Abstract
Hepatitis E virus (HEV), a non-enveloped, positive-sense, single-stranded RNA virus, is a major cause of enteric hepatitis. Classified into the family Hepeviridae, HEV comprises four genotypes (genotypes 1-4), which belong to a single serotype. We describe a monoclonal antibody (mAb), 8G12, which equally recognizes all four genotypes of HEV, with ∼2.53–3.45 nM binding affinity. The mAb 8G12 has a protective, neutralizing capacity, which can significantly block virus infection in host cells. Animal studies with genotypes 1, 3 and 4 confirmed the cross-genotype neutralizing capacity of 8G12 and its effective prevention of hepatitis E disease. The complex crystal structures of 8G12 with the HEV E2s domain (the most protruded region of the virus capsid) of the abundant genotypes 1 and 4 were determined at 4.0 and 2.3 Å resolution, respectively. These structures revealed that 8G12 recognizes both genotypes through the epitopes in the E2s dimerization region. Structure-based mutagenesis and cell-model assays with virus-like particles identified several conserved residues (Glu549, Lys554 and Gly591) that are essential for 8G12 neutralization. Moreover, the epitope of 8G12 is identified as a key epitope involved in virus-host interactions. These findings will help develop a common strategy for the prevention of the most abundant form of HEV infection.
Keywords: HEV, cross-genotype neutralizing antibody, immunodominant, epitope
Introduction
Hepatitis E is increasingly becoming an important public health concern in both developing and developed countries1. Hepatitis E is transmitted primarily via the fecal-oral route2 and is caused by HEV, a small non-enveloped, positive-sense, single-stranded RNA icosahedral virus, of the genus Hepevirus3. The infection-to-mortality ratio of HEV is small; however, this significantly increases in infants under the age of 2 years, in pregnant women, and in adults over the age of 56 years (5%-25%)4,5,6. Four major genotypes of HEV have been identified to date, with a single serotype: genotypes 1 and 2 are human viruses that circulate only in humans as an epidemic form, whereas genotypes 3 and 4 are zoonotic, swine viruses that infect a wide range of mammalian species as well as humans in sporadic cases1,7.
The 7.5-kb genome of HEV contains three open reading frames8, of which ORF2 encodes a single, viral capsid protein of 660 amino acids (aa). The capsid protein is responsible for virion assembly, host interaction and immunogenicity and is formed by homodimeric subunits, which comprise a protrusion domain9,10 (E2s: aa 455-60211 or its N-terminal extension version E2: aa 394-60612). These dimers project from the surface of the virus and initiate infection by interacting with host cells.
We have previously shown that a truncated form of this capsid protein, p239 (aa 368-606) can self-assemble into virus-like particles (VLPs) of ∼23 nm in diameter and subsequently penetrate susceptible cells, similar to a live virus12. We later produced the recombinant vaccine, HEV239, and employed this in a randomized, double-blind, placebo-controlled, phase 3 clinical trial involving more than 100 000 volunteers13. Moreover, we introduced a commercial p239/E2-based IgG and IgM kit for HEV surveillance and worldwide clinical identification14,15.
Immunologically, there is a major, predominant antigenic determinant among the different HEV genotypes. Structural comparisons of the E2s (aa 455-602) of genotype 111 (resolution of 2.0 Å) with that of VLPs (T=1; aa 112-608) for genotype 39 and genotype 410 (3.5 Å) have revealed that the HEV capsid is formed by capsomeres comprising a homodimeric ORF2, and that virus infection is initiated through the interaction of this protruding homodimer with host cells. Cryo-electron microscopy studies16 have additionally confirmed these structural findings.
To understand the neutralization mechanism and immunological nature of HEV, we previously determined the crystal structure of the antigen-binding fragment (Fab) of a genotype-specific neutralizing (herein referred to as genotype 1-preferred) monoclonal antibody (mAb), 8C11, in complex with HEV E2sHEV-1 domain (aa 455-602; superscript refers to genotype 1) at 1.9 Å resolution. We identified the epitope of 8C11 on E2sHEV-1 and, using mutational and cell-model assays, showed that Arg512 was the most crucial residue for 8C11-E2sHEV-1 interaction17. In addition, our structural and functional studies on E2s genotype 4 (E2sHEV-4) identified the essential role of residue 497 in the epitope of 8C11 in its recognition of different genotypes17.
As a continuation of our efforts to understand the neutralization mechanism of HEV, here we report the characterization of a novel mAb with high immunodominance, 8G12, as a cross-genotype, neutralizing antibody of HEV. Through structural studies, we investigated the blocking profile and epitope function of 8G12 in serum samples taken from experimental Macaques rhesus monkeys challenged with HEVs of various genotypes. Overall, we demonstrate that 8G12 effectively inhibits all genotype HEV infections in vitro and in vivo. These findings are important for the improvement of the current vaccine and in the future development of any antibody-based drugs that protect against infection from all genotype HEVs as well as the onset and progression of subsequent hepatitis.
Results
Identification of mAb 8G12 and its cross-genotype neutralization of HEV
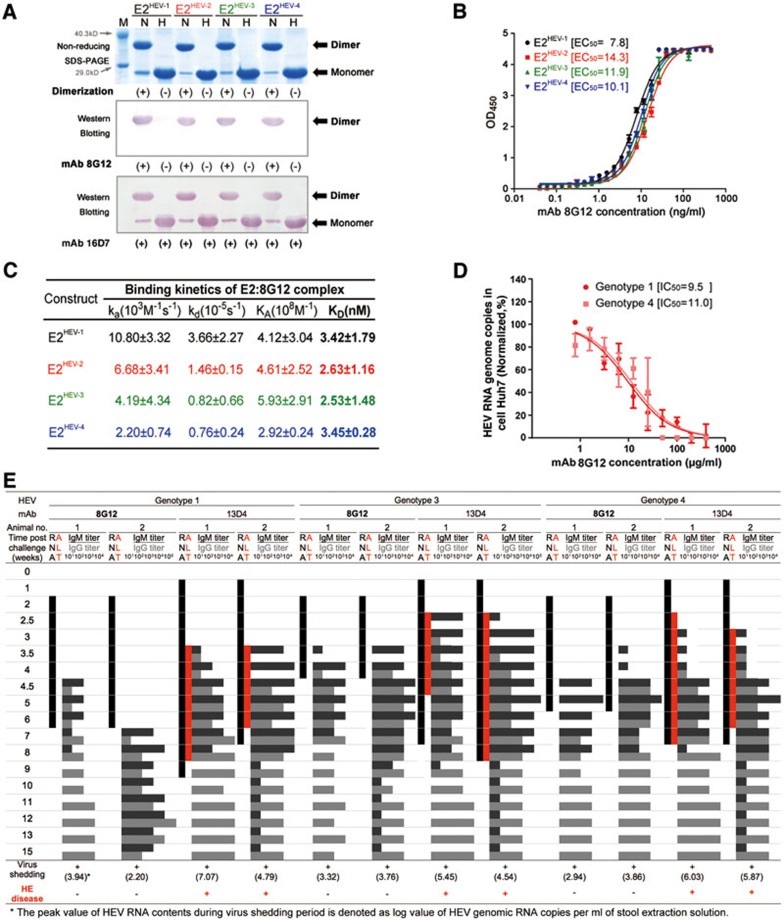
The 8G12 antibody was isolated from several murine mAbs raised against the E2 domain, as described by us previously18. We found that 8G12 reacted only with dimeric E2, but not with monomeric E2, while the previously known mAb 16D7 reacted with both monomeric and dimeric E219 (Figure 1A). The western blotting confirmed that 8G12 recognized the dimeric E2 of all four HEV genotypes, without any discrimination. This was further verified by enzyme-linked immunosorbent assay (ELISA), where serial dilution of the 8G12 antibody showed a similar E2 detection profile for all four genotypes (Figure 1B). These findings were again confirmed using a Biacore 3000 SPR, where the 8G12 antibody demonstrated a high binding affinity (∼2.53 to 3.45 nM) for the E2 domain of all four HEV genotypes (Figure 1C and Supplementary information, Figure S1). For comparison purposes, mAb 8C11 (which preferentially binds HEV genotype 1) had an affinity of 5.89 nM for the E2 of genotype 1 and a 100× lower affinity (0.646 μM) for the E2 of genotype 417.
Figure 1.
