Abstract
F-box and WD repeat domain-containing 7 (FBW7) is the substrate recognition component of the Skp1-Cul1-F-box (SCF) ubiquitin ligase complex and functions as a major tumor suppressor by targeting various oncoproteins for degradation. Genomic deletion or mutation of FBW7 has frequently been identified in many human cancers but not in pancreatic ductal adenocarcinoma. Thus it is important to know how the tumor suppressive function of FBW7 is impaired in pancreatic cancer. In this study, we first observed that low FBW7 expression correlated significantly with ERK activation in pancreatic cancer clinical samples, primarily due to KRAS mutations in pancreatic cancer. We further showed that ERK directly interacted with FBW7 and phosphorylated FBW7 at Thr205, which sequentially promoted FBW7 ubiquitination and proteasomal degradation. Furthermore, the phospho-deficient T205A FBW7 mutant is resistant to ERK activation and could significantly suppress pancreatic cancer cell proliferation and tumorigenesis. These results collectively demonstrate how the oncogenic KRAS mutation inhibits the tumor suppressor FBW7, thus revealing an important function of KRAS mutations in promoting pancreatic cancer progression.
Keywords: KRAS, pancreatic cancer, ERK, FBW7
Introduction
Pancreatic ductal adenocarcinoma (PDAC), the most common form of pancreatic cancer, is a devastating disease with a 5-year overall survival rate of < 5%, the highest fatality rate among all cancers1,2. The incidence of pancreatic cancer remains equal to its mortality rate. Due to a lack of practical early diagnostic means and a high metastatic rate at initial diagnosis, only 15%-20% of patients are resectable3. Moreover, the 5-year survival rate among radically operated patients is only ∼18%, due to early local or metastatic recurrence4. Therefore, the improvement of clinical practice is a pressing need, which requires a better understanding of the mechanisms underlying the occurrence and development of PDAC.
Genetically, PDAC progresses as a complex result of the activation of oncogenes and inactivation of tumor suppressors. For example, oncogenic mutations in genes such as KRAS and loss-of-function mutation of tumor suppressors, such as p53, CDNK2A/p16, DPC4/SMAD4 and BRCA2, are frequently observed in PDAC5,6. Mutations in KRAS were found in > 90% of patient specimens and were proposed to be initiators of PDAC. In addition, inactivating mutations of CDKN2A were detected in almost 95% of PDAC cases, making this gene the most frequently mutated tumor suppressor. In addition, mutations in p53, DPC4 and BRCA2 were identified in 75%, 50% and 5%-10% of PDAC cases, respectively5,6. Consistent with the genetic data, mouse models with activating KRAS mutations combined with p53 or DPC4 deletion exhibited accelerated PDAC development7, supporting the idea of a complex synergistic pro-tumor effect involving the alteration of both oncogenes and tumor suppressor genes.
F-box and WD repeat domain-containing 7 (FBW7) is the substrate recognition component of the Skp1-Cul1-F-box (SCF) ubiquitin ligase complex and is located within 4q32, a chromosomal region that is frequently deleted in cancers8,9. It has been well established that FBW7 functions as a tumor suppressor by targeting multiple oncoprotein substrates for ubiquitination and degradation, including cyclin E, c-Myc, c-Jun, Notch-1, SREBP1 and Mcl-18,10, thus regulating cell proliferation, apoptosis and metabolism. Hence, loss of function of FBW7 has been proposed to drive the progression of cancer. Indeed, deletion and mutation of FBW7 have been frequently identified in various human malignancies, such as gastric cancer11, colon cancer12, breast carcinoma13, esophageal squamous cell carcinoma14 and intrahepatic cholangiocarcinoma15. Low expression of FBW7 is significantly correlated with poor prognosis11,12,13,14,15. Overall, ∼6% of human tumors harbor mutations in FBW78. Emerging evidence has also demonstrated that FBW7 is regulated by multiple protein factors, such as p53, Pin1, Hes-5 and Numb4, as well as microRNAs16. However, the function and regulation of FBW7 is poorly studied in the occurrence and progression of PDAC. Importantly, elevated KRAS activity stimulates many downstream signaling pathways. Evidence accumulated over the last few decades has demonstrated that the Ras-Raf-MEK-ERK signaling pathway plays an important role in pancreatic cancer development and progression by contributing to cell cycle regulation, differentiation, proliferation, survival, migration and angiogenesis17,18,19. Previously, it was indicated that Ras activity may regulate cyclin E degradation via the FBW7 pathway20. This observation prompted us to investigate whether the Ras-Raf-MEK-ERK pathway plays a role in FBW7 degradation in PDAC.
Herein, we report that FBW7 is rarely mutated and the protein level is significantly downregulated in PDAC, through a mechanism involving an activated Ras-Raf-MEK-ERK signaling pathway. We demonstrate that ERK kinase directly interacts with and phosphorylates FBW7 at Thr205, which leads to the ubiquitination and proteosomal degradation of FBW7 itself and the elevation of multiple FBW7 substrates, such as c-Myc, thus facilitating the proliferation and survival of pancreatic cancer cells. Furthermore, overexpression of a phospho-deficient mutant of FBW7 (Thr205Ala) significantly reduced the tumorigenesis potential of PDAC cells both in vitro and in vivo. Through our experiments, we established a novel link between the oncogenic KRAS-mutant/MAPK/ERK signaling axis and the tumor suppressor FBW7 and revealed the mechanism underlying the impairment of FBW7 function in PDAC. These findings may help develop new therapeutic strategies to treat pancreatic cancer.
Results
FBW7 is downregulated in pancreatic cancer
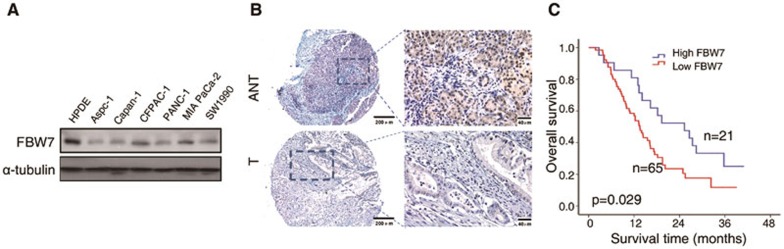
To investigate the role of FBW7 in PDAC progression, we first determined the expression level of FBW7 in various human pancreatic cancer cell lines with KRAS mutations (Aspc-1, Capan-1, CFPAC-1, PANC-1, MIA PaCa-2 and SW1990). We found that FBW7 expression levels were significantly downregulated in six human pancreatic cancer cell lines compared with normal human pancreatic ductal epithelium (HPDE) cells (Figure 1A). We then analyzed FBW7 expression in PDAC clinical samples by immunohistochemistry (IHC) staining of tissue microarrays (TMAs), which contain 86 pairs of tumor and adjacent normal tissues. Based on staining intensity, we grouped the pancreatic tumor specimens according to FBW7 expression level as negative/weak, moderate, and strong (Supplementary information, Figure S1). FBW7 was consistently downregulated in the tumor tissues compared with adjacent normal tissues (Figure 1B). Next, we explored the relationship between FBW7 expression and the clinicopathological features of PDAC. We found that decreased IHC signal of FBW7 correlated with poor tumor differentiation (Supplementary information, Table S1; P = 0.017). However, no significant correlation was observed between FBW7 staining and tumor size, grade or lymph node metastasis (Supplementary information, Table S1; P > 0.05). The Kaplan-Meier survival curves and log rank test showed that high FBW7 expression significantly correlated with better overall survival (OS) in PDAC (Figure 1C; n = 86, P = 0.029). Thus, low expression of FBW7 is associated with high malignancy and poor prognosis in PDAC cases.
Figure 1.
