kDa) and large (230 kDa) | Periosteal cells, osteoblast enriched cultures and endochondral bone. | Used universal cDNA probe to detect tenascin-C in osteogenic and chondrogenic regions. Full length tenascin-C present only in osteogenic regions and expressed by osteoblasts. Periosteal cultures express 3 isoforms but enriched osteoblast cultures express Tn230 only | Mackie and Tucker.45 | |
| Small (190 kDa) | Embryonic chick brain | Identified Tn190 as ligand to contactin/F11. Contactin/F11 binds FNIII-5,6 via first 3 Ig-domains and binding efficacy is reduced by incorporation of alternatively spliced FNIII | Zisch et al.58 | |
| Tn190, 200, 230 | Embryonic knee cartilage | IHC revealed Tn190 present in E11 articular cartilage. Peripheral articular cartilage and fibrocartilage expresses Tn200, and Tn230 is expressed in perichondrium | Pacifici et al.51 | |
| Small and large | Whole chick embryos | ISH revealed large and small tenascin-C abundantly expressed in embryo at E3–7. From E10–15 expression was spatially regulated; chord glia, Bergmann glia, endoderm-derived epithelium at growing tips of lung bronchioles, endothelium of major vessels, and osteogenic regions predominantly express large isoform. Small isoform associated with cartilage deposition and chondrocyte proliferation e.g. surrounding E10 notohord | Tucker.46 | |
| Small and large | Embryonic aorta and adjacent mesenchyme | Identified 3 isoforms containing 0, 1 or 3 alternatively spliced FNIII repeats, in haematopoietic progenitor/primordial germ cell migratory pathways of which the smallest was most prominent | Anstrom and Tucker.52 | |
| Mouse | Small (210 kDa) and large (260 kDa) | Gut mesenchyme | Small isoform predominant at E14, but after birth abundance of large isoform increases | Aufderheide and Ekblom.53 |
| Small (190 kDa) and large (220 kDa) | Embryonic intestine, brain | 190 kDa isoform more prevalent than 220 kDa isoform in mouse ileum, but relative concentration is the same from E14 to adulthood. Adult brain expresses single 160 kDa isoform. Developmental appearance of increasing concentration gradient of all tenascin-C from crypt to villus in ECM at epithelial-mesenchyme interface. Proposed to facilitate epithelial shedding in the villus | Probstmeier et al.227 | |
| Small (5.5 kb) and Large(7 kb) | Brain, submandibular gland, thymus, lung, heart, spleen, kidney, liver, pancreas, esophagus, stomach, intestine, bladder, skin and skeletal muscle | Two isoforms observed between E17 and P6 in skeletal muscle, stomach, cerebellum, bladder, duodenum, jejunum, ileum, and colon. Large (7 kb) isoform observed in lung, kidney and cerebrum. Small isoform observed in thymus and skin. Expression of all variants was decreased at P32, but small (5.5 kb) message continued to be transcribed in thymus, colon and cerebellum | Saga et al.34 | |
| Small (6 kb) and large (8 kb) | Kidney, intestine | Large variant predominant in newborn mouse kidney but postnatally small variant increases in abundance. E13 intestines express small isoform, and by birth the larger isoform is predominant | Weller et al.40 | |
| Small (190 kDa, 200 kDa/ 6 kb) and large (225 kDa, 240 kDa/ 8 kb) | Cerebellum | Expression of larger isoforms down regulated faster than smaller isoforms from P0 to >P60 | Bartsch et al.57 | |
| Small (200 kDa/ 6 kb) and large (230 kDa/ 8 kb) | Thymus, spleen, lymph nodes, lung, skin, cerebellum | Small isoform abundantly expressed in adult thymus, weaker expression in cerebellum, skin and none in spleen, testes, skeletal muscle, liver, kidney and heart. WB revealed 200 kDa tenascin in adult spleen and lymph nodes while thymus also contained 230 kDa isoform | Ocklind et al.35 | |
| Small (200 kDa) and large (250 kDa) | NIH-3T3 cells | TGFβ1 and FGF induce expression of small and large isoforms respectively in mouse embryonic fibroblast cell line | Tucker et al.47 | |
| Rat | Small (6.5 kb) and large (7.2 kb/ 280 kDa) | Lung | 7.5 kb mRNA more abundant than 6.5 kb in developing rat lung. Corresponds with prominent 280 kDa isoform detected from E17. All tenascin-C expression increases at early postnatal age and decreases to levels found in adult by P21. Bacterially expressed FNIII-6,8 peptide inhibited lung branching morphogenesis though only slightly more than FNIII1–5 and A-D | Young et al.54 |
| Small (6.4 kb/ 180 kDa) and large (7.3 kb/ 230 kDa) | Lung explant culture | TGFβ preferentially induces expression of 180 kDa isoform containing 1 alternatively spliced FNIII, over 230 kDa isoform containing 5 in dose dependent manner. Strong expression of both occurs from P2 to P30, and decreases from P30 into adulthood, although 230 kDa isoform more abundant from E19 onwards. | Zhao and Young.55 | |
| Small (180 kDa) and large (230 kDa) | Cultured lung epithelial, fibroblast and endothelial cells | The conditioned medium of lung fibroblasts and endothelium both expressed both 180 and 230 kDa variants, whereas lung alveolar cells expressed very little total tenascin-C | Zhao and Young.228 | |
| Range from small (190 kDa) to large (280 kDa) | Cortex, thalamus and cerebellum | E16 and P7 cortex, thalamus, cerebellum. All tissues expressed isoforms ranging from 190–280 kDa. Large variant is most prominent at E16 and P7 in all tissues. Expression of total tenascin-C increased most in P7 cortex and thalamus, but no major shift in the ratio of isoform expression was observed | Götz et al., 1997 79 |
Large versus small
A good example of this derives from an elegant series of studies in the developing chick which revealed 4 main isoforms; Tn260, Tn230, Tn200 and Tn190, which have apparent Mr of 260, 230, 200 and 190 kDa respectively.36,37 Tn260 was rarely expressed in contrast to Tn230, Tn200 and Tn190, which were widely expressed during embryonic and early postnatal development.38,39 High Mr variants (Tn230/200) were associated with regions of active tissue remodeling, cell migration and cell division; evidenced by their presence in epithelial substratum for migrating neurons, embryonic skin fibroblasts, whole brain, cerebellum, chord glia, Bergmann glia, endoderm-derived epithelium at developing lung bronchioles, growing wing bud tips, base of feather buds, major blood vessel endothelium, kidney, lung, osteoblasts and regions of osteogenesis; where the expression of tenascin-C is only required transiently.22,34,40-49 In these regions, large Mr tenascin-C is expressed by migrating glia and Bergmann glia in the developing chick spinal chord and cerebellum respectively; expression in the latter of which facilitates granule cell migration.46 In contrast, lower Mr isoforms (Tn190/200) are observed to be expressed more stably in dense connective tissue in areas such as gizzard tendons, intramuscular connective tissue, aortic mesenchyme, articular cartilage, inner layer of perichondrium and zones of active chondrocyte proliferation, where cell condensation and differentiation are more prevalent.22,39,41,44-47,50-52
Similar patterns of tenascin-C expression are observed in mice and rats, although significantly less is known about the presence of alternatively spliced tenascin-C during human fetal development. WB and real-time PCR (RT-PCR) techniques revealed that smaller Mr tenascin-C variants are the predominant isoforms expressed in E14 mouse gut mesenchyme,53 and in the developing thymus and skin from E17 to P6 respectively.35 Furthermore, smaller isoforms are observed to be persistently expressed into adulthood in thymus, colon, cerebellum, lymph nodes and splenic tissues.34,35 In contrast, the larger Mr isoforms are the predominant variants expressed in embryonic mouse kidney and in developing rat lung.40,54,55 Small isoforms are also implicated in regulating development of rat lungs, where the expression of a small variant is shown to be preferentially induced by TGFβ in explant cultures,55 and low Mr isoforms have the effects of inhibiting lung branching morphology and aveolarization.54
The splicing pattern within developing tissues was also observed to vary over time, as well as by location within the organism. For example, there is a shift in the relative abundance of splices away from large isoforms, in favor of smaller ones with increasing age in developing mouse kidney, cerebellum, skeletal muscle, stomach, bladder, duodenum, ileum, jejunum and colon.34,40,56,57 In embryonic chicken brain and gizzard, the relative proportion of smaller variants also increases with developmental age,22,43 as is also true in embryonic mouse intestine, gut mesenchyme, and chick cerebellum.40,44,53
The smallest tenascin-C isoform, with no alternatively spliced FNIII included, is known to bind strongly to FN and the glycosylphosphatidylinositol (GPI)-anchored neural cell adhesion molecule contactin/F1158-60 and to promote cell attachment and the formation of focal adhesions. This is in contrast to larger tenascin-C isoforms, which prevent focal adhesion formation and drive cell migration.61 These data imply that predominance of the larger isoform at sites of active tissue remodeling aid cell migration and dynamic tissue organization, while the smaller, pro-adhesive isoforms mediate stability of newly formed tissues toward the end of development and into adulthood. Indeed, in the prenatal chick brain, larger isoforms are abundantly expressed from E6 although the relative occurrence of smaller isoforms increases from E6 to E15;43 and at postnatal day 3 only a single 7.2 Kda message encoding a 220 kDa peptide is detected.44 Increased cell migration in the developing CNS was found to correlate with accumulation of long tenascin-C, but not short tenascin-C isoforms;50,57,62,63 indicating that long tenascin-C splices facilitate neurite motility in development.64
These studies provided a wealth of information about tenascin-C splicing during development, but it is worth mentioning that 2 isoforms containing a different repertoire of alternatively spliced FNIII repeats can still exhibit the same molecular mass, as each alternatively spliced FNIII repeat is approximately the same size (89–92 amino acids), with a mass of ∼10 kDa each. Moreover, analysis of tenascin-C forms based purely on Mr does not allow distinction between tenascin-C that has been alternatively spliced and tenascin-C that has been proteolytically clipped. As it became apparent that each alternatively spliced FNIII repeat had unique functions, analysis of the precise FNIII repeats that make up each isoform became increasingly important.
Analysis of specific FNIII repeats
The pattern of expression of specific FNIII domains in developing tissues and the functional consequences of the expression of these different isoforms are described below and summarized in Tables 4 and 5 respectively. In both the text and tables individual splice variants separated by (,) are included in the same transcript, while those separated by (/) are not.
Table 4.
Associations between specific alternatively spliced FIII repeats and developing tissues. Individual splice variants separated by (,) are included in the same transcript, while those separated by (/) are not
| Cell/Tissue Type | Alternatively spliced FNIII repeats | Associations | Method of Identification | Reference |
|---|---|---|---|---|
| Chick embryo | A,B | FNIII-A,B containing tenascin-C synthesized by migrating glia and osteoblasts at sites of epithelial-mesenchymal interactions in feather buds, kidney, bronchiole tips and tendons | ISH, IHC, RT-PCR | Tucker et al.46 |
| Embryonic mouse cerebellum | A1,A2,A4,B,D/ A1,A2,A4,D/ D/ no FNIII | Splices contained all FNIII or excluded C, B-C, A1-C or A1-D. Expression of isoforms containing 6, 5, 4, 1, 0 alternatively spliced FNIII decreased from E14 to adulthood, although expression of larger isoforms decreased faster than shorter isoforms | ISH, RT-PCR, Northern Blot (NB), sequencing, Southern blotting (SB) | Dörries and Schachner.56 |
| Rat aortic smooth muscle cells (ASMCs) | Full length/ D/ none | Treatment with PDGF-BB subunit homodimer or Angiotensin II induced expression of mRNAs containing all, one or no variable FNIII repeats. Tenascin-C also inhibited cell adhesion of ASMCs to FN | Radiolabelling, WB, cell adhesion assays, RT-PCR | LaFleur et al.229 |
| Embryonic chick spinal cord, tendons, base of feather buds, bronchiole tips, skin fibroblasts. | A/ B/ C/ A,B/ AD2/ AD1 | First report of FNIII-AD1/ AD2/ C in chicken. ISH cDNA probes for FNIII-A/ B/ C hybridize in E7 bronchiole tips, ligamentum flavum, kidney mesenchyme, FNIII-A,B in E7 aorta endothelium, spinal-chord ependyma and E10 spinal chord, tendons and base of feather buds. FNIII-C absent in spinal chord. Identified FNIII-AD1 and AD2 in E11 skin fibroblasts by RT-PCR | ISH, WB, IHC, RT-PCR | Tucker et al.29 |
| Chick embryonic lung bud tips, feather buds, bone | AD1/ AD2 | AD2 observed in E10 bronchiole bud tips. AD1 expression more widespread and abundant in developing bone (where 85% tenascin-C contained AD1) | Quantitative ISH, WB, IHC, RT-PCR and SB | Derr et al.49 |
| Human Fetal Membranes | Predominant isoforms D/ A1,A2,A3,A4/A1,A2,A4 | Identified 8 mRNA isoforms associated with processes analogous to tissue remodeling and wound response prior to labor and delivery in normal membranes Speculated that inclusion of FNIII A3 provides substrate for MMP-2, 3, 7 digestion prior to membrane rupture | Sequencing, SB, RT-PCR | Bell et al.96 |
| Mouse postnatal day 6 cerebellum | 27 isoforms ranging from 1–6 FNIII repeats | Identified 27 tenascin-C isoforms (22 of which were novel) in P6 cerebellum. Cerebellum confirmed as major expresser of tenascin-C in P6 brain. Only splice containing FNIII-D found in adult brain | RT-PCR, SB | Joester and Faissner.31 |
| Mouse embryonic whole tooth | D/ A1,D/ B,D/ B,C,D/ A1,A2,A4,B,D | E13 whole tooth expresses multiple alternatively spliced isoforms | ISH with cDNA probes, RT-PCR, IHC | Sahlberg et al.80 |
| Mouse embryonic dental papilla mesenchyme | D/ B,D/ A1,A2,A4,B,D | E12 dental mesenchyme expresses FNIII-D following induction by FGF-4 and TGFβ. TGFβ induces expression of long mRNA containing 5 alternatively spliced FNIII. Propose mesenchyme becomes sensitive to epithelial induction of tenascin-C during E11 | ISH with cDNA probes, RT-PCR, IHC | Sahlberg et al.80 |
| Mouse NSCs | 20 isoforms identified, novel A1,A4,B,D | Identified 20 isoforms in embryonic forebrain derived NSCs. Transcription factor Pax6 overexpression induced isoforms with 4, 5, and 6 alternatively spliced FNIII repeats, but downregulated smaller ones | RT-PCR, gene overexpression | Von Holst et al.69 |
| Mouse embryonic NSCs | Full length | Identified N-and O-linked HNK-1 epitopes expressed in NSC rich regions is almost exclusively expressed on full length tenascin-C. Small interfering RNA (siRNA) knockdown showed HNK-1 on full length tenascin-C promotes NSC proliferation via modulating EGF-R expression | Immunocytochemistry (ICC), WB, siRNA knockdown, RT-PCR, liquid chromatography MS/MS | Yagi et al.71 |
| Rat Hippocampal Neurons | AD1, various others. | FNIII AD1 detected in VZ and area dentate of rat brain. Observable shift from short to long isoform expression in rat hippocampus from embryonic day 16, to postnatal day 5 | ICC, RT-PCR, ISH | Garwood et al.32 |
Table 5.
Functional consequences of specific alternatively spliced FIII repeats during development. Individual splice variants separated by (,) are included in the same transcript, while those separated by (/) are not
| Cell/Tissue Type | Alternatively spliced FNIII repeats inc. | Function | Method of Identification | Reference |
|---|---|---|---|---|
| Chick embryonic skin fibroblasts | A,B,C/ C/ none | FNIII-A,B,C/ C/ none containing fusion protein had no effect on promoting cell adhesion. FNIII-7,8 promoted adhesion as efficiently as full-length tenascin-C evidenced by perturbation with mAb Tn68 | Generated fusion proteins, cell attachment assays, WB, electron microscopy, antibody perturbation experiments | Spring et al.38 |
| Embryonic rat hippocampal and mesencephalon neurons | B,D | Promotes neurite outgrowth and cell adhesion to substratum. Effects inhibited by anti FNIII-B,D mAb J1/tn2 | Antibody perturbation, cell substrate adhesion assay, ICC, rotary shadowing and electron microscopy | Lochter et al.92 |
| Bovine aortic endothelial cells | A3/ D | Antibody perturbation revealed FNIII-A3/ D mediate loss of focal adhesions. Confirmed by addition of recombinant protein containing FNIII-A1,A2,A3,A4,B,C,D | Antibody perturbation experiment, focal adhesion assays | Murphy-Ullrich et al.127 |
| Early postnatal mouse cerebellar cortex | A1,A2,A4/ B,D | FNIII-A1,A2,A4 promotes P6 granule cell neuron migration, but not outgrowth. B,C has no effect on migration but promotes outgrowth and increases the proportion of neurite bearing cells | Antibody perturbation experiment, cell migration assays, neurite outgrowth assays | Husmann et al.77 |
| Rat Lung (Fetal/ Postnatal) | A-D | Inhibits lung branching morphogenesis and aveolarization | Produced spliced domain anti-serums, IHC | Young et al.54 |
| Adult mouse mammary gland | A1,A2,A4,B,C,D | FNIII 1–3/ A1-D/ all (1–8) inhibit β-caesin expression and milk production during involution of mammary gland | Generated recombinant FNIII fragments, ICC, NB, WB | Jones et al.81 |
| Chicken PNS and CNS neuron cultures | A | FNIII-A promotes PNS and CNS neuron adhesion, increasing proportion of cells with extending neurites. Adhesion effects inhibited by anti-β1 integrin antibodies | Generated FNIII-A fusion protein, cell adhesion assays, antibody perturbation experiments | Phillips et al.85 |
| Embryonic and Postnatal Mouse and Rat CNS Neurons | A1,A2,A4,B,D/ A1,A2,A3/ B,D/ D,6 | FNIII-A1,A2,A4,B,D supported initial attachment in E18 rat and P6 mouse neurons. FNIII-A1,A2,A4 was repulsive to neurons, while B,D/ D,6/ and 6 promoted neurite outgrowth in E18 rat hippocampal and P0 mouse dorsal root ganglia explants | Cell binding assays, repulsion assays, neurite outgrowth assays, WB | Götz et al.76 |
| Chicken Embryos | AD2/AD1/C | Decreased cell attachment and actin microfilament bundle organization on cells adherent to FN. Increased adhesion on AD2/AD1/C containing substrata without focal adhesion | Cell adhesion assays, immunofluorescence (IF), RT-PCR, SB, IHC, in-situ hybridization | Fischer et al.86 |
| Rat cortical and thalamic explants | 4,5/ A1,A2,A4/ D | Tenascin-C IHC and western blot staining identified isoforms ranging from 190–280 kDa in E16-P7 cortical tissue. mAb perturbation with J1/Tn1, J1/Tn2/ J1/Tn4 inhibited axon outgrowth by binding FNIII-A1,A2,A4/ D and 4,5 respectively. J1/Tn3 to EGF-L had no effect | IHC, ISH, mAb perturbation, WB | Götz et al.79 |
| Rat embryonic cerebral cortical and hypothalamic neuronal cells | D/ A1,A4/ A4 | Surface bound long and short isoforms promote E17 neurite process extension. Soluble long and short variants have no effect, or inhibit outgrowth respectively. FNIII-A1,A4/ D/ 6 are permissive and 6–8 are inhibitory. Different sites are masked/exposed when surface bound | Neurite outgrowth assays, antibody perturbation, generating recombinant FNIII proteins, ICC, WB | Meiners and Geller.64 |
| Embryonic rat kidney | A1,A2,A4/ B,D | Tenascin-C expressed in kidneys from E14 past birth, strongest expression in cortical regions at newest growth. No alternatively spliced FNIII are implicated in kidney development in vitro | Antibody perturbation experiments (used 11 antibodies, 6 of which were novel) | Talts et al.230 |
| Rat embryonic cerebral cortical neurons, rat cerebral cortical astrocytes, baby hamster kidney cells (BHKs) | Full/ A1,A2,A3,A4/ B,C,D/ D | Bound human FNIII-A-D promotes neurite outgrowth. mAb perturbation revealed FNIII-D as outgrowth permissive region on FNIII A-D bound to astrocytes, and A1–4/ D as permissive on A-D bound BHKs. Bacterially expressed FNIII A1-A4 and B,C,D promoted astrocyte outgrowth on BHK cells | mAb perturbation, neurite outgrowth assays, binding assay, WB | Meiners et al.93 |
| Rat embryonic cerebellar granule neurons | Full/ C/ D | Discovered FNIII-C mRNA in early postnatal rat cerebellum. FNIII-D permits neurite extension, C regulates orientation and growth | RT-PCR, neurite guidance assays, mAb perturbation, IF, WB | Meiners et al.90 |
| Rat Embryonic Hippocampal Neurons | B,D/ D | FNIII-B,D/ D promote neurite process extension and outgrowth. The B,D effect was contactin/F3 dependent | Expressed hybrid-fusion proteins, Neurite outgrowth assays, RT-PCR | Rigato et al.91 |
| Early postnatal rat and mouse cerebellar granule neuronal cultures | D | Unique amino acid sequence VFDNFVLK within FNIII-D promotes neurite outgrowth in α7/ β1 integrin subunit dependent manner | Antibody perturbation, synthesized recombinant wild type and mutant FNIII-D, neurite outgrowth assay, ICC, affinity chromatography, WB | Mercado et al.78 |
| Embryonic rat hippocampal neurons | D | FNIII-D mediated E18 hippocampal neurite outgrowth in Ca2+, PLC, contactin and β1 integrin dependent manner | Neurite outgrowth assay, antibody perturbation, inhibitor experiments, pull down assay, WB, video microscopy | Michele and Faissner.x95 |
| Embryonic rat retinal explant | B,D/ D,6/ A1,A2/ A1,D | FNIII-B,D fusion protein promoted strongest fiber outgrowth in E18 retinal explants, followed by A1,D. FNIII-A1,A2 is inhibitory. The FNIII-D responsible for outgrowth with effects modulated by neighboring FNIII | Generated alternatively spliced FNIII fusion proteins to human Ig-Fc fragment, antibody perturbation, axon/ neurite outgrowth assay | Siddiqui et al.94 |
Developing mouse
Tenascin-C is abundantly expressed in the mouse olfactory bulb, ventricular zone (VZ) and sub ventricular zone (SVZ) of the CNS during development and postnatally65-67 where it has been shown to orchestrate the development of niches permitting the development of neural stem cells (NSC) altering their response to FGF-2 and BMP-4.68 Embryonic and postnatal day 6 mouse cerebellum express 4 and 27 different major mRNA isoforms respectively, of which the largest decrease in expression faster than the shorter variants during neural development.31,56 Similarly, NSCs express 20 isoforms which are differentially regulated by Pax6 and Sam68 transcription factors.69,70 In NSCs, expression of the largest isoform comprising FNIII-A1,A2,A4,B,C,D was demonstrated to contain N- and O-linked natural killer-1 (HNK-1) carbohydrate epitopes;71 first described on tenascin-C by Kruse et al. 72 HNK-1 epitopes; otherwise known as CD57 are named after the HNK-1 mAb, and are found on the surface of a subset of T-lymphocytes and natural killer (NK) cells, but also on a number of cell adhesion molecules distributed throughout the nervous system including myelin-associated glycoprotein,73 neural cell adhesion molecule,74 and L1 cell adhesion molecule.75 HNK-1 has functions in cell-cell and cell-substrate interactions, and promotes NSC proliferation via modulating the expression of the EGF-R.71
Further functional studies demonstrated how tenascin-C splice variants are capable of modulating the response of neurons during CNS development. In E18 rat and P6 mouse neurons, FNIII-A1,A2,A4,B,D coated plates, supported initial attachment.76 FNIII-B,D/ D,6 and 6 containing isoforms facilitate neurite outgrowth in P6 granule cell neurons and E18 rat hippocampal, P0 mouse dorsal root ganglia explants respectively, and even increase the proportion of neurite bearing cells in culture.76,77 Mercado et al.78 demonstrated that the FNIII-D repeat is strongly inductive of neurite outgrowth on account of 2 short sequences within the VFDNFVLK amino acid sequence, which permit interactions with α7 and β1 subunit containing integrins. The FNIII-A1,A2,A4 region in contrast is anti-adhesive and was demonstrated to promote neuronal migration and repulsion in rat E18 and P0 mouse root dorsal ganglia neurons,76,77 and E16 rat cortical and thalamic axonal outgrowth.79
Non-neural sites exhibiting a diverse expression of splice variants include the developing tooth, which at E13 expresses D/ A1,D/ B,D/ B,C,D/ and A1,A2,A4,B,D.80 Nearby dental mesenchyme papilla expressed FNIII-D following induction by FGF-4 and TGFβ, the latter of which also induced the expression of a long mRNA variant containing 5 alternatively spliced FNIII repeats. Interestingly, the response of dental mesenchyme papilla to growth factor induction always occurred in E12 but not always in E11, suggesting that the dental mesenchyme becomes responsive to the induction of tenascin-C at some point during E11.80 In the mammary gland, full length tenascin-C isoforms are highly prevalent during involution, and recombinant proteins containing FNIIIA1-D inhibit milk protein synthesis, suggestive of a role for tenascin-C in facilitating the cessation of lactation in mammals.81
Developing chicken
In Situ hybridization (ISH) experiments using cDNA probes complementary to FNIII AD2 or C revealed a strict spatial pattern of tenascin-C splicing in the developing chick. These probes did not hybridize to chondrogenic or osteogenic regions; while AD2 cDNA probes did hybridize at regions of epithelial-mesenchymal interactions including lung bronchioles and the base of feather buds.28,48 In contrast probes for AD1 and A,B hybridize strongly in almost all regions where tenascin-C is detected in the E10 chick embryo, such as feather buds, lung bronchioles, tendons, ligamentum flavum, cartilage and bone; with exceptions including spinal cord glia and sternal perichondrium.28,48 In developing bone, up to 85% of the tenascin-C transcripts detected via quantitative immunohistochemistry (IHC) included the AD1 repeat.49 FNII-C localizes in a pattern which exclusively overlaps the expression pattern for FNIII-A,B, with the exception that FNIII-C cDNA probes never hybridize in the spinal cord, or aorta endothelium, indicating a more tightly regulated pattern of expression in this location.29
Functionally, tenascin-C was shown to promote adhesion and outgrowth of E3 spinal cord, E8 sensory, E8–11 sympathetic and E6 retinal ganglion cell neurons in chickens.82-84 In CNS neuron neurons this effect was mediated by FNIII-A, but in peripheral nervous system (PNS) neurons was mediated by FNIII-A and 3. Cell adhesion to FNIII-A and 3 was inhibited by anti- β1-integrin antibodies, and arginylglycylaspartic (RGD) peptides respectively, illustrating that the adhesive effects of FNIII-A on neurons is β1-integrin dependent.85
In chicken embryos, the expression of isoforms containing FNIII-AD2/AD1/ and C are dramatically up-regulated at sites of active tissue remodeling and FN expression within the developing feather-bud and sternum. In vitro these variants have been shown to decrease cell attachment and organization of actin microfilament bundles in myoblasts cultured on FN.86 However, cells bound to a FNIII-AD2/AD1/C containing substrata developed stronger adhesions than those bound on tenascin-C containing no alternatively spliced FNIII, but did not form focal adhesions as is the case on FN. Instead they formed F-actin microfilament bundles at non-uniform adhesion points, giving the cells an irregular shape much like when adherent cells are cultured on thrombospondin-1.86,87 The introduction of alternatively spliced FNIII modules may conversely disrupt adhesions between tenascin-C and other ECM molecules, as was demonstrated by FNIII A/B/C splices binding less strongly to FN and the GPI-anchored immunoglobin-superfamily neural cell adhesion molecule contactin/F11.58 This is because in contactin/F11, the first 3-Ig domains preferentially bind to the uninterrupted FNIII-5,6 region;59 as a result, contactin/F11-tenascin-C binding is attenuated by the inclusion of alternatively spliced FNIII in the tenascin-C molecule which interrupt this binding region.58 Larger chick tenascin-C isoforms possess annexin II binding ability, and hence promote proliferation, cell migration and induce loss of focal adhesions hence encouraging cells cultured on high Mr tenascin-C to become motile.88,89 In addition, contactin/F11 binding to FNIII-5,6 was shown to be inhibited by heparin sulfate and dermatan sulfate.60 All in all, the inclusion of adhesion modulating FNIII repeats such as AD2/AD1/C in the developing chick embryo create an environment conducive to tissue remodeling by permitting changes in motility, adhesion and cell shape.