Comparable reactivity and neutralization nature of mAb 8G12 is observed among various HEV genotypes. (A) E2 proteins with four genotypes were subjected to non-reducing SDS-PAGE and western blot analysis with the neutralizing mAb 8G12 to investigate their reactivity in association with each genotype. The lanes marked with H indicate heated samples under reducing conditions (samples were heated up to 100 °C for 3 min) whereas the lanes marked with N indicate samples under non-reducing conditions (0.1% SDS, in the absence of β-mercaptoethanol (BME) and not heated). (+) denotes dimerization or reactivity with 8G12, (−) denotes monomer or loss of the respective property. (B) The interaction of E2 proteins with four genotypes against mAb 8G12 was tested by ELISA and EC50 was calculated by sigmoid trend fitting. (C) The affinity constants of the interaction of E2 proteins with four genotypes against mAb 8G12 were measured by SPR in Biacore 3000. (D) Inhibition profiles of mAb 8G12 blocking authentic HEV from infecting the host cell. HEV genotypes 1 or 4 show 1 × 105 genomic RNA copies in infected vulnerable Huh7 cells in the presence of serial dilutions of mAb 8G12. The inhibition profiles of HEV RNA genome copies vs. concentration of mAb 8G12 were fitted to a sigmoid trend to generate the IC50 value. (E) Neutralization of HEV infection rate by mAb 8G12 in a Macaques rhesus monkey model. HEV of genotypes 1, 3 and 4 was mixed with mAb 8G12 or mAb 13D4 (negative control) and used to inoculate monkeys. Serum and stool samples were taken before and weekly after infection to monitor serum levels of alanine aminotransferase (ALT; red bar), shedding of the HEV RNA in stool samples (black bar) and HEV IgG/IgM antibody titer (gray histogram). (+) denotes occurrence of virus shedding or Hepatitis E disease, (−) denotes a negative event. Also refer to Supplementary information, Figure S5.
We next sought to characterize the cross-genotype neutralizing capability of mAb 8G12 against HEV infection in human hepatoma Huh7 cells in vitro. The blocking efficiency of the overloaded mAb 8G12 was 94.56% for HEV genotype 1 and 91.62% for genotype 4, with comparable IC50 concentrations of 9.5 μg/ml (95% confidence interval: 6.7–13.4 μg/ml) and 11.0 μg/ml (95% confidence interval: 6.8–17.9 μg/ml) for genotypes 1 and 4, respectively (Figure 1D and Supplementary information, Figure S2A). In addition to 8C11 and 8G12, we also tested other mAbs (12E11 and 12E9) for their neutralization efficacy. We found that 8C11 and 12E11 showed distinct neutralization specificities: 8C11 preferred genotype 1 over genotype 4 (IC50 of 9.3 μg/ml for genotype 1; 30.9 μg/ml for genotype 4; Supplementary information, Figure S2B), as expected, whereas 12E11 preferred genotype 4 over genotype 1 (Supplementary information, Figure S2C). Although mAb 12E9 binds with HEV, it has no neutralization capacity (Supplementary information, Figure S2D). Moreover, the cross-blocking of 8G12 and 8C11 against genotype 1 (p239 VLP) shows the specificity of 8G12 over 8C11 (Supplementary information, Figures S3 and S4). These results demonstrate the potent cross-genotype inhibitory potential of 8G12 against HEV infection.
Protective role of 8G12 against HEV in a rhesus monkey animal model
We next evaluated the cross-genotype neutralizing capability of mAb 8G12 against HEV in a disease animal model, as previously described14,18,19. Briefly, two rhesus monkeys were used for each of the HEV genotypes 1, 3 and 4 and their respective control groups (12 monkeys in total). In the experimental groups, rhesus monkeys were challenged with a dose of 2-5 × 107 genomic equivalent of HEV genotypes 1, 3 or 4, which were pre-incubated with 5 mg of mAb 8G12. In the control group, monkeys were also injected with HEV genotypes 1, 3 or 4 that were instead pretreated with a broad neutralizing antibody for bird flu virus, mAb 13D420, which should have no neutralization capacity against HEV. For all monkeys, blood samples were collected before and at regular intervals after infection to measure alanine aminotransferase (ALT), anti-HEV IgM and IgG serum levels.
In the control group, hepatitis developed toward the end of week 3/early week 4, as determined by the elevation in ALT levels (values over 40IU/l21). All six pathogenic monkeys in the control group acquired Hepatitis E within 26.5 ± 8.5 days, with a peak ALT of 120.5 ± 1.4 IU/l as compared with that before virus challenge (32.7 ± 6.0 IU/l). In animals challenged with 8G12-pretreated HEV, ALT levels remained at normal levels throughout the assay period. In the control monkeys, the development of hepatitis was concomitant with an increase in anti-HEV antibody levels, indicating that mAb 13D4 had no neutralization capacity. Moreover, we monitored the serum IgM/IgG seroconversion rate and found that all 8G12-pretreated monkeys showed an ∼1-week delay in E2 IgG seroconversion as compared with 13D4-pretreated samples: 8G12-pretreated monkeys showed seroconversion at 3½ weeks after infection with genotypes 3 and 4, and at 4½ weeks with genotype 1.
In parallel, stool samples were harvested, and RT-PCR was used to determine the duration of virus excretion as a measure of the viral replication speed. For most 8G12-pretreated monkeys, stool virus shedding was first detected at 11 days after infection and persisted for 15-31 days. Comparing 8G12-pretreated and 13D4-pretreated cases, the onset time point of viral detection was delayed by 1 week with 8G12 pretreatment, the duration time shortened by ∼2-6 weeks and the average peak value of shed HEV RNA content lowered to 103.34 ± 0.67 copies versus 105.63 ± 0.92 copies (Figure 1E and Supplementary information, Figure S5A-S5F). These observations further confirmed the efficacy of 8G12 in inhibiting viral replication in these animals. Taken together, 8G12 treatment delays the onset of serological features and virus excretion, and thus offers a protective effect against HEV in Macaques rhesus monkeys.
mAb 8G12 blocks the binding of naturally acquired antibodies
Viruses undergoing high gene variation will typically silence their broad neutralizing sites when infiltrating the immune system of the host, as exemplified by HIV22 and Influenza virus23,24,25. We next sought to determine the predominance of 8G12-like antibodies in the anti-HEV pools during the immune responses elicited by HEV infection and vaccine inoculation. Previously, we obtained sera from volunteers participating in our phase-3 clinical trial (NCT01014845)13. These subjects were randomly assigned to vaccine or placebo groups, and sera were obtained at various intervals after inoculation. We also obtained sera from patients with native HEV acute infections. These hepatitis patients were identified through Hepatitis E surveillance tests that we implemented prior to our clinical trial13 in order to ascertain the prevalence of Hepatitis E and its onset rate in the general population. Sera from both the pre-screening and clinical trial were subjected to well-defined anti-HEV IgG titration according to the HEV WHO antibody standards13. In addition to these samples, we used antibodies raised in rhesus monkeys experimentally challenged with a dose of 2-5 × 105 genomic equivalent of HEV genotype 1, 3 or 4 (Supplementary information, Figure S5G-S5I).
To first calculate the blocking ratio, we examined whether binding of the HEV-reactive sera to p239 could be inhibited by mAb 8G12 in a linear, reactive range. In blocking assays, we found that mAb 8G12 showed immunodominance over human antibodies acquired from previous infections or through vaccination. In addition, mAb 8G12 dominated over the antibodies generated in the sera obtained from challenged rhesus monkeys. We found that blocking was more effective in acute sera from both human and monkey samples (mean interquartile blocking of 81% for human samples; 85%-90% for monkey samples) as compared with that obtained from vaccinated and placebo-treated subjects obtained at 7 and 31 months after the first inoculation (mean interquartile blocking of 68%-70%; Figure 2).
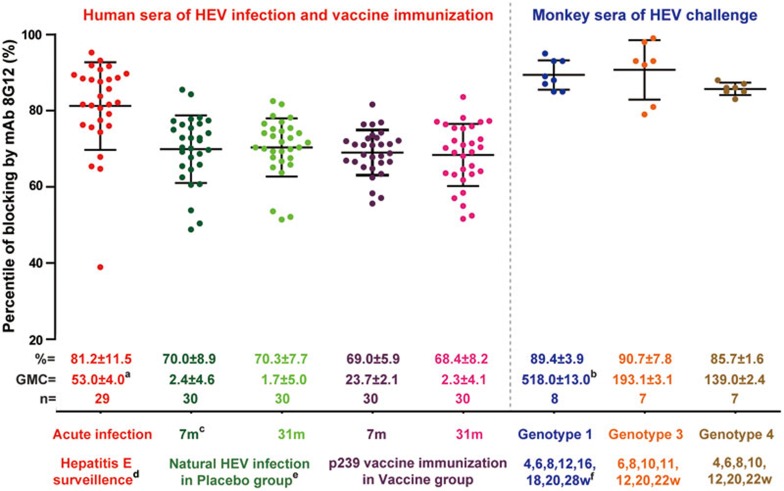
Figure 2.