FBW7 is downregulated in pancreatic cancer. (A) Immunoblot analysis of the indicated human pancreatic cancer cell lines. The HPDE cell line was included as a positive control for the detection of endogenous FBW7 expression, and α-tubulin was used as a loading control. (B) IHC staining of human pancreatic adenocarcinoma tissue arrays using specific antibodies for FBW7. T, tumor; ANT, adjacent normal tissues; scale bar, 200 μm; magnification scale bar, 40 μm. (C) Kaplan-Meier analysis of overall survival rate of pancreatic cancer patients according to the expression of FBW7 (n = 86, P = 0.029, log-rank test).
Expression of FBW7 inversely correlates with ERK activation
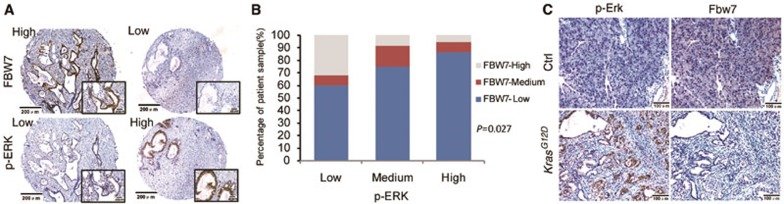
KRAS mutation and the consequential activation of the MAPK kinase axis are regarded as major driving forces of PDAC progression. To explore the potential association of MAPK signaling and FBW7 expression, we first analyzed the levels of FBW7 and phospho-ERK (p-ERK), which represents the activation of MAPK signaling, in human pancreatic cancer cell lines. Western blotting showed a general inverse correlation between the levels of FBW7 and p-ERK in all six pancreatic cancer cell lines examined (Supplementary information, Figure S2). Consistently, IHC staining also indicated an inverse correlation between the levels of FBW7 and p-ERK in human pancreatic cancer samples (Figure 2A and 2B; n = 86, P = 0.027). To further support this point, an inverse correlation between the levels of Fbw7 and p-Erk was detected on slides containing samples derived from KrasG12D transgenic mice by IHC staining (Figure 2C). Taken together, these results suggest that the abundance of FBW7 inversely correlates with ERK activation in PDAC.
Figure 2.
Expression of FBW7 inversely correlates with ERK activation. (A) Representative IHC staining of FBW7 and p-ERK in pancreatic adenocarcinoma tissue arrays (scale bar, 200 μm; inset scale bar, 40 μm). (B) Statistical analysis of the correlation between the levels of FBW7 and p-ERK (P = 0.027; P value was obtained using a Pearson χ2 test). (C) Representative IHC staining of Fbw7 and p-Erk in KrasG12D transgenic mice. Control slides were obtained from the same mouse strain, which was kept in the same conditions and sacrificed at the same age as the transgenic mice (scale bar, 100 μm).
ERK negatively regulates FBW7 stability
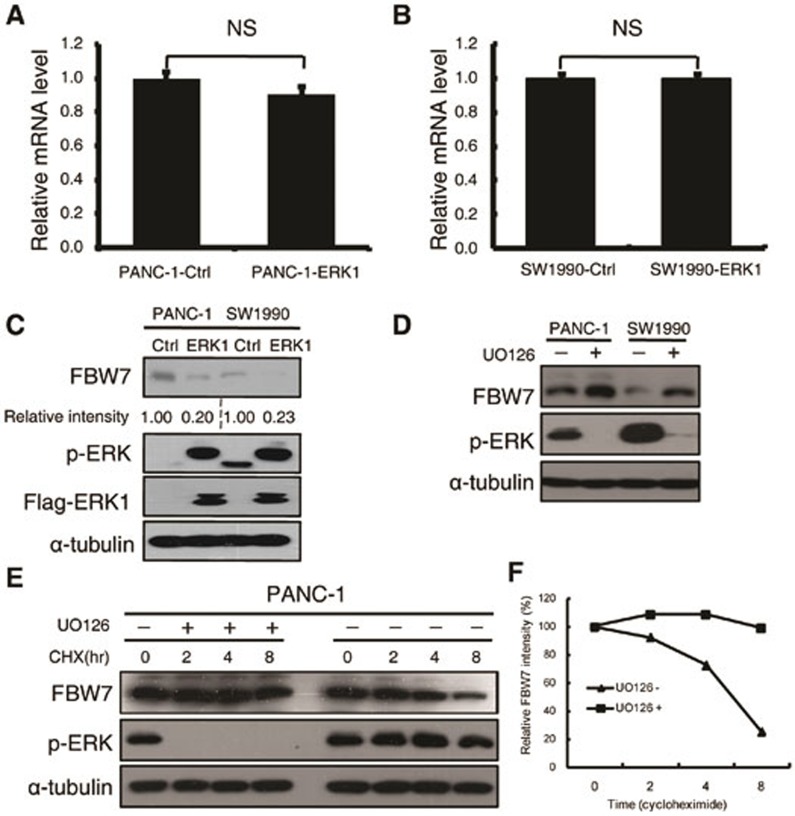
To test the hypothesis that the activation of MAPK signaling negatively regulates FBW7, we examined the expression of FBW7 at both the mRNA and protein levels by q-PCR and immunoblotting, respectively, upon overexpression of ERK1 kinase in two pancreatic cancer cell lines. Notably, the FBW7 mRNA level was barely affected (Figure 3A and 3B). Besides, DNA methylation of FBW7 promoter region was also analyzed. There were 16 CpG sites at the promoter region of FBW7/hCDC4-α (Supplementary information, Figure S3A). Bisulfate sequencing results demonstrated that CpG sites were seldom methylated. Moreover, overexpression of ERK had no detectable influence on the methylation status of CpG sites in the promoter region (Supplementary information, Figure S3B), which is consistent with the transcriptional analysis of FBW7 upon ERK1 overexpression, indicating that FBW7 was regulated mostly via post-transcriptional modifications. As expected, the FBW7 protein level was significantly decreased upon overexpression of wild-type ERK1 but not the kinase-dead mutant (Figure 3C and Supplementary information, Figure S3C). As constitutive activation of ERK is observed in almost all pancreatic cell lines, we pretreated the pancreatic cancer cell lines with the MEK/ERK inhibitors (UO126, AZD6244, trametinib and SCH772984) to suppress ERK activity before harvesting and then determined FBW7 protein level. FBW7 protein levels significantly increased in response to inhibition of MAPK-ERK signaling in pancreatic cells (Figure 3D and Supplementary information, Figure S3D). Similarly, downregulation of endogenous ERK1 by shRNA constructs significantly increased the abundance of FBW7 in SW1990 cells (Supplementary information, Figure S3E). Furthermore, we performed the cycloheximide (CHX) chase experiment to determine the FBW7 half-life with or without UO126 treatment in PANC-1 cell lines. As expected, inhibition of the MAPK/ERK pathway dramatically extended the FBW7 half-life (Figure 3E and 3F). These results suggest that activation of the Ras/Raf/MEK/ERK pathway negatively regulates FBW7 stability in pancreatic cancer cells.
Figure 3.
ERK negatively regulates FBW7 stability. (A, B) qPCR analysis of FBW7 expression in PANC-1 and SW1990 cells infected with empty vector or ERK1-expressing virus. GAPDH mRNA expression was used as an internal control (n = 3, independent t-test, P > 0.05). (C) Western blot analysis of FBW7 expression in PANC-1 and SW1990 cells infected with empty vector or ERK1-expressing virus. The band intensity was quantified with Image J software. (D) Western blot analysis of FBW7 expression in PANC-1 and SW1990 cells in the presence and absence of the MEK-1 inhibitor (UO126, 10 μM). DMSO was used as the control. (E) PANC-1 cells were treated with 20 μg/ml CHX in the presence or absence of 10 μM UO126, and whole-cell lysates (WCL) were collected at the indicated time points for immunoblot analysis. (F) Semi-quantification with α-tubulin as a loading control and relative FBW7 levels at time 0 were set as 100%.