Developing rat
AD1 containing variants are found in the highly plastic SVZ and dentate gyrus of the developing rat brain, with an observable shift in expression from shorter to longer isoforms as development progresses from embryonic day 1 to postnatal day 5.32 An early mAb perturbation study revealed that FNIII-4,5/A1,A2,A4/ D but not the EGF-L was required for axonal outgrowth from E16 cortical and thalamic explants.79 In vitro, FNIII-C facilitates the orientation of rat cerebellar granular neurons, but has no effect on outgrowth.90 The variable rat FNIII B-D promotes contactin/F3 dependent neurite outgrowth when FNIII-C is excluded from the transcript; as its inclusion disrupts the contactin/F3 binding site formed between the adjacent FNIII-B and D.91 FNIII-B,D containing tenascin-C is widely reported to promote neuron outgrowth in embryonic rat hippocampal neurons, mesencephalic neurons, cortical astrocytes and retinal neurons.76,91-94 This repeat has a conserved function in P6 mouse cerebellar neurons, where it does not alter the rate of cell proliferation, but does increase the proportion of neurite bearing cells in culture.77 Similarly to mouse neurites, FNIII-D also promotes rat neurite outgrowth in a manner dependent on Ca2+, PLC, protein kinase-C and contactin.95 The outgrowth of E18 rat hippocampal neurons by FNIII-D was also inhibited by addition of anti-α7/ β1 integrin mAbs, caffeine, thapsigargin, inositol triphosphate receptor, ryanodine, 3,4,5-trimethylbenzoic acid 8-(diethylamino)octyl ester and proved dependent upon Ca2+ mobilization from the endoplasmic reticulum.95
Interestingly the abundance of FNIII-B,C in regions of high cell motility and turnover led some investigators to assess any potential functions of the repeat in developing organs outside of the CNS. However, despite a high abundance of tenascin-C in the E14 rat kidney mesenchyme, antibody perturbation of FNIII-B,C and A1,A2,A4 failed to exhibit any negative effect on the development of the kidney, suggesting that in an in vivo model at least, the presence of alternatively spliced FNIII in these tissues have no developmental function.95 The model did not take into account the potential roles of variable FNIII repeats on in vivo specific processes in development, such as vascularization which would make good considerations for the future.
Developing human
A wealth of tenascin-C splice variants was found within human fetal membranes which encapsulate the developing fetus and amniotic fluid. Bell et al.96 identified 8 different tenascin-C splice variants containing between 0 and 7 spliced FNIII repeats, which were speculated to promote spontaneous membrane rupture during labor by contributing to tissue remodeling processes in a manner analogous to a wound response. The predominant isoforms detected by RT-PCR contained FNIII-D/ and A1-A4, either with or without FNIII-A3.96 The selective inclusion/exclusion of tenascin-C isoforms containing repeat FNIII A3, may provide functional significance with regard to mediating the susceptibility of tenascin-C to proteolytic degradation by matrix metalloproteinase (MMP) -2 and 3; both of which can cleave tenascin-C at a site located within the FNIII A3. MMP-7 also exhibits protease activity, and can digest tenascin-C between FNIII repeats A3-D;96,97 perhaps facilitating fetal membrane rupture prior to birth. In this way, ‘long’ tenascin-C isoforms would be susceptible to MMP-2, 3 and 7 mediated fragmentations, whereas ‘small’ tenascin-C isoforms lacking A3 would be resistant. This process may generate a novel regulatory pathway where FNIII A3 containing isoform dependent processes can be regulated by MMP-2 and MMP-3.96
Tenascin-C splicing - cancer
Tenascin-C is abundantly expressed in the stroma of many solid tumors (reviewed in 1,98,3). In addition to elevated tenascin-C in tumors, the splicing pattern often differs compared to healthy tissues. Studies examining ‘long’ and ‘short’ tenascin-C isoforms are summarized in Table 6, while Table 7 details specific alternatively spliced FNIII detected in tumors and Table 8 the functional consequences of particular FNIII repeats on tumor cell biology.
Table 6.
Association of ‘long’ and ‘short’ tenascin-C splice variants with cancer
| Species | Alternatively Spliced FNIII Repeats (or size of splice variant if known) | Cell or Tissue Type | Features of Study | Reference |
|---|---|---|---|---|
| Human | Small (210 kDa) and large (230 kDa) | U-251MG Glioma | First identification of tenascin-C then called glioma-mesenchymal extracellular matrix antigen (GMEM). Identified major 230 kDa isoform and minor 210 kDa isoform. Did not specify whether difference in size of isoform is due to proteolysis or molecular heterogeneity | Bourdon et al.231 |
| Large (220, 230, 280 kDa) Large (320 kDa) | U-251 MG Glioma | Purified hexabrachions from conditioned media. Identified hexabrachions of different sizes, created by incorporation of different sized tenascin-C isoforms. In glioma, 220, 230 and 280 kDa tenascin-C isoforms were in roughly equal abundance | Aukhil et al.232 | |
| Small (180 kDa) and large (250 kDa) | Ductal and lobular breast carcinomas | Assessed distribution using mAbs in breast tissue from fetal, adult resting, lactating, aging parenchyma, fibrocystic, fibroadenomas, cystosarcoma phylloides and ductal and locular carcinomas. Total expression increased during fetal growth, gestation, hyperplasia, dysplasia, benign tumors, and much increased in infiltrating and intraductal breast carcinomas | Howeedy et al.99 | |
| Small (0 AS-FNIII), large (7 AS FNIII – 1.9 kb insertion between FNIII 5- 6 | Lung cancer tissues | 6 of 10 and 3 of 3 adenocarcinomas and SCC respectively, exhibited elevated expression of large isoforms relative to small ones. Variant containing 7 FNIII comprised 27%, 24%, 54% and 42% of the total tenascin-C in adenocarcinoma, SCC, large cell carcinoma and small cell carcinoma respectively | Oyama et al.101 | |
| Small (∼6 kb/190 kDa) and large (∼8 kb/330 kDa) | Normal, hyperplastic and neoplastic breast tissue | Invasive carcinoma expressed 6 and 8 kb tenascin-C mRNA isoforms. Small variant accounts for 85% tenascin-C in healthy tissues, and < 40% total in malignant ones. Intermediate sized variants detected in 3 of 16 invasive carcinomas. In fibroadenomas, 8 kb isoform associated with high stromal cellularity, findings supported by intermediate protein bands also detected from these donors | Borsi et al.100 | |
| Small (190 kDa) and large (280 kDa and 330 kDa) | Fibrosarcoma (HT-1080), Rhabdomyosarcoma (RD) and SV40 transformed fibroblast cell lines (WI-38-VA and AG-280), Melanoma (SK-MEL-28) | In WI-38-VA and SK-MEL-28 cell lines, only 330 kDa isoform visible in conditioned media. RD cells expressed low amounts of 330 kDa isoform in conditioned media and faintly in cell extract. HT-1080 and AG-280 expressed no detectable tenascin-C. Normal fibroblasts expressed 190 and 280 kDa isoforms in cell extracts and conditioned media | Carnemolla et al.233 | |
| Small (190 kDa) and large (280 kDa) | Transfected BHKs with long and short tenascin-C cDNAs using λgt11 vector. Tenascin-C purified from SK-MEL-28 cells | Characterized binding locations of 11 mAbs to regions of tenascin-C, and found that mAbs BC-2, α-A2, α-A3, α-B and α-D bind to alternatively spliced FNIII repeats A1 and A4, A2, A3, B and D respectively | Balza et al.234 | |
| Small and large | Prostatic hyperplasia and carcinoma | WB and RT-PCR analysis identified small and large isoforms of tenascin-C protein | Ibrahim et al.235 | |
| Small (190 kDa) and large (250 kDa) | Invasive CRC | Tenascin-C absent in normal tissues. Variants purified from invasive CRC were 190 and 250 kDa in size, identical to sizes those found in human fetal fibroblasts | Sakai et al.103 | |
| Large (330 kDa) | CRC and colorectal adenomas | Large tenascin-C splice variant (330 kDa) was detected in 7 of 15 carcinomas. Presence correlated with expression of FN extra domain-B (ED-B) | Hauptmann et al.105 | |
| Small (190 kDa) | Renal cell carcinoma (RCC) and Oncocytoma | Small 190 kDa isoform is predominantly expressed in RCC. Large isoform is almost absent | Lohi et al.102 | |
| Small and large | Malignantly transformed fibroblasts | Normal human fibroblasts predominantly express smaller or larger tenascin-C isoforms when cultured in more acidic or basic culture medium respectively (pH 6.8–7.2) Malignantly transformed cells are resistant to external pH regulation of splicing and predominantly express large variant because a more basic cytosolic pH is maintained | Borsi et al.166 | |
| Small (284 bp PCR product), and large (490, 556, 750, 1651 and 1924 bp) A4/ B/ C containing isoforms | Malignant and benign ovarian tumors | The smallest splice variant mRNA fragment (284 bp) found in all tumors tested. FNIII-B expression was widespread in all except smallest variant. Larger variants also expressed A4 and C. Nine/12 malignant compared to 1/6 benign tumors exhibited increase in intensity of larger ∼490 and 556 bp products relative to smallest 284 bp | Wilson et al.236 | |
| Small and large | Endometrial adenocarcinoma | Total tenascin-C expression induced by 20 ng/ml TGFβ. Many individual splice isoforms identified, the most abundant contained none, 1 or 7 alternatively spliced FNIII repeats | Vollmer et al.237 | |
| Small (190 kDa/ 5.5 kb) and large (220, 250 kDa/ 7.5 kb) | Lung cancer | All 30 lung cancers tested expressed 190 kDa isoform (5.5 kb mRNA), and also in 28 cases a larger 250 kDa band (7.5 kb mRNA); sometimes accompanied by a 220 kDa band. Normal lung only expressed 5.5 kb mRNA transcript, but 190 and 250 kDa tenascin-C could be very weakly detected by WB | Kusagawa et al.238 | |
| Small (5.8 kb) and large (7.5 kb) | Skin (dermal keratinocytes and fibroblasts) – non cancerous | In keratinocytes treatment with IFNγ and TNFα slightly increased mRNA expression of large and small isoforms respectively, while IL-4 increased both small and large isoforms equally | Latijnhouwers et al.204 | |
| Small (220 kDa) and large (320 kDa) | Chondrosarcoma Clinical Specimens, and Cell Line JJ012 | High small:large isoform ratio found in normal human articular chondrocytes. Low small:large isoform ratio found in chondrosarcomas and correlates with low survival. Determined via Semi-quantitative RT-PCR, IHC, survival analysis | Ghert et al.239 | |
| D (∼250 kDa) | NSCLC | 18-fold increase in large isoform expression observed in recurrent NSCLC compared to non-recurrent NSCLC, determined via quantitative RT-PCR and WB | Parekh et al.104 | |
| Large isoforms (unspecified) | Neurofibromatomas, plexiform neurofibromas and malignant peripheral nerve sheath tumors (MPNSTs) | Relative expression levels of total tenascin-C in neurofibromatomas, plexiform neurofibromas and MPNSTs were 1: 2.98: 4.95, with larger spliced variants accounting for 27.6%, 54.1% and 60.3% respectively. Determined by RT-PCR | Lévy et al.240 | |
| Small (210 kDa) and large (260 kDa) | Amdc-s cells (NIH-3T3 transfected with human AdoMetDC), Ras-E4 (transfected with c-Ha-rasVal12 oncogene) and Odc-n (NIH-3T3 cells transformed by overexpression of human ODC) | S-adenosylmethionine decarboxylase overexpressed in NIH-3T3 cells produced aggressive transformed cells. Amdc-s express 9.2-fold more tenascin-C than non-transformed cells, with the 260 kDa isoform abundantly expressed in Amdc-s, and the only isoform expressed in Ras-E4 cells; while Odc-n cells additionally expressed 210 kDa isoform | Paasinen-Sohns et al.241 | |
| Rat | Small (180 kDa) and large (220 kDa, very weak 280 kDa) | Hepatic and sarcoma derived cell linesx | Two hepatic and one sarcoma-derived cell lines shown to express major isoform 220 kDa isoform. Cell lines explanted into nude mice, epithelial sarcoma induced tenascin-C expression in stromal mouse tissue but no-longer expressed tenascin-C themselves. After transplantation the stromal hepatic-derived cell lines still prominently expressed tenascin-C | Sakai et al.242 |
Table 7.