Blocking of naturally acquired anti-HEV by 8G12. The blocking activity of 8G12 against anti-HEV was determined as described in Supplementary information, Figure S6. Samples included acute serum samples from 29 patients with confirmed diagnosis of hepatitis E; 30 samples each from healthy subjects participating in the control and vaccine groups of a phase 3 clinical trial (7 and 31 months enrolment in the trial); and acute serum samples from rhesus monkeys challenged with 5.55 × 105, 2.69 × 105, 4.73 × 105 genomic doses of HEV genotype 1 (n = 8), genotype 3 (n = 7) or genotype 4 (n = 7), respectively. Each sample was diluted to yield an OD value of 1.0 before use. Blocking activity of the mAb against each serum sample (dots) was calculated as 1 − (OD value of mAb treated with sera/OD value of the concurrent untreated control) × 100%. The horizontal and vertical lines indicate the median and interquartile range of blocking activity against the different groups of samples. Note: (a) anti-HEV IgG levels of the human sera were quantified and expressed in WHO units per mL (WU/ml), as previously described13, and calculated to express the GMC (Geometric Mean Concentration) with standard deviations; (b) anti-HEV IgG titer of the monkey sera was defined by maximum dilution titer to make detectability in IgG kit; (c) number of months after first p239 vaccine inoculation with a 0, 1 and 6 month immunization program; (d) hepatitis E surveillance was implemented before the p239 vaccine phase-3 clinical trial; the subjects acquiring hepatitis E were verified by abnormal ALT and simultaneous IgM/IgG seroconversion; (e) subjects in the placebo group who received hepatitis B vaccine and presented significantly positive IgG; (f) number of weeks after the challenge with HEV in monkeys. mAb 8G12 blocks HEV reactive primate sera to p239 by more than 70%, demonstrating the formation of a dominant immune complex in the immune response. Refer to Supplementary information, Figure S6 for details of the 8G12 Fab and 8G12-escape p239 mutated proteins.
A comparison of the blocking activity among 8G12, 8C11 and 12A10 using the sera from experimental HEV-challenged monkeys shows that mAb 8G12 efficiently blocks the binding of anti-HEV antibodies that are produced in response to infection with HEV genotypes 1, 3 or 4, with comparable blocking values of 89.4%, 90.7% and 85.7%, respectively. Although the epitope of 8C11 is immunodominant in naturally acquired HEV antibodies (mean interquartile blocking of 77.8%, 73.8% and 67.5% for genotype 1, 3 and 4, respectively), it shows a significantly lower blocking rate for genotypes 3 and 4 as compared with 8G12 (Supplementary information, Figure S6). The substantially lower blocking activity for 12A10, however, indicates that the epitope of 12A10 is not immunogenic in natural HEV infections (mean interquartile blocking of 35%-38%), which is consistent with the findings of others26. Blocking by the 8G12 Fab fragment (mean interquartile blocking of 29%-37%) was also markedly reduced as compared with 8G12; this could be due to its reduced spatial hindrance of blocking as compared with the whole mAb. We also used the reduced activity of the 8G12-espcape p239 mutant with respect to the wild-type p239, to assess the exact epitope of binding. The mean epitope reactivity of 17%-27% by this mutant (E549A and G591A), indicates the dominance of 8G12's epitope over other HEV antibody binding sites. From these results, we can infer that mAb 8G12 most likely binds to a dominant, immunogenic site of HEV, which may constitute a major target for antibody responses to infection and vaccination in humans and primates.
Crystal structure of immune complexes of genotypes 1 and 4
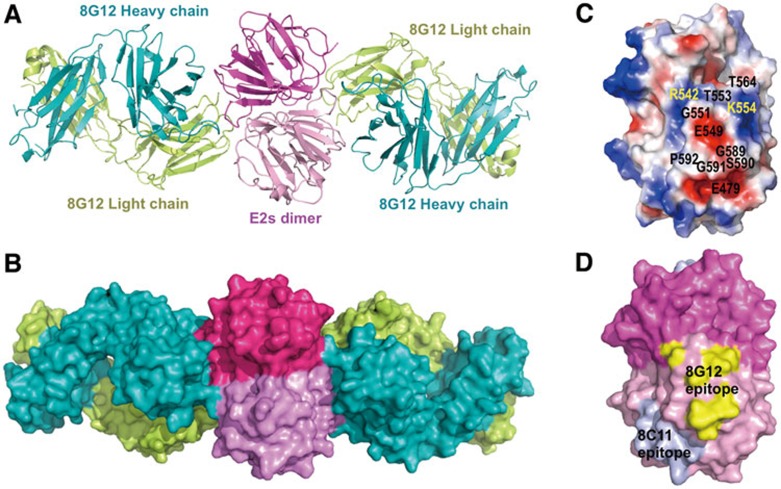
Next, we sought to study the details of the 8G12's epitope on E2s at the atomic level to decipher its mechanism of cross-genotype neutralization. The crystal structure of the immune complex of 8G12 Fab with the E2s of genotype 4 (E2sHEV-4) was determined at 2.3 Å resolution and refined to an R-factor of 18.7 % (Rfree = 24.2 %; Table 1). In the structure, one dimeric E2sHEV-4 bound with two 8G12 Fab molecules (Figure 3), which is consistent with the analytical ultracentrifugation results (Supplementary information, Figure S7). The E2sHEV-4 dimer had a well-defined electron density map that was similar to the E2sHEV-4 structure in the absence of the antibody. The superposition of E2sHEV-4 in the complex with free E2sHEV-4 yielded an rmsd of 0.4 Å for all Cα atoms.
Table 1. Diffraction data collection and refinement statistics for 8G12 Fab in complex with E2s.
| E2sHEV-1: 8G12 Fab | E2sHEV-4: 8G12 Fab | |
|---|---|---|
| Data collection | ||
| Cell parameters (Å, °) | a=54.2, b=192.0, c=192.6 | a=56.8, b=89.5, c=137.7 |
| β=90.0 | β=95.9 | |
| Space group | P21 | P21 |
| Resolution rangea (Å) | 50.0-4.00 (4.07-4.00) | 30.0-2.30 (2.38-2.30) |
| Total reflections | 101,538 | 349,821 |
| Unique reflections | 31,821 | 61,168 |
| Redundancy | 3.2 (2.6) | 5.7 (4.7) |
| Completeness (%) | 90.5 (81.9) | 99.8 (99.6) |
| Overall (I/sI) | 5.8 (1.2) | 14.3 (2.4) |
| Rsymb (%) | 13.1 (64.1) | 8.6 (65.2) |
| Refinement | ||
| Resolution range (Å) | 34.0-4.0 | 30.0-2.3 |
| Reflections | 30,302 | 60,870 |
| Rfactorc | 26.4 | 18.7 |
| Rfreed | 29.6 | 24.2 |
| RMSD bond lengths (Å) | 0.006 | 0.008 |
| RMSD bond angles (°) | 1.6 | 1.2 |
| Wilson B-factor (Å2) | 92.3 | 34.7 |
| Average B-factors (Å2) | 137.4 | 53.5 |
| E2s domain | 105.9 | 41.1 |
| Variable domain of heavy chain | 118.8 | 48.6 |
| Constant domain of heavy chain | 159.9 | 59.0 |
| Variable domain of light chain | 131.5 | 56.4 |
| Constant domain of light chain | 192.5 | 71.9 |
| Ramachandran Plot | ||
| Favored and allowed region (%) | 99.0 | 99.8 |
| Generously allowed regions (%) | 0.5 | 0.2 |
| Disallowed regions (%) | 0.5 | 0 |
aNumbers in parentheses refer to the highest resolution shell.
bRsym = Σh Σi|I1(h) − <I(h)| / Σh Σi I1(h)
cRfactor = Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|.
dRfree is calculated using the same equation as that for R factor but 6.2% for E2sHEV-1:8G12 Fab and 3.27% for E2sHEV-4:8G12 Fab of reflections were chosen randomly and omitted from the refinement.
Figure 3.
Crystal structure of the Fab of cross-genotype neutralizing mAb 8G12 in complex with HEV capsid protein E2s domain. (A) Cartoon representation shows the dimeric E2sHEV-4:8G12 Fab complex. 8G12 heavy chain (H) and light chain (L) are depicted in cyan and yellow, respectively. In the dimeric E2s, one monomer is shown in dark pink and the other in light pink. (B) Surface representation for the E2sHEV-4:8G12 Fab complex. (C) Image depicts the electrostatic surface potential of the dimeric E2sHEV-4 (red, negative; blue, positive; gray, neutral) with the key residues of the 8G12's epitope are marked. The dimeric E2sHEV-4 is shown in an orientation in which most of the epitopes are visible. More epitopes are from one E2s monomer while only two residues (R542 and T564) from the other monomer (D) Surface representation shows the mapping of the epitopes of 8G12 and 8C11 on E2sHEV-1/4. Structure-related figures were prepared using the program PyMol37.