ERK interacts with FBW7
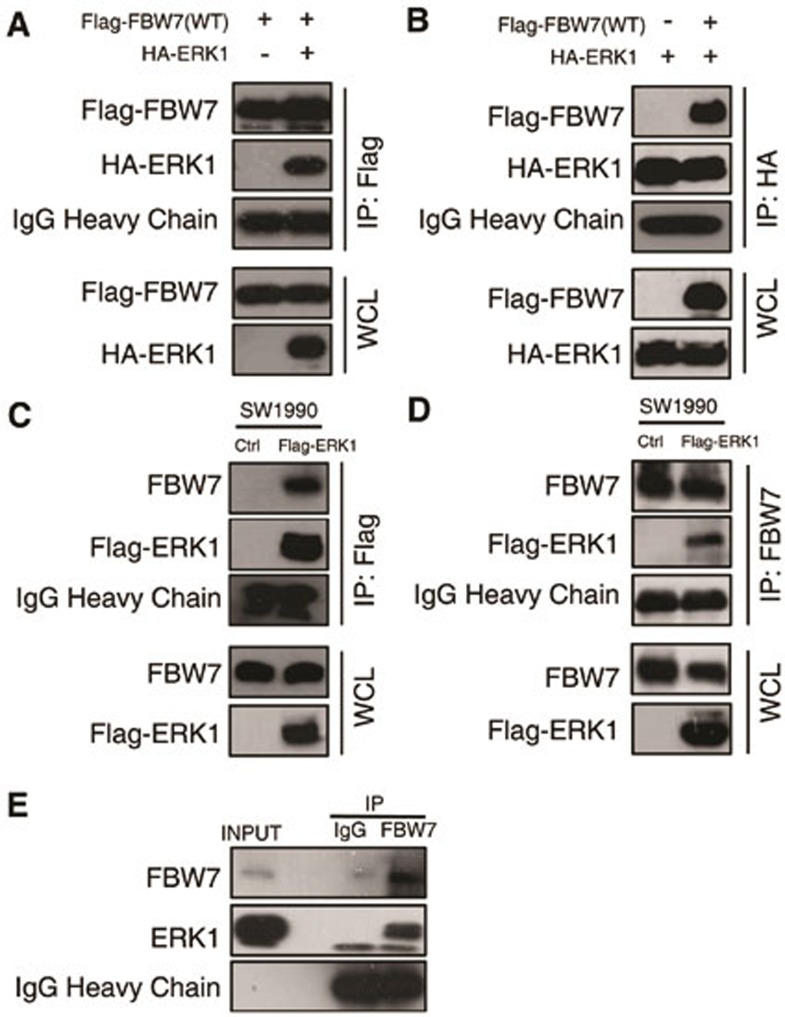
To explore the mechanism by which ERK kinase modulates FBW7 stability, we investigated whether there is a direct interaction between FBW7 and ERK1. In a co-immunoprecipitation assay with ectopically expressed ERK1 and FBW7, we found that Flag-tagged FBW7 could precipitate HA-ERK1 from transfected 293T cells (Figure 4A). Conversely, HA-ERK1 could also pull down Flag-FBW7 (Figure 4B). In addition, the physical interaction between FBW7 and ERK1 was supported by experiments conducted on pancreatic cancer cell line SW1990 stably expressing ERK1, which showed that ERK1 was able to co-immunoprecipitate with endogenous FBW7 and vice versa (Figure 4C and 4D). Furthermore, endogeneous interaction between FBW7 and ERK1 was validated by co-immunoprecipitation with FBW7 antibody (Figure 4E).
Figure 4.
ERK interacts with FBW7. (A, B) Immunoblot (IB) analysis of whole-cell lysates (WCL) and immunoprecipitates (IP) from 293T cells transfected with Flag-WT-FBW7, or HA-ERK1 or together. Thirty hours post-transfection, cells were pretreated with 15 μM MG132 for 10 h before harvesting. (C, D) Immunoblot analysis of WCL and immunoprecipitates from the indicated SW1990 cells stably expressing the Flag-ERK1 construct. Cells were pretreated with 15 μM MG132 for 10 h before harvesting. (E) SW1990 cell extracts were immunoprecipitated with an antibody against FBW7 or control IgG and analyzed by immunoblot analysis.
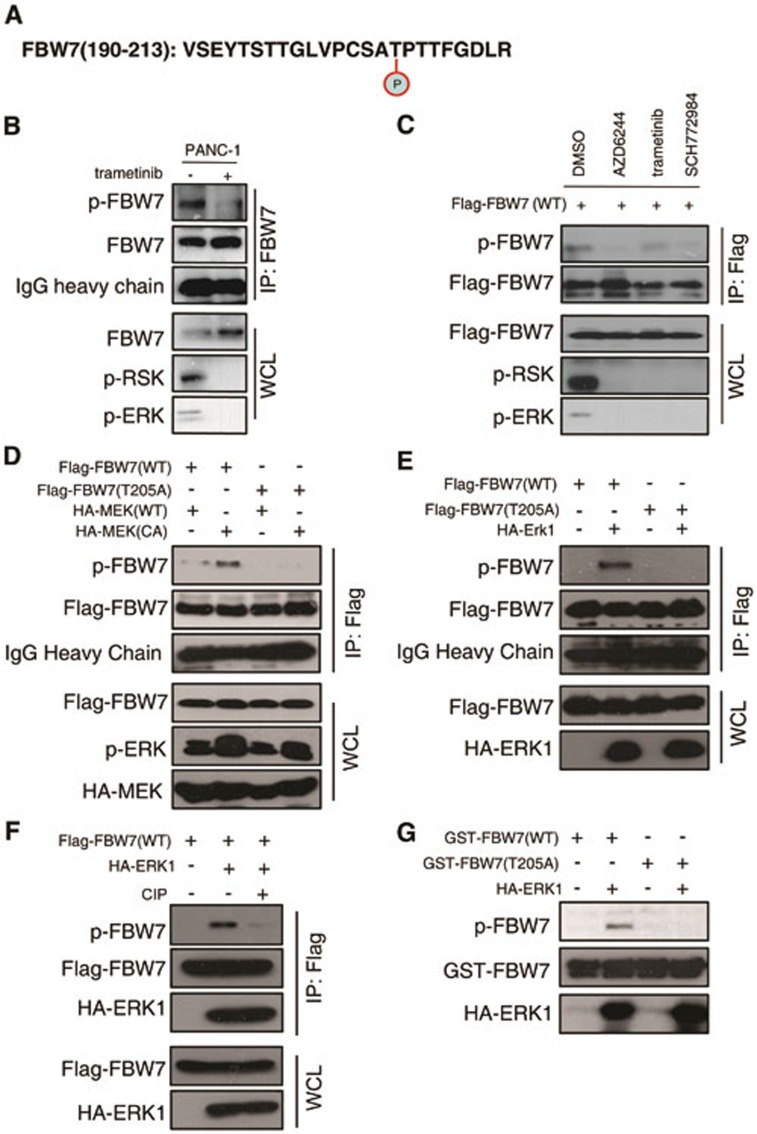
ERK phosphorylates FBW7 at T205
According to a recent study, Threonine205-Proline206 is the only TP motif of FBW7, and this TP motif has been shown to be involved in FBW7 stability control21. Thus, we first validated T205 phosphorylation of endogenous FBW7 expressed in SW1990 cell line after MG132 treatment by mass spectrometry analysis and peptides containing phosphorylated T205 were successfully recovered (Figure 5A and Supplementary information, Figure S4A). Next, we investigated whether ERK kinase directly phosphorylates FBW7, as they physically interact. We chose CFPAC-1 cells, which has a relatively higher FBW7 expression level. After UO126 treatment, the phosphorylation of endogenous FBW7 in the PDAC cell line CFPAC-1 was significantly reduced when detected with a phospho-Threonine-Proline (TP) monoclonal antibody (p-Thr-Pro-101) that specifically recognizes the phosphorylated TP motif (Supplementary information, Figure S4B). To further understand how ERK kinase phosphorylates FBW7 in vivo, we developed a specific antibody that only recognizes FBW7 phosphorylated at the T205 site. To validate the specificity of this antibody, we performed a dot blot assay and found that the phospho-T205-FBW7 (p-FBW7) antibody preferentially detected the phosphorylated peptide but not the unmodified peptide (Supplementary information, Figure S4C). We treated PANC-1 cells with trametinib, a MEK inhibitor, and the phosphorylation of endogenous FBW7 in PANC-1 cells was also significantly reduced when detected with the specific p-FBW7 antibody (Figure 5B). Similarly, after treatment with multiple MEK/ERK inhibitors, the phosphorylation of FBW7 in 293T cells all significantly decreased (Figure 5C). Next, we conducted a series of experiments to determine whether T205 is indeed an ERK phosphorylation site. Notably, co-transfection of a constitutively active form of MEK1 (MEK1-CA) or ERK1 kinase dramatically increased the phosphorylation of wild-type FBW7 but not the T205A mutant, suggesting that MAPK kinase signaling indeed controls FBW7 phosphorylation status (Figure 5D and 5E). Additionally, the phosphorylation of FBW7 by ERK was abolished by calf intestinal phosphatase (CIP) treatment (Figure 5F), indicating that ERK kinase modulates FBW7 in a phosphorylation-dependent manner. To prove that ERK kinase directly phosphorylates FBW7, glutathione S-transferase (GST)-fused recombinant FBW7 proteins, both wild type and T205A mutant, were incubated with purified HA-ERK1 kinase in the presence of ATP and detected with the p-FBW7 antibody. As shown in Figure 5G, ERK kinase could also specifically phosphorylate FBW7 at T205 in vitro. Furthermore, we performed hot kinase assay by incubating purified ERK kinase with recombinant FBW7 proteins in the presence of gamma-32P-ATP and indeed the incorporation of 32P was much reduced in T205A-FBW7 protein compared with WT-FBW7 (Supplementary information, Figure S4D). Thus, these data together suggest that ERK kinase directly phosphorylates FBW7 at the T205 site.