Associations of specific tenascin-C splice variants, or individual alternatively spliced FNIII repeats in cancer. Individual splice variants separated by (,) are included in the same transcript, while those separated by (/) are not
| Species | Alternatively Spliced FNIII Repeats (or size of splice variant if known) | Cell or Tissue Type | Features of Study | Reference |
|---|---|---|---|---|
| Human | None/ D/ A4,B,D/ A1,A2,A3,A4/ A1,A2,A3,A4,B,D/ A1,A2,A3,A4,B, AD1/ A1,A2,A3,A4,B,C,D | U-251MG and U87-MG glioblastoma cell lines, MG-63 human osteosarcoma cell line, SK-N-SH neuroblastoma cell line, IMR-90 human lung fibroblast cell line, and HUVECs | Identified the novel FNIII-AD1 repeat located between FNIII-B and C, via RT-PCR and sequencing. Identified other large splice variants also by RT-PCR | Sriramarao and Bourdon et al.27 |
| A1,A2,A3,A4,B,C,D | U-251MG Glioblastoma cell line | Utilized radio-ligand binding assay to identify cell surface annexin II as a high affinity receptor for the whole alternatively spliced FNIII A-D region of tenascin-C (at the time this excluded FNIII AD2 and AD1) | Chung and Erickson.88 | |
| Human | AD1 | Hs578T breast ductal carcinoma, SK-MEL-24 melanoma, WERI retinoblastoma and BCC basal cell carcinoma, A431 epidermoid carcinoma cell line | Identified the presence of AD1in human cell lines Hs578T, SK-MEL-24, WERI, BCC but not in A431 epidermoid carcinoma cells | Derr et al.49 |
| Chicken | Small (190 kDa) and large (200 kDa, 230 kDa) QT6 contained AD1,D/ A,AD1,C/ A,AD1,D/ AD1,C,D/ A,AD1,C,D/ A,B,AD1,D/ A,B,AD2,AD1,D | QT6 quail fibrosarcoma, SL-29 chick embryo fibroblast | SL-29 express 190, 200 and 230 kDa isoforms encoding 0, 1 and 3 alternatively spliced FNIII in equal ratios. QT6 fibrosarcoma expressed predominantly 230 kDa isoform containing 3 spliced FNIII but has minor bands containing 0, 1, 2 and 4 alternatively spliced FNIII. In QT6 cells splices containing more than 3 alternatively spliced FNIII are rare in protein form | Derr et al.49 |
| AD2 | Malignant oral mucosae | Discovered human xFNIII-AD2 repeat. Possesses 70% amino acid and 55% amino acid sequence similarity with chicken-AD2. Identified in 2/10 oral cancers, but was absent in 40/40 normal, reactive, pre-malignant and other oral mucosae specimens | Mighell et al.28 | |
| Large A1,A2,A3,A4,B,AD1,C,D | Melanoma SK-MEL-28 | Large isoform is predominant isoform expressed by SK-MEL-28 although others are expressed in lesser abundance. Identified via sequencing, SB, RT-PCR | Bell et al.96 | |
| C | Astrocytoma and glioblastoma | FNIII-C associated with high grade (III) astrocytoma and glioblastoma tumors, blood vessels and proliferating cells. Absent in healthy tissues and barely detected in meningioma, low grade xastrocytoma, breast, lung and gastric carcinomas. Used TN-11 (mAb for FNIII-C) | Carnemolla et al.116 | |
| D/ A1,A2,A4 containing large isoforms. | CRC, ulcerative colitis and liver metastases | IHC with mAbs K8 (small isoform), 19H12, 201A, J1/tn1 (A1,A2,A4 region) and J1/tn2 (FNIII-D region). J1/tn2 stained strongly in stage I/II CRC but less so in stage III/IV tumors and liver metastases. The 19H12 staining was much stronger in metastases than early stage tumors | Dueck et al.125 | |
| Small and large (A1,A4 containing variants) | Oral SCC | Large tenascin-C variant stained strongly in tumor stroma, and expressed in single positive layer of cells at the tumor-stroma interface near invading cells, illustrated by ISH and IHC with mAb BC-2. Higher expression of large variant associated with increasing tumor stage | Hindermann et al.114 | |
| D, B/D | Breast Cancer cells MDA-MB 231, MDA-MB 468, MCF-7, T47D | Associated with metastasis and elevated invasion risk. Via RT-PCR, SB, sequencing. These variants are synthesized by stromal fibroblasts in malignant tissue, and by periductal fibroblasts, and myoepithelial cells in DCIS | Adams et al.121 | |
| C containing large isoforms | Cerebral cavernomas | Total tenascin-C localized to vascular walls and in interspace between blood cavities in cavernomas. FNIII-C localized to sub-endothelium of blood vessels in cavernomas and white matter surrounding the lesion sites. No FNIII-C was found in normal brain tissue. FNIII-C associated to tumor blood vessels in brain cancer | Viale et al.117 | |
| B containing large isoforms | Breast cancer (intraductal cancers) | Recombinant full length FNIII region of tenascin-C promoted in vitro migration and mitotic activity, effects perturbed by adding mAb 4C8MS (against FNIII-B). Large tenascin-C variants localized to invasion front of intraductal and ductal cancers. Positive correlation between large tenascin-C isoform expression and proliferation rate | Tsunoda et al.122 | |
| A1,A4/ A3, A4, B | Prostatic Adenocarcinoma | IHC revealed normal prostate tissue devoid of FNIII-A1,A4 containing tenascin-C, but staining is observed in tumor stroma with strong deposition around neoplastic glands. ISH revealed FNIII-A3,A4,B abundant in tumor cytosol and is associated with tumor invasion front and loss of cell adhesion in all adenocarcinomas tested | Katenkamp et al.115 | |
| A1, B, D, AD1 | UCC | A1, B, D Restricted to invasive tumors, tumor blood vessels and destructed muscle. AD1 associated with compact invasion pattern. Studied via Semi-quantitative IHC and RT-PCR correlated to tumor stage | Berndt et al.120 | |
| A1/ D | U87 Glioblastoma | Generated recombinant human mAbs to FNIII A1 and D (F16 and P12 respectively). F16 selectively localized at tumor site in U87 glioblastoma, and was rapidly cleared from other organs. F16 identified for potential antibody-based pharmaceutical development | Brack et al.154 | |
| Small (322bp fragment) and large (2243bp fragment A1,A2,A3,A4,B,C,D) | Cutaneous SCC and actinic keratosis (AK) | Large isoform found in 5%, 63% and 88% of normal, AK and SCC samples respectively. In SCC, tenascin-C associated with basal cells at invasion front and papillary/reticular dermis | Dang et al.243 | |
| A1 and A4 containing large isoforms | Immortalized human corneal epithelial cells (HCEs) | Co-deposited laminin-332 and large tenascin-C variant in plaque beneath adhering cells and was Golgi dependent. HCE adhesion to laminin and large variant tenascin-C was dependent on α3β1 integrin | Katz et al.244 | |
| C | Lung cancer | Generated mAb G11 specific to FNIII-C and via IHC observed expression in majority of lung cancers, in vascular and stromal pattern. G11 demonstrated preferential localization to tumor site in rats grafted with U87 gliomas, so may have application in delivery of imaging or therapeutics to glioma or lung tumors | Silacci et al.118 | |
| None and A1,A2,A3,A4,B,AD2,AD1,C,D | Thyroid carcinoma cell lines. TT medullary carcinoma, ARO/ FRO anaplastic carcinoma, WRO follicular carcinoma, BHP 2/ BHP 5/ BHP 7/ BHP 10/ BHP 14/ BHP 17/ BHP 18, BHP 19 and SW579 papillary carcinoma | RT-PCR illustrated large transcript fragment containing full alternatively spliced cassette (variable splice region 1 - VS1) was more prevalent than the small variant in all cancers tested, except for medullary carcinoma cell line TT | Tseleni-Balafouta et al.245 | |
| A1 and A4 containing large isoforms | Atypical oral brush epithelium | Specificity and sensitivity of conventional HE staining for atypical oral brush biopsies increased from 96–99% and 78–95% respectively when combined with BC-2 immunostaining for large FNIII-A1/A4 containing tenascin-C isoforms | Driemel et al.126 | |
| B containing large variants | Oral SCC | IF with BC-3 mAb (for FNIII-B). No large tenascin-C variant identified in normal oral basement membrane. In dysplastic and neoplastic oral mucosa long tenascin-C variant containing FNIII-B was co-localized to laminin-5/gamma-2 in the basement membrane region. Extent of reorganization of large variant in basement membrane correlated with malignancy grade | Franz et al.119 | |
| C | CRC | Serum levels of large-tenascin-C variants containing FNIII-C quantified by ELISA. Primary CRC patients 5260 ± 3243.3 pg/ml, recurrent CRC patients 4106 ± 2261.1 pg/ml and healthy donors 2364 ± 749.6 pg/ml. Sensitivity for detecting CRC via serum levels was 56.6%, exceeding conventional tumor markers CEA at 40.1% and CA19–9 at 23.6% | Takeda et al.146 | |
| C | Hepatic recurrence of CRC | High serum levels of large tenascin-C splice variants in 2 patients; as detected via ELISA sensitive to FNIII-C domain, were associated with hepatic recurrence of CRC | Takeda et al.147 | |
| B, C | UCC | Measured urine concentration of FNIII-B, C in 104 UBC patients, 11 patients with cystitis and 15 healthy donors. Increased urinary FNIII-B concentration correlates with tumor progression in UBC. Proteolytic fragmentation of tenascin-C was also observed in urine from invasive tumor patients | Richter et al.148 | |
| AD1, AD2 | Breast cancer DCIS, ductal carcinomas, lobular carcinomas, fibroadenoma | AD1 and AD2 present in 34.9% and 23.1% of invasive breast carcinomas respectively. AD1 and AD2 not tumor specific but expression is increased in carcinomas from younger women. AD1 localized to tumor cells and myoepithelium of normal breast ducts. AD1 associated with ER negative and grade III tumors | Guttery et al.123 | |
| A1 | Melanoma | 24 primary and 29 metastatic melanoma lesions stained with F8, L19 and F16 mAbs for extra domain-A containing FN (EDA-FN), EDB-FN and FNIII-A1 respectively. F16 strongly stained basal lamina and deeper layers of tissue compared to others. F16-IL2 could therefore be useful as therapeutic for malignant melanoma | Frey et al.246 | |
| B | UBC | Determined urine levels of FNIII-B/ C containing tenascin-C in 35 patients via ELISA. FNIII-B could predict cases without tumor recurrence, or with tumor existence. Could also predict whether UBC was muscle/or non-muscle invasive | Gecks et al.149 | |
| A1/ C | Clear cell RCC (ccRCC), papillary (pRCC), chromophobe-primary RCC (chRCC) | Detected FNIII A1/C in RCC samples. FNIII A1 in addition to FN ED-A/B was associated with vascular structures. By contrast FNIII-C was absent in ccRCC, strongly expressed in 80% of pRCC and was widely expressed in chRCC | Galler et al.247 | |
| Mouse, human | Large isoforms | NIH-3T3, human melanoma cells | Overexpression of SRSF6 transcription factor in mice induces hyperplasia of sensitized skin, and in melanoma is associated with increases in expression of full length tenascin-C isoforms found in invasive carcinomas; Knockdown of SRSF6 in NIH-3T3 down-regulated large tenascin-C variant. SRSF6 associates with exons 10–14 (encoding FNIII-5,A1,A2,A3,A4) | Jensen et al.168 |
Table 8.
Functional consequences of tenascin-C alternative splicing in cancer. Individual splice variants separated by (,) are included in the same transcript, while those separated by (/) are not
| Species | Alternatively Spliced FNIII Repeats (or size of splice variant if known) | Cell or Tissue Type | Features of Study | Reference |
|---|---|---|---|---|
| Human | none and A1,A2,A3,A4,B,C,D | U-251MG cell line, and BHKs transfected with human tenascin-C | Small isoform tenascin-C binds purified and mixed FN substrate via FNIII-3 and a region within FNIII-6,7,8. Solid phase binding assay demonstrated stronger binding of small rather than full length tenascin-C to FN. FNIII-A-D fusion protein showed minimal displacement of bound FN-Tenascin-C complexes compared to FNIII-3/3–5/1–5 | Chung et al.129 |
| Human | Small and large (220 and 320 kDa isoforms respectively) | Chondrosarcoma Cell Line JJ012 | Small isoform of tenascin-C binds FN and promotes adhesion when bound to plastic. Large isoform does not bind FN and fails to promote cell attachment. Determined via ELISA, cell attachment assays, antibody blocking experiments | Ghert et al.61 |
| Human | D (∼250 kDa) | Non-Small Cell Lung Carcinoma | Inhibits CD3-dependent lymphocyte proliferation and INFγ secretion in tumor-invading lymphocytes. Observed via Lymphocyte proliferation assay, ELISPOT assay and antibody perturbation. Eighteen-fold increase in large isoform expression | Parekh et al.104 |
| Human | A2 | SV40-transformed human embryonic lung fibroblasts (WI-38-VA-13 cell line) | FNIII-A2 domain contains syndecan-4 binding cryptic site exposed by MMP-2 digestion. Stimulates β1 integrin mediated cell adhesion to FN. Suggests the extracellular portion of syndecan-4 binds FNIII-A2 and subsequently promotes β1 integrin clustering and activation | Saito et al.132 |
| Human | D/ B,D | Breast cancer (MCF-7, T47D, MDA-MD-231, MDA-MB-468, GI101) and fibroblasts | Overexpression of full length tenascin-C increased mean invasion index (MII) in MDA-MD-231 and T47D cells, but overexpression of FNIII- D and B,D by tumor cells enhanced proliferation and invasion significantly. Fibroblasts overexpressing full length, or FNIII-B/ B,D also promoted invasion of tumor cells. Tenascin-C upregulated expression of MMP-13 and TIMP-3 | Hancox et al.130 |
| Human | A1,A2,A3,A4,B,AD2,AD1,C,D | Pancreatic cancer | Recombinant FNIII-A-D bound cell surface annexin II, and suppressed gemcitabine mediated cytotoxicity in pancreatic cancer cells in dose dependent manner. This interaction increased intracellular phosphorylation status of PI3K, Akt, IKKα/β and NF-kB. NF-kB inhibition by siRNA restored gemcitabine cytotoxicity | Gong et al.135 |
| Human | B,AD1,D | Breast cancer cell lines (MCF-7, T-47 D, ZR-75–1, MDA-MB-231 and GI-101) | Overexpression of FNIII B,AD1,D enhanced tumor cell invasion and growth relative to baseline levels | Guttery et al.123 |
| Human | Small (220 kDa) and large (320 kDa) | Chondrosarcoma | Exogenous addition of 320 kDa isoform stimulates twice the levels of MMP-1 expression observed when adding 220 kDa isoform. Thirty-fold activation of MMP-1 promoter by 320 kDa isoform, compared to 220 kDa isoform. Furthermore, collagenase and invasion activity of chondrosarcoma increased 3-fold in 320 kDa variant treated cells | Galoian et al.128 |
Tissue levels of large tenascin-C variants
What little tenascin-C is present in healthy or benign tissues generally consists of the small variant containing no alternatively spliced FNIII repeats (Mr 180–190 kDa). In contrast, a range of larger isoforms with Mr 210, 220, 230, 250, 260, 280, 320 and 330 kDa, which would be predicted to contain one or more alternatively spliced FNIII repeats, have been described to be deposited in the ECM of a variety of human tumors including breast, colon, bladder, ovaries, prostate, pancreas, kidney, liver, uterus, brain, mouth, lung, skin, cartilage, connective tissues and peripheral nervous system (Table 6).
For example, total tenascin-C expression was observed to dramatically increase in invasive and intraductal breast carcinomas relative to normal breast tissues, with 2 major isoforms of 180 kDa and 250 kDa detectable via WB.99 Subsequent analysis revealed that 85% of tenascin-C in normal breast tissue is a small 180 kDa isoform, but in malignancy the larger 330 kDa variant becomes predominant, with the small isoform always accounting for less than 40% of the total tenascin-C.100 Similarly, elevated expression of large tenascin-C isoforms relative to smaller ones in lung cancer, was reported by Oyama et al.101 who found elevated mRNA isoforms encoding a large tenascin-C variant with 7 alternatively spliced FNIII repeats in 6 of 10 adenocarcinomas, and 3 of 3 squamous cell carcinomas (SCCs) tested.
However there are exceptions to this rule; the expression of tenascin-C in cancer is highly polymorphic, depending on the type of cell involved, and the stage of disease. This is evidenced by examples of cancers where the smallest tenascin-C isoform is predominantly expressed, such as in some renal, colon and breast carcinomas.99,102,103
Association of large isoforms with disease progression
A number of studies have suggested that large tenascin-C isoforms may correlate with prognosis, invasion and cancer progression in the bladder, brain, colon, lung, breast, B-cell non-Hodgkin's lymphoma, and adipocytic tumors. Furthermore, the presence of these large isoforms in some cases can also indicate disease recurrence and chemotherapy resistance (Table 6).
In the case of non-small cell lung carcinoma (NSCLC), an 18-fold increase in the expression of large tenascin-C variants was predictive of disease recurrence.104 Similarly, large 8 kb mRNA variants correlated with high stromal cellularity in breast cancer, and 330 kDa protein variants with extra domain-B containing FN (EDB-FN) positive colon cancer; linking large tenascin-C isoform expression with a tumor permissive stroma and the presence of invasive and angiogenic tumor marker protein EDB-FN, the expression of which is otherwise absent in healthy tissues.100,105 Likewise, in normal and benign breast cancer, the 190 kDa isoform was predominantly expressed, with a shift to expression of large 330 kDa variant in invasive disease. Furthermore in 3 of 16 cases of invasive disease, a number of intermediate splice variants were identified which suggested that while stromal cells are capable of synthesizing tenascin-C of 2 major isoforms, 190 and 330 kDa, epithelial cells can express tenascin-C isoforms of intermediate mass.100
Again though, while elevated tenascin-C expression in tumors is often associated with poor prognosis and patient outcome, there are many examples of tumors where there is no such correlation. There is no correlation between tenascin-C expression and prognosis or disease stage in stomach adenocarcinoma;106 metastasis, invasion or survival in gastric carcinoma;107,108 survival in oral and pharyngeal SCC;109 survival, clinical stage or metastasis in pancreatic carcinoma.110 There are also cases where tenascin-C expression is absent in poor prognosis tumors, such as in cases of cervical carcinoma.111 Other studies found no associations between tenascin-C expression pattern and a number of clinical features such as nodal metastasis, tumor necrosis or blood vessel invasion in 32 invasive breast carcinomas.112 Interestingly, one study found that in colorectal carcinoma (CRC) low expression of tenascin-C is associated with lymph node metastasis, perhaps indicating that in some cancers tenascin-C is capable of enforcing some anti-metastasis mechanisms.113
Specific FNIII repeats and their association with tumors
Technological advances in the field now permit the identification of specific spliced tenascin-C variants in cancer, and many studies have used these tools to identify subsets of tenascin-C isoforms, or single alternatively spliced FNIII repeats in tumors. While a systematic analysis of every splice variant in specific tumors is for the most part still lacking, these studies illustrate the distribution of specific FNIII repeats in malignancy, and shown that in some cases distinct FNIII repeats are associated with increased tumor grade, invasion, migration and metastases (Table 6).
For example, many IHC studies have utilized mAbs specific to regions FNIII-A1/A4, such as BC-2; showing that FNIII A1/A4 containing variants are not detected in normal cells, but are found at the tumor invasion front with apparent associations with migrating tumor cells. In oral SCC FNIII-A1, A4 containing variants are localized to the tumor stroma at the interface near invading cells, with greater expression correlating with higher disease stage.114 Similarly, FNIII-A1,A4 and A3,A4,B are absent in normal prostate, yet strongly localized to neoplastic glands and the tumor invasion front in prostatic adenocarcinoma by IHC and ISH respectively.115 In addition, FNIII-C containing tenascin-C isoforms were identified in brain cancer tissues via IHC using the mAb TN-11,116 and subsequently using a catalog ISH probes and RT-PCR techniques. This study found strong associations of FNIII-C with high grade astrocytoma and glioblastoma, cerebral cavernomas and lung cancers where the distribution of FNIII-C was consistently associated with tumor stroma, tumor blood vessels and proliferating cells in these tissues.116-118 Likewise, FNIII-B containing tenascin-C isoforms co-localize to laminin-5/γ2 in the basement membrane region of oral SCC, where increased expression positively correlates with malignancy grade.119 Another study revealed by IHC and RT-PCR that invasive urothelial cell carcinoma (UCC) exhibits restricted expression of FNIII-A1/ B or D which is closely associated with tumor blood vessels, invasive tumor stroma and damaged muscle. Moreover, the same study identified an association between AD1 and a compact invasion pattern.120
Similarly, the preferential expression of tenascin-C isoforms containing FNIII repeats B/ D/ AD1/ AD2/ B,D and B,AD1,D is well characterized in breast cancers including ductal carcinoma in situ (DCIS) and is known to occur at the tumor invasion front, and in cancers with elevated risk of metastasis.49,121-124 These isoforms were originally thought to be tumor specific; although a more recent study identified FNIII-AD1 and AD2 expression in the myoepithelium of larger normal ducts in the breast, while also detecting their presence in 34.9% and 23.1% of invasive breast carcinomas respectively.123 There is also a clear correlation between expression of AD1 and AD2 containing tenascin-C isoforms and invasive estrogen receptor negative breast cancer in younger women (≤40 y of age), however no such correlation was observed between FNIII-AD1 and AD2 prevalence with human epidermal growth factor receptor 2 expression. Since one of the primary chemotherapeutics for the treatment of breast cancer is the ER antagonist tamoxifen, the prevalence of FNIII AD1 and AD2 may be suitable as predictor for tamoxifen-resistant breast cancer, or breast cancer which is not yet tamoxifen resistant; yet may become so. In this way, surveillance of the tenascin-C isoform expression pattern could positively impact patient treatment.123
Other IHC studies illustrated how the pattern of tenascin-C splicing changes dynamically during disease progression. For example, anti-FNIII-D mAbs stained strongly in early stage I/II CRC, but less so in later stage III/IV tumors and liver metastases. However, FNIII-A1,A2,A4 containing isoforms were much more abundant in metastases then early stage tumors demonstrating a shift in expression from FNIII-D to A1,A2,A4 during the development of CRC.125
These observations may prove useful in cancer patient diagnosis or stratification. For example, Driemel et al. demonstrated the diagnostic potential of combining hematoxylin and eosin staining with BC-2 IHC, in atypical oral brush biopsies, which would allow for the additional detection of FNIII-A1/ A4 containing isoforms. This combination protocol increased the sensitivity and specificity of conventional HE staining from 96 to 99% and 78 to 95% respectively.126
Consequences of FNIII repeat expression in cancer
Tenascin-C possesses the ability to modulate cell behavior in normal, tumor and tumor-associated cells in a manner which is cell-type specific, and which may either directly or indirectly affects cancer initiation and progression. Functional studies have identified roles for large tenascin-C variants in promoting a tumor supportive tissue microenvironment, with early studies identifying roles in the down-regulation of focal adhesion activity,127 promotion of cell migration,50 up-regulation of ECM degrading enzymes128 and prevention of binding to FN;39,61,129 processes which would favor the development of malignancy by reducing adhesion and facilitating cell migration (Table 8).
Large tenascin-C isoforms may promote the destruction of the local tissue environment during tumorigenesis and metastasis by inducing the expression of ECM degrading enzymes such as MMP-1 in chondrosarcoma128 and MMP-13 in breast cancer. Although here MMP-13 expression is accompanied by increased levels of TIMP-3, and was not tenascin-C isoform specific.130 The degradation of the ECM and the driving of an epithelial-mesenchymal transition is crucial in the process of tissue invasion as observed in metastasis. In chondrosarcoma, addition of the large 320 kDa tenascin-C isoform conferred a 3-fold increase in collagenase and invasion activity via modulation of MMP-1 expression.128
Functional studies in human cells have also served to elucidate the role of specific tenascin-C isoforms in malignancy, and these have been best characterized in breast cancer (reviewed in124). AD1 containing isoforms promoted cell growth and invasion above control levels in breast cancer cells.123 These data are supported by previous findings of FNIII-AD1 expression in breast ductal carcinoma, melanoma, retinoblastoma, basal cell carcinoma cell lines and in highly invasive Japanese quail fibrosarcoma cells (QT6 cell line).49 QT6 predominantly express a major 230 kDa isoform containing 3 alternatively spliced FNIII, although minor isoforms containing 0, 1, 2 and 4 spliced FNIII are detected in low abundance. FNIII-AD1 is abundantly expressed in QT6 cells, and correlates with these cells exhibiting invasive capabilities as demonstrated in a Boyden chamber assay where they passed through a FN coated filter.49 Other studies have also identified AD1 containing tenascin-C isoforms at sites of high cell motility.131 Together, these findings may suggest that AD1 containing tenascin-C isoforms modulate cell-FN interactions and contribute to a tissue microenvironment that facilitates changes in cell morphology, proliferation and migration in cancer.49,123,131
Large tenascin-C isoforms have been directly implicated in the induction of proliferation, migration and control of cell spreading in vitro. MMP-2 mediated proteolytic cleavage of large tenascin-C variants reveals a cryptic syndecan-4 binding site within the FNIII-A2 domain. Here, a 22mer fusion peptide containing an adhesive YTITIRG sequence bound to the heparin sulfated side chains of the β1 integrin co-receptor synecan-4, hence inducing cell adhesion and spreading on FN.132 This study found that mutation of the cytosolic region of syndecan-4 had no effect on promoting the inhibition of β1 integrin action, suggesting the extracellular portion of syndecan-4 binds FNIII-A2 and hence promotes β1 integrin clustering and activation, rather than mediating this effect via downstream targets of syndecan-4. Elevated levels of both activated MMP-2 and degraded tenascin-C were identified in NSCLC, with proteolytically degraded tenascin-C serving as a good indicator of stage-1 NSCLC recurrence.133 It is therefore possible to speculate a role for MMP-2 in degrading tenascin-C in lung cancer, and that FNIII-A2 cryptic site interaction with syndecan-4 in lung tissues play a role in mediating early stage recurrence of the disease.
A follow up study identified that sustained activation of α5β1 by the FNIII-A2 cryptic site caused non-transformed mouse embryonic fibroblasts NIH-3T3 to become resistant to serum deprivation-induced anoiksis by activating the pro-survival Akt/Bcl-2 pathway, and enhanced PDGF-dependent proliferation of NIH-3T3 cells via promoting the association between PDGF receptor β (PDGF-Rβ) and the activated molecular complex that comprises of α5β1 and syndecan-4.134 This extended the activation of the PDGF-Rβ and its downstream Ras/MAPK pathways. Furthermore, the study suggested that tenascin-C may de-regulate cell growth processes by triggering the continual activation of α5β1 by the FNIII-A2 cryptic site; this mechanism was implied by the observation that confluent monolayers of fibroblasts continued to proliferate in an α5β1 dependent manner, and had apparently overridden their innate cell-density mediated cell cycle arrest mechanisms.134 This demonstrates a highly significant role for MMP-2 cleaved tenascin-C in the promotion of cancer.