The HEV genotypes 1 and 4 are the most abundant genotypes but with different host ranges27. Hence, in addition to genotype 4, we also sought to determine the complex structure of E2sHEV-1:8G12 to verify the conservation of the epitope of 8G12 in these genotypes. However, the complex crystals of E2sHEV-1:8G12 diffracted only up to 4.0 Å. The structure was determined and refined to a final R-value of 26.4 % (Rfree = 29.6%). Nonetheless, the complex was reasonably well defined in the electron density map, with good stereochemical parameters (Table 1). In both complex structures, the loop region 138-141H of the 8G12 Fab (superscripted H and L represent heavy and light chain, respectively) at the opposite side of the antibody-antigen interface was not well defined in the electron density map, a region that is often found disordered in the Fab structures28. The overall structure of the immune complexes for genotypes 1 and 4 were very similar, and the superposition of the genotype 1 complex onto the genotype-4 complex yielded an rmsd of ∼0.7 Å for 1 146 Cα atoms (one E2s dimer and two Fab molecules). The interaction details discussed in the following sections are based on the high-resolution genotype-4 complex structure, unless otherwise mentioned.
Interactions between the HEV E2s and 8G12 antibody
In the E2sHEV-4:8G12 structure, ∼844 Å2 of the E2s dimer surface was buried by the binding of one 8G12 Fab molecule (calculated by PISA5). The 8G12's binding region consisted of the complementarity determining region (CDR) loops L3, H2 and H3, whereas the other CDR loops (H1, L1 and L2) were not involved in the interaction. Key interactions between the antibody (8G12) and antigen (E2s) were mediated by 17 hydrogen-bonding contacts (≤ 3.2 Å) involving the main-chain and side-chain atoms (Supplementary information, Table S1). In addition, several van der Waals interactions were observed between E2s and 8G12 Fab. Notably, the heavy chain of the antibody was mainly involved in these interactions, which accounted for ∼58% of the total buried area of E2s at the interface.
The immune complex structures of genotypes 1 and 4 revealed identical 8G12-interacting residues. The 8G12's epitope is located at the E2s dimerization region (Figure 3), which is consistent with the cross-neutralization property of 8G12 for all genotypes. 8G12 Fab interacts with both monomers of the E2s dimer. It shows more interactions with one monomer, and interacts with the second monomer through two hydrogen-bonding contacts and hydrophobic/van der Waals interactions (Supplementary information, Table S1 and Figure S4C). Notably, we have shown that E2s dimerization is essential for HEV infection11, and it is also worth mentioning that the epitope of the genotype 1-preferred neutralizing mAb 8C11 on E2sHEV-1 is located in the groove region, an entirely different site17 (Figure 3D).
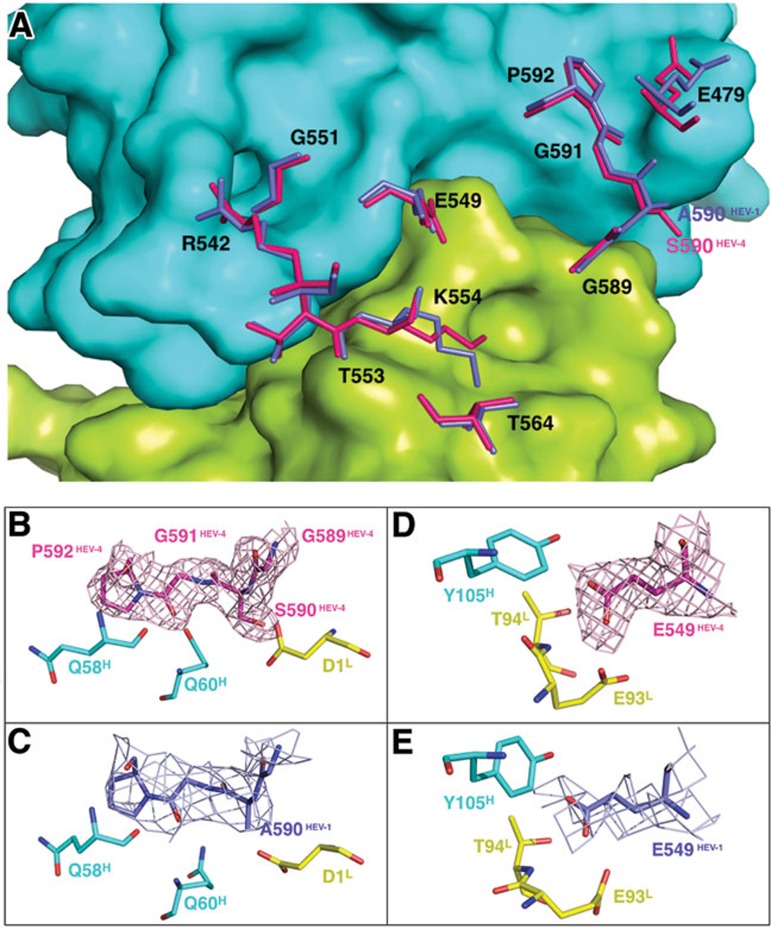
Following sequence analysis, we found that the 8G12's epitope was highly conserved among all four genotypes, with the exception of residue 590, which is a serine residue in genotype 4 but an alanine residue in the other genotypes (Figure 4A and Supplementary information, Figure S8). It is worth noting that the residue at this position makes hydrogen-bonding contacts with the light chain of mAb 8G12 through the main-chain amide group. Thus, the serine/alanine substitution in this position is unlikely to alter the interaction. The overall high structural and sequence conservation at the E2S dimerization region, and the observed interactions with the mAb 8G12 provide the structural basis for cross-genotype neutralization by 8G12.
Figure 4.
Comparison of the 8G12's epitope for genotypes 1 and 4. (A) 8G12's epitope for genotype 1 (blue) and 4 (fluorescent pink) are superimposed with the background of the 8G12 molecular surface. (B) and (C) show enlargements of the E2s loop region (aa 589-592) for genotypes 4 and 1, respectively, with interacting residues from 8G12. (D) and (E) show an enlargement of the region surrounding residue E549 for genotype 4 and 1 respectively. Sample electron density (2Fo-Fc) maps contoured at 1σ above the mean are shown for E2sHEV-1/4 residues.
Validation of the key interactions
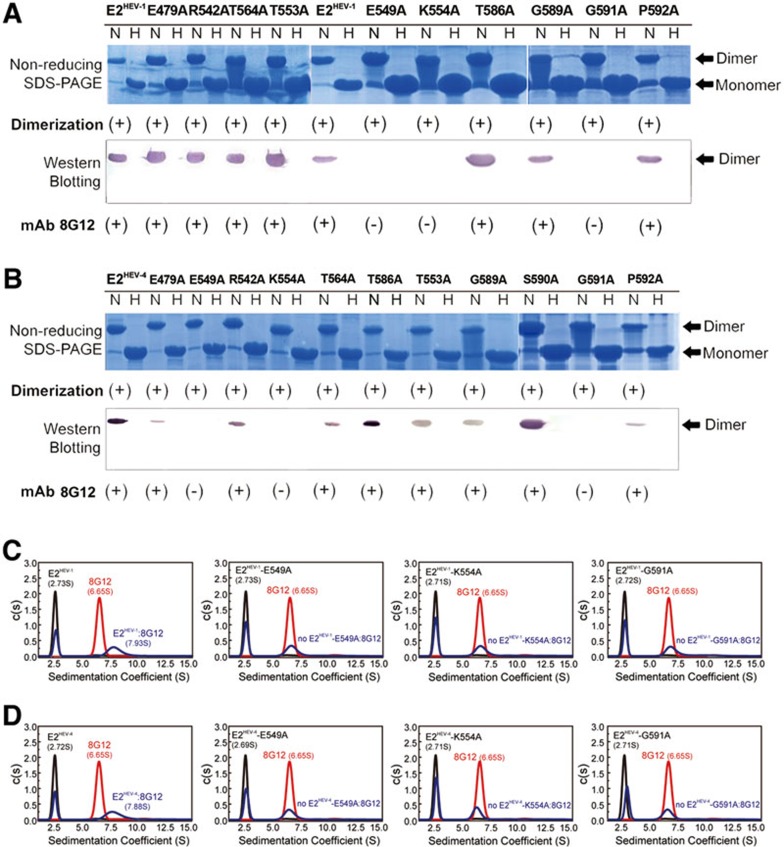
We next sought to verify the importance of key interacting residues of the E2 domain using structure-based mutagenesis for both genotypes 1 and 4. Six residues — E479, R542, E549, T553, K554, T564 — were mutated and binding with mAb 8G12 was determined by western blotting and analytical ultracentrifuge (AUC) experiments. We observed that point mutations E549A and K554A in the E2 domain from both genotypes completely abolished its binding with mAb 8G12, with no detectable bands on the immunostained membranes (Figure 5A and 5B), and no formation of immune complexes in the AUC analysis (Figure 5C, 5D and Supplementary information, Figure S9).