Figure 5.
ERK phosphorylates FBW7 at T205. (A) Detection of in vivo FBW7 T205 phosphorylation by mass spectrometry analysis. (B) PANC-1 cells were pretreated with the proteasome inhibitor MG132 and trametinib, as indicated, overnight before harvest. Endogenous FBW7 phosphorylation status was examined by immunoblot analysis after immunoprecipitates (IP). (C) 293T cells were transfected with Flag-WT-FBW7. Thirty hours posttransfection, cells were pretreated with MG132 and various MEK/ERK inhibitors overnight before harvesting. FBW7 phosphorylation status was examined by immunoblot analysis after immunoprecipitation. (D-F) 293T cells were transfected with Flag-WT-FBW7 and Flag-T205A-FBW7, together with HA-MEK1 and HA-ERK1 constructs, respectively. Wild-type (WT) and the constitutively active (CA) form of MEK1 were used as indicated. Thirty hours post-transfection, cells were pretreated with MG132 overnight before harvesting. FBW7 phosphorylation status was examined by immunoblot analysis after immunoprecipitation. Immunoprecipitates were treated with CIP for 20 min at room temperature, and the reaction was stopped by the addition of SDS loading buffer (F). (G) Recombinant GST-WT-FBW7 and GST-T205A-FBW7 proteins were incubated with HA-ERK1 kinase in the presence of ATP and the kinase reaction buffer. Thirty minutes later, the reaction was stopped by the addition of the loading buffer. The kinase reaction products were resolved by SDS-PAGE, and FBW7 phosphorylation was detected by the specific p-FBW7 antibody.
ERK destabilizes FBW7 by promoting its ubiquitination
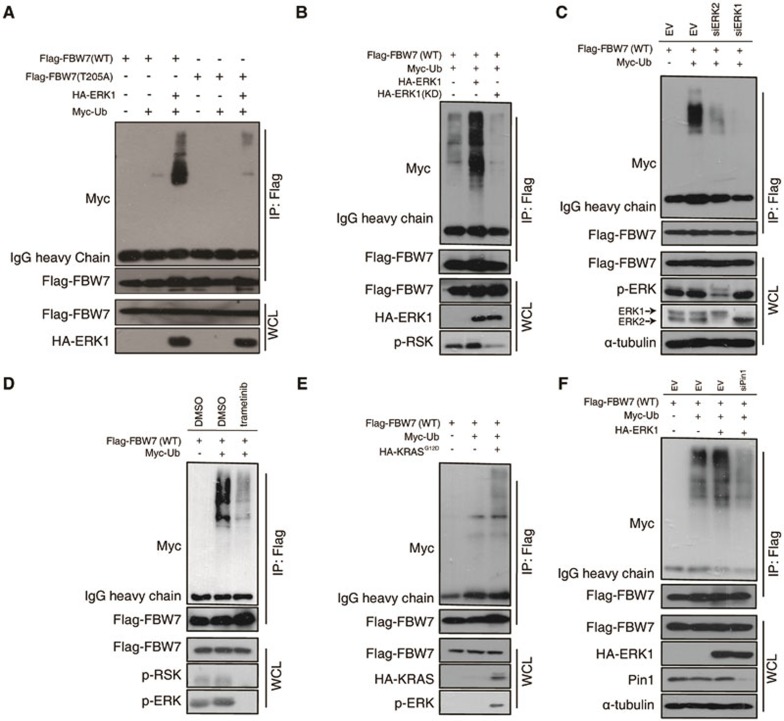
FBW7 is the substrate-binding unit of the SCF E3 ubiquitin ligase complex that targets a group of phosphorylated proteins for ubiquitination and degradation. Interestingly, it has been previously reported that FBW7 itself undergoes post-translational regulations through ubiquitination-dependent proteosomal degradation when phosphorylated at the T205 site21. To test whether ERK kinase promotes FBW7 degradation in a phosphorylation-dependent manner, we first treated cells with MG132 to block the proteasome pathway. Notably, although MG132 treatment led to an increase in total FBW7 protein, the upregulation of phosphorylated FBW7 was much more dramatic (Supplementary information, Figure S5), suggesting that phosphorylated FBW7 is indeed highly unstable. Next, we investigated whether ERK kinase regulates FBW7 ubiquitination. First, we observed that wild-type HA-ERK1, but not kinase-dead mutant, greatly increased the poly-ubiquitination of WT-FBW7 but not the T205A mutant (Figure 6A and 6B). Moreover, depletion of endogenous ERK1 and ERK2 by siRNA transfection or trametinib treatment significantly reduced the ubiquitination of FBW7 (Figure 6C and 6D), indicating an important role for ERK kinase in regulating FBW7 ubiquitination. Furthermore, co-transfection of KrasG12D also significantly promoted the ubiquitination of FBW7 (Figure 6E). Previously, it was reported that peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) regulates FBW7 ubiquitination when T205 is phosphorylated21. Thus, we hypothesized that ERK kinase promotes FBW7 ubiquitination in a Pin1-dependent manner. Indeed, depletion of endogenous Pin1 by siRNA drastically reversed the ERK-promoted poly-ubiquitination of FBW7 (Figure 6F). Together, these data strongly suggest that ERK kinase promotes FBW7 ubiquitination in a T205 phosphorylation- and Pin1-dependent manner.
Figure 6.