Chung and Erickson.,88 first showed that the large tenascin-C isoform containing the full alternatively spliced FNIII cassette is a high affinity binding partner of the cell surface located calcium-dependent phospholipid-binding protein annexin II, binding of which reduces focal adhesions, enhances mitogenesis and promotes cell migration.89,127 Similarly, addition of full length tenascin-C to pancreatic cancer cells ameliorated the cytotoxic response to chemotherapeutic agent gemcitabine, in a manner which was dependent upon binding between FNIII A1-D and cell surface annexin II.135 Addition of recombinant FNIII A1-D increased the intracellular phosphorylation status of protein kinase B signal cascade constituents including PI3k, Akt, IKKα and NF-kB.135 Although in this study the effects of FNIII A1-D were not assessed in the context of promoting cell survival, proliferation or growth, such phosphoproteins are central mediators of these processes; for example, phospho-Akt is a major promoter of cell survival and effects this via phosphorylating and hence inactivating a number of pro-apoptotic target proteins including Bad, Bax, Bim and Forkhead transcription factors (FOXOs).136
The anti-FNIII-B mAb 4C8MS attenuated in vitro migration and mitotic activity in intraductal breast cancers.122 Functional overexpression of FNIII-D/ B,D and B,AD1,D in human breast cancer cell lines or FNIII B and B,D in fibroblasts significantly increased proliferative and invasive capabilities of breast cancer cells compared to vector controls in vitro; in addition to inducing a 2- and 4-fold increase in expression of MMP-13 and TIMP-3 respectively.123,130 Interestingly, during embryonic development, FNIII B and D promote neurite outgrowth in a manner dependent upon interactions with the F3/contactin and α7β1 integrin,78,91 permitting speculation that conserved mechanisms promoting cell migration in embryogenesis are being re-activated in tumorigenesis.
A further antibody perturbation study determined that specific splice variants of tenascin-C containing FNIII-D reduced immune surveillance in solid tumors, evidenced by the large tenascin-C variant inhibiting CD3-FN mediated lymphocyte proliferation and interferon-γ (IFNγ) production in tumor invading lymphocytes.104 The presence of FNIII-D would therefore be expected to be accompanied by increased tumor recurrence rates on account of an impaired immune clearance mechanism. Interestingly, Parekh et al. 104 found that large 250 kDa isoform expression is increased 18-fold in recurrent NSCLC compared to non-recurrent NSCLC; which if presuming FNIII-D expression was increased also, would be consistent with this hypothesis.
Serum levels of tenascin-C
Without taking into account individual splice isoforms, global serum levels of tenascin-C have been shown to have either diagnostic or prognostic functions in pancreatic adenocarcinoma,137 CRC (healthy control 3.2+/−1.7 μg/ml, CRC mean 5.0+/−4.2 μg/ml, CRC without metastases 4.3+/−3 μg/ml
CRC with metastases 6.8+/−6 μg/ml),138 inflammatory bowel disease (healthy control 3.2+/−1.7 μg/ml, prior to surgery for ulcerative collitis (UCC) 17.2+/− 4.6 μg/ml, ulcerative collitis + ongoing medical treatment, median 5.3 μg/ml, Crohn's disease + ongoing medical treatment, median 4.9 μg/ml),139 melanoma (healthy control 444+/− 171 ng/ml, stage I/II melanoma 419+/−103.3, stage IV melanoma 782.6+/−629.5),140 and auto-immune bullous diseases of the skin,.141 In NSCLC patients tenascin-C is cited as a predictive biomarker of angiogenesis, and levels above 96 ng/ml in patient serum inversely correlate with patient survival.142 However in diseases like breast cancer and ovarian cancer where serum levels of tenascin-C are elevated from 137 ± 26.8 pg/ml and 90.1 pg/ml to 344.1 ± 42.4 pg/ml and 130.5 pg/ml respectively, there is no predictive or prognostic function in relation to patient survival;143,144 with another study identifying elevated serum tenascin-C levels as a dubious tumor biomarker as serum levels also increased in during infection and as a consequence of inflammation.145
Two studies identified elevated serum levels of FNIII-C as a suitable tumor biomarker for CRC (healthy control 2364.3+/−749.6 pg/ml, primary CRC 5260+/−3243.3 pg/ml, recurrent CRC 4160+/−2261.1 pg/ml), where it exceeded the sensitivity of conventional CRC diagnostic tests for serum markers carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19–9) at 40.1% and 23.6% respectively, and diagnosed CRC with 56.6% sensitivity,146 furthermore elevated serum levels of FNIII-C were associated with hepatic recurrence of CRC metastases.147
Urine levels of FNIII-B from patients with UCC and urothelial carcinoma of the urinary bladder (UBC) have been quantified in several studies using commercially available ELISA kits (FNIII-B containing isoforms: healthy control 200 ng/ml, cystitis 650 ng/ml, pTa grade UBC ∼1000 ng/ml, pT3–4 grade UBC ∼8500 ng/ml, FNIII-C containing isoforms: healthy control ∼60 ng/ml, cystitis ∼200 ng/ml, pTa grade UBC ∼200 ng/ml, pT3–4 grade UBC ∼990 ng/ml) (FNIII-B containing isoforms: non-muscle invasive UBC ∼2.4 ng/ml. non-muscle invasive relapse ∼2.9 ng/ml, muscle invasive relapse ∼8 ng/ml).148,149 While proteolytic fragmentation of tenascin-C was observed in the urine from invasive UCC patients, and was indicative of enhanced protease activity and tissue destructive disease,148 FNIII-B concentration in UBC patient urine could predict existence or recurrence of a tumor, and also whether the disease was muscle invasive.149 The first study also quantified urine levels of FNIII-C containing isoforms, although these were found not to be indicative of disease stage in UCC patients.148
However, the number of cancers where serum levels of tenascin-C sufficiently correlate with disease progression is low and tenascin-C would appear an unsuitable universal cancer biomarker as had perhaps once been hoped. Moreover, the serum levels of tenascin-C detected in healthy controls vary vastly from one study to another, which may partly be explained by use of different ELISA kits and sample preparation, but not wholly, as many of these studies use comparable methodology. Therefore at the current time, in some specific disease types, serum or urine tenascin-C surveillance may improve diagnosis when combined with already existing techniques. Interestingly, tenascin-W exhibits an even more restricted expression pattern in normal tissues than tenascin-C and has been proposed as a superior biomarker for malignancy.150
Therapeutic exploitation of tenascin-C splice variants
The strong association between the expression of alternatively spliced FNIII repeats of tenascin-C with tumors, and in many cases very aggressive tumors, combined with their reduced expression in normal healthy tissues has raised interest in pharmacological targeting of these modules. These studies are extensively reviewed in,3 and are therefore only summarized below.
Most studies to date have focused on using antibodies to alternatively spliced FNIII domains to deliver cytotoxic or anti-inflammatory factors direct to the tumor, but given the active role these domains play in driving tumor cell biology, there also exists the potential for developing antagonists of these domains for use in cancer treatment. Antibodies specific for FNIII-A1 (F16) and FNIII-C/D (81C6) have been best characterized, although others are in development. 131I-labeled 81C6 has been trialled as a therapeutic in malignant glioma where it targets residual cancer cells following brain surgery.151 131I-81C6 increased median survival of glioblastoma multiforme patients, although correlations were observed between irreversible neurotoxicity and decreased survival. Nevertheless, 131I-labeled 81C6 delivers highly targeted radio-isotope therapy and is now undergoing phase III trials with the trade name Neuradiab.152,153 Three other anti-tenascin-C antibody conjugates are currently under development, all of which use the FNIII A1 targeting antibody F16.154 F16 is conjugated with interleukin (IL) -2, 131I, and 124I for the therapy of lung and breast cancer, Hodgkin's lymphoma and various tumors respectively.153 F16-IL-2 is reportedly a highly efficacious and safe immunotherapeutic when administered in combination with the chemotherapeutic temozolomide to BALB/c nude mice with sub-cutaneous and intracranial human glioblastoma tumors, F16-IL-2 induced a complete remission in these animals, and promoted leukocyte infiltration of the tumors. Following complete remission, the mice remained tumor free for 160 d155 The latest instalment in this exciting story is published in this issue in the article by Catania et al, who report data from a phase Ib/II trial administering F16-IL2 cytokine fusion proteins in combination with doxorubicin to patients with solid tumors and metastatic breast cancer.
Alternative splicing beyond development and cancer
Alternative splicing of tenascin-C is not restricted to developing tissues and tumors, but is more likely a universal feature of tenascin-C expression during tissue remodeling. Indeed 22 different tenascin-C splice isoforms were identified in pseudophakic and aphakic bullous keratopathy corneas, with the predominant isoforms containing one, 2, both or neither of the FNIII A1/D modules.156,157 These data suggest that a detailed and systematic examination of tenascin-C splicing will reveal important information about the role of this molecule in different locations. Using well-established PCR techniques requiring the generation of primers to flank the alternatively spliced region and subsequent ISH, cloning and sequencing of the expanded PCR products,31,69 or exploiting the ability of deep sequencing technology to identify all mRNA species present within cells of different tissues and lineages, will help to reveal the full picture of this post transcriptional modification of tenascin-C at the mRNA level.
Regulation of tenascin-C splicing
As described above, tenascin-C splicing is regulated in a very distinct spatio-temporal manner, which is clearly observed during embryogenesis and in numerous pathological conditions. The mechanisms by which this complex process is controlled in such a highly orchestrated time and location dependent manner are beginning to emerge.
The regulation of tenascin-C expression is mediated by growth factors and cytokines in a situation-specific response. During embryogenesis in humans and mice, tenascin-C expression is upregulated by TGFβ.47,158 TGFβ-1 provides the most significant upregulation of tenascin-C although expression is also responsive to TGFβ-2, -3 and -5.159 TGFβ-1 is also observed to induce preferential expression of the small tenascin-C isoform lacking FNIII A1-D in endometrial adenocarcinoma and NIH-3T3 cell lines.160 In vivo, tenascin-C expression closely correlates with expression of TGFβ in the heart, bone marrow megakaryocytes, breast cancer, ovarian cancer and inflamed joints.161,162 FGF is also a potent inducer of tenascin-C expression, with the expression pattern of FGF2 in the mouse CNS corresponding with that of tenascin-C.47 In addition to FGF up-regulating tenascin-C expression on the whole, FGF1 and FGF2 has been shown to preferentially induce the expression of large tenascin-C splice variants in the lateral ventricles of the rat brain, and the NIH-3T3 fibroblast cell line respectively.47,160,163
The expression of tenascin-C is influenced by the presence of pro-inflammatory cytokines, which also can either up-regulate the expression of all tenascin-C isoforms, or induce the preferential expression of long (tenascin-C isoforms containing one or more alternatively spliced FNIII), or short isoforms (containing no alternatively spliced FNIII). This effect has been investigated in keratinocytes, where IL-4 has been shown to induce the expression of short and long mRNA transcripts equally, with a relative abundance of 1:1.5. In contrast, the potent immune-activator tumor necrosis factor-α (TNFα) preferentially induces the expression of short tenascin-C transcripts, while IFNγ favors the expression of long transcripts.164
Another important regulator of tenascin-C splicing is intracellular pH (pHi). In cell cultures of lung and dermal fibroblast cell-lines, a physiological pH ∼7.0 results in the expression of small tenascin-C splice isoforms lacking the alternatively spliced FNIII domains A1-D. However when cells are exposed to more basic pH ∼7.30–7.50 as is the case in artificial culture experiments, fetal tissues and aggressive tumors, tenascin-C expression switches to the longer-isoform with one or more alternatively spliced FNIII repeats included in the transcript.153,165,166 pH regulation of tenascin-C splicing may therefore contribute to the diverse range of tenascin-C splice variants which are found in human fetal membranes.96 Similarly, malignantly transformed fibroblasts principally express large tenascin-C isoforms regardless of the extracellular pH in vitro (pHo); interestingly, this is because these cells maintain a basic pHi throughout a wide range of pHo.166 Bumke et al.,167 confirmed that the large isoform is preferentially expressed at basic pHo, but that serum starvation in fibroblasts reduces expression of the large tenascin-C isoform; independent of culture media pH.
Most recently, the splicing factor protein SRSF6 has been identified as a regulator of tenascin-C alternative splicing. SRSF6 induction in transgenic mice resulted in expression of full-length tenascin-C isoforms, which were absent in normal uninduced skin. The presence of the full-length tenascin-C isoform accompanied hyperplasia of keratinocytes, although this effect and the abundance of full-length tenascin-C could be reverted following removal of SRSF6 induction.168 SRSF6 mediated alternative splicing regulation of tenascin-C results in the excessive proliferation of keratinocytes in response to tissue injury, and therefore SRSF6 can modulate tissue homeostasis in vivo by promoting the expression of long tenascin-C isoforms in preference to shorter ones.
Post Translational Regulation of Tenascin-C
In addition to control of tenascin-C function at the transcriptional and post transcriptional level, the impact of tenascin-C on cell phenotype is profoundly affected by modifications to the protein after translation.
Tenascin-C biosynthesis
Following translation, tenascin-C assembles intracellularly into a 1.1–2.0 MDa 6-armed hexabrachion structure comprised of 6 monomers.169 This process is largely determined by N-terminal residues which are arranged into 3 cysteine-rich heptad repeats which form a coiled-coil region (amino acids 118–145 in human tenascin-C);38,170 a feature common to other oligomeric matricellular proteins such as thrombospondin and cartilage oligomeric matrix protein-1.171 Coiled-coils have a repeat pattern of hydrophobic and charged amino acids arranged as hxxhcxc (denoted a-g) called a heptad. In tenascin-C, these repeats produce short α-helices in which residues a and d form a 3,4-hydrophobic repeat, creating a stable non-polar core buried at the center of the helix. This stable arrangement projects the charged Cysteine side-chains at positions e and g out from the helix, where they can promote the oligomerization of tenascin-C subunits by forming inter-chain di-sulfide bonds.170
Two models for tenascin-C oligomerization have been proposed. In one model this event is a rapid co-translational process where 6 monomers are simultaneously assembled into a single hexabrachion in as little as 60 minutes following translation initiation. These data are based on pulse-chase analysis which revealed an absence of any intermediate oligomers suggesting that assembly is initiated at the N-terminus and completed in a single step, before translation is completed.171 This contrasts with models of assembly described for other large ECM proteins such as collagen, FN and vonWillebrand factor which are all assembled post-translationally from C-terminal assembly start points. In this model, 6 ribosome-associated polypeptide chains would have to multimerize simultaneously during translation. It has been suggested that the conformation of poly-ribosomes on the surface of the endoplasmic reticulum may be conducive to this: these ribosomes have been observed by electron microscopy to be 35 nm apart. Taking in to account the length of the N-terminal region of the TA domain and the ER membrane-spanning region in the newly translated peptide, any newly synthesized peptide must be 55 nm in length to permit N-terminal binding with other ribosome associated transcripts. As human tenascin-C transcripts have been recorded at 86.6 nm in length when folded,172 it is argued that the N-terminal heptad repeats on newly synthesized tenascin-C transcripts could engage each other and form the hexabrachion in a co-translational manner.171 In concordance with this, tenascin-R is shorter than tenascin-C and has previously been observed to form only dimers and trimers, but not hexamers; perhaps because the polypeptide chain is too short to allow interactions with 6 other transcripts simultaneously.
In the second model, the hexabrachion has been described as a composite of 2 trimers; each of which are stabilized by coiled-coil interactions prior to dimerization of these 2 subunits into the hexabrachion.170 This description concurs with preliminary electron micrograph images of the hexabrachion by Erickson and Inglesias, which revealed the presence of a single central globular domain tethered by 2 T-junctions, each in turn bound to 3-arms. In this model, hexabrachion assembly is described as a 2-step process dependent upon key Cysteine residues and a conserved glutamic acid residue. Site-specific mutagenesis was utilized to create numerous N-terminal tenascin-C polypeptide fragments which were then analyzed by far-UV circular dichroism for signs of secondary structure development. In this model the tenascin-C monomers form homotrimer intermediates stabilized at the N-terminal heptad repeats by a parallel 3-stranded α-helical coiled-coil (Fig. 3). This conformation clusters the 3 TA domains from each homotrimer together (Fig. 3), allowing 2 homotrimers to bind via their respective TA domains by a high concentration of weak homophillic interactions.170
Figure 3.
Hexabrachion assembly is a 2-step process. Multimerization of the N-terminal region of tenascin-C during hexabrachion assembly. 1. The N-terminal region of 3 tenascin-C monomers. Black cylinders represent the N-terminal heptad repeat residues 118–145, and gray circles represent TA domains. 2. The N-terminal heptads contain 3 cysteine-rich heptad repeats with hydrophobic (h) and charged (c) amino acid residues arranged in the conformation hxxhcxc. These monomers form an intermediary trimer which is stabilized by α-helical coiled-coil interactions between the N-terminal domains of the monomers. Three. The oligomerization of the adjacent TA domains increases homophillic binding affinity between the 2 trimers, which bind to form the hexabrachion. Di-sulfide bonds stabilize the hexabrachion but are not required for its formation (Adapted from Kammerer et al.170).
The substitution of the amino acid residues present within or nearby the N-terminal heptad repeats can significantly alter the hexabrachion assembly process. Substitution of the N-terminal C-64 residue with glycine results in the formation of trimeric tenascin-C intermediates suggesting a functional role for C-64 in joining trimeric intermediates into hexabrachions, perhaps by di-sulfide bonding. No other N-terminal Cysteine residue is considered essential for hexabrachion assembly, because site directed mutagenesis of C other than C-64 within the N-terminal heptad repeat region (C-111, 113, 146, 147) still results in the secretion of hexamers from the cell regardless of whether conducted on individual Cysteine or in pairwise combinations.173 Substitution of all Cysteine residues simultaneously at positions 111, 113, 140, 146 and 147 did not prevent hexabrachion formation, with the majority of oligomers containing over 4 subunits. However subsequent mutation of C-64 resulted in tenascin-C monomers, compounding the importance of C-64 in tenascin-C oligomerization via di-sulfide bonds.173
Substitution of a conserved glutamic acid-130 with alanine or leucine at heptad position e, abolishes the interaction between the residue at this position and hydrophobic residues at positions a and d. This allows for more efficient packing of hydrophobic residues within the heptad to increase the number of α-helices which can join the coiled-coil from 3 to 4; forming a 4 stranded α-helical coiled-coil.170 Normally, E130 forms a salt-bridge linkage with arginine-125 in a preceding heptad on an adjacent tenascin-C polypeptide chain.
While hexabrachions are often formed as homotypic molecules comprised of identical subunits, there are instances where heterotypic molecules can be formed. Kammerer et al.170 demonstrated that 2 triplets of tenascin-C trimers; one missing both the FNIII and FBG domain, were able to heterodimerize forming a heterotypic hexabrachion. Furthermore, smaller oligomers comprising of 2–5 tenascin-C molecules have also been identified, although it is not known whether these smaller isoforms are deliberately assembled in this manner, or if they are merely remnants of hexabrachions degraded by proteases.172
Post-translational modification of tenascin-C
The assembly of the hexabrachion from tenascin-C monomers reportedly takes only 5 minutes, with a rate limiting step in tenascin-C biosynthesis coming at the point of transport from the endoplasmic reticulum to the golgi apparatus.171 This is indicative of substantial post-translational modification of tenascin-C and indeed this molecule has been shown to be glycosylated.
Human tenascin-C possesses 26 potential N-glycosylation sites, of which 2 are in the TA domain, 2 are in the EGF-L repeat region, 21 are in the FNIII domain, and 1 is in the FBG domain (Fig. 4).23,174 Analysis of the human tenascin-C cDNA sequence revealed a cluster of 10 potential N-linked glycosylation sites within the alternatively spliced FNIII repeats A1-A4, within 2 clusters given as NLT-X11-NWT-X23-NLT-X63-NWT-X23-NLT. Three further putative N-glycosylation sites were identified in human FNIII B,19,23 and further analysis with the NetNGlyc 1.0 Server revealed 4 more predicted N- glycosylation sites in the FNIII AD1 and AD2 domains.174 In chicken tenascin-C, 17 potential N-glycosylation sites were identified by gene analysis, 6 of which were located in the FNIII AD1 and AD2 domains.38
Figure 4.
Predicted N- and O-glycosylation sites within tenascin-C. Tenascin-C possesses 26 putative N- glycosylation sites,19,23,174 and 34 putative O-linked glycosylation sites;175 the locations of which are represented on the tenascin-C stick diagram by triangles and circles respectively.
Analysis of the human tenascin-C peptide sequence with the NetOGlyc 4.0 Server tool; which predicts mucin type GalNAc O-glycosylation sites in mammalian proteins, predicts 34 putative O-glycosylation sites within tenascin-C.175 Of these, 8 were located in the TA domain, 2 within the EGF-L repeat region and 24 within the FNIII domain, while none were located in the FBG domain (Fig. 4).175At first glance, the distribution of potential O-glycosylation sites within tenascin-C clearly contrasts with that of N-glycosylation sites. O-glycosylation sites are densely arranged within the TA domain and FNIII 1–5, perhaps suggestive of a role in modulating macromolecular assembly and cell adhesion processes. By contrast, N-glycosylation sites are predominantly found within the alternatively spliced FNIII and may therefore be implicated in mechanisms related to development and tumorigenesis.
Confirmation of tenascin-C glycosylation derived from studies where purified human or chicken tenascin-C was treated with glycoside hydrolase enzymes and revealed changes in apparent Mr when subject to reducing-PAGE. Treatment of affinity purified chick embryonic fibroblast tenascin-C with endoglycosidase-F, was shown in 2 studies to result in a ∼20 kDa,176 and ∼10 kDa,50 reduction in apparent Mr of all 3 tenascin-C splice variants detected. Additionally, treatment with neuraminidase resulted in a 10 kDa reduction in Mr of all gradient purified human glioma and chicken embryonic fibroblast tenascin-C variants detected.169 All together, these data suggest that carbohydrates contribute significantly to the mass of tenascin-C in humans and chickens, and that the most active glycosylation sites must exist within the constitutively expressed portion of the molecule, as in each study the Mr of all tenascin-C splice variants was affected equally by endoglycosidase-F and neuraminidase treatment. Not much is known about the tissue specific expression patterns and the functional significance of tenascin-C glycosylation beyond studies performed on the HNK-1 epitope.72 The HNK-1 epitope, is presented on either an N- or O- linked oligosaccharide and believed to play a role in mediating cell adhesion and substrate binding processes. HNK-1 is distributed on a range of neural cell adhesion molecules;73-75 and also in some lymphocyte subsets. In tenascin-C this epitope is found in isoforms containing FNIII-A1,A2,A4,B,C,D and which promoted NSC proliferation via modulating the expression of the EGF-R.71 In this way the presence of glycosylation sites on alternatively spliced FNIII subunits confers greater control of protein function which can be regulated by alternative splicing.