Figure 5.
Mutagenesis of the E2s:8G12 interaction interface. (A-B) The mutants and wild-type E2 (A, genotype 1; B, genotype 4) were subjected to non-reducing SDS-PAGE and western blotting with the neutralizing mAb 8G12 to study the effects of these mutations on E2s-8G12 interaction. The lanes marked with H indicate heated samples under reducing conditions (samples were heated up to 100 °C for 3 min) whereas the lanes marked with N indicate samples under non-reducing conditions (samples were diluted in 0.1% SDS, in the absence of β-mercaptoethanol (BME) and were not heated). (+) denotes dimerization or reactivity with 8G12, (−) denotes monomer or loss of the respective property. (C-D) Sedimentation velocity (SV) was utilized to detect the mAb 8G12 binding of E2HEV-1 and E2HEV-4. The c(s) profile of E2HEV-1/E2HEV-4 and its mutants, mAb 8G12 alone, and the antigen-antibody mixture are denoted as black, red and cyan solid curves, respectively. The molar ratio of E2HEV-1/E2HEV-4 or its mutant versus mAb 8G12 was 5:1, so that the antigen was in surplus.
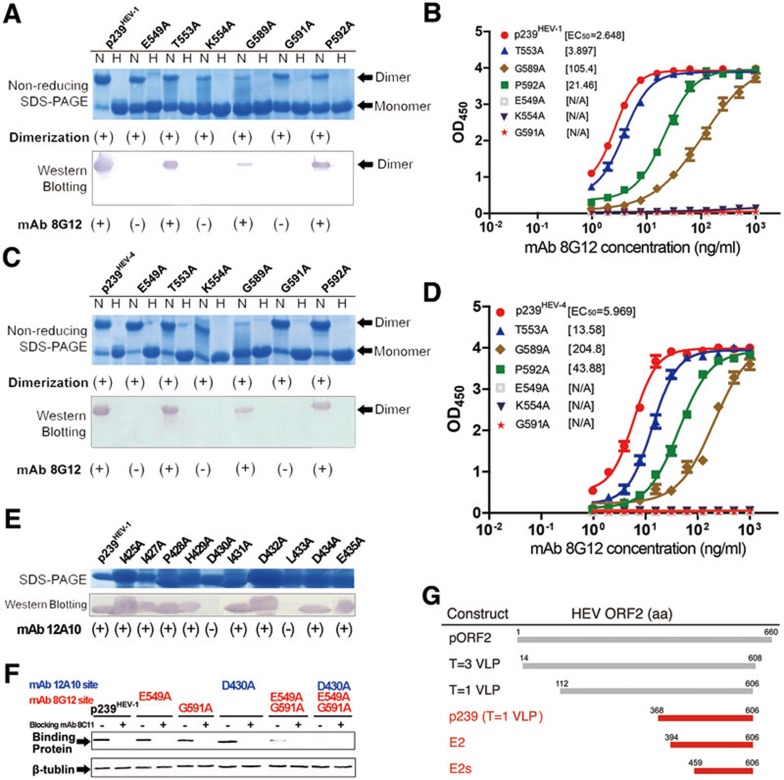
Next, we investigated the role of flexibility versus rigidity at a loop region (G589-S590-G591-P592) that lies in close contact with 8G12 by main-chain interactions (Figure 4B and 4C). This was accomplished by mutating flexible Gly residues to Ala (G589A and G591A), which is small but rigid, and by mutating the rigid Pro residue to the relatively less rigid Ala (P592A). We found that G589A and P592A mutants showed reduced binding to 8G12 whereas G591A mutant showed a complete loss of binding (Figure 6A and 6C); this loss-of-binding might be due to the differences in flexibility or systematic conformational changes. It has been previously shown that G591 and P592 are essential epitopes for the neutralizing antibody MAB272, and the double mutant, G591A and P592A, in the context of HEV VLPHEV-3 exhibits no binding to Huh7 cells9.
Figure 6.
Mutagenesis of the p239:8G12 and p239:12A10 interaction interfaces, and key mutations of p239 affecting binding to Huh7 cells. (A, C, E) The mutants and wild-type p239 (A and E, genotype 1; C, genotype 4) were subjected to non-reducing SDS-PAGE and western blotting with the neutralizing mAb 8G12 or mAb 12A10 to study the effects of these mutations on p239:8G12 and p239:12A10 interactions. The 12A10 mAb was previously shown to recognize the linear sequence KGIAIPHDIDLGESR (aa 423-438) of HEV ORF229. The lanes marked with H indicate heated samples under reducing conditions (samples were heated up to 100 °C for 3 min) while the lanes marked with N indicate samples under non-reducing conditions (samples were diluted in 0.1% SDS, in the absence of β-mercaptoethanol (BME) and were not heated). All lanes in E were run under reducing conditions. (+) denotes dimerization or reactivity with 8G12 or 12A10, (−) denotes monomer or loss of the respective property. (B, D) Interaction of the mutants and wild-type p239 (B, genotype 1; D, genotype 4) against mAb 8G12 was tested by enzyme-linked immunosorbent assay and EC50 was calculated by sigmoid trend fitting. (F) Binding of p239 and its mutants to Huh7 cells, as detected by western blotting. Endogenous β-tubulin served as a loading control. Single mutants p239-E549A, p239-G591A and p239-D430A retained the cell binding as compared with the wild-type p239. However, the double mutant p239-E549A-G591A caused a significant decrease in cell binding, and the triple mutant p239-D430A-E549A-G591A completely abrogated binding. (−) denotes the absence of the blocking mAb (8C11), (+) denotes the presence of this blocking mAb. Please also see Supplementary information, Figure S10. (G) Schematic bar representation of various ORF2 constructs.
Subsequently, we created mutations on 8G12's epitope of HEV VLP p239HEV-1 or p239HEV-4 and tested their binding to 8G12 by serial dilution and ELISA. The ELISA profiles of the wild-type (WT) p239HEV-1/4 were similar, and demonstrated the cross-genotype reactivity of 8G12 determinants. Three mutants — T553A, G589A and P592A — showed decreased reactivity against mAb 8G12, with higher EC50 values as compared with the WT p239. Consistent with the results observed for mutations on E2 (see Figure 5), three other mutants — E549A, K554A and G591A — were almost undetectable by 8G12, even at high Ab concentrations (Figure 6B and 6D), indicating that these residues play an essential role in the antibody-antigen interaction (Supplementary information, Table S1); this is also consistent with our structural observations. Of these important epitopes, E549 is the only residue that is in close contact with both the heavy and light chains of 8G12, and makes four hydrogen-bonding contacts (≤ 3.2 Å) (Figure 4D and 4E; Supplementary information, Table S1) and several van der Waals interactions; K554, on the other hand, forms salt bridges with the light chain residue E93L.
Cell binding analysis of HEV virus-like particle (VLP) mutants
The ORF2 construct p239 (aa 368-606) forms VLPs that specifically absorb and penetrate the host cell. There are two important sites on p239 that are involved in virus attachment to the host cell: (i) aa 423-438, the linear epitope of mAb 12A1029; and (ii) Arg512, the most important residue of the conformational epitope of mAb 8C1117. Previously, we reported that the combined mutation of these two sites, Δ12A10-8C11 (R512A), causes almost complete abrogation of host cell penetration17. To further identify the key residues of the 8G12's epitope, we generated p239 mutants harboring combined mutations in the 8G12 and 12A10 binding sites and tested their penetration and entry into vulnerable Huh7 cells. We selected residues for which the mutants showed no binding to their respective antibody, including E549, K554 and G591 of 8G12's epitope (Figure 6A-6D) and D430 and L433 of 12A10's epitope (Figure 6E). Notably, the double mutant p239 (E549A and G591A), as well as the triple mutant p239 (E549A, K554A and G591A) showed significantly reduced binding to the host cell (Supplementary information, Figure S10A). Further, when we introduced the D430A mutation of 12A10's epitope (such as 12A10 (D430A)-8G12(E549A-G591A); Figure 6F and Supplementary information, Figure S10B), we observed complete disruption of host cell penetration. Thus, combining mutations in 8C11's or 8G12's epitope with those of the 12A10's epitope completely abrogates host-cell penetration.
Discussion
Viral entry is the earliest step in virus infection, with some viruses making contact with host cells through virus attachment or virus adsorption to cell-surface membrane receptors. The virus adsorption region (AR) is defined as the virus surface region that directly interacts with its complementary receptor on host cells. Previously, it was reported that dimerization of the protruding region (E2s domain) is a prerequisite for HEV virulence and plays an important role in the host-virus interaction9,10,11,17. Therefore, it seems reasonable to propose that the AR is located, at least in part, near the dimerization region and includes the identified epitope region of these mAbs. Thus, not surprisingly, the mAb 8G12 targets this dimerization region and neutralizes HEV across all genotypes.
Here we also revealed that all three epitope sites (of mAb 8C11, 8G12 and 12A10) are essential for the interaction of the virus with host cells. Mutating the 12A10's epitope in combination with either 8G12's or 8C11's epitope can abolish the interaction with the host and prevent infection. Although the 12A10's epitope is important, we found that its mutation alone is insufficient to prevent host cell penetration. Thus, to completely prevent infection, a combination of 12A10 with 8G12 or 8C11 is likely required.
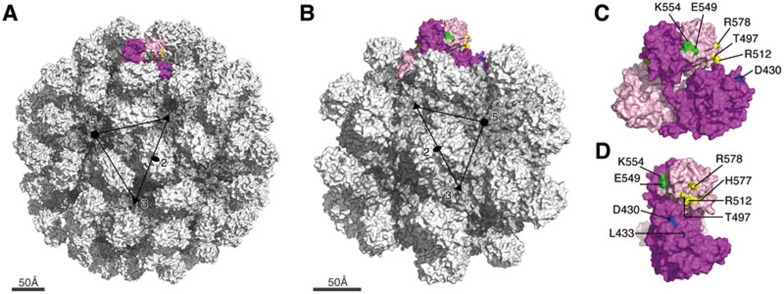
Previous studies have explored the receptor binding site of pORF2 of HEV, although the cell receptor for HEV has not yet been identified10,29. The E2 domain of HEV is homologous to the receptor-binding domain of Norovirus in the Caliciviridae family, for which the blood-group trisaccharides act as cell receptors10. The superposition of these two structures suggests that trisaccharides might locate on the top of the HEV protruding spike (E2 domain) between the loops created by aa 549-566 and aa 583-593. Notably, these two loop regions are potential sugar-binding sites, implicating that they might have important functional roles in cell receptor binding. In the present study, we show that mutations at residues on p239 within this potential receptor-binding region (E549A, K554A and G591A) lead to a significant loss of binding to host cells (Supplementary information, Figure S10). In addition, mutation of residue N562 in this region to Gln has been shown to result in loss of HEV infectivity in both cultured cells and rhesus macaques30. Meanwhile, this residue was also reported to be one of the several essential residues for host cell interaction9. To have a better spatial view of all the key epitope sites, i.e., the potential receptor binding sites, we mapped all the residues that have been shown to be involved in binding to the host cell (Figure 7, Table 2 and Supplementary information, Figure S11) on the surface of T = 3 virion-sized particle31 (Figure 7A) and T = 1 HEV VLP9,10,32 (Figure 7B). Interestingly all of these residues within E2 domain are located either in the dimerization region or in the groove region. It suggests the involvement and importance of these two regions of E2 domain in the virus-host interactions. In addition the linear epitope of 12A10 especially the residue D430 present in the M domain also plays a key role in the virus-host interaction.
Figure 7.
Putative host-binding site on the surface of HE virus-like particle (VLP). (A) the molecular surface representation of the T = 3 virion-sized particle (PDB code 3IYO) and (B) the T = 1 VLP (PDB code 2ZTN). The 2-fold, 3-fold and 5-fold axes of the icosahedral lattice are denoted as solid oval, triangle and pentagon, respectively. (C) The front view and (D) the side view of a dimeric subunit of the construct (aa 112-608) is shown. The monomers are colored in dark and light pink. The key residues on the epitopes of 12A10, 8C11 and 8G12, which are crucial for the viral infection, are labeled and highlighted in blue, yellow and green, respectively.
Table 2. The amino acids recognized by the neutralizing antibodies and those confirmed to be involved in virus-host interactions.
| mAb | Key aa of the epitope region | Method used to identify | Location of the epitope | Genotype | Reference |
|---|---|---|---|---|---|
| Based on mutation/truncation studies to test antibody binding | |||||
| 8C11 | E479, Y485, D496, R512, K534, H577, R578 | Crystal Structure combined with mutation assay | Groove region | 1 | 17 |
| 8G12 | E549,K554,G591 | Crystal Structure combined with mutation assay | Dimerization region | 1 & 4 | this study |
| 12A10 | Linear epitope 423-438 | Mutation assay | M domain | 1 | 29 |
| 8H3 | E479,Y485,I529, K534 | Mutation assay | Groove region | 1 | 11 |
| MAB1323 | S487, S488, T489, P491, N562,T564 | Density fractionation assay | Dimerization region | 3 | 9 |
| MAB272 | D496, T497, G591, P592 | Density fractionation assay | Groove region | 3 | 9 |
| Fab224 | E479,K534,Y485, I529 | Mutation assay | Groove region | 1 | 32 |
| HEV#4 and HEV#31 | Linear epitope 578-607 | Truncation binding assay | Dimerization region and groove region | 1 | 33 |
| Based on cell binding assay | |||||
| 4 mAbs | T489, P491, Y561, N562, T564, T585, T586 | Binding to host cell model assay | Dimerization region | 3 | 9 |
| 8C11 | T497, R512, H577, R578 | Binding to host cell model assay | Groove region | 1 | 17 |
| 8G12 | E549, K554, G591 | Binding to host cell model assay | Dimerization region | 1 & 4 | this study |
| 12A10 | D430 and L433 | Binding to host cell model assay | M domain | 1 | this study |
Here we show that the mAb 8G12 neutralizes HEV infection in the cell model and confers HE disease protection in our rhesus monkey model. The neutralization in vivo manifested as a postponement of the onset of virus shedding, a decrease in the virus amount, and, presumably, protection against the subsequent development of acute hepatitis, although the HEV infection was not completely suppressed at the high-dose virus challenge (HE disease model). This observation is similar to our previous studies with mAb 8C11-pretreatment18 and p239 vaccination19, which also did not completely block the HEV infection in HE disease model. However the 8C11 pretreatment and p239 vaccination exhibited complete HEV-infection protection against a 3-log lower virus challenge (HEV infection model). Collectively, our findings suggest that the mAb 8G12 could completely block the infection of HEV genotypes 1 and 4 in an infection model used in this study and the disease model study will be the subject of future analyses.
Our structural studies of mAb 8G12 identified the dimerization region of the E2s domain as one of the key interacting regions of the antibody. Indeed, we show this region to be immune predominant and a site for cross-genotype neutralization. In HEV biology, dominant antibodies are elicited by the conserved 8G12 epitope during the immune response, and this dominance may account for there being one major serotype of HEV. The current p239HEV-1 vaccine only targets genotype 1, although it showed protection against heterogeneous HEV-induced disease in the clinical trial data13. The 8G12-interacting region could be exploited, preserved and/or combined with epitopes for other neutralizing mAbs such as 8C11 and 12A10 for the development of more potent HEV vaccines (Figure 7, Table 2 and Supplementary information, Figure S11). Moreover, the mAb 8G12 binding assay will benefit the quality assessment of the current vaccine p239HEV-1 and the surveillance of potential 8G12-escape HEVs. This might also be adopted to monitor any immune escape from the current vaccine protection. Overall, the current study advances our understanding of effective immune responses against HEV and provides important information for vaccine design and improvement.
Materials and Methods
Construction and antibody preparation
The E2s, E2 and p239 genes of HEV were cloned as previously described12,17. All mutated constructs were generated with site-directed PCR reactions. pTO-T7 expression plasmid and E. Coli ER2566 strain were used for protein expression. The 8G12 Fab was obtained by papain digestion and purified with DEAE-5PW (TOSOH Bioscience, Tokyo, Japan). A schematic bar diagram of HEV ORF2 constructs is shown in Figure 6G.
Protein purification, crystallization and structure determination
The HEV E2s were purified as previously described11. The E2s:8G12 (ratio of 1:1.5M) was kept at 37 °C for 2 h and purified by G5000PWxl (TOSOH) and concentrated to 8 mg/ml. The crystallization condition for E2sHEV-1:8G12 was 12% PEG 5000 and 0.05 M NaH2PO4, whereas that for E2sHEV-4:8G12 was 14% PEG 1500. Crystals were grown using hanging-drop vapor diffusion method at 21 °C. For both crystals, 30% glycerol supplemented with reservoir condition was used as a cryo-protectant and data was collected at 100 K at Shanghai Synchrotron beam line 17U. Data sets were processed by HKL200034. The structures were solved by molecular replacement with PHASER35. The models were built using COOT36, refined by CNS37 and PHENIX38, and analyzed by PROCHECK39. For E2sHEV-1:8G12 structure, the NCS restrains were applied during refinement. There are 0.5% residues of E2sHEV-1:8G12 complex in the disallowed region such as E2s residues A477, V503, A575 and H577 and the rest from 8G12, most of them located in the loop regions. This might be due to the limitation of the data set. The structure-related figures in this manuscript were prepared using the program PyMol40.