ERK destabilizes FBW7 by promoting its ubiquitination. (A) Immunoblot (IB) analysis of whole-cell lysates (WCL) and anti-Flag immunoprecipitates of 293T cells transfected with HA-ERK1 together with Myc-Ub and various Flag-FBW7 constructs. Thirty hours posttransfection, cells were treated with MG132 overnight before harvest. (B) Immunoblot (IB) analysis of WCL and anti-Flag immunoprecipitates of 293T cells transfected with wild-type HA-ERK1 or kinase-dead (KD) mutant together with Myc-Ub and Flag-WT-FBW7 constructs. Thirty hours posttransfection, cells were treated with MG132 overnight before harvest. (C) Immunoblot analysis of WCL and anti-Flag immunoprecipitates of Hela cells transfected with Flag-WT-FBW7 together with Myc-Ub and siRNA against ERK1/2. Thirty hours posttransfection, cells were treated with MG132 overnight before harvest. (D) Immunoblot analysis of WCL and anti-Flag immunoprecipitates of 293T cells transfected with Flag-WT-FBW7 and Myc-Ub constructs. Thirty hours posttransfection, cells were treated with MG132 and trametinib (where indicated) overnight before harvest. (E) Immunoblot analysis of WCL and anti-Flag immunoprecipitates of 293T cells transfected with HA-KRASG12D together with Myc-Ub and Flag-WT-FBW7 constructs. Thirty hours posttransfection, cells were treated with MG132 overnight before harvest. (F) Immunoblot analysis of WCL and anti-Flag immunoprecipitates of 293T cells transfected with Flag-WT-FBW7 and HA-ERK1 together with Myc-Ub and siRNA oligos against Pin1. Thirty hours posttransfection, cells were treated with MG132 overnight before harvest.
T205 phosphorylation inhibits FBW7 tumor suppressor function in pancreatic cancer
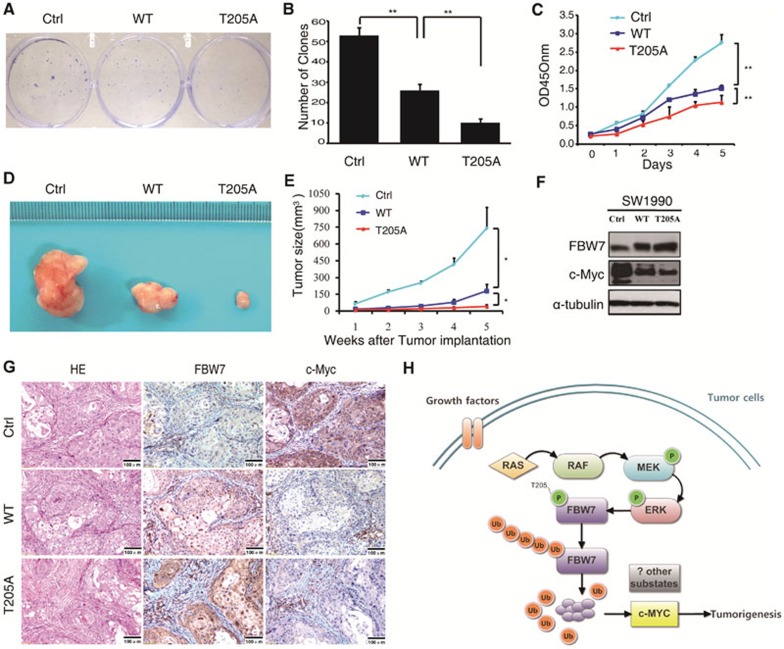
Given the critical role of Thr205 phosphorylation in controlling FBW7 stability, we continued to assess how this phosphorylation inhibits FBW7 tumor suppressor function in PDAC. First, we established SW1990 stable cell lines ectopically expressing wild-type FBW7 and a T205A phospho-deficient mutant, designated as SW1990/FBW7WT and SW1990/FBW7T205A respectively. In an anchorage-dependent growth assay, SW1990/FBW7T205A cells formed fewer and smaller colonies compared with its mock and SW1990/FBW7WT counterparts (Figure 7A and 7B). Moreover, SW1990/FBW7T205A also displayed much lower proliferation potential than SW1990 control and SW1990/FBW7WT cells (Figure 7C). Furthermore, we also found that SW1990/FBW7T205A cells displayed a significant decrease in cell migration ability (Supplementary information, Figure S6A and S6B). To further confirm the in vitro phenotype of FBW7T205A, we performed xenograft studies. In total, 4 × 106 cells of each cell line were subcutaneously inoculated into the backs of nude mice, and tumor growth was determined. As shown in Figure 7D and 7E, although FBW7WT displayed a dramatic effect in tumor growth inhibition, which clearly demonstrated that the expression level of FBW7 is critical for blocking tumor progression, FBW7T205A still displayed a significantly stronger effect in reducing tumor volume than wild-type FBW7. Taken together, these results suggested that the FBW7 T205A mutant conferred a growth disadvantage on pancreatic cancer cells both in vitro and in vivo. FBW7 has been reported to target various oncoproteins for ubiquitination and degradation, such as cyclin E, c-Myc, c-Jun, Notch-1, SREBP1 and Mcl-1. To determine the major downstream substrate that FBW7 might target in PDAC, we next examined changes in the known substrates responding to FBW7 overexpression by western blot. Notably, among the 8 known substrates examined, only c-Myc showed a significant reduction in response to overexpression of FBW7 (Figure 7F and Supplementary information, Figure S6C-S6E). By contrast, other known FBW7 substrates, including cyclin E, c-Jun, Notch-1, SREBP1, PGC-1α, KLF5 and Mcl-1, were barely affected. Consistently, western blotting results also showed a general inverse correlation between the levels of FBW7 and c-Myc in all six pancreatic cancer cell lines examined (Supplementary information, Figure S2). Furthermore, the pathological correlation between FBW7 and c-Myc was further validated in specimens from xenograft tumors (Figure 7G). Next, we examined whether the ERK/FBW7/c-Myc axis identified in PDAC cells is also valid in clinical samples. Correlation studies showed that a high p-ERK level correlated with low FBW7 expression and high c-Myc expression in PDAC specimens. Conversely, a low p-ERK level correlated with high FBW7 expression and low c-Myc expression (Supplementary information, Figure S6F). These results suggest that ERK kinase may exert its oncogenic function partially through elevating c-Myc expression by inhibiting FBW7 tumor suppressor function.
Figure 7.
Phosphorylation at the T205 site inhibits FBW7 tumor suppressor function in pancreatic cancer. (A, B) Representative images of clone formation on plastic plates with SW1990 cell lines expressing FBW7 (wild type and T205A mutant) or empty vector. Quantification of cell growth is shown in B (n = 3, independent t-test, **P < 0.01). (C) Cell growth curve assay with SW1990 cell lines expressing FBW7 (wild type and T205A mutant) or empty vector (n = 3, independent t-test, **P < 0.01). (D, E) SW1990 cells stably expressing WT-FBW7, T205A-FBW7, or empty vector were injected into nude mice. At the indicated times, tumors were measured with Vernier calipers (mean ± SEM; n = 5, independent t-test, *P < 0.05). (F) Western blot analysis for c-Myc in SW1990 cell lines stably expressing Flag-WT-FBW7 or Flag-T205A-FBW7 (with empty vector as a negative control). (G) Representative images of tumor tissue from staining with antibodies against FBW7 and c-Myc. Scale bar, 100 μm. (H) Proposed model of how the Ras/Raf/MEK/ERK pathway regulates FBW7 stability to influence its tumor suppressive functions in pancreatic cancer.
Discussion
Generally, loss of function of tumor suppressors in cancer is frequently caused by mutation or deletion22. FBW7 is ranked among the most important tumor suppressors, with a relatively high mutation rate in cancer exceeded only by p53 and PTEN. Mutation of FBW7 has previously been reported in several types of cancer11,12,13,14,15. Overall, ∼6% of tumors exhibited mutations in FBW7, with the highest mutation rates found in cholangiocarcinoma (35%) and T-cell acute lymphoblastic leukemia (31%)8. However, in PDAC, FBW7 mutation was only reported in a study with limited samples (1 out of 11 samples)23. In a recent whole-exome sequencing analysis of 99 pancreatic cancer specimens, the top 16 most frequently mutated genes were identified, including KRAS and TP5324, but the FBW7 gene mutation status was not included. Similarly, our own sequencing results from 60 individual patients showed that < 2% of specimens exhibited FBW7 mutation (1 out of 60 samples; Supplementary information, Table S2). These findings together suggest that FBW7 mutation is a rare event in PDAC. Interestingly, we found that FBW7 is expressed at a low level in PDAC and is tightly associated with cancer progression. FBW7 protein expression was significantly reduced in six pancreatic cancer cell lines compared with normal HPDE cells. The expression level of FBW7 in PDAC patients' TMAs suggested that the expression of FBW7 was downregulated in PDAC samples compared with paracancerous tissues. Importantly, low expression of FBW7 significantly correlates with poor OS in PDAC cases. Therefore, there must be a mechanism that inhibits FBW7 in PDAC without affecting FBW7 genetic status.