Glycosylation of tenascin-C is likely to significantly impact its ability to modulate cell adhesive and protein binding functions. This may occur via the blocking of receptor and protein binding sites by large branched carbohydrate chains, which obscure tenascin-C from potential binding partners. It is perhaps via this mechanism that the collagen binding affinity of vitronectin is reduced upon glycosylation,177 and that fibroblast cell adhesion and spreading is improved following a lack of FN glycosylation.178 In contrast, the addition of new carbohydrate epitopes to tenascin-C may enhance protein and cell interactions, for example sialylation of vitronectin is required for rat hepatic stellate cell spreading,179 and O-glycosylated oncofetal FN induces epithelial to mesenchymal transition in lung cancer cells in synergy with TGFβ.180
Tenascin-C glycosylation may confer a strategy by which the protein is protected from proteolytic degradation. An early study on rat FN revealed that less glycosylated variants exhibit a greater rate of protease degradation,181 indicating early on that FN glycosylation protects the protein from fragmentation; therefore a similar protective mechanism may be conserved in tenascin-C. The abundance of putative N–glycosylation sites within the alternatively spliced FNIII A1-D closely associate with the FNIII domains which are susceptible to protease degradation by MMP-1, -2, -3 and -7;96,97 and may therefore exist to improve the half-life of tenascin-C.
Glycosylation of tenascin-C may offer a mechanism by which macromolecular assembly of the hexabrachion can be regulated by the expression of glycosyltransferase enzymes. The impact of glycosylation on multimer assembly is observed in vitronectin, where a reduced glycosylation state is associated with increased multimer size.177 It is therefore possible that the prevalence of N- and O-glycosylation sites within in the TA domain of tenascin-C is so high in order to function in a manner designed to interfere with macromolecular assembly of the hexabrachion, which may therefore indirectly affect the ability of the HXB to integrate into the ECM, although this is just speculation.
Despite a wealth of evidence revealing that tenascin-C is glycosylated, specific glycosylation sites have not yet been characterized in the protein, and as a result it is not known how many of the putative N- and O-glycosylation sites are active. Although glycosylation of tenascin-C is understood to modulate cell adhesion, substrate binding and cell proliferation processes, there are likely many more functions yet to be discovered. Further studies are required to advance our understanding of the exciting role played by tenascin-C glycosylation in modifying cell behavior.
Assembly of tenascin-C into a fibrillar matrix
Once suitably modified, the export of glycosylated tenascin-C from the golgi is rapid, with hexabrachions taking as little as 45 minutes to be detected in the conditioned culture medium of U-138 glioma cells after addition of a labeling medium.171
Tenascin-C is incorporated into the ECM in a manner which is dependent upon the presence of heparan sulfate proteoglycans (HSPGs). Chung and Erickson., generated Chinese hamster ovary cell mutants with a total block in heparin sulfate and chondroitin sulfate synthesis; this blockade resulted in the failure of the cells to assemble a FN matrix, and no tenascin-C was incorporated into the ECM as a result.182 The generation of a tenascin-C rich ECM has previously been shown to be dependent upon a template of FN and α5, αv and β1 integrins.183 It may be that once incorporated into the ECM, tenascin-C stabilizes FN as tenascin-C knockout mice generally exhibit reduced FN deposition.15,184
Tenascin-C may bind to proteoglycan molecules which are themselves bound to FN fibrils; in support of this an analysis revealed a co-localization between perlecan, FN and tenascin-C. Perlecan binds the constitutive FNIII 3–5 of tenascin-C, and this binding is enhanced in the short tenascin-C isoform (∼190 kDa). The insertion of alternatively spliced FNIII repeats after FNIII 5 results in an attenuation of perlecan-tenascin-C binding;182 and could possibly reduce adhesion in cells expressing longer isoforms of tenascin-C, as is observed in cancer. Another factor in matrix formation is the ECM molecule periostin, which has been shown to aid incorporation of tenascin-C into the ECM, albeit through a currently unknown mechanism.185
It is not clear how tenascin-C may be affected by incorporation into a 3-dimensional matrix, a process very likely to change the conformation of this molecule, perhaps exposing or concealing specific domains and binding sites. Moreover, it is not known how tenascin-C signals as part of a multicomponent ECM, where interaction with other matrix residents may affect the availability of binding sites and where molecules can synergistically regulate cell behavior. What is clear is that the conformation of tenascin-C does have profound effects on its function; this is evidenced by data showing that soluble tenascin-C binds readily to FN in glycerol gradients assays,129 where the extended conformation of both proteins appear to favor interaction.186 Conversely, FN does not bind to tenascin-C that has been immobilized onto ELISA plates, but tenascin-C can bind to immobilized FN. 39,129,187,188 As such the context and conformation of tenascin-C should be considered major players in defining ECM function.
Proteolytic processing of tenascin-C
One other means of regulating tenascin-C function is proteolytic cleavage. Fragments of tenascin-C have been reported in swabs from individuals with gingivitis,189 chronic venous leg ulcer exudates,164 and atherosclerotic plaques190 among other locations.
Mice lacking MMP-7191 or MMP-9192 exhibited lower levels of tenascin-C and tenascin-C fragments after myocardial infarction, suggesting that these metalloproteases can degrade tenascin-C in vivo. In contrast, MMP-19 null mice exhibited elevated tenascin-C protein during a model of asthma, indicating that MMP-19 may prevent the accumulation of tenascin-C.193 Tenascin-C is also susceptible to proteolytic degradation by MMPs-1, 2, 3 and 7 in vitro with the majority of MMP cleavage sites located within the alternatively spliced region of the protein, making long tenascin-C isoforms more susceptible to fragmentation than shorter isoforms.97 MMP-7 cleaves all tenascin-C isoforms by removing the 16 kDa N-terminal knob, but it also digests FNIII A3-D within the alternatively spliced region; its ability to digest AD1 and AD2 is not yet known.97 However, in chronic wounds tenascin-C fragmentation was not inhibited by EDTA and E64 ruling out a role for MMPs and cysteine proteases. It was subsequently confirmed that tenascin-C was digested by leucocyte elastase and possibly other serine proteases.164 Furthermore, a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) -5 has demonstrated an ability to cleave tenascin-C into fragments of which some correspond to the N-terminal TA domain,194 with specific tenascin-C fragments identified as inducers of cartilage matrix degradation.195
Degradation of tenascin-C results in modulation of tissue protein levels, a process key to limiting tenascin-C expression in situations where transient expression is vital, for example in the developing embryo and in healing wounds. The reduction over time of longer variants compared to shorter variants fits well with data showing that longer isoforms are more readily cleaved. Proteolysis also releases smaller fragments from large multimodular proteins that exhibit quite different functions to the intact molecule. For example, fragmented EGF-L domains of tenascin-C induce apoptosis in cultured smooth muscle cells (SMCs) in contrast to full length tenascin-C,190 and FN matrix assembly is unaffected by full-length tenascin-C, but is inhibited by the FNIII domains or the FBG domain of tenascin-C.196 Similarly, cleavage of tenascin-C within the FNIII A3 subunit by MMP-2 generates a cryptic binding site within FNIII A2 which permits interaction with heparan sulfated side-chains of syndecan-4 to promote β1 integrin clustering and cell spreading.97,132 Finally, full-length tenascin-C is found to be anti-adhesive and to prevent FN-mediated cell spreading of T98G glioblastoma cells, but this effect was lost following cleavage with the metalloprotease meprin β.197
Summary and Conclusions
Following tenascin-C through its life cycle, from the switching on of TNC, through transcription, translation, and protein modification, all the way though to its degradation, reveals a number of mechanisms by which different forms of this molecule are created. Moreover, it is clear that the spatial and temporal control of these events enables the expression of precise tenascin-C forms with distinct functions designed to carry out specific tasks at the right time and place. The molecular events that enable different stimuli to induce the expression of specific spliced variants of tenascin-C remain to be elucidated. However, the way in which the inclusion of alternatively spliced FNIII domains with unique interaction sites confers new binding capabilities or susceptibilities to proteolytic cleavage to tenascin-C is exemplified in many tissues during development and in tumors. Conversely, the inclusion of extra FNIII repeats that interrupt existing binding sites, causing loss of a particular function, is also observed at these locations. Post translational modification of tenascin-C has not yet been extensively studied but already data suggest that glycosylation can have a profound effect on its function. Moreover, it is emerging that the architecture of tenascin-C matters; its assembly into a fibrillar matrix at the cell surface impacts the conformation of this molecule, potentially exposing cryptic binding sites or covering exposed binding sites, and it will be of great interest to determine precisely how tenascin-C acts within the context of complex, 3D tissue environments. Finally, even proteolytic destruction of tenascin-C creates collateral damage that confers novel functions to this molecule. The destruction of existing binding sites and generation of smaller fragments with new binding sites can drive entirely novel processes compared to the intact molecule. It seems that the more we learn about this extraordinary matrix glycoprotein, the more complex and intricate the story becomes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The authors are funded by the Kennedy Trust for Rheumatology Research and Arthritis Research UK.
References
- 1. Midwood KS, Orend G. The role of tenascin-C in tissue injury and tumorigenesis. J Cell Commun Signal 2009; 3:287-310; PMID:19838819; http://dx.doi.org/ 10.1007/s12079-009-0075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiquet-Ehrismann R, Orend G, Chiquet M, Tucker RP, Midwood KS. Tenascins in stem cell niches. Matrix Biol 2014; 37C:112-23; http://dx.doi.org/ 10.1016/j.matbio.2014.01.007 [DOI] [PubMed] [Google Scholar]
- 3.Orend G, Saupe F, Schwenzer A, and Midwood, KS. The extracellular matrix and cancer: regulation of tumor cell biology by tenascin-C. iConcept Press 2014. ISBN: 978-1-922227-25-6. [Google Scholar]
- 4. Forsberg E, Hirsch E, Frohlich L, Meyer M, Ekblom P, Aszodi A, Werner S, Fassler R. Skin wounds and severed nerves heal normally in mice lacking tenascin-C. Proc Nat Acad Sci USA 1996; 93:6594-9; PMID:8692862; http://dx.doi.org/ 10.1073/pnas.93.13.6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fukamauchi F, Wang YJ, Mataga N, Kusakabe M. Effects of cholecystokinin-B receptor antagonist on dopamine system in tenascin mutant mice. Neuroreport 1997; 8:3919-22; PMID:9462466; http://dx.doi.org/ 10.1097/00001756-199712220-00015 [DOI] [PubMed] [Google Scholar]
- 6. Nakao N, Hiraiwa N, Yoshiki A, Ike F, Kusakabe M. Tenascin-C promotes healing of Habu-snake venom-induced glomerulonephritis: studies in knockout congenic mice and in culture. Am J Pathol 1998; 152:1237-45; PMID:9588892 [PMC free article] [PubMed] [Google Scholar]
- 7. Ohta M, Sakai T, Saga Y, Aizawa S, Saito M. Suppression of hematopoietic activity in tenascin-C-deficient mice. Blood 1998; 91:4074-83; PMID:9596652 [PubMed] [Google Scholar]
- 8. Talts JF, Wirl G, Dictor M, Muller WJ, Fassler R. Tenascin-C modulates tumor stroma and monocyte/macrophage recruitment but not tumor growth or metastasis in a mouse strain with spontaneous mammary cancer. J Cell Sci 1999; 112(Pt 12):1855-64; PMID:10341205 [DOI] [PubMed] [Google Scholar]
- 9. Van Obberghen-Schilling E, Tucker RP, Saupe F, Gasser I, Cseh B, Orend G. Fibronectin and tenascin-C: accomplices in vascular morphogenesis during development and tumor growth. Int J Dev Biol 2011; 55:511-25; PMID:21769776; http://dx.doi.org/ 10.1387/ijdb.103243eo [DOI] [PubMed] [Google Scholar]
- 10. Saupe F, Schwenzer A, Jia Y, Gasser I, Spenle C, Langlois B, Kammerer M, Lefebvre O, Hlushchuk R, Rupp T, et al. Tenascin-C downregulates wnt inhibitor dickkopf-1, promoting tumorigenesis in a neuroendocrine tumor model. Cell Rep 2013; 5:482-92; PMID:24139798; http://dx.doi.org/ 10.1016/j.celrep.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 11. Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G, et al. Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nat Med 2009; 15:774-80; PMID:19561617; http://dx.doi.org/ 10.1038/nm.1987 [DOI] [PubMed] [Google Scholar]
- 12. El-Karef A, Yoshida T, Gabazza EC, Nishioka T, Inada H, Sakakura T, Imanaka-Yoshida K. Deficiency of tenascin-C attenuates liver fibrosis in immune-mediated chronic hepatitis in mice. J Pathol 2007; 211:86-94; PMID:17121418; http://dx.doi.org/ 10.1002/path.2099 [DOI] [PubMed] [Google Scholar]
- 13. Nakahara H, Gabazza EC, Fujimoto H, Nishii Y, D'Alessandro-Gabazza CN, Bruno NE, Takagi T, Hayashi T, Maruyama J, Maruyama K, et al. Deficiency of tenascin C attenuates allergen-induced bronchial asthma in the mouse. Eur J Immunol 2006; 36:3334-45; PMID:17125141; http://dx.doi.org/ 10.1002/eji.200636271 [DOI] [PubMed] [Google Scholar]
- 14. Chiquet-Ehrismann R, Tucker RP doi: 10.1101/cshperspect.a004960. Tenascins and the importance of adhesion modulation. Cold Spring Harb Perspect Biol 2011; 3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mackie EJ, Tucker RP. The tenascin-C knockout revisited. J Cell Sci 1999; 112(Pt 22):3847-53; PMID:10547346 [DOI] [PubMed] [Google Scholar]
- 16. Mokone GG, Gajjar M, September AV, Schwellnus MP, Greenberg J, Noakes TD, Collins M. The guanine-thymine dinucleotide repeat polymorphism within the tenascin-C gene is associated with achilles tendon injuries. Am J Sports Med 2005; 33:1016-21; PMID:15983124; http://dx.doi.org/ 10.1177/0363546504271986 [DOI] [PubMed] [Google Scholar]
- 17. Zhao Y, Zhao F, Zong L, Zhang P, Guan L, Zhang J, Wang D, Wang J, Chai W, Lan L, et al. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PloS One 2013; 8:e69549; PMID:23936043; http://dx.doi.org/ 10.1371/journal.pone.0069549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuda A, Hirota T, Akahoshi M, Shimizu M, Tamari M, Miyatake A, Takahashi A, Nakashima K, Takahashi N, Obara K, et al. Coding SNP in tenascin-C Fn-III-D domain associates with adult asthma. Hum Mol Genet 2005; 14:2779-86; PMID:16115819; http://dx.doi.org/ 10.1093/hmg/ddi311 [DOI] [PubMed] [Google Scholar]
- 19. Gulcher JR, Alexakos MJ, Le Beau MM, Lemons RS, Stefansson K. Chromosomal localization of the human hexabrachion (tenascin) gene and evidence for recent reduplication within the gene. Genomics 1990; 6:616-22; PMID:1692804; http://dx.doi.org/ 10.1016/0888-7543(90)90495-G [DOI] [PubMed] [Google Scholar]
- 20. Rocchi M, Archidiacono N, Romeo G, Saginati M, Zardi L. Assignment of the gene for human tenascin to the region q32-q34 of chromosome 9. Hum Genet 1991; 86:621-3; PMID:1709136 [DOI] [PubMed] [Google Scholar]
- 21. Udalova IA, Ruhmann M, Thomson SJ, Midwood KS. Expression and immune function of tenascin-C. Crit Rev Immunol 2011; 31:115-45; PMID:21542790; http://dx.doi.org/ 10.1615/CritRevImmunol.v31.i2.30 [DOI] [PubMed] [Google Scholar]
- 22. Jones FS, Burgoon MP, Hoffman S, Crossin KL, Cunningham BA, Edelman GM. A cDNA clone for cytotactin contains sequences similar to epidermal growth factor-like repeats and segments of fibronectin and fibrinogen. Proc Nat Acad Sci U S A 1988; 85:2186-90; PMID:2451243; http://dx.doi.org/ 10.1073/pnas.85.7.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gulcher JR, Nies DE, Marton LS, Stefansson K. An alternatively spliced region of the human hexabrachion contains a repeat of potential N-glycosylation sites. Proc Nat Acad Sci U S A 1989; 86:1588-92; PMID:2466295; http://dx.doi.org/ 10.1073/pnas.86.5.1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones FS, Jones PL. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev Dyn 2000; 218:235-59; PMID:10842355; http://dx.doi.org/ 10.1002/(SICI)1097-0177(200006)218:2%3c235::AID-DVDY2%3e3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 25. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413-5; PMID:18978789; http://dx.doi.org/ 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 26. Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature 2014; 509:575-81; PMID:24870542; http://dx.doi.org/ 10.1038/nature13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sriramarao P, Bourdon MA. A novel tenascin type III repeat is part of a complex of tenascin mRNA alternative splices. Nucleic Acids Res 1993; 21:163-8; PMID:7680113; http://dx.doi.org/ 10.1093/nar/21.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mighell AJ, Thompson J, Hume WJ, Markham AF, Robinson PA. Human tenascin-C: identification of a novel type III repeat in oral cancer and of novel splice variants in normal, malignant and reactive oral mucosae. Int J Cancer 1997; 72:236-40; PMID:9219826; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19970717)72:2%3c236::AID-IJC6%3e3.0.CO;2-S [DOI] [PubMed] [Google Scholar]
- 29. Tucker RP, Spring J, Baumgartner S, Martin D, Hagios C, Poss PM, Chiquet-Ehrismann R. Novel tenascin variants with a distinctive pattern of expression in the avian embryo. Development 1994; 120:637-47; PMID:7512896 [DOI] [PubMed] [Google Scholar]
- 30. Gulcher JR, Nies DE, Alexakos MJ, Ravikant NA, Sturgill ME, Marton LS, Stefansson K. Structure of the human hexabrachion (tenascin) gene. Proc Nat Acad Sci U S A 1991; 88:9438-42; PMID:1719530; http://dx.doi.org/ 10.1073/pnas.88.21.9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joester A, Faissner A. Evidence for combinatorial variability of tenascin-C isoforms and developmental regulation in the mouse central nervous system. J Biol Chem 1999; 274:17144-51; PMID:10358070; http://dx.doi.org/ 10.1074/jbc.274.24.17144 [DOI] [PubMed] [Google Scholar]
- 32. Garwood J, Theocharidis U, Calco V, Dobbertin A, Faissner A. Existence of tenascin-C isoforms in rat that contain the alternatively spliced AD1 domain are developmentally regulated during hippocampal development. Cell Mol Neurobiol 2012; 32:279-87; PMID:21968644; http://dx.doi.org/ 10.1007/s10571-011-9759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joester A, Faissner A. The structure and function of tenascins in the nervous system. Matrix Biol 2001; 20:13-22; PMID:11246000; http://dx.doi.org/ 10.1016/S0945-053X(00)00136-0 [DOI] [PubMed] [Google Scholar]
- 34. Saga Y, Tsukamoto T, Jing N, Kusakabe M, Sakakura T. Murine tenascin: cDNA cloning, structure and temporal expression of isoforms. Gene 1991; 104:177-85; PMID:1717349; http://dx.doi.org/ 10.1016/0378-1119(91)90248-A [DOI] [PubMed] [Google Scholar]
- 35. Ocklind G, Talts J, Fassler R, Mattsson A, Ekblom P. Expression of tenascin in developing and adult mouse lymphoid organs. J Histochem Cytochem 1993; 41:1163-9; PMID:7687262; http://dx.doi.org/ 10.1177/41.8.7687262 [DOI] [PubMed] [Google Scholar]
- 36. Chiquet-Ehrismann R, Mackie EJ, Pearson CA, Sakakura T. Tenascin: an extracellular matrix protein involved in tissue interactions during fetal development and oncogenesis. Cell 1986; 47:131-9; PMID:2428505; http://dx.doi.org/ 10.1016/0092-8674(86)90374-0 [DOI] [PubMed] [Google Scholar]