Neutralization assay on HEV-infection cell model
Approximately 1 × 105 genomic equivalent of genotype 1 or 4 virus was mixed with serially diluted aliquots of mAbs or PBS at 37 °C for 30 min and added to triplicate cultures of Huh7 cells. The infected cultures were incubated at 37 °C for 60 min, and then washed three times with PBS. The cells were harvested and then tested for the HEV RNA content. The HEV RNA genome was extracted by a robot Biomen NX (Beckman, CA, USA). Real-time RT-PCR was performed with the following primers designed using the sequence with the DDBJ accession number D11092: JVHEVF (forward; 5261-5278 nt of HEV genome), 5′-GGTGGTTTCTGGGGTGAC-3′ JVHEVP (probe 1; 5283-5300 nt) FAM-TGATTCTCAGCCCTTCGC-TAMRA; and JVHEVR (reverse, 5311-5328 nt) 3′-AGGGGTTGGTTGGATGAA-5′. RNA content in cultures infected with the mAb-treated virus was expressed as a percentage of that in the control cultures infected with untreated virus. The inhibition profiles were fitted to a sigmoid trend to generate the IC50 value. The IC50 for mAb 8G12 was used to estimate the applied amount of mAbs to pretreat HEV in the subsequent animal experiments.
Animal study
A total of 15 Macaques rhesus monkeys were used during screening. The monkeys showed no detectable HEV IgG/IgM in serum at 1:10 dilution using ELISA kits (Wantian, Beijing, China); no ALT abnormality in the serum, as determined using an ALT-ELISA kit (Maker Biotechnology, SC, China); and no detectable RNA in stool samples, as determined by RT-PCR.
Two monkeys per group were inoculated with six combinations of HEV from genotypes 1, 3 and 4 that had been mixed and pre-incubated at 37 °C for 2 h with mAb 8G12 or mAb 13D4 in a 1:2 (v/v) ratio. The mAb 13D4 served as a control as it is directed against the H5N1 flu and therefore should be non-neutralizing against HEV genotypes. Each monkey intravenously received 1 500 μl (500 μl virus extraction solutions, 1 000 μl mAb of 5 mg/ml) of the corresponding inoculant mixture. The virus stocks were purified from stool and bile samples from each challenged monkey. The final copies of the RNA genome for genotypes 1, 3 and 4 were 5.55 × 107/500 μl, 2.69 × 107/500 μl and 4.73 × 107/500 μl, respectively. Because HEV genotype 2 mainly prevails in Africa and Mexico, we were unable to work with this genotype due to the geographic limitations. However, we used a recombinant E2 protein of HEV genotype 2 in the antigenicity assay. Notably, E2s genotype 2 shares 86%-92% sequence identity with other genotypes (Supplementary information, Figure S8).
Prior to the experiment, three monkeys were inoculated with 500 μl inoculum of 5.55 × 105, 2.69 105, 4.73 × 105 genomic RNA copies of HEV genotype 1, 3 or 4, respectively, to check the virulence of the virus in developing Hepatitis E. The sera from these three monkeys were serially diluted to measure the blocking profile of mAbs 12A10, 8C11 and 8G12, as well as the 8G12 Fab and the 8G12's epitope reactivity of the 8G12-escape p239 mutant (harboring two point mutations at E549A and G591A) as compared with the WT p239 along the virus life cycle; this included assessment of infection, Hepatitis E development and convalescence. Stool samples were collected before inoculation and twice per week for 15 weeks after inoculation. Samples were assayed for HEV virus genomes by above-mentioned RT-PCR and levels of ALT, anti-HEV IgM or IgG by ELISA.
BIAcore biosensor analysis
CM-5 sensor chips were coated with a carboxylated dextran polymer matrix, where the goat anti-mouse antibody Fc fragment (GAM-Fc) was amine coupled (BIAcore 3000, GE). The carboxyl groups on the dextran surface were activated with 35 μl of a 1:1 (v/v) solution of N-hydroxysuccinimide and N-ethyl-N-(3-diethylarninopropyl) carbodiimide for 7 min. The GAM-Fc was diluted with 10 mM sodium acetate (pH 5.0) to a final concentration of 60 μg/ml for coupling. Eventually, one flow cell of a chip was coated with 13 000 RU (resonance units) of the GAM-Fc, whereas the other flow cell was left uncoated and blocked as a control. The affinity measurements of mAb 8G12 binding with different antigens were initiated by passing HBS (10 mM HEPES, pH 7.4 and 150 mM NaCl) over the sensor surface for 100 s at 10 μl/min, followed by injection of 10 μg/ml of mAb 8G12 at 30 μl/min for 3.3 min, and then injections of serially diluted antigens at 30 μl/min for 3.3 min. Every measurement on the BIAcore 3000 biosensor was performed three times and the individual values were used for affinity constant fitting to produce the mean affinity constant with measure error evaluation.
ELISA assay
ELISA was performed to weight the genotype-common reactive nature of mAb 8G12, to monitor the IgG/IgM response in experimental monkeys, and to monitor the inhibition ratio of mAb 8G12 against human/monkey serum, based on genotype-specific antigens. In brief, the antigen (E2, p239, or 8G12-escape p239 mutant harboring E549A and G591A mutations; 100 ng per well) was coated in a 96-well microplate and then incubated at 37 °C for 30 min along with (i) serial dilutions of mAb 8G12 for 8G12 reactivity evaluation; (ii) serial dilutions of the sera of monkeys for antibody response monitoring; (iii) 10 μg per well of mAb 12A10, 8C11, 8G12 and 8G12 Fab with a predetermined dilution of human/monkey serum (producing 1 OD reading in a serial diluted pre-test ELISA) for 30 min at 37 °C for mAb blocking ratio against pAb test; or (iv) serial dilutions of the free-form of mAb 8C11 or 8G12 for cross-blocking ratio calculations. Wells were then incubated with HRP-conjugated goat anti-mouse (GAM; for mAb), goat anti-human (GAH; for human/monkey serum) secondary antibody (DAKO, Glostrup, Denmark) or HRP-conjugated mAb 8C11/8G12 (producing 1 OD reading in a serial diluted pre-test ELISA) for 30 min at 37 °C. Subsequently, 100 μl tetramethylbenzidine substrate was added and the plates incubated for 10 min at 37 °C. The reaction was stopped by the addition of 50 μl of 2 M H2SO4, and the OD was measured using the microplate reader (Sunrise, Tecan) at 450 nm with a reference wavelength of 620 nm (measurement range: 0–4.0 OD at 400 to 750 nm). IC50 or EC50 was calculated at the half-point of the curve by polynomial fitting. The blocking ratio of the mAbs or Fab against serum was the resultant OD ratio of the sera binding to coated antigens with and without pretreatment. The percentage of 8G12's epitope reactivity among monkey sera was calculated by the increase in the EC50 for the 8G12-escape p239 mutant as compared with that of WT p239; i.e., % 8G12 reactivity = (1-EC50[wild-type]/EC50[8G12-escape]) × 100%.
Analytical Ultra-Centrifugation (AUC)
Sedimentation velocity was used to monitor the binding of the antigen and mAb in a neutral solution, as previously described17. The experiments were conducted at 20 °C on a Beckman XL-A AUC equipped with absorbance optics and an An60-Ti rotor. All samples were diluted to ∼1.0 OD at 280 nm in a 1.2-cm light path in a buffer comprising 20 mM HEPES, pH 7.5, and 150 mM NaCl. The rotor speed was set to 50 000 rpm for E2 protein, 40 000 rpm for mAb 8G12, and 30 000 rpm for the immune complex. E2 or mutated proteins and mAb 8G12 were mixed at a 5:1 molar ratio and incubated at 37 °C for 1 h to permit the interaction to occur, if existent. The sedimentation coefficient was obtained with c(s) method using the Sedfit software kindly provided by Dr Schuck P (National Institutes of Health, Bethesda, MA, USA).
Binding assay on VLP-based cell model
Human hepatoma Huh7 cells were incubated at 37 °C for 1 h in the presence of (i) p239 or its mutants (20 μg) and (ii) p239 or its mutants (20 μg) pre-incubated with neutralizing mAb 8C11 (200 μg) as a blocking mAb control. The cells were then harvested and lysed with lysis buffer (20 mM KOH-HEPES buffer, pH 8.0, 0.2 mM EDTA, 5% glycerol, 250 mM NaCl, 0.5% NP-40, 0.25% sodium deoxycholate, 1 mM DTT and protease inhibitors). The samples were separated by SDS-PAGE and transferred onto nitrocellulose membranes, as previously described. After transfer, the membranes were immersed for 30 min in blocking solution (5% non-fat milk in PBS, pH 7.45) and washed in PBS containing 0.1% Tween 20 (PBST). Subsequently, membranes were incubated with primary antibody (4A6, which recognizes the linear epitope HEV ORF2 aa 443-457) and washed three times with PBST. Alexa Fluor 680-conjugated mouse anti-β-tubulin and rabbit anti-α-tubulin (Invitrogen) or HRP-conjugated GAM (for mAb; DAKO) were then added and the reaction signals were examined by Image Quant LAS4000mini (GE).
Accession numbers
The coordinates and structure factors for both E2sHEV-1:8G12 and E2sHEV-4:8G12 complexes have been deposited in the Protein Data Bank (accession numbers 4PLK and 4PLJ).
Acknowledgments
The data sets were collected at the beam line 17U of Shanghai Synchrotron Radiation Facility (SSRF), China. SL and NX acknowledge funding support from the Chinese government: Fujian Provincial Science Fund for Distinguished Young Scholars (Grant 2011J06015), National Natural Science Foundation (Grants 81172885 and 81371818), International Science and Technology Collaborative Program (Grant 2011DFG33050) and Key Program in New Drug R&D (Grants 2012ZX09101316 and 2014AA021302). We thank Dr Tom Terwilliger (Los Alamos National Laboratory) for the suggestions on refinement with PHENIX program. JS and CL-H acknowledge funding support from Academic Research Fund Tier 1 (Grant R154000616112), the National University of Singapore (NUS), Singapore.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
E2 and mAb 8G12 binding curves in Surface Plasma Resonance (SPR).
Inhibition profiles of mAb 8G12, 8C11, 12E11 and 12E9 blocking authentic HEV from infecting the host cell.
Cross blocking of mAbs 8G12 and 8C11 against p239 VLP (genotype 1).
Cross-genotype neutralizing mAb 8G12 binds to the E2s dimeric interface region.
Neutralization of HEV infection rates by mAb 8G12 in Macacus rhesus monkey model.
Comparison of the blocking ratio of mAb 12A10, 8C11 and 8G12 against monkey sera.
Stoichiometry of E2sHEV-4:8G12 complex in solution.
Structure-based sequence alignment among E2s genotype 1, 3 and 4 in combination with sequence-based alignment for E2s genotype 2 are shown.
SV binding test of the E2HEV-1 and E2HEV-4 constructs via Ala-scanning mutagenesis on the 8G12 binding interface.
Binding of p239 and its mutants disturbing mAb 8G12 and 12A10 binding sites in Huh7 cells was detected by western blotting.
The potential virus-host binding region.
Hydrogen bonding contacts (≤ 3.2 Å) between the E2s epitope and the 8G12 Fab
References
- Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Tessari P, Sofia A, Saffioti S, et al. Effects of chronic metabolic acidosis on splanchnic protein turnover and oxygen consumption in human beings. Gastroenterology. 2010;138:1557–1565. doi: 10.1053/j.gastro.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Petit MA, Capel F, Dubanchet S, Mabit H. PreS1-specific binding proteins as potential receptors for hepatitis B virus in human hepatocytes. Virology. 1992;187:211–222. doi: 10.1016/0042-6822(92)90309-d. [DOI] [PubMed] [Google Scholar]
- Jaiswal SP, Jain AK, Naik G, Soni N, Chitnis DS. Viral hepatitis during pregnancy. Int J Gynaecol Obstet. 2001;72:103–108. doi: 10.1016/s0020-7292(00)00264-2. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Deng Q, Zhai JW, Michel ML, et al. Identification and characterization of peptides that interact with hepatitis B virus via the putative receptor binding site. J Virol. 2007;81:4244–4254. doi: 10.1128/JVI.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N, Renou C, Di Liberto G, Boutrouille A, Eloit M. Hepatitis E: a curious zoonosis. Front Biosci. 2008;13:7172–7183. doi: 10.2741/3219. [DOI] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Mori Y, Miyazaki N, et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci USA. 2009;106:12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guu TS, Liu Z, Ye Q, et al. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci USA. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tang X, Seetharaman J, et al. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 2009;5:e1000537. doi: 10.1371/journal.ppat.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SW, Zhang J, He ZQ, et al. Mutational analysis of essential interactions involved in the assembly of hepatitis E virus capsid. J Biol Chem. 2005;280:3400–3406. doi: 10.1074/jbc.M410361200. [DOI] [PubMed] [Google Scholar]
- Zhu FC, Zhang J, Zhang XF, et al. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet. 2010;376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ge SX, Huang GY, et al. Evaluation of antibody-based and nucleic acid-based assays for diagnosis of hepatitis E virus infection in a rhesus monkey model. J Med Virol. 2003;71:518–526. doi: 10.1002/jmv.10523. [DOI] [PubMed] [Google Scholar]
- Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- Xing L, Kato K, Li T, et al. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology. 1999;265:35–45. doi: 10.1006/viro.1999.0005. [DOI] [PubMed] [Google Scholar]
- Tang X, Yang C, Gu Y, et al. Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc Natl Acad Sci USA. 2011;108:10266–10271. doi: 10.1073/pnas.1101309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gu Y, Ge SX, et al. Analysis of hepatitis E virus neutralization sites using monoclonal antibodies directed against a virus capsid protein. Vaccine. 2005;23:2881–2892. doi: 10.1016/j.vaccine.2004.11.065. [DOI] [PubMed] [Google Scholar]
- Li SW, Zhang J, Li YM, et al. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine. 2005;23:2893–2901. doi: 10.1016/j.vaccine.2004.11.064. [DOI] [PubMed] [Google Scholar]
- Chen Y, Qin K, Wu WL, et al. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J Infect Dis. 2009;199:49–58. doi: 10.1086/594374. [DOI] [PubMed] [Google Scholar]
- Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BS. Advances in antiviral vaccine development. Immunol Rev. 2013;255:230–242. doi: 10.1111/imr.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Torresi J, Locarnini SA, et al. Amino-terminal epitopes are exposed when full-length open reading frame 2 of hepatitis E virus is expressed in Escherichia coli, but carboxy-terminal epitopes are masked. J Med Virol. 1997;52:289–300. doi: 10.1002/(sici)1096-9071(199707)52:3<289::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med. 2012;367:1237–1244. doi: 10.1056/NEJMra1204512. [DOI] [PubMed] [Google Scholar]
- Fransson J, Teplyakov A, Raghunathan G, et al. Human framework adaptation of a mouse anti-human IL-13 antibody. J Mol Biol. 2010;398:214–231. doi: 10.1016/j.jmb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- He S, Miao J, Zheng Z, et al. Putative receptor-binding sites of hepatitis E virus. J Gen Virol. 2008;89:245–249. doi: 10.1099/vir.0.83308-0. [DOI] [PubMed] [Google Scholar]
- Graff J, Zhou YH, Torian U, et al. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J Virol. 2008;82:1185–1194. doi: 10.1128/JVI.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Li TC, Mayazaki N, et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Wang JC, Li TC, et al. Spatial configuration of hepatitis E virus antigenic domain. J Virol. 2011;85:1117–1124. doi: 10.1128/JVI.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield DJ, Glamann J, Emerson SU, Purcell RH. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J Virol. 2000;74:5548–5555. doi: 10.1128/jvi.74.12.5548-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr. Pt A 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK – a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Delano W.The PyMOL molecular graphics system. San Carlos, CA: Schrödinger, available freomhttp://www.pymol.org/ . 2002
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E2 and mAb 8G12 binding curves in Surface Plasma Resonance (SPR).
Inhibition profiles of mAb 8G12, 8C11, 12E11 and 12E9 blocking authentic HEV from infecting the host cell.
Cross blocking of mAbs 8G12 and 8C11 against p239 VLP (genotype 1).
Cross-genotype neutralizing mAb 8G12 binds to the E2s dimeric interface region.
Neutralization of HEV infection rates by mAb 8G12 in Macacus rhesus monkey model.
Comparison of the blocking ratio of mAb 12A10, 8C11 and 8G12 against monkey sera.
Stoichiometry of E2sHEV-4:8G12 complex in solution.
Structure-based sequence alignment among E2s genotype 1, 3 and 4 in combination with sequence-based alignment for E2s genotype 2 are shown.
SV binding test of the E2HEV-1 and E2HEV-4 constructs via Ala-scanning mutagenesis on the 8G12 binding interface.
Binding of p239 and its mutants disturbing mAb 8G12 and 12A10 binding sites in Huh7 cells was detected by western blotting.
The potential virus-host binding region.
Hydrogen bonding contacts (≤ 3.2 Å) between the E2s epitope and the 8G12 Fab