KRAS mutation has been regarded as the most frequent oncogenic event in PDAC. Overall, oncogenic KRAS signaling in PDAC is through three major pathways: Raf/MEK/ERK, PI3K/Pdk1/Akt and the Ral guanine nucleotide exchange factor pathway25. As the best characterized Ras downstream pathway, the MAPK signaling axis is of particular importance in PDAC development17. The pathway has been reported to play a central role in diverse cellular responses, such as proliferation, differentiation, and survival. Briefly, activated Ras binds to Raf, thereby directly activating MEK kinase via phosphorylation. MEK1/2 subsequently phosphorylates residues in the activation loops of ERK kinase, resulting in their activation. Upon activation, ERK kinase can phosphorylate and activate both cytosolic and nuclear targets18. Previously, Minella et al.20 indicated that Ras stabilized cyclin E via the activation of the MAPK pathway and inhibition of FBW7-dependent cyclin E ubiquitination in vivo. However, the underlying mechanism was not further clarified. Inspired by their findings, we speculated that cross-talk may occur between FBW7 and the MAPK pathway. As expected, in the current study, we found that FBW7 levels are inversely correlated with ERK activation in PDAC. On one hand, ectopic expression of ERK1 significantly reduced the endogenous FBW7 protein level. On the other hand, inhibition of the MAPK pathway dramatically increased endogenous FBW7 abundance. By contrast, inhibition of AKT kinase dramatically shortened Fbw7 half-life, indicating the PI3K/AKT pathway may play a different role in regulating Fbw7 stability (Supplementary information, Figure S3F and S3G). Furthermore, we found that ERK directly interacts with FBW7 and phosphorylates the T205 site to destabilize FBW7. Before our study, a report demonstrated that Pin1, the only peptidyl-prolyl cis/trans isomerase, interacts with FBW7 in a phospho-T205-dependent manner and promotes FBW7 self-ubiquitination and protein degradation21. However, the signaling to trigger T205 phosphorylation, which is important for understanding the physiological regulation, was largely left untouched in that study. Hence, our research not only filled the missing upstream signaling that controls FBW7 stability via the T205 site but also identified new functions of the KRAS-MAPK kinase pathways in PDAC. Interestingly, a similar regulation paradigm between other oncogenic kinase and tumor suppressing proteins has also been reported. For example, decreased p27 expression levels in tumor cells mostly occur due to enhanced proteolysis, given that mutation of the p27 gene is a rare event in cancer. This low expression level is well explained by the finding that cylin A/CDK2 phosphorylates the CDK inhibitor p27 and triggers its destruction by the oncoprotein SCF-SKP2 in many cancer types26, such as oral squamous cell carcinoma27. Another example is that the oncoprotein NEDD4-1 negatively regulates the stability of the tumor suppressor PTEN by catalyzing PTEN polyubiquitination and destruction in lung cancer28,29. Thus, in addition to genetic mutations, accelerated protein turnover upon certain post-translational modification is another important means by which oncoproteins inactivate tumor suppressors during tumorigenesis. To dissect a possible role of self-ubiquitination in ERK-mediated FBW7 degradation, we generated a FBW7 R465H mutant that could not ubiquitinate substrates based on a previous report21. As shown in Supplementary information, Figure S7A, R465H is more heavily ubiquitinated than WT FBW7, which is consistent with the previous report. Co-transfected ERK kinase could still promote poly-ubiquitination of the R465H mutant, which possibly indicates the involvement of other E3 ligases in FBW7 stability control, but the possibility of self-ubiquitination can not be formally excluded. Furthermore, we generated T205E FBW7 construct. As shown in Supplementary information, Figure S7B, T205E mutant failed to form homo-dimer as WT and T205A FBW7 did. We also tested the ubiquitination potential of T205E, and interestingly T205E are less ubiquitinated than WT (Supplementary information, Figure S7C). Although the T→E mutation generated a negative charge at aa205, which is similar to what a phosphor group does after phosphorylation, Pin1 isomerase does not interact with EP motif and could not perform the cis-trans conversions. Thus, the T205E mutant may not be appropriate to be used to evaluate the regulatory effect involving Pin1. Overall, our study established an important regulatory function of FBW7 by the activated RAS-MAPK-ERK signaling pathway, although more efforts are required to dissect how FBW7 is ubiquitinated after its phosphorylation.
Previous studies have shown that FBW7 functions as a tumor suppressor by targeting multiple well-known oncoproteins for degradation. However, the functional role of FBW7 in PDAC progression has not yet been well studied, although a recent stem cell study reported that FBW7 deletion in the adult pancreatic ducts induces the direct conversion of ductal cells into β cells30. In our study, by overexpressing the phospho-deficient mutant form of FBW7 (FBW7T205A mutant), we observed a remarkable growth disadvantage in pancreatic cancer cells both in vitro and in vivo. To uncover the underlying molecular mechanism, we examined the changes in many known FBW7 substrates, such as cyclin E31, c-Myc32, c-Jun33, Notch-134, SREBP135, PGC-1α36, KLF-537 and Mcl-138. Intriguingly, only the level of c-Myc decreased significantly, suggesting a central role for c-Myc in FBW7-mediated PDAC inhibition. c-Myc is one of the most frequently dysregulated oncogenes in human malignancies, and it regulates multiple processes including uncontrolled cell proliferation, cell growth, genomic instability and tumor cell metabolism39,40. Previous studies have also demonstrated that high levels of c-Myc are frequently detected in pancreatic cancer and promote the progression of pancreatic cancer41,42,43. Recent studies have shown that the expression levels of mutant KRAS and c-Myc share equal importance in the initiation and maintenance of pancreatic cancer44,45. Therefore, through the phosphorylation and inhibition of FBW7, the KRAS-MAPK axis may mainly rescue c-Myc expression to trigger a major alteration in the transcription profile towards malignancy and cell growth advantage. However, further studies are required to determine whether other substrates, in addition to c-Myc, contribute to this phenotype in PDAC.
In summary, by exploring signaling pathways unique to PDAC and absent from other types of cancer, we demonstrated for the first time that the oncogenic KRAS-MAPK signaling cascade, one of the major driving forces for PDAC progression, phosphorylates the tumor suppressor FBW7 at the T205 site and promotes FBW7 ubiquitination and destruction. Thus, the dysregulation of FBW7 substrates, such as c-Myc, promotes tumorigenesis and poor clinical prognosis (Figure 7H). These findings have revealed important aspects of the functional roles of oncogenic KRAS signaling in PDAC and may contribute to designing new therapeutic strategies to treat pancreatic cancer.
Materials and Methods
Cells and reagents
The human pancreatic cancer cell lines SW1990, PANC-1, CFPAC-1, Capan-1, MIA PaCa-2 and AsPC-1 with KRAS mutations were obtained from the American Type Culture Collection. The SW1990 cells were cultured in L-15 medium supplemented with 10% fetal bovine serum (FBS). PANC-1 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS. CFPAC-1 cells were cultured in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% FBS. Capan-1 cells were cultured in IMDM supplemented with 20% FBS. MIA PaCa-2 cells were cultured in DMEM supplemented with 10% FBS and 2.5% horse serum. AsPC-1 cells were cultured in Roswell Park Memorial Institute medium supplemented with 10% FBS. The HPDE cells were cultured in complete keratinocyte serum-free medium supplemented with 50 μg/mL bovine pituitary extract and 5 ng/mL epidermal growth factor (Gibco). All of the cell culture media contained 100 U/mL penicillin and 100 mg/mL streptomycin. UO126, AZD6244, trametinib, SCH772984 and MG132 were purchased from Selleck. CHX, CIP and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich.
Tissue specimens
The clinical tissue samples used in this study were histopathologically and clinically diagnosed at Fudan University Shanghai Cancer Center from 2010 to 2011. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained. The clinical information on the samples is presented in Supplementary information, Table S1. Tissue slides derived from P48-Cre;KrasLSL-G12D;Ink4a/ArfF/F mice were provided by Chiao PJ. The histological grading and pathological annotation were performed by two independent pathologists at our center. The correlation between p-ERK and FBW7 was analyzed using the χ2 test. The use of human PDAC tissue specimens was evaluated and approved by the Ethical Committee of Fudan University Shanghai Cancer Center, and written informed consent was obtained from all participants or their appropriate surrogates.
Plasmids
The Flag-tagged coding sequence of human FBW7 or the relevant T205A mutant was cloned into the lentiviral vector pCDH-CMV-MCS-EF1-puro (SBI, USA) to generate FBW7 expression plasmids. The first 300 amino acids of the human FBW7 protein and the T205A mutant were cloned into the pGEX-4T-1 vector to generate bacterial expression constructs. The expression vectors for HA-tagged MEK1 (WT-MEK) and constitutively active MEK1 (CA-MEK; in which the Raf1-dependent regulatory phosphorylation sites S218 and S222 were substituted by aspartic residues) were constructed as previously described46.
shRNA and siRNA treatments
pLKO.1-TRC cloning vector (Addgene:10878) was used to express shRNAs against ERK1. In brief, 21-bp oligos targeting ERK1 was ligated into the pLKO.1-TRC cloning vector to generate the pLKO.1-shERK1 construct. pLKO.1-shScr (Addgene:1864) containing scrambled non-target shRNA was used as control. siRNA duplexes against ERK1, ERK2 and Pin1 were transfected into 293T cells using Lipofectamine 2000 (Invitrogen). The shRNA oligo and siRNA duplex sense sequences were listed in Supplementary information, Table S3.
Cell proliferation assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8; Dojindo) as previously described47.
Colony formation assay
Five hundred cells were placed in each well of a 6-well plate and maintained in media containing 10% FBS for 10 days. Colonies were fixed with methanol and stained with 0.1% crystal violet in 20% methanol for 15 min. The number of colonies was counted using an inverted microscope.
RNA isolation and quantitative real-time PCR
Total RNA was prepared using TRIzol reagent (Invitrogen, USA), and cDNA was obtained by reverse transcription using a TaKaRa PrimeScript RT reagent Kit. The expression status of candidate genes and GAPDH were determined by quantitative real-time PCR using an ABI 7900HT Real-Time PCR system (Applied Biosystems, USA). All reactions were run in triplicate.
Primers used:
Human FBW7: 5′-CCACTGGGCTTGTACCATGTT-3′ (forward); 5′-CAGATGTAATTCGGCGTCGTT-3′ (reverse).
Human GAPDH: 5′-CGACCACTTTGTCAAGCTCA-3′ (forward); 5′-AGGGGAGATTCAGTGTGGTG-3′ (reverse).
Western blot
The proteins were extracted using EBC lysis buffer (50 mM, Tris pH 8.0, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Selleck). The protein concentrations were measured using a Bradford Protein Assay Kit (Bio-Rad). Total proteins (20 μg) were separated by SDS-PAGE and blotted onto polyvinylidene fluoride membranes (Bio-Rad). After blocking with 5% non-fat milk in tris-buffered saline with tween at room temperature for 30 min, the membranes were probed with antibodies. The antibody against FBW7 was purchased from Bethyl, and p-FBW7 was custom-ordered from Abclonal Technology. The PGC1α, ERK1, HA and tubulin antibodies were purchased from Santa Cruz, and p-ERK, Mcl-1 and the phospho-TP monoclonal antibodies were purchased from Cell Signaling Technology, while c-Myc, cyclin E, Notch1, c-Jun, p-AKT and p-RSK antibodies were all purchased from Abcam. The GST, Myc, SREBP1 and KLF-5 antibodies were purchased from Proteintech. The Flag antibody was purchased from Sigma-Aldrich. All secondary antibodies were purchased from Dako. After incubation with primary and secondary antibodies, immunoblots were incubated with an enhanced chemiluminescence detection kit (Millipore) and visualized in a dark room.
In vivo ubiquitination analysis
Cells were transfected with a plasmid encoding Flag-FBW7 along with Myc-tagged ubiquitin. The HA-ERK1 vector was co-transfected to assess its effect on FBW7 ubiquitination. Thirty-six hours after transfection, 15 μM MG132 was added to block proteasome degradation, and cells were harvested in EBC buffer containing protease inhibitors. Two milligrams of whole-cell lysates was incubated with Flag beads (Sigma-Aldrich) for 4 h, followed by washing 5 times with NETN buffer (0.5% NP-40, 1 mM EDTA, 20 mM Tris, pH 8.0 and 100 mM NaCl). The washed pellet was boiled in SDS-containing lysis buffer, resolved by SDS-PAGE, and detected by immunoblotting.
In vitro kinase assay
Cells from the 293T line were transfected with HA-ERK1. Forty-eight hours later, ERK1 was immunoprecipitated using an HA-matrix (Roche) and then incubated with 1 μg of GST-FBW7 protein (wild type and T205A mutant) in the presence of 200 μM cold ATP in kinase reaction buffer for 30 min. The reaction was stopped by the addition of SDS-containing lysis buffer, and the proteins were resolved by SDS-PAGE and detected by immunoblotting.
Tumorigenesis study
BALB/c-nu mice (5-6 weeks of age, 18-20 g, Shanghai SLAC Laboratory Animal Co., Ltd.) were housed in sterile filter-capped cages. A total of 4 × 106 cells in 100 μl phosphate-buffered saline were injected subcutaneously into the backs of the mice. Tumor size was measured every week with calipers from the time of the formation of palpable tumors. Tumor volume was calculated by the formula of length × width2 × 0.5248. Six weeks postimplantation, the mice were euthanized, and the tumors were surgically dissected. The tumor specimens were fixed in 4% paraformaldehyde. Samples were then processed for histopathological examination. All animal experiments were performed according to the guidelines for the care and use of laboratory animals and were approved by IACUC of Fudan University and the University of Texas, MD Anderson Cancer Center.
Mass Spectrometry analysis to detect FBW7 T205 phosphorylation in vivo
SW1990 cells were pretreated with MG132 overnight. Whole-cell lysates were collected to perform immunoprecipitation with FBW7 antibody. The phosphorylation status of FBW7 were analyzed by mass spectrometry according to the protocols reported previously49.
Statistics
Unless otherwise indicated, data are expressed as the mean ± SD, and the differences between any two groups were compared by t-tests. The χ2 test was used to analyze the relationship between FBW7 expression and clinicopathological characteristics. The survival curve was plotted using the Kaplan-Meier method and compared by the log-rank test. All statistical analyses were conducted using the SPSS 19.0 statistical software package. A P value of < 0.05 was considered significant.
Other methods are described in the Supplementary information, Data S1.
Acknowledgments
We thank HuanYu Xia for assistance in collecting the patient data. We thank Fengqun Zhang, Min Chen, Xiangling Chen, Liyan Gong, Binghai Cui, Huafang Ouyang and Xiaoduo Xie for technical support. This work was supported by the National Natural Science Foundation of China (81372651, 81201900, 81172276 and 81101565), the Sino-German Center (GZ857), and the PhD Programs Foundation of the Ministry of Education of China (20120071120104).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Representative micrographs showing negative, weak, moderate, or strong expression of FBW7 and p-ERK in pancreatic cancer tissues (scale bar: 100μm).
Immunoblot analysis of FBW7, p-ERK, c-Myc, CyclinE and Pin1 expression level in 6 pancreatic cancer cells.
ERK negatively regulates FBW7.
ERK phosphorylates FBW7 at T205.
293T cells were transfected with Flag-WT-FBW7 and Flag-T205A-FBW7 constructs.
Overexpression of FBW7 is significantly correlated with decreased expression levels of c-Myc in pancreatic cancer.
(A) Immunoblot (IB) analysis of whole cell lysates (WCL) and anti-Flag immunoprecipitates of 293T cells transfected with Flag-WT-FBW7 and Flag-R465H-FBW7 constructs together with HA-ERK1 and Myc-Ub constructs.
Clinicopathological features and correlation of Fbw7 expression in in PDAC.
Alterational Rates of KRAS and Fbw7 in 60 selected patients.
List of shRNA oligos and siRNA duplex sense sequence
Materials and Methods
References
- Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poruk KE, Firpo MA, Adler DG, Mulvihill SJ. Screening for pancreatic cancer: why, how, and who. Ann Surg. 2013;257:17–26. doi: 10.1097/SLA.0b013e31825ffbfb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–1210. doi: 10.1016/j.gassur.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, Hruban RH. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 2012;18:4257–4265. doi: 10.1158/1078-0432.CCR-12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–1094. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142:1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Knuutila S, Aalto Y, Autio K, et al. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, et al. SCF (FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori T, Mimori K, Iwatsuki M, et al. P53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788–3794. doi: 10.1158/0008-5472.CAN-08-2846. [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Mimori K, Ishii H, et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int J Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- Ibusuki M, Yamamoto Y, Shinriki S, Ando Y, Iwase H. Reduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patients. Cancer Sci. 2011;102:439–445. doi: 10.1111/j.1349-7006.2010.01801.x. [DOI] [PubMed] [Google Scholar]
- Kurashige J, Watanabe M, Iwatsuki M, et al. Overexpression of microRNA-223 regulates the the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhbold C, Utsunomiya T, Morine Y, et al. Loss of FBXW7 expression is associated with poor prognosis in intrahepatic cholangiocarcinoma. Hepatol Res. 2014;44:E346–E352. doi: 10.1111/hepr.12314. [DOI] [PubMed] [Google Scholar]
- Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:2000–2015. doi: 10.18632/oncotarget.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Neuzillet C, Hammel P, Tijeras-Raballand A, Couvelard A, Raymond E. Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev. 2013;32:147–162. doi: 10.1007/s10555-012-9396-2. [DOI] [PubMed] [Google Scholar]
- Collisson EA, Trejo CL, Silva JM, et al. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minella AC, Welcker M, Clurman BE. Ras activity regulates cyclin E degradation by the FBW7 pathway. Proc Natl Acad Sci USA. 2005;102:9649–9654. doi: 10.1073/pnas.0503677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SH, Lau AW, Lee TH, et al. Negative regulation of the stability and tumor suppressor function of FBW7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Okano K, Muraki S, et al. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann Surg. 2013;258:336–346. doi: 10.1097/SLA.0b013e3182827a65. [DOI] [PubMed] [Google Scholar]
- Calhoun ES, Jones JB, Ashfaq R, et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Kudo Y, Ogawa I, et al. Role of Cks1 overexpression in oral squamous cell carcinomas: cooperation with Skp2 in promoting p27 degradation. Am J Pathol. 2004;165:2147–2155. doi: 10.1016/S0002-9440(10)63264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, et al. NEDD4–1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio N, Scrima M, Palaia L, et al. Oncogenic role of the E3 ubiquitin ligase NEDD4–1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am J Pathol. 2010;177:2622–2634. doi: 10.2353/ajpath.2010.091075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Gruber R, Gu G, Behrens A. Loss of FBW7 reprograms adult pancreatic ductal cells into α, δ, and β cells. Cell Stem Cell. 2014;15:139–153. doi: 10.1016/j.stem.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFBW7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein FBW7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the FBW7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Punga T, Bengoechea-Alonso MT, Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J Biol Chem. 2006;281:25278–25286. doi: 10.1074/jbc.M604983200. [DOI] [PubMed] [Google Scholar]
- Trausch-Azar JS, Abed M, Orian A, Schwartz AL. Isoform-specific SCFFbw7 ubiquitination mediates differential regulation of PGC-1α. J Cell Physiol. 2015;230:842–852. doi: 10.1002/jcp.24812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Li H, Li S, et al. The FBW7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretones G, Delgado MD, León J.Myc and cell cycle control Biochim Biophys Acta 2014. Apr 1. pii: S1874–9399(14)00073-X. doi: 10.1016/j.bbagrm.2014.03.013 [DOI] [PubMed]
- Wahlström T, Arsenian Henriksson M.Impact of MYC in regulation of tumor cell metabolism Biochim Biophys Acta 2014. Jul 17. pii: S1874–9399(14)00192–8. doi: 10.1016/j.bbagrm.2014.07.004 [DOI] [PubMed]
- Skoudy A, Hernandez-Munoz I, Navarro P. Pancreatic ductal adenocarcinoma and transcription factors: role of c-Myc. J Gastrointest Cancer. 2011;42:76–84. doi: 10.1007/s12029-011-9258-0. [DOI] [PubMed] [Google Scholar]
- Grippo PJ, Sandgren EP. Acinar-to-ductal metaplasia accompanies c-myc-induced exocrine pancreatic cancer progression in transgenic rodents. Int J Cancer. 2012;131:1243–1248. doi: 10.1002/ijc.27322. [DOI] [PubMed] [Google Scholar]
- He C, Jiang H, Geng S, et al. Expression of c-Myc and Fas correlates with perineural invasion of pancreatic cancer. Int J Clin Exp Pathol. 2012;5:339–346. [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Rajbhandari N, Liu C, et al. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res. 2013;73:1821–1830. doi: 10.1158/0008-5472.CAN-12-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Rajbhandari N, Wagner KU. Cancer cell dormancy in novel mouse models for reversible pancreatic cancer: a lingering challenge in the development of targeted therapies. Cancer Res. 2014;74:2138–2143. doi: 10.1158/0008-5472.CAN-13-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, et al. Transformation of mammalian cells by constitutively active MAP kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Huang Z, Huang S, Wang Q, et al. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;71:2582–2589. doi: 10.1158/0008-5472.CAN-10-3032. [DOI] [PubMed] [Google Scholar]
- Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- Gao D, Wan L, Inuzuka, et al. Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction. Mol Cell. 2010;39:797–808. doi: 10.1016/j.molcel.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative micrographs showing negative, weak, moderate, or strong expression of FBW7 and p-ERK in pancreatic cancer tissues (scale bar: 100μm).
Immunoblot analysis of FBW7, p-ERK, c-Myc, CyclinE and Pin1 expression level in 6 pancreatic cancer cells.
ERK negatively regulates FBW7.
ERK phosphorylates FBW7 at T205.
293T cells were transfected with Flag-WT-FBW7 and Flag-T205A-FBW7 constructs.
Overexpression of FBW7 is significantly correlated with decreased expression levels of c-Myc in pancreatic cancer.
(A) Immunoblot (IB) analysis of whole cell lysates (WCL) and anti-Flag immunoprecipitates of 293T cells transfected with Flag-WT-FBW7 and Flag-R465H-FBW7 constructs together with HA-ERK1 and Myc-Ub constructs.
Clinicopathological features and correlation of Fbw7 expression in in PDAC.
Alterational Rates of KRAS and Fbw7 in 60 selected patients.
List of shRNA oligos and siRNA duplex sense sequence
Materials and Methods