- 37. Chiquet M, Vrucinic-Filipi N, Schenk S, Beck K, Chiquet-Ehrismann R. Isolation of chick tenascin variants and fragments. A C-terminal heparin-binding fragment produced by cleavage of the extra domain from the largest subunit splicing variant. Eur J Biochem 1991; 199:379-88; PMID:1712728 [DOI] [PubMed] [Google Scholar]
- 38. Spring J, Beck K, Chiquet-Ehrismann R. Two contrary functions of tenascin: dissection of the active sites by recombinant tenascin fragments. Cell 1989; 59:325-34; PMID:2478295; http://dx.doi.org/ 10.1016/0092-8674(89)90294-8 [DOI] [PubMed] [Google Scholar]
- 39. Chiquet-Ehrismann R, Matsuoka Y, Hofer U, Spring J, Bernasconi C, Chiquet M. Tenascin variants: differential binding to fibronectin and distinct distribution in cell cultures and tissues. Cell Regul 1991; 2:927-38; PMID:1725601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weller A, Beck S, Ekblom P. Amino acid sequence of mouse tenascin and differential expression of two tenascin isoforms during embryogenesis. J Cell Biol 1991; 112:355-62; PMID:1703162; http://dx.doi.org/ 10.1083/jcb.112.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chiquet M, Fambrough DM. Chick myotendinous antigen. I. A monoclonal antibody as a marker for tendon and muscle morphogenesis. J Cell Biol 1984; 98:1926-36; PMID:6725406; http://dx.doi.org/ 10.1083/jcb.98.6.1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiquet M, Fambrough DM. Chick myotendinous antigen. II. A novel extracellular glycoprotein complex consisting of large disulfide-linked subunits. J Cell Biol 1984; 98:1937-46; PMID:6202699; http://dx.doi.org/ 10.1083/jcb.98.6.1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoffman S, Crossin KL, Edelman GM. Molecular forms, binding functions, and developmental expression patterns of cytotactin and cytotactin-binding proteoglycan, an interactive pair of extracellular matrix molecules. J Cell Biol 1988; 106:519-32; PMID:2448317; http://dx.doi.org/ 10.1083/jcb.106.2.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prieto AL, Jones FS, Cunningham BA, Crossin KL, Edelman GM. Localization during development of alternatively spliced forms of cytotactin mRNA by in situ hybridization. J Cell Biol 1990; 111:685-98; PMID:1696267; http://dx.doi.org/ 10.1083/jcb.111.2.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mackie EJ, Tucker RP. Tenascin in bone morphogenesis: expression by osteoblasts and cell type-specific expression of splice variants. J Cell Sci 1992; 103(Pt 3):765-71; PMID:1282516 [DOI] [PubMed] [Google Scholar]
- 46. Tucker RP. The in situ localization of tenascin splice variants and thrombospondin 2 mRNA in the avian embryo. Development 1993; 117:347-58; PMID:7693413 [DOI] [PubMed] [Google Scholar]
- 47. Tucker RP, Hammarback JA, Jenrath DA, Mackie EJ, Xu Y. Tenascin expression in the mouse: in situ localization and induction in vitro by bFGF. J Cell Sci 1993; 104(Pt 1):69-76; PMID:7680659 [DOI] [PubMed] [Google Scholar]
- 48. Tucker RP, McKay SE. The expression of tenascin by neural crest cells and glia. Development 1991; 112:1031-9; PMID:1718677 [DOI] [PubMed] [Google Scholar]
- 49. Derr LB, Chiquet-Ehrismann R, Gandour-Edwards R, Spence J, Tucker RP. The expression of tenascin-C with the AD1 variable repeat in embryonic tissues, cell lines and tumors in various vertebrate species. Differentiation 1997; 62:71-82; PMID:9404002; http://dx.doi.org/ 10.1046/j.1432-0436.1997.6220071.x [DOI] [PubMed] [Google Scholar]
- 50. Kaplony A, Zimmermann DR, Fischer RW, Imhof BA, Odermatt BF, Winterhalter KH, Vaughan L. Tenascin Mr 220,000 isoform expression correlates with corneal cell migration. Development 1991; 112:605-14; PMID:1724418 [DOI] [PubMed] [Google Scholar]
- 51. Pacifici M, Iwamoto M, Golden EB, Leatherman JL, Lee YS, Chuong CM. Tenascin is associated with articular cartilage development. Dev Dyn 1993; 198:123-34; PMID:7508293; http://dx.doi.org/ 10.1002/aja.1001980206 [DOI] [PubMed] [Google Scholar]
- 52. Anstrom KK, Tucker RP. Tenascin-C lines the migratory pathways of avian primordial germ cells and hematopoietic progenitor cells. Dev Dyn 1996; 206:437-46; PMID:8853992; http://dx.doi.org/ 10.1002/(SICI)1097-0177(199608)206:4%3c437::AID-AJA9%3e3.0.CO;2-J [DOI] [PubMed] [Google Scholar]
- 53. Aufderheide E, Ekblom P. Tenascin during gut development: appearance in the mesenchyme, shift in molecular forms, and dependence on epithelial-mesenchymal interactions. J Cell Biol 1988; 107:2341-9; PMID:2461951; http://dx.doi.org/ 10.1083/jcb.107.6.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young SL, Chang LY, Erickson HP. Tenascin-C in rat lung: distribution, ontogeny and role in branching morphogenesis. Dev Biol 1994; 161:615-25; PMID:7508872; http://dx.doi.org/ 10.1006/dbio.1994.1057 [DOI] [PubMed] [Google Scholar]
- 55. Zhao Y, Young SL. TGF-β regulates expression of tenascin alternative-splicing isoforms in fetal rat lung. Am J Physiol 1995; 268:L173-80; PMID:7532367 [DOI] [PubMed] [Google Scholar]
- 56. Dorries U, Schachner M. Tenascin mRNA isoforms in the developing mouse brain. J Neurosci Res 1994; 37:336-47; PMID:7513764; http://dx.doi.org/ 10.1002/jnr.490370306 [DOI] [PubMed] [Google Scholar]
- 57. Bartsch S, Bartsch U, Dorries U, Faissner A, Weller A, Ekblom P, Schachner M. Expression of tenascin in the developing and adult cerebellar cortex. J Neurosci 1992; 12:736-49; PMID:1372043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zisch AH, D'Alessandri L, Ranscht B, Falchetto R, Winterhalter KH, Vaughan L. Neuronal cell adhesion molecule contactin/F11 binds to tenascin via its immunoglobulin-like domains. J Cell Biol 1992; 119:203-13; PMID:1382076; http://dx.doi.org/ 10.1083/jcb.119.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vaughan L, Weber P, D'Alessandri L, Zisch AH, Winterhalter KH. Tenascin-contactin/F11 interactions: a clue for a developmental role? Perspect Dev Neurobiol 1994; 2:43-52; PMID:7530143 [PubMed] [Google Scholar]
- 60. Weber P, Ferber P, Fischer R, Winterhalter KH, Vaughan L. Binding of contactin/F11 to the fibronectin type III domains 5 and 6 of tenascin is inhibited by heparin. FEBS Lett 1996; 389:304-8; PMID:8766721; http://dx.doi.org/ 10.1016/0014-5793(96)00609-6 [DOI] [PubMed] [Google Scholar]
- 61. Ghert MA, Qi WN, Erickson HP, Block JA, Scully SP. Tenascin-C splice variant adhesive/anti-adhesive effects on chondrosarcoma cell attachment to fibronectin. Cell Struct Funct 2001; 26:179-87; PMID:11565810; http://dx.doi.org/ 10.1247/csf.26.179 [DOI] [PubMed] [Google Scholar]
- 62. Crossin KL, Hoffman S, Tan SS, Edelman GM. Cytotactin and its proteoglycan ligand mark structural and functional boundaries in somatosensory cortex of the early postnatal mouse. Dev Biol 1989; 136:381-92; PMID:2479585; http://dx.doi.org/ 10.1016/0012-1606(89)90264-9 [DOI] [PubMed] [Google Scholar]
- 63. Steindler DA, Cooper NG, Faissner A, Schachner M. Boundaries defined by adhesion molecules during development of the cerebral cortex: the J1/tenascin glycoprotein in the mouse somatosensory cortical barrel field. Dev Biol 1989; 131:243-60; PMID:2462518; http://dx.doi.org/ 10.1016/S0012-1606(89)80056-9 [DOI] [PubMed] [Google Scholar]
- 64. Meiners S, Geller HM. Long and short splice variants of human tenascin differentially regulate neurite outgrowth. Mol and Cell Neurosci 1997; 10:100-16; PMID:9361291; http://dx.doi.org/ 10.1006/mcne.1997.0643 [DOI] [PubMed] [Google Scholar]
- 65. Garcion E, Faissner A, ffrench-Constant C. Knockout mice reveal a contribution of the extracellular matrix molecule tenascin-C to neural precursor proliferation and migration. Development 2001; 128:2485-96; PMID:11493565 [DOI] [PubMed] [Google Scholar]
- 66. Gates MA, Thomas LB, Howard EM, Laywell ED, Sajin B, Faissner A, Gotz B, Silver J, Steindler DA. Cell and molecular analysis of the developing and adult mouse subventricular zone of the cerebral hemispheres. J Comp Neurol 1995; 361:249-66; PMID:8543661; http://dx.doi.org/ 10.1002/cne.903610205 [DOI] [PubMed] [Google Scholar]
- 67. Treloar HB, Ray A, Dinglasan LA, Schachner M, Greer CA. Tenascin-C is an inhibitory boundary molecule in the developing olfactory bulb. J Neurosci 2009; 29:9405-16; PMID:19641104; http://dx.doi.org/ 10.1523/JNEUROSCI.2356-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garcion E, Halilagic A, Faissner A, ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development 2004; 131:3423-32; PMID:15226258; http://dx.doi.org/ 10.1242/dev.01202 [DOI] [PubMed] [Google Scholar]
- 69. von Holst A, Egbers U, Prochiantz A, Faissner A. Neural stem/progenitor cells express 20 tenascin C isoforms that are differentially regulated by Pax6. J Biol Chem 2007; 282:9172-81; PMID:17264084; http://dx.doi.org/ 10.1074/jbc.M608067200 [DOI] [PubMed] [Google Scholar]
- 70. Moritz S, Lehmann S, Faissner A, von Holst A. An induction gene trap screen in neural stem cells reveals an instructive function of the niche and identifies the splicing regulator sam68 as a tenascin-C-regulated target gene. Stem Cells 2008; 26:2321-31; PMID:18617690; http://dx.doi.org/ 10.1634/stemcells.2007-1095 [DOI] [PubMed] [Google Scholar]
- 71. Yagi H, Yanagisawa M, Suzuki Y, Nakatani Y, Ariga T, Kato K, Yu RK. HNK-1 epitope-carrying tenascin-C spliced variant regulates the proliferation of mouse embryonic neural stem cells. J Biol Chem 2010; 285:37293-301; PMID:20855890; http://dx.doi.org/ 10.1074/jbc.M110.157081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kruse J, Keilhauer G, Faissner A, Timpl R, Schachner M. The J1 glycoprotein–a novel nervous system cell adhesion molecule of the L2/HNK-1 family. Nature 1985; 316:146-8; PMID:2409452; http://dx.doi.org/ 10.1038/316146a0 [DOI] [PubMed] [Google Scholar]
- 73. McGarry RC, Helfand SL, Quarles RH, Roder JC. Recognition of myelin-associated glycoprotein by the monoclonal antibody HNK-1. Nature 1983; 306:376-8; PMID:6196641; http://dx.doi.org/ 10.1038/306376a0 [DOI] [PubMed] [Google Scholar]
- 74. Kruse J, Mailhammer R, Wernecke H, Faissner A, Sommer I, Goridis C, Schachner M. Neural cell adhesion molecules and myelin-associated glycoprotein share a common carbohydrate moiety recognized by monoclonal antibodies L2 and HNK-1. Nature 1984; 311:153-5; PMID:6206400; http://dx.doi.org/ 10.1038/311153a0 [DOI] [PubMed] [Google Scholar]
- 75. Nieke J, Schachner M. Expression of the neural cell adhesion molecules L1 and N-CAM and their common carbohydrate epitope L2/HNK-1 during development and after transection of the mouse sciatic nerve. Differentiation 1985; 30:141-51; PMID:2420673; http://dx.doi.org/ 10.1111/j.1432-0436.1985.tb00525.x [DOI] [PubMed] [Google Scholar]
- 76. Gotz B, Scholze A, Clement A, Joester A, Schutte K, Wigger F, Frank R, Spiess E, Ekblom P, Faissner A. Tenascin-C contains distinct adhesive, anti-adhesive, and neurite outgrowth promoting sites for neurons. J Cell Biol 1996; 132:681-99; PMID:8647898; http://dx.doi.org/ 10.1083/jcb.132.4.681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Husmann K, Faissner A, Schachner M. Tenascin promotes cerebellar granule cell migration and neurite outgrowth by different domains in the fibronectin type III repeats. J Cell Biol 1992; 116:1475-86; PMID:1371773; http://dx.doi.org/ 10.1083/jcb.116.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mercado ML, Nur-e-Kamal A, Liu HY, Gross SR, Movahed R, Meiners S. Neurite outgrowth by the alternatively spliced region of human tenascin-C is mediated by neuronal alpha7beta1 integrin. J Neurosci 2004; 24:238-47; PMID:14715956; http://dx.doi.org/ 10.1523/JNEUROSCI.4519-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gotz M, Bolz J, Joester A, Faissner A. Tenascin-C synthesis and influence on axonal growth during rat cortical development. Eur J Neurosci 1997; 9:496-506; PMID:9104592; http://dx.doi.org/ 10.1111/j.1460-9568.1997.tb01627.x [DOI] [PubMed] [Google Scholar]
- 80. Sahlberg C, Aukhil I, Thesleff I. Tenascin-C in developing mouse teeth: expression of splice variants and stimulation by TGFbeta and FGF. Eur J Oral Sci 2001; 109:114-24; PMID:11347655; http://dx.doi.org/ 10.1034/j.1600-0722.2001.00990.x [DOI] [PubMed] [Google Scholar]
- 81. Jones PL, Boudreau N, Myers CA, Erickson HP, Bissell MJ. Tenascin-C inhibits extracellular matrix-dependent gene expression in mammary epithelial cells. Localization of active regions using recombinant tenascin fragments. J Cell Sci 1995; 108(Pt 2):519-27; PMID:7539436 [DOI] [PubMed] [Google Scholar]
- 82. Wehrle B, Chiquet M. Tenascin is accumulated along developing peripheral nerves and allows neurite outgrowth in vitro. Development 1990; 110:401-15; PMID:1723942 [DOI] [PubMed] [Google Scholar]
- 83. Perez RG, Halfter W. Tenascin in the developing chick visual system: distribution and potential role as a modulator of retinal axon growth. Dev Biol 1993; 156:278-92; PMID:7680630; http://dx.doi.org/ 10.1006/dbio.1993.1076 [DOI] [PubMed] [Google Scholar]
- 84. Bartsch S, Husmann K, Schachner M, Bartsch U. The extracellular matrix molecule tenascin: expression in the developing chick retinotectal system and substrate properties for retinal ganglion cell neurites in vitro. Eur J Neurosci 1995; 7:907-16; PMID:7542126; http://dx.doi.org/ 10.1111/j.1460-9568.1995.tb01078.x [DOI] [PubMed] [Google Scholar]
- 85. Phillips GR, Edelman GM, Crossin KL. Separate cell binding sites within cytotactin/tenascin differentially promote neurite outgrowth. Cell Adhes Commun 1995; 3:257-71; PMID:8846026; http://dx.doi.org/ 10.3109/15419069509081291 [DOI] [PubMed] [Google Scholar]
- 86. Fischer D, Tucker RP, Chiquet-Ehrismann R, Adams JC. Cell-adhesive responses to tenascin-C splice variants involve formation of fascin microspikes. Mol Biol Cell 1997; 8:2055-75; PMID:9348542; http://dx.doi.org/ 10.1091/mbc.8.10.2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Adams JC, Lawler J. Cell-type specific adhesive interactions of skeletal myoblasts with thrombospondin-1. Mol Biol Cell 1994; 5:423-37; PMID:7519904; http://dx.doi.org/ 10.1091/mbc.5.4.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chung CY, Erickson HP. Cell surface annexin II is a high affinity receptor for the alternatively spliced segment of tenascin-C. J Cell Biol 1994; 126:539-48; PMID:7518469; http://dx.doi.org/ 10.1083/jcb.126.2.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chung CY, Murphy-Ullrich JE, Erickson HP. Mitogenesis, cell migration, and loss of focal adhesions induced by tenascin-C interacting with its cell surface receptor, annexin II. Mol Biol Cell 1996; 7:883-92; PMID:8816995; http://dx.doi.org/ 10.1091/mbc.7.6.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Meiners S, Mercado ML, Nur-e-Kamal MS, Geller HM. Tenascin-C contains domains that independently regulate neurite outgrowth and neurite guidance. J Neurosci 1999; 19:8443-53; PMID:10493745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rigato F, Garwood J, Calco V, Heck N, Faivre-Sarrailh C, Faissner A. Tenascin-C promotes neurite outgrowth of embryonic hippocampal neurons through the alternatively spliced fibronectin type III BD domains via activation of the cell adhesion molecule F3/contactin. J Neurosci 2002; 22:6596-609; PMID:12151539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lochter A, Vaughan L, Kaplony A, Prochiantz A, Schachner M, Faissner A. J1/tenascin in substrate-bound and soluble form displays contrary effects on neurite outgrowth. J Cell Biol 1991; 113:1159-71; PMID:1710226; http://dx.doi.org/ 10.1083/jcb.113.5.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Meiners S, Powell EM, Geller HM. Neurite outgrowth promotion by the alternatively spliced region of tenascin-C is influenced by cell-type specific binding. Matrix Biol 1999; 18:75-87; PMID:10367733; http://dx.doi.org/ 10.1016/S0945-053X(98)00008-0 [DOI] [PubMed] [Google Scholar]
- 94. Siddiqui S, Horvat-Brocker A, Faissner A. The glia-derived extracellular matrix glycoprotein tenascin-C promotes embryonic and postnatal retina axon outgrowth via the alternatively spliced fibronectin type III domain TNfnD. Neuron Glia Biol 2008; 4:271-83; PMID:19508743; http://dx.doi.org/ 10.1017/S1740925X09990020 [DOI] [PubMed] [Google Scholar]
- 95. Michele M, Faissner A. Tenascin-C stimulates contactin-dependent neurite outgrowth via activation of phospholipase C. Mol and Cell Neurosci 2009; 41:397-408; PMID:19394429; http://dx.doi.org/ 10.1016/j.mcn.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 96. Bell SC, Pringle JH, Taylor DJ, Malak TM. Alternatively spliced tenascin-C mRNA isoforms in human fetal membranes. Mol Hum Reprod 1999; 5:1066-76; PMID:10541570; http://dx.doi.org/ 10.1093/molehr/5.11.1066 [DOI] [PubMed] [Google Scholar]
- 97. Siri A, Knauper V, Veirana N, Caocci F, Murphy G, Zardi L. Different susceptibility of small and large human tenascin-C isoforms to degradation by matrix metalloproteinases. J Biol Chem 1995; 270:8650-4; PMID:7536739; http://dx.doi.org/ 10.1074/jbc.270.15.8650 [DOI] [PubMed] [Google Scholar]
- 98. Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett 2006; 244:143-63; PMID:16632194; http://dx.doi.org/ 10.1016/j.canlet.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 99. Howeedy AA, Virtanen I, Laitinen L, Gould NS, Koukoulis GK, Gould VE. Differential distribution of tenascin in the normal, hyperplastic, and neoplastic breast. Lab Invest 1990; 63:798-806; PMID:1701508 [PubMed] [Google Scholar]
- 100. Borsi L, Carnemolla B, Nicolo G, Spina B, Tanara G, Zardi L. Expression of different tenascin isoforms in normal, hyperplastic and neoplastic human breast tissues. Int J Cancer 1992; 52:688-92; PMID:1385335; http://dx.doi.org/ 10.1002/ijc.2910520504 [DOI] [PubMed] [Google Scholar]
- 101. Oyama F, Hirohashi S, Shimosato Y, Titani K, Sekiguchi K. Qualitative and quantitative changes of human tenascin expression in transformed lung fibroblast and lung tumor tissues: comparison with fibronectin. Cancer Res 1991; 51:4876-81; PMID:1716516 [PubMed] [Google Scholar]
- 102. Lohi J, Tani T, Laitinen L, Kangas L, Lehto VP, Virtanen I. Tenascin and fibronectin isoforms in human renal cell carcinomas, renal cell carcinoma cell lines and xenografts in nude mice. Int J Cancer 1995; 63:442-9; PMID:7591246; http://dx.doi.org/ 10.1002/ijc.2910630324 [DOI] [PubMed] [Google Scholar]
- 103. Sakai T, Kawakatsu H, Hirota N, Yokoyama T, Sakakura T, Saito M. Specific expression of tenascin in human colonic neoplasms. Br J Cancer 1993; 67:1058-64; PMID:7684238; http://dx.doi.org/ 10.1038/bjc.1993.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Parekh K, Ramachandran S, Cooper J, Bigner D, Patterson A, Mohanakumar T. Tenascin-C, over expressed in lung cancer down regulates effector functions of tumor infiltrating lymphocytes. Lung Cancer 2005; 47:17-29; PMID:15603851; http://dx.doi.org/ 10.1016/j.lungcan.2004.05.016 [DOI] [PubMed] [Google Scholar]
- 105. Hauptmann S, Zardi L, Siri A, Carnemolla B, Borsi L, Castellucci M, Klosterhalfen B, Hartung P, Weis J, Stocker G, et al. Extracellular matrix proteins in colorectal carcinomas. Expression of tenascin and fibronectin isoforms. Lab Invest 1995; 73:172-82; PMID:7543628 [PubMed] [Google Scholar]
- 106. Zirbes TK, Baldus SE, Moenig SP, Schmitz K, Thiele J, Holscher AH, Dienes HP. Tenascin expression in gastric cancer with special emphasis on the WHO-, Lauren-, and Goseki-classifications. Int J Mol Med 1999; 4:39-42; PMID:10373635 [DOI] [PubMed] [Google Scholar]
- 107. Ikeda Y, Mori M, Kajiyama K, Haraguchi Y, Sasaki O, Sugimachi K. Immunohistochemical expression of tenascin in normal stomach tissue, gastric carcinomas and gastric carcinoma in lymph nodes. Br J Cancer 1995; 72:189-92; PMID:7541237; http://dx.doi.org/ 10.1038/bjc.1995.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ilunga K, Iriyama K. Expression of tenascin in gastric carcinoma. Br J Surg 1995; 82:948-51; PMID:7544220; http://dx.doi.org/ 10.1002/bjs.1800820730 [DOI] [PubMed] [Google Scholar]
- 109. Atula T, Hedstrom J, Finne P, Leivo I, Markkanen-Leppanen M, Haglund C. Tenascin-C expression and its prognostic significance in oral and pharyngeal squamous cell carcinoma. Anticancer Res 2003; 23:3051-6; PMID:12926160 [PubMed] [Google Scholar]
- 110. Juuti A, Nordling S, Louhimo J, Lundin J, Haglund C. Tenascin C expression is upregulated in pancreatic cancer and correlates with differentiation. J Clin Pathol 2004; 57:1151-5; PMID:15509674; http://dx.doi.org/ 10.1136/jcp.2003.015818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pilch H, Schaffer U, Schlenger K, Lautz A, Tanner B, Hockel M, Knapstein PG. Expression of tenascin in human cervical cancer–association of tenascin expression with clinicopathological parameters. Gynecol Oncol 1999; 73:415-21; PMID:10366470; http://dx.doi.org/ 10.1006/gyno.1999.5405 [DOI] [PubMed] [Google Scholar]
- 112. Moch H, Torhorst J, Durmuller U, Feichter GE, Sauter G, Gudat F. Comparative analysis of the expression of tenascin and established prognostic factors in human breast cancer. Pathol Res Pract 1993; 189:510-4; PMID:7690952; http://dx.doi.org/ 10.1016/S0344-0338(11)80357-2 [DOI] [PubMed] [Google Scholar]
- 113. Sugawara I, Hirakoshi J, Masunaga A, Itoyama S, Sakakura T. Reduced tenascin expression in colonic carcinoma with lymphogenous metastasis. Invasion Metastasis 1991; 11:325-31; PMID:1726610 [PubMed] [Google Scholar]
- 114. Hindermann W, Berndt A, Borsi L, Luo X, Hyckel P, Katenkamp D, Kosmehl H. Synthesis and protein distribution of the unspliced large tenascin-C isoform in oral squamous cell carcinoma. J Pathol 1999; 189:475-80; PMID:10629546; http://dx.doi.org/ 10.1002/(SICI)1096-9896(199912)189:4%3c475::AID-PATH462%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 115. Katenkamp K, Berndt A, Hindermann W, Wunderlich H, Haas KM, Borsi L, Zardi L, Kosmehl H. mRNA expression and protein distribution of the unspliced tenascin-C isoform in prostatic adenocarcinoma. J Pathol 2004; 203:771-9; PMID:15221936; http://dx.doi.org/ 10.1002/path.1589 [DOI] [PubMed] [Google Scholar]
- 116. Carnemolla B, Castellani P, Ponassi M, Borsi L, Urbini S, Nicolo G, Dorcaratto A, Viale G, Winter G, Neri D, et al. Identification of a glioblastoma-associated tenascin-C isoform by a high affinity recombinant antibody. Am J Pathol 1999; 154:1345-52; PMID:10329587; http://dx.doi.org/ 10.1016/S0002-9440(10)65388-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Viale GL, Castellani P, Dorcaratto A, Pau A, Sehrbundt E, Siri A, Biro A, Zardi L. Occurrence of a glioblastoma-associated tenascin-C isoform in cerebral cavernomas and neighboring vessels. Neurosurgery 2002; 50:838-42; discussion 42; PMID:11904036; http://dx.doi.org/ 10.1097/00006123-200204000-00028 [DOI] [PubMed] [Google Scholar]
- 118. Silacci M, Brack SS, Spath N, Buck A, Hillinger S, Arni S, Weder W, Zardi L, Neri D. Human monoclonal antibodies to domain C of tenascin-C selectively target solid tumors in vivo. Protein Eng Des Sel 2006; 19:471-8; PMID:16928692; http://dx.doi.org/ 10.1093/protein/gzl033 [DOI] [PubMed] [Google Scholar]
- 119. Franz M, Hansen T, Borsi L, Geier C, Hyckel P, Schleier P, Richter P, Altendorf-Hofmann A, Kosmehl H, Berndt A. A quantitative co-localization analysis of large unspliced tenascin-C(L) and laminin-5/gamma2-chain in basement membranes of oral squamous cell carcinoma by confocal laser scanning microscopy. J Oral Pathol Med 2007; 36:6-11; PMID:17181735; http://dx.doi.org/ 10.1111/j.1600-0714.2006.00492.x [DOI] [PubMed] [Google Scholar]
- 120. Berndt A, Anger K, Richter P, Borsi L, Brack S, Silacci M, Franz M, Wunderlich H, Gajda M, Zardi L, et al. Differential expression of tenascin-C splicing domains in urothelial carcinomas of the urinary bladder. J Cancer Res Clin Oncol 2006; 132:537-46; PMID:16788848; http://dx.doi.org/ 10.1007/s00432-006-0106-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Adams M, Jones JL, Walker RA, Pringle JH, Bell SC. Changes in tenascin-C isoform expression in invasive and preinvasive breast disease. Cancer Res 2002; 62:3289-97; PMID:12036947 [PubMed] [Google Scholar]
- 122. Tsunoda T, Inada H, Kalembeyi I, Imanaka-Yoshida K, Sakakibara M, Okada R, Katsuta K, Sakakura T, Majima Y, Yoshida T. Involvement of large tenascin-C splice variants in breast cancer progression. Am J Pathol 2003; 162:1857-67; PMID:12759243; http://dx.doi.org/ 10.1016/S0002-9440(10)64320-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Guttery DS, Hancox RA, Mulligan KT, Hughes S, Lambe SM, Pringle JH, Walker RA, Jones JL, Shaw JA. Association of invasion-promoting tenascin-C additional domains with breast cancers in young women. Breast Cancer Res 2010; 12:R57; PMID:20678196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Guttery DS, Shaw JA, Lloyd K, Pringle JH, Walker RA. Expression of tenascin-C and its isoforms in the breast. Cancer Metastasis Rev 2010; 29:595-606; PMID:20814719; http://dx.doi.org/ 10.1007/s10555-010-9249-9 [DOI] [PubMed] [Google Scholar]
- 125. Dueck M, Riedl S, Hinz U, Tandara A, Moller P, Herfarth C, Faissner A. Detection of tenascin-C isoforms in colorectal mucosa, ulcerative colitis, carcinomas and liver metastases. Int J Cancer 1999; 82:477-83; PMID:10404058; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19990812)82:4%3c477::AID-IJC2%3e3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 126. Driemel O, Dahse R, Berndt A, Pistner H, Hakim SG, Zardi L, Reichert TE, Kosmehl H. High-molecular tenascin-C as an indicator of atypical cells in oral brush biopsies. Clin Oral Investig 2007; 11:93-9; PMID:17111122; http://dx.doi.org/ 10.1007/s00784-006-0086-8 [DOI] [PubMed] [Google Scholar]
- 127. Murphy-Ullrich JE, Lightner VA, Aukhil I, Yan YZ, Erickson HP, Hook M. Focal adhesion integrity is downregulated by the alternatively spliced domain of human tenascin. J Cell Biol 1991; 115:1127-36; PMID:1720121; http://dx.doi.org/ 10.1083/jcb.115.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Galoian KA, Garamszegi N, Garamszegi SP, Scully SP. Molecular mechanism of tenascin-C action on matrix metalloproteinase-1 invasive potential. Exp Biol Med 2007; 232:515-22; PMID:17392487 [PubMed] [Google Scholar]
- 129. Chung CY, Zardi L, Erickson HP. Binding of tenascin-C to soluble fibronectin and matrix fibrils. J Biol Chem 1995; 270:29012-7; PMID:7499434; http://dx.doi.org/ 10.1074/jbc.270.48.29012 [DOI] [PubMed] [Google Scholar]
- 130. Hancox RA, Allen MD, Holliday DL, Edwards DR, Pennington CJ, Guttery DS, Shaw JA, Walker RA, Pringle JH, Jones JL. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res: BCR 2009; 11:R24; PMID:19405959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Tucker RP. Quantitative in situ localization of tenascin-C alternatively spliced transcripts in the avian optic tectum. Mol Vis 1998; 4:18; PMID:9756954 [PubMed] [Google Scholar]
- 132. Saito Y, Imazeki H, Miura S, Yoshimura T, Okutsu H, Harada Y, Ohwaki T, Nagao O, Kamiya S, Hayashi R, et al. A peptide derived from tenascin-C induces beta1 integrin activation through syndecan-4. J Biol Chem 2007; 282:34929-37; PMID:17901052; http://dx.doi.org/ 10.1074/jbc.M705608200 [DOI] [PubMed] [Google Scholar]
- 133. Cai M, Onoda K, Takao M, Kyoko IY, Shimpo H, Yoshida T, Yada I. Degradation of tenascin-C and activity of matrix metalloproteinase-2 are associated with tumor recurrence in early stage non-small cell lung cancer. Clin Cancer Res 2002; 8:1152-6; PMID:11948127 [PubMed] [Google Scholar]
- 134. Tanaka R, Seki Y, Saito Y, Kamiya S, Fujita M, Okutsu H, Iyoda T, Takai T, Owaki T, Yajima H, et al. Tenascin-C-derived peptide TNIIIA2 highly enhances cell survival and platelet-derived growth factor (PDGF)-dependent cell proliferation through potentiated and sustained activation of integrin alpha5beta1. J Biol Chem 2014; 289:17699-708; PMID:24808173; http://dx.doi.org/ 10.1074/jbc.M113.546622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gong XG, Lv YF, Li XQ, Xu FG, Ma QY. Gemcitabine resistance induced by interaction between alternatively spliced segment of tenascin-C and annexin A2 in pancreatic cancer cells. Biol Pharm Bull 2010; 33:1261-7; PMID:20686216; http://dx.doi.org/ 10.1248/bpb.33.1261 [DOI] [PubMed] [Google Scholar]
- 136. Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochimi Biophys Acta 2011; 1813:1978-86; PMID:21440011; http://dx.doi.org/ 10.1016/j.bbamcr.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 137. Esposito I, Penzel R, Chaib-Harrireche M, Barcena U, Bergmann F, Riedl S, Kayed H, Giese N, Kleeff J, Friess H, et al. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J Pathol 2006; 208:673-85; PMID:16450333; http://dx.doi.org/ 10.1002/path.1935 [DOI] [PubMed] [Google Scholar]
- 138. Riedl S, Bodenmuller H, Hinz U, Holle R, Moller P, Schlag P, Herfarth C, Faissner A. Significance of tenascin serum level as tumor marker in primary colorectal carcinoma. Int J Cancer 1995; 64:65-9; PMID:7545144; http://dx.doi.org/ 10.1002/ijc.2910640113 [DOI] [PubMed] [Google Scholar]
- 139. Riedl S, Tandara A, Reinshagen M, Hinz U, Faissner A, Bodenmuller H, Buhr HJ, Herfarth C, Moller P. Serum tenascin-C is an indicator of inflammatory bowel disease activity. Int J Colorectal Dis 2001; 16:285-91; PMID:11686525; http://dx.doi.org/ 10.1007/s003840100312 [DOI] [PubMed] [Google Scholar]
- 140. Burchardt ER, Hein R, Bosserhoff AK. Laminin, hyaluronan, tenascin-C and type VI collagen levels in sera from patients with malignant melanoma. Clin Exp Dermatol 2003; 28:515-20; PMID:12950343; http://dx.doi.org/ 10.1046/j.1365-2230.2003.01326.x [DOI] [PubMed] [Google Scholar]
- 141. Schenk S, Bruckner-Tuderman L, Chiquet-Ehrismann R. Dermo-epidermal separation is associated with induced tenascin expression in human skin. Br J Dermatol 1995; 133:13-22; PMID:7545419; http://dx.doi.org/ 10.1111/j.1365-2133.1995.tb02486.x [DOI] [PubMed] [Google Scholar]
- 142. Ishiwata T, Takahashi K, Shimanuki Y, Ohashi R, Cui R, Takahashi F, Shimizu K, Miura K, Fukuchi Y. Serum tenascin-C as a potential predictive marker of angiogenesis in non-small cell lung cancer. Anticancer Res 2005; 25:489-95; PMID:15816617 [PubMed] [Google Scholar]
- 143. Didem T, Faruk T, Senem K, Derya D, Murat S, Murat G, Oznur K. Clinical significance of serum tenascin-c levels in epithelial ovarian cancer. Tumour Biol 2014; 35:6777-82; PMID:24722824; http://dx.doi.org/ 10.1007/s13277-014-1923-z [DOI] [PubMed] [Google Scholar]
- 144. Tastekin D, Tas F, Karabulut S, Duranyildiz D, Serilmez M, Guveli M, Vatansever S. Clinical significance of serum tenascin-C levels in breast cancer. Tumour Biol 2014; 35:6619-25; PMID:24696262; http://dx.doi.org/ 10.1007/s13277-014-1875-3 [DOI] [PubMed] [Google Scholar]
- 145. Schenk S, Muser J, Vollmer G, Chiquet-Ehrismann R. Tenascin-C in serum: a questionable tumor marker. Int J Cancer 1995; 61:443-9; PMID:7538974; http://dx.doi.org/ 10.1002/ijc.2910610402 [DOI] [PubMed] [Google Scholar]
- 146. Takeda A, Otani Y, Iseki H, Takeuchi H, Aikawa K, Tabuchi S, Shinozuka N, Saeki T, Okazaki Y, Koyama I. Clinical significance of large tenascin-C spliced variant as a potential biomarker for colorectal cancer. World J Surg 2007; 31:388-94; PMID:17219282; http://dx.doi.org/ 10.1007/s00268-006-0328-6 [DOI] [PubMed] [Google Scholar]
- 147. Takeda A, Otani Y, Hirooka E, Okada K, Torii T, Shinozuka N, Koyama I. Plasma large Tenascin-C spliced variant as a possible biomarker for the prediction of hepatic recurrence in colorectal cancer. Surgery 2007; 141:124-5; PMID:17188181; http://dx.doi.org/ 10.1016/j.surg.2006.07.044 [DOI] [PubMed] [Google Scholar]
- 148. Richter P, Tost M, Franz M, Altendorf-Hofmann A, Junker K, Borsi L, Neri D, Kosmehl H, Wunderlich H, Berndt A. B and C domain containing tenascin-C: urinary markers for invasiveness of urothelial carcinoma of the urinary bladder? J Cancer Res Clin Oncol 2009; 135:1351-8; PMID:19326143; http://dx.doi.org/ 10.1007/s00432-009-0576-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Gecks T, Junker K, Franz M, Richter P, Walther M, Voigt A, Neri D, Kosmehl H, Wunderlich H, Kiehntopf M, et al. B domain containing Tenascin-C: a new urine marker for surveillance of patients with urothelial carcinoma of the urinary bladder? Clin Chim Acta 2011; 412:1931-6; PMID:21763295; http://dx.doi.org/ 10.1016/j.cca.2011.06.038 [DOI] [PubMed] [Google Scholar]
- 150. Brellier F, Martina E, Degen M, Heuze-Vourc'h N, Petit A, Kryza T, Courty Y, Terracciano L, Ruiz C, Chiquet-Ehrismann R. Tenascin-W is a better cancer biomarker than tenascin-C for most human solid tumors. BMC Clin Pathol 2012; 12:14; PMID:22947174; http://dx.doi.org/ 10.1186/1472-6890-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Akabani G, Reardon DA, Coleman RE, Wong TZ, Metzler SD, Bowsher JE, Barboriak DP, Provenzale JM, Greer KL, DeLong D, et al. Dosimetry and radiographic analysis of 131I-labeled anti-tenascin 81C6 murine monoclonal antibody in newly diagnosed patients with malignant gliomas: a phase II study. J Nucl Med 2005; 46:1042-51; PMID:15937318 [PubMed] [Google Scholar]
- 152. Cokgor I, Akabani G, Kuan CT, Friedman HS, Friedman AH, Coleman RE, McLendon RE, Bigner SH, Zhao XG, Garcia-Turner AM, et al. Phase I trial results of iodine-131-labeled antitenascin monoclonal antibody 81C6 treatment of patients with newly diagnosed malignant gliomas. Jo Clin Oncol 2000; 18:3862-72; PMID:11078500 [DOI] [PubMed] [Google Scholar]
- 153. Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov 2011; 10:767-77; PMID:21921921; http://dx.doi.org/ 10.1038/nrd3554 [DOI] [PubMed] [Google Scholar]
- 154. Brack SS, Silacci M, Birchler M, Neri D. Tumor-targeting properties of novel antibodies specific to the large isoform of tenascin-C. Clin Cancer Res 2006; 12:3200-8; PMID:16707621; http://dx.doi.org/ 10.1158/1078-0432.CCR-05-2804 [DOI] [PubMed] [Google Scholar]
- 155. Pedretti M, Verpelli C, Marlind J, Bertani G, Sala C, Neri D, Bello L. Combination of temozolomide with immunocytokine F16-IL2 for the treatment of glioblastoma. Br J Cancer 2010; 103:827-36; PMID:20736949; http://dx.doi.org/ 10.1038/sj.bjc.6605832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Saghizadeh M, Khin HL, Bourdon MA, Kenney MC, Ljubimov AV. Novel splice variants of human tenascin-C mRNA identified in normal and bullous keratopathy corneas. Cornea 1998; 17:326-32; PMID:9603390; http://dx.doi.org/ 10.1097/00003226-199805000-00014 [DOI] [PubMed] [Google Scholar]
- 157. Ljubimov AV, Saghizadeh M, Spirin KS, Khin HL, Lewin SL, Zardi L, Bourdon MA, Kenney MC. Expression of tenascin-C splice variants in normal and bullous keratopathy human corneas. Invest Ophthalmol Vis Sci 1998; 39:1135-42; PMID:9620072 [PubMed] [Google Scholar]
- 158. Pearson CA, Pearson D, Shibahara S, Hofsteenge J, Chiquet-Ehrismann R. Tenascin: cDNA cloning and induction by TGF-β. EMBO J 1988; 7:2977-82; PMID:2460335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Chimal-Monroy J, Diaz de Leon L. Expression of N-cadherin, N-CAM, fibronectin and tenascin is stimulated by TGF-beta1, beta2, beta3 and beta5 during the formation of precartilage condensations. Int J Dev Biol 1999; 43:59-67; PMID:10213083 [PubMed] [Google Scholar]
- 160. Tucker RP, Chiquet-Ehrismann R. The regulation of tenascin expression by tissue microenvironments. Biophys Acta 2009; 1793:888-92; PMID:19162090; http://dx.doi.org/ 10.1016/j.bbamcr.2008.12.012 [DOI] [PubMed] [Google Scholar]
- 161. Akhurst RJ, Lehnert SA, Faissner A, Duffie E. TGF β in murine morphogenetic processes: the early embryo and cardiogenesis. Development 1990; 108:645-56; PMID:1696875 [DOI] [PubMed] [Google Scholar]
- 162. Soini Y, Kamel D, Apaja-Sarkkinen M, Virtanen I, Lehto VP. Tenascin immunoreactivity in normal and pathological bone marrow. J Clin Pathol 1993; 46:218-21; PMID:7681854; http://dx.doi.org/ 10.1136/jcp.46.3.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Suzuki S, Li AJ, Ikemoto M, Imamura T. Expression of tenascin-C long isoforms is induced in the hypothalamus by FGF-1. Neuroreport 2002; 13:1041-5; PMID:12060805; http://dx.doi.org/ 10.1097/00001756-200206120-00013 [DOI] [PubMed] [Google Scholar]
- 164. Latijnhouwers MA, de Jongh GJ, Bergers M, de Rooij MJ, Schalkwijk J. Expression of tenascin-C splice variants by human skin cells. Arch Dermatol Res 2000; 292:446-54; PMID:11000288; http://dx.doi.org/ 10.1007/s004030000152 [DOI] [PubMed] [Google Scholar]
- 165. Borsi L, Balza E, Gaggero B, Allemanni G, Zardi L. The alternative splicing pattern of the tenascin-C pre-mRNA is controlled by the extracellular pH. J Biol Chem 1995; 270:6243-5; PMID:7534307; http://dx.doi.org/ 10.1074/jbc.270.11.6243 [DOI] [PubMed] [Google Scholar]
- 166. Borsi L, Allemanni G, Gaggero B, Zardi L. Extracellular pH controls pre-mRNA alternative splicing of tenascin-C in normal, but not in malignantly transformed, cells. Int J Cancer 1996; 66:632-5; PMID:8647625; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19960529)66:5%3c632::AID-IJC9%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 167. Bumke MA, Neri D, Elia G. Modulation of gene expression by extracellular pH variations in human fibroblasts: a transcriptomic and proteomic study. Proteomics 2003; 3:675-88; PMID:12748947; http://dx.doi.org/ 10.1002/pmic.200300395 [DOI] [PubMed] [Google Scholar]
- 168. Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat Struct Mol Biol 2014; 21:189-97; PMID:24440982; http://dx.doi.org/ 10.1038/nsmb.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Taylor HC, Lightner VA, Beyer WF, Jr., McCaslin D, Briscoe G, Erickson HP. Biochemical and structural studies of tenascin/hexabrachion proteins. J Cell Biochem 1989; 41:71-90; PMID:2482292; http://dx.doi.org/ 10.1002/jcb.240410204 [DOI] [PubMed] [Google Scholar]
- 170. Kammerer RA, Schulthess T, Landwehr R, Lustig A, Fischer D, Engel J. Tenascin-C hexabrachion assembly is a sequential two-step process initiated by coiled-coil α-helices. J Biol Chem 1998; 273:10602-8; PMID:9553121; http://dx.doi.org/ 10.1074/jbc.273.17.10602 [DOI] [PubMed] [Google Scholar]
- 171. Redick SD, Schwarzbauer JE. Rapid intracellular assembly of tenascin hexabrachions suggests a novel cotranslational process. J Cell Sci 1995; 108(Pt 4):1761-9; PMID:7542260 [DOI] [PubMed] [Google Scholar]
- 172. Erickson HP, Inglesias JL. A six-armed oligomer isolated from cell surface fibronectin preparations. Nature 1984; 311:267-9; PMID:6482952; http://dx.doi.org/ 10.1038/311267a0 [DOI] [PubMed] [Google Scholar]
- 173. Luczak JA, Redick SD, Schwarzbauer JE. A single cysteine, Cys-64, is essential for assembly of tenascin-C hexabrachions. J Biol Chem 1998; 273:2073-7; PMID:9442046; http://dx.doi.org/ 10.1074/jbc.273.4.2073 [DOI] [PubMed] [Google Scholar]
- 174. Gupta R, Jung E, Brunak S Prediction of N-glycosylation sites in human proteins. (in preparation 2004). [Google Scholar]
- 175. Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 2013; 32:1478-88; PMID:23584533; http://dx.doi.org/ 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Vaughan L, Huber S, Chiquet M, Winterhalter KH. A major, six-armed glycoprotein from embryonic cartilage. EMBO J 1987; 6:349-53; PMID:3582363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Sano K, Asanuma-Date K, Arisaka F, Hattori S, Ogawa H. Changes in glycosylation of vitronectin modulate multimerization and collagen binding during liver regeneration. Glycobiology 2007; 17:784-94; PMID:17369286; http://dx.doi.org/ 10.1093/glycob/cwm031 [DOI] [PubMed] [Google Scholar]
- 178. Jones GE, Arumugham RG, Tanzer ML. Fibronectin glycosylation modulates fibroblast adhesion and spreading. J Cell Biol 1986; 103:1663-70; PMID:2946699; http://dx.doi.org/ 10.1083/jcb.103.5.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Sano K, Miyamoto Y, Kawasaki N, Hashii N, Itoh S, Murase M, Date K, Yokoyama M, Sato C, Kitajima K, et al. Survival signals of hepatic stellate cells in liver regeneration are regulated by glycosylation changes in rat vitronectin, especially decreased sialylation. J Biol Chem 2010; 285:17301-9; PMID:20335177; http://dx.doi.org/ 10.1074/jbc.M109.077016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Alisson-Silva F, Freire-de-Lima L, Donadio JL, Lucena MC, Penha L, Sa-Diniz JN, Dias WB, Todeschini AR. Increase of O-glycosylated oncofetal fibronectin in high glucose-induced epithelial-mesenchymal transition of cultured human epithelial cells. PloS One 2013; 8:e60471; PMID:23593224; http://dx.doi.org/ 10.1371/journal.pone.0060471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kasbaoui L, Harb J, Bernard S, Meflah K. Differences in glycosylation state of fibronectin from two rat colon carcinoma cell lines in relation to tumoral progressiveness. Cancer Res 1989; 49:5317-22; PMID:2766300 [PubMed] [Google Scholar]
- 182. Chung CY, Erickson HP. Glycosaminoglycans modulate fibronectin matrix assembly and are essential for matrix incorporation of tenascin-C. J Cell Sci 1997; 110(Pt 12):1413-9; PMID:9217327 [DOI] [PubMed] [Google Scholar]
- 183. Ramos DM, Chen B, Regezi J, Zardi L, Pytela R. Tenascin-C matrix assembly in oral squamous cell carcinoma. Int J Cancer 1998; 75:680-7; PMID:9495234; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19980302)75:5%3c680::AID-IJC4%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 184. Matsuda A, Yoshiki A, Tagawa Y, Matsuda H, Kusakabe M. Corneal wound healing in tenascin knockout mouse. Invest Ophthalmol Vis Sci 1999; 40:1071-80; PMID:10235540 [PubMed] [Google Scholar]
- 185. Kii I, Nishiyama T, Li M, Matsumoto K, Saito M, Amizuka N, Kudo A. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J Biol Chem 2010; 285:2028-39; PMID:19887451; http://dx.doi.org/ 10.1074/jbc.M109.051961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Rocco M, Carson M, Hantgan R, McDonagh J, Hermans J. Dependence of the shape of the plasma fibronectin molecule on solvent composition. Ionic strength and glycerol content. J Biol Chem 1983; 258:14545-9; PMID:6643501 [PubMed] [Google Scholar]
- 187. Chiquet-Ehrismann R, Kalla P, Pearson CA, Beck K, Chiquet M. Tenascin interferes with fibronectin action. Cell 1988; 53:383-90; PMID:2452695; http://dx.doi.org/ 10.1016/0092-8674(88)90158-4 [DOI] [PubMed] [Google Scholar]
- 188. Hoffman S, Edelman GM. A proteoglycan with HNK-1 antigenic determinants is a neuron-associated ligand for cytotactin. Proc Nat Acad Sci U S A 1987; 84:2523-7; PMID:2436234; http://dx.doi.org/ 10.1073/pnas.84.8.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Ruggiero S, Cosgarea R, Potempa J, Potempa B, Eick S, Chiquet M. Cleavage of extracellular matrix in periodontitis: gingipains differentially affect cell adhesion activities of fibronectin and tenascin-C. Biochimi Biophys Acta 2013; 1832:517-26; PMID:23313574; http://dx.doi.org/ 10.1016/j.bbadis.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Wallner K, Li C, Shah PK, Wu KJ, Schwartz SM, Sharifi BG. EGF-Like domain of tenascin-C is proapoptotic for cultured smooth muscle cells. Arterioscler Thromb Vasc Biol 2004; 24:1416-21; PMID:15178565; http://dx.doi.org/ 10.1161/01.ATV.0000134299.89599.53 [DOI] [PubMed] [Google Scholar]
- 191. Chiao YA, Zamilpa R, Lopez EF, Dai Q, Escobar GP, Hakala K, Weintraub ST, Lindsey ML. In vivo matrix metalloproteinase-7 substrates identified in the left ventricle post-myocardial infarction using proteomics. J Proteome Res 2010; 9:2649-57; PMID:20232908; http://dx.doi.org/ 10.1021/pr100147r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Zamilpa R, Lopez EF, Chiao YA, Dai Q, Escobar GP, Hakala K, Weintraub ST, Lindsey ML. Proteomic analysis identifies in vivo candidate matrix metalloproteinase-9 substrates in the left ventricle post-myocardial infarction. Proteomics 2010; 10:2214-23; PMID:20354994; http://dx.doi.org/ 10.1002/pmic.200900587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Gueders MM, Hirst SJ, Quesada-Calvo F, Paulissen G, Hacha J, Gilles C, Gosset P, Louis R, Foidart JM, Lopez-Otin C, et al. Matrix metalloproteinase-19 deficiency promotes tenascin-C accumulation and allergen-induced airway inflammation. Am J Respir Cell Mol Biol 2010; 43:286-95; PMID:19843707; http://dx.doi.org/ 10.1165/rcmb.2008-0426OC [DOI] [PubMed] [Google Scholar]
- 194. Zhen EY, Brittain IJ, Laska DA, Mitchell PG, Sumer EU, Karsdal MA, Duffin KL. Characterization of metalloprotease cleavage products of human articular cartilage. Arthritis Rheum 2008; 58:2420-31; PMID:18668564; http://dx.doi.org/ 10.1002/art.23654 [DOI] [PubMed] [Google Scholar]
- 195. Sofat N, Robertson SD, Hermansson M, Jones J, Mitchell P, Wait R. Tenascin-C fragments are endogenous inducers of cartilage matrix degradation. Rheumatol Int 2012; 32:2809-17; PMID:21874326; http://dx.doi.org/ 10.1007/s00296-011-2067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. To WS, Midwood KS. Cryptic domains of tenascin-C differentially control fibronectin fibrillogenesis. Matrix Biol 2010; 29:573-85; PMID:20708078; http://dx.doi.org/ 10.1016/j.matbio.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 197. Ambort D, Brellier F, Becker-Pauly C, Stocker W, Andrejevic-Blant S, Chiquet M, Sterchi EE. Specific processing of tenascin-C by the metalloprotease meprinbeta neutralizes its inhibition of cell spreading. Matrix Biol 2010; 29:31-42; PMID:19748582; http://dx.doi.org/ 10.1016/j.matbio.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 198. Iyer AK, Tran KT, Borysenko CW, Cascio M, Camacho CJ, Blair HC, Bahar I, Wells A. Tenascin cytotactin epidermal growth factor-like repeat binds epidermal growth factor receptor with low affinity. J Cell Physiol 2007; 211:748-58; PMID:17311283; http://dx.doi.org/ 10.1002/jcp.20986 [DOI] [PubMed] [Google Scholar]
- 199. Swindle CS, Tran KT, Johnson TD, Banerjee P, Mayes AM, Griffith L, Wells A. Epidermal growth factor (EGF)-like repeats of human tenascin-C as ligands for EGF receptor. J Cell Biol 2001; 154:459-68; PMID:11470832; http://dx.doi.org/ 10.1083/jcb.200103103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Rodrigues M, Yates CC, Nuschke A, Griffith L, Wells A. The matrikine tenascin-C protects multipotential stromal cells/mesenchymal stem cells from death cytokines such as FasL. Tissue Eng Part A 2013; 19:1972-83; PMID:23541003; http://dx.doi.org/ 10.1089/ten.tea.2012.0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. De Laporte L, Rice JJ, Tortelli F, Hubbell JA. Tenascin C promiscuously binds growth factors via its fifth fibronectin type III-like domain. PloS One 2013; 8:e62076; PMID:23637968; http://dx.doi.org/ 10.1371/journal.pone.0062076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Schumacher S, Stube EM. Regulated binding of the fibrinogen-like domains of tenascin-R and tenascin-C to the neural EGF family member CALEB. J Neurochem 2003; 87:1213-23; PMID:14622101; http://dx.doi.org/ 10.1046/j.1471-4159.2003.02112.x [DOI] [PubMed] [Google Scholar]
- 203. Goh FG, Piccinini AM, Krausgruber T, Udalova IA, Midwood KS. Transcriptional regulation of the endogenous danger signal tenascin-C: a novel autocrine loop in inflammation. J Immunol 2010; 184:2655-62; http://dx.doi.org/ 10.4049/jimmunol.0903359 [DOI] [PubMed] [Google Scholar]
- 204. Latijnhouwers MA, Pfundt R, de Jongh GJ, Schalkwijk J. Tenascin-C expression in human epidermal keratinocytes is regulated by inflammatory cytokines and a stress response pathway. Matrix Biol 1998; 17:305-16; PMID:9749946; http://dx.doi.org/ 10.1016/S0945-053X(98)90083-X [DOI] [PubMed] [Google Scholar]
- 205. Jinnin M, Ihn H, Asano Y, Yamane K, Trojanowska M, Tamaki K. Upregulation of tenascin-C expression by IL-13 in human dermal fibroblasts via the phosphoinositide 3-kinase/Akt and the protein kinase C signaling pathways. J Invest Dermatol 2006; 126:551-60; PMID:16374482; http://dx.doi.org/ 10.1038/sj.jid.5700090 [DOI] [PubMed] [Google Scholar]
- 206. Nakoshi Y, Hasegawa M, Sudo A, Yoshida T, Uchida A. Regulation of tenascin-C expression by tumor necrosis factor-α in cultured human osteoarthritis chondrocytes. J Rheumatol 2008; 35:147-52; PMID:18061975 [PubMed] [Google Scholar]
- 207. Jinnin M, Ihn H, Asano Y, Yamane K, Trojanowska M, Tamaki K. Tenascin-C upregulation by transforming growth factor-β in human dermal fibroblasts involves Smad3, Sp1, and Ets1. Oncogene 2004; 23:1656-67; PMID:15001984; http://dx.doi.org/ 10.1038/sj.onc.1207064 [DOI] [PubMed] [Google Scholar]
- 208. Jinnin M, Ihn H, Asano Y, Yamane K, Trojanowska M, Tamaki K. Platelet derived growth factor induced tenascin-C transcription is phosphoinositide 3-kinase/Akt-dependent and mediated by Ets family transcription factors. J Cell Physiol 2006; 206:718-27; PMID:16245312; http://dx.doi.org/ 10.1002/jcp.20527 [DOI] [PubMed] [Google Scholar]
- 209. Yamamoto K, Dang QN, Kennedy SP, Osathanondh R, Kelly RA, Lee RT. Induction of tenascin-C in cardiac myocytes by mechanical deformation. Role of reactive oxygen species. J Biol Chem 1999; 274:21840-6; PMID:10419501; http://dx.doi.org/ 10.1074/jbc.274.31.21840 [DOI] [PubMed] [Google Scholar]
- 210. Asparuhova MB, Ferralli J, Chiquet M, Chiquet-Ehrismann R. The transcriptional regulator megakaryoblastic leukemia-1 mediates serum response factor-independent activation of tenascin-C transcription by mechanical stress. FASEB J 2011; 25:3477-88; PMID:21705668; http://dx.doi.org/ 10.1096/fj.11-187310 [DOI] [PubMed] [Google Scholar]
- 211. Scherer C, Pfisterer L, Wagner AH, Hodebeck M, Cattaruzza M, Hecker M, Korff T. Arterial wall stress controls NFAT5 activity in vascular smooth muscle cells. J Am Heart Assoc 2014; 3:e000626; PMID:24614757; http://dx.doi.org/ 10.1161/JAHA.113.000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Jones FS, Chalepakis G, Gruss P, Edelman GM. Activation of the cytotactin promoter by the homeobox-containing gene Evx-1. Proc Nat Acad Sci U S A 1992; 89:2091-5; PMID:1372434; http://dx.doi.org/ 10.1073/pnas.89.6.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Chiquet-Ehrismann R, Tannheimer M, Koch M, Brunner A, Spring J, Martin D, Baumgartner S, Chiquet M. Tenascin-C expression by fibroblasts is elevated in stressed collagen gels. J Cell Biol 1994; 127:2093-101; PMID:7528751; http://dx.doi.org/ 10.1083/jcb.127.6.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Gherzi R, Carnemolla B, Siri A, Ponassi M, Balza E, Zardi L. Human tenascin gene. Structure of the 5'-region, identification, and characterization of the transcription regulatory sequences. J Biol Chem 1995; 270:3429-34; PMID:7531707; http://dx.doi.org/ 10.1074/jbc.270.7.3429 [DOI] [PubMed] [Google Scholar]
- 215. Copertino DW, Edelman GM, Jones FS. Multiple promoter elements differentially regulate the expression of the mouse tenascin gene. Proc Nat Acad Sci U S A 1997; 94:1846-51; PMID:9050867; http://dx.doi.org/ 10.1073/pnas.94.5.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Gherzi R, Briata P, Boncinelli E, Ponassi M, Querze G, Viti F, Corte G, Zardi L. The human homeodomain protein OTX2 binds to the human tenascin-C promoter and trans-represses its activity in transfected cells. DNA Cell Biol 1997; 16:559-67; PMID:9174161; http://dx.doi.org/ 10.1089/dna.1997.16.559 [DOI] [PubMed] [Google Scholar]
- 217. Mettouchi A, Cabon F, Montreau N, Dejong V, Vernier P, Gherzi R, Mercier G, Binetruy B. The c-Jun-induced transformation process involves complex regulation of tenascin-C expression. Mol Cell Biol 1997; 17:3202-9; PMID:9154819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Jones PL, Jones FS, Zhou B, Rabinovitch M. Induction of vascular smooth muscle cell tenascin-C gene expression by denatured type I collagen is dependent upon a beta3 integrin-mediated mitogen-activated protein kinase pathway and a 122-base pair promoter element. J Cell Sci 1999; 112(Pt 4):435-45; PMID:9914156 [DOI] [PubMed] [Google Scholar]
- 219. Shirasaki F, Makhluf HA, LeRoy C, Watson DK, Trojanowska M. Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene 1999; 18:7755-64; PMID:10618716; http://dx.doi.org/ 10.1038/sj.onc.1203360 [DOI] [PubMed] [Google Scholar]
- 220. Jones FS, Meech R, Edelman DB, Oakey RJ, Jones PL. Prx1 controls vascular smooth muscle cell proliferation and tenascin-C expression and is upregulated with Prx2 in pulmonary vascular disease. Circ Res 2001; 89:131-8; PMID:11463719; http://dx.doi.org/ 10.1161/hh1401.093582 [DOI] [PubMed] [Google Scholar]
- 221. McKean DM, Sisbarro L, Ilic D, Kaplan-Alburquerque N, Nemenoff R, Weiser-Evans M, Kern MJ, Jones PL. FAK induces expression of Prx1 to promote tenascin-C-dependent fibroblast migration. J Cell Biol 2003; 161:393-402; PMID:12741393; http://dx.doi.org/ 10.1083/jcb.jcb.200302126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Ghatnekar A, Trojanowska M. GATA-6 is a novel transcriptional repressor of the human Tenascin-C gene expression in fibroblasts. Biochimi Biophys Acta 2008; 1779:145-51; PMID:18177748; http://dx.doi.org/ 10.1016/j.bbagrm.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Sivasankaran B, Degen M, Ghaffari A, Hegi ME, Hamou MF, Ionescu MC, Zweifel C, Tolnay M, Wasner M, Mergenthaler S, et al. Tenascin-C is a novel RBPJkappa-induced target gene for Notch signaling in gliomas. Cancer Res 2009; 69:458-65; PMID:19147558; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-2610 [DOI] [PubMed] [Google Scholar]
- 224. Erickson HP, Taylor HC. Hexabrachion proteins in embryonic chicken tissues and human tumors. J Cell Biol 1987; 105:1387-94; PMID:3654758; http://dx.doi.org/ 10.1083/jcb.105.3.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Matsuoka Y, Spring J, Ballmer-Hofer K, Hofer U, Chiquet-Ehrismann R. Differential expression of tenascin splicing variants in the chick gizzard and in cell cultures. Cell Differ Dev 1990; 32:417-23; PMID:1711921; http://dx.doi.org/ 10.1016/0922-3371(90)90058-5 [DOI] [PubMed] [Google Scholar]
- 226. Koch M, Wehrle-Haller B, Baumgartner S, Spring J, Brubacher D, Chiquet M. Epithelial synthesis of tenascin at tips of growing bronchi and graded accumulation in basement membrane and mesenchyme. Exp Cell Res 1991; 194:297-300; PMID:1709104; http://dx.doi.org/ 10.1016/0014-4827(91)90368-5 [DOI] [PubMed] [Google Scholar]
- 227. Probstmeier R, Martini R, Tacke R, Schachner M. Expression of the adhesion molecules L1, N-CAM and J1/tenascin during development of the murine small intestine. Differentiation 1990; 44:42-55; PMID:1701406; http://dx.doi.org/ 10.1111/j.1432-0436.1990.tb00535.x [DOI] [PubMed] [Google Scholar]
- 228. Zhao Y, Young SL. Tenascin in rat lung development: in situ localization and cellular sources. Am J Physiol 1995; 269:L482-91; PMID:7485520 [DOI] [PubMed] [Google Scholar]
- 229. LaFleur DW, Fagin JA, Forrester JS, Rubin SA, Sharifi BG. Cloning and characterization of alternatively spliced isoforms of rat tenascin. Platelet-derived growth factor-BB markedly stimulates expression of spliced variants of tenascin mRNA in arterial smooth muscle cells. J Biol Chem 1994; 269:20757-63; PMID:7519614 [PubMed] [Google Scholar]
- 230. Talts JF, Eng H, Zhang HY, Faissner A, Ekblom P. Characterization of monoclonal antibodies against tenascin-C: no apparent effect on kidney development in vitro. Int J Dev Biol 1997; 41:39-48; PMID:9074936 [PubMed] [Google Scholar]
- 231. Bourdon MA, Wikstrand CJ, Furthmayr H, Matthews TJ, Bigner DD. Human glioma-mesenchymal extracellular matrix antigen defined by monoclonal antibody. Cancer Res 1983; 43:2796-805; PMID:6342760 [PubMed] [Google Scholar]
- 232. Aukhil I, Joshi P, Yan Y, Erickson HP. Cell- and heparin-binding domains of the hexabrachion arm identified by tenascin expression proteins. J Biol Chem 1993; 268:2542-53; PMID:7679097 [PubMed] [Google Scholar]
- 233. Carnemolla B, Borsi L, Bannikov G, Troyanovsky S, Zardi L. Comparison of human tenascin expression in normal, simian-virus-40-transformed and tumor-derived cell lines. Eur J Biochem 1992; 205:561-7; PMID:1374030 [DOI] [PubMed] [Google Scholar]
- 234. Balza E, Siri A, Ponassi M, Caocci F, Linnala A, Virtanen I, Zardi L. Production and characterization of monoclonal antibodies specific for different epitopes of human tenascin. FEBS Lett 1993; 332:39-43; PMID:7691659; http://dx.doi.org/ 10.1016/0014-5793(93)80479-E [DOI] [PubMed] [Google Scholar]
- 235. Ibrahim SN, Lightner VA, Ventimiglia JB, Ibrahim GK, Walther PJ, Bigner DD, Humphrey PA. Tenascin expression in prostatic hyperplasia, intraepithelial neoplasia, and carcinoma. Hum Pathol 1993; 24:982-9; PMID:7504654; http://dx.doi.org/ 10.1016/0046-8177(93)90112-T [DOI] [PubMed] [Google Scholar]
- 236. Wilson KE, Langdon SP, Lessells AM, Miller WR. Expression of the extracellular matrix protein tenascin in malignant and benign ovarian tumours. Br J Cancer 1996; 74:999-1004; PMID:8855965; http://dx.doi.org/ 10.1038/bjc.1996.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Vollmer G, Tan MI, Wunsche W, Frank K. Expression of tenascin-C by human endometrial adenocarcinoma and stroma cells: heterogeneity of splice variants and induction by TGF-β. Biochem Cell Biol 1997; 75:759-69; PMID:9599665 [PubMed] [Google Scholar]
- 238. Kusagawa H, Onoda K, Namikawa S, Yada I, Okada A, Yoshida T, Sakakura T. Expression and degeneration of tenascin-C in human lung cancers. Br J Cancer 1998; 77:98-102; PMID:9459152; http://dx.doi.org/ 10.1038/bjc.1998.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Ghert MA, Jung ST, Qi W, Harrelson JM, Erickson HP, Block JA, Scully SP. The clinical significance of tenascin-C splice variant expression in chondrosarcoma. Oncology 2001; 61:306-14; PMID:11721178; http://dx.doi.org/ 10.1159/000055338 [DOI] [PubMed] [Google Scholar]
- 240. Levy P, Ripoche H, Laurendeau I, Lazar V, Ortonne N, Parfait B, Leroy K, Wechsler J, Salmon I, Wolkenstein P, et al. Microarray-based identification of tenascin C and tenascin XB, genes possibly involved in tumorigenesis associated with neurofibromatosis type 1. Clin Cancer Res 2007; 13:398-407; PMID:17202312; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-0182 [DOI] [PubMed] [Google Scholar]
- 241. Paasinen-Sohns A, Kaariainen E, Yin M, Jarvinen K, Nummela P, Holtta E. Chaotic neovascularization induced by aggressive fibrosarcoma cells overexpressing S-adenosylmethionine decarboxylase. Int J Biochem Cell Biol 2011; 43:441-54; PMID:21134486; http://dx.doi.org/ 10.1016/j.biocel.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 242. Sakai T, Kawakatsu H, Hirota N, Yokoyama T, Takoka T, Sakakura T, Saito M. Tenascin expression in vitro and in vivo: comparison between epithelial and nonepithelial rat cell lines. Exp Cell Res 1993; 206:244-54; PMID:7684694; http://dx.doi.org/ 10.1006/excr.1993.1144 [DOI] [PubMed] [Google Scholar]
- 243. Dang C, Gottschling M, Roewert J, Forschner T, Stockfleth E, Nindl I. Tenascin-C patterns and splice variants in actinic keratosis and cutaneous squamous cell carcinoma. Br J Dermatol 2006; 155:763-70; PMID:16965426; http://dx.doi.org/ 10.1111/j.1365-2133.2006.07401.x [DOI] [PubMed] [Google Scholar]
- 244. Katz S, Hukkanen M, Lounatmaa K, Rousselle P, Tervo T, Virtanen I. Cooperation of isoforms of laminin-332 and tenascin-CL during early adhesion and spreading of immortalized human corneal epithelial cells. Exp Eye Res 2006; 83:1412-22; PMID:16963023; http://dx.doi.org/ 10.1016/j.exer.2006.07.021 [DOI] [PubMed] [Google Scholar]
- 245. Tseleni-Balafouta S, Gakiopoulou H, Fanourakis G, Voutsinas G, Balafoutas D, Patsouris E. Tenascin-C protein expression and mRNA splice variants in thyroid carcinoma. Exp Mol Pathol 2006; 80:177-82; PMID:16259977; http://dx.doi.org/ 10.1016/j.yexmp.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 246. Frey K, Fiechter M, Schwager K, Belloni B, Barysch MJ, Neri D, Dummer R. Different patterns of fibronectin and tenascin-C splice variants expression in primary and metastatic melanoma lesions. Exp Dermatol 2011; 20:685-8; PMID:21649738; http://dx.doi.org/ 10.1111/j.1600-0625.2011.01314.x [DOI] [PubMed] [Google Scholar]
- 247. Galler K, Junker K, Franz M, Hentschel J, Richter P, Gajda M, Gohlert A, von Eggeling F, Heller R, Giavazzi R, et al. Differential vascular expression and regulation of oncofetal tenascin-C and fibronectin variants in renal cell carcinoma (RCC): implications for an individualized angiogenesis-related targeted drug delivery. Histochem Cell Biol 2012; 137:195-204; PMID:22075565; http://dx.doi.org/ 10.1007/s00418-011-0886-z [DOI] [PubMed] [Google Scholar]
- 248. Nies DE, Hemesath TJ, Kim JH, Gulcher JR, Stefansson K. The complete cDNA sequence of human hexabrachion (Tenascin). A multidomain protein containing unique epidermal growth factor repeats. J Biol Chem 1991; 266:2818-23; PMID:1704365 [PubMed] [Google Scholar